Summary
SNARE-mediated synaptic exocytosis is orchestrated by facilitatory and inhibitory mechanisms. Genetic ablations of Complexins, a family of SNARE complex–binding proteins, in mice and Drosophila cause apparently opposite effects on neurotransmitter release, leading to contradictory hypotheses of Complexin function. Reconstitution experiments with different fusion assays and Complexins also yield conflicting results. We therefore performed cross-species rescue experiments to compare the functions of murine and Drosophila Complexins in both mouse and fly synapses. We found that murine and Drosophila Complexins employ conserved mechanisms to regulate exocytosis despite their strikingly different overall effects on neurotransmitter release. Both Complexins contain distinct domains that facilitate or inhibit synaptic vesicle fusion, and the strength of each facilitatory or inhibitory function differs significantly between murine and Drosophila Complexins. Our results show that a relative shift in the balance of facilitatory and inhibitory functions results in differential regulation of neurotransmitter release by murine and Drosophila Complexins in vivo, reconciling previous incompatible findings.
Introduction
SNARE (soluble N-ethylmaleimide–sensitive factor–attachment protein receptor)-mediated synaptic vesicle exocytosis is tightly controlled by a large number of regulatory proteins to ensure the exquisite temporal and spatial precision of neurotransmitter release at synapses (Brunger, 2005; Jahn and Scheller, 2006). Complexins constitute a family of small and highly charged proteins that bind to the assembled SNARE complex (Ishizuka et al., 1995; McMahon et al., 1995; Reim et al., 2005; Takahashi et al., 1995). They generally contain a central α-helix and an accessory α-helix in the middle portion of the protein, and the N- and C-terminal sequences that are probably largely unstructured (Pabst et al., 2000). Complexins bind to the SNARE complex with high affinity (Bowen et al., 2005; Li et al., 2007; Pabst et al., 2002). The central α-helix of Complexins interacts with the SNARE motifs of Syntaxin-1 and Synaptobrevin-2 within the SNARE complex in an anti-parallel fashion (Bracher et al., 2002; Chen et al., 2002). Complexins can also bind to the target-SNAREs (Syntaxin-1 and SNAP-25) heterodimer with a lower affinity (Guan et al., 2008; Weninger et al., 2008; Yoon et al., 2008).
Biophysical and physiological studies have indicated diverse functions for Complexins in vesicle fusion, some of which are incompatible (Brose, 2008). Complexins have been shown to inhibit SNARE-mediated cell fusion (Giraudo et al., 2006) and proteoliposome fusion in bulk ensemble assay (Schaub et al., 2006), and this inhibition is released by the Ca2+ sensor Synaptotagmin-1 and Ca2+. Biochemically, Synaptotagmin-1 competes with Complexins for the SNARE complex binding and displaces Complexins from the SNARE complex in a Ca2+-dependent manner (Tang et al., 2006). These studies suggest a fusion clamp model for Complexin function, in which Complexins inhibit the transfer of the force generated by the SNARE complex assembly onto the fusing membranes and arrest synaptic vesicle fusion before Ca2+ influx. Upon Ca2+ binding, Synaptotagmin-1 displaces Complexins from the SNARE complex to release this inhibition and triggers exocytosis (Giraudo et al., 2006; Maximov et al., 2009; Schaub et al., 2006; Tang et al., 2006). However, Complexins have also been shown to stimulate proteoliposome fusion in both single-vesicle fusion assay (Yoon et al., 2008) and bulk ensemble assay (Malsam et al., 2009), indicating a facilitatory role. These in vitro results are further confounded by in vivo genetic studies. Genetic knockout of Complexins in mice leads to a reduction in both evoked and spontaneous release at multiple glutamatergic and GABAergic synapses in cultures and in acute brain slices (Reim et al., 2001; Strenzke et al., 2009; Xue et al., 2008b), and a decrease in Ca2+-triggered exocytosis in adrenal chromaffin cells (Cai et al., 2008), supporting a stimulatory function for Complexins. In contrast, genetic deletion of Complexin in fruit fly Drosophila melanogaster greatly enhances spontaneous release but decreases Ca2+-evoked release (Huntwork and Littleton, 2007), favoring the fusion clamp model. Moreover, knockdown of Complexins by RNA interference in mass-cultured mouse cortical neurons decreases evoked release and increases spontaneous release at glutamatergic synapses (Maximov et al., 2009). To explain the discrepancy between this result and those obtained previously from Complexin knockout mice, the authors (Maximov et al., 2009) suggest that this is due to the different preparations used (autaptic cultures for knockout studies (Reim et al., 2001; Xue et al., 2008b) versus mass cultures for knockdown study (Maximov et al., 2009)), disavowing the fact that the knockout studies also employed mass cultures and acute brain slices (Xue et al., 2008b), and found similar results to those obtained from autaptic cultures. Hence, many studies seem at odds with each other and the precise in vivo role of Complexins in exocytosis is still unclear.
An in vivo structure-function analysis of murine Complexin I (CplxI) in Complexin I/II double knockout mouse neurons indicates that the SNARE complex binding is essential for CplxI function, and that the N terminus of CplxI facilitates release, whereas an accessory α-helix between the N terminus and the central α-helix inhibits release (Xue et al., 2007). A biophysical study reveals that CplxI inhibits SNARE complex formation, but strongly stimulates membrane fusion after the assembly of the SNARE complex in vitro (Yoon et al., 2008). These studies indicate that Complexins play both facilitatory and inhibitory roles in exocytosis, but they still do not explain why genetic deletions of Complexins in two model organisms, mouse and fly, have such different effects on neurotransmitter release. Furthermore, the amino acid sequence homology is low between murine and Drosophila Complexins except for the central α-helix that is essential for the binding to the SNARE complex, and part of the N terminus (Figure S1; (Huntwork and Littleton, 2007; Reim et al., 2005)). Thus, the dramatic difference in loss-of-function phenotypes of Complexin deficient mice and flies lead to the conclusion that Complexin function must differ between mice and flies.
To test whether the functions of murine and Drosophila Complexins are conserved in synaptic vesicle exocytosis, and to gain insight into their functional and structural differences, it is essential to compare murine and Drosophila Complexins in the same experimental in vivo systems. A detailed structure-function analysis is also necessary because a complete removal of Complexins is unlikely to reveal all aspects of their function (Xue et al., 2007). We therefore undertook a systematic cross-species rescue approach to compare the functions of murine and Drosophila Complexins at both mouse and fly synapses. We find that both murine and Drosophila Complexins contain distinct functional domains and play dual roles in neurotransmitter release. They facilitate and inhibit release via similar domains, but the facilitatory or inhibitory strength of a given domain varies between murine and Drosophila Complexins. Thus, both murine and Drosophila Complexins utilize conserved mechanisms in release process, but the integration of facilitation and inhibition differs substantially between them, leading to an apparently opposite overall effect on exocytosis. Our results reveal conserved functions of Complexins between species and indicate that the interplay of dual functions orchestrates neurotransmitter release.
Results
Drosophila Cplx arrests neurotransmitter release in Cplx-TKO mouse neurons
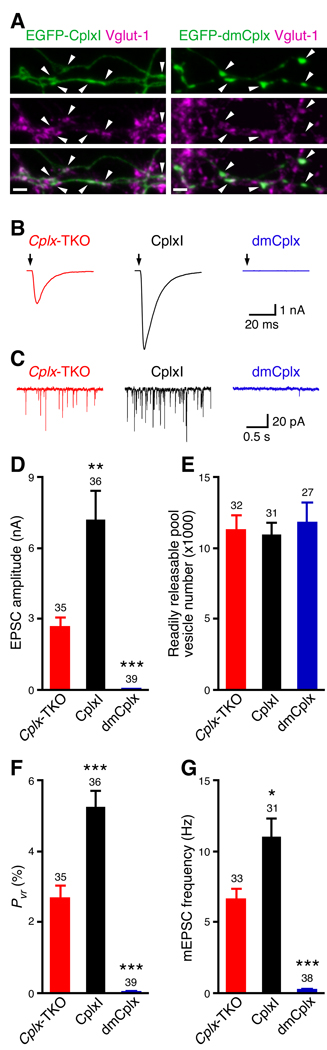
Prior to examining Complexin function at mouse synapses, we assessed Complexin protein localization by immunostaining and confocal micrscopy. The antibodies against Drosophila Complexin (dmCplx) or murine Complexin I (CplxI) show some nonspecific labeling in immunostaining of mouse neurons, we therefore generated enhanced green fluorescent protein (EGFP) tagged dmCplx and CplxI. We transfected cultured hippocampal neurons with EGFP-dmCplx and EGFP-CplxI, and immunolabeled the neurons with antibodies against EGFP and a presynaptic marker, vesicular glutamate transporter-1 (Vglut-1). EGFP-CplxI is distributed diffusely in axonal processes and synapses (Figure 1A), consistent with previous subcellular localization studies (McMahon et al., 1995; Reim et al., 2005). In contrast, EGFP-dmCplx shows a punctate pattern along axons and is enriched at presynaptic varicosities (Figure 1A). The synaptic localization of EGFP-dmCplx is similar to the punctate distribution of murine CplxIII and CplxIV, which is mediated by farnesylation of their C termini ((Reim et al., 2005) and see below). Nevertheless, both EGFP-dmCplx and EGFP-CplxI are present at presynaptic termini that are labeled by Vglut-1, indicating that both CplxI and dmCplx reach synapses in mouse neurons (Figure 1A).
Figure 1.
Murine CplxI facilitates, whereas Drosophila Cplx inhibits neurotransmitter release in Cplx-TKO hippocampal neurons. (A) In cultured hippocampal neurons, EGFP-CplxI and EGFP-dmCplx are present at presynaptic termini labeled by a presynaptic marker, Vglut1. White arrowheads indicate the synaptic varicosities containing Vglut1 and EGFP-CplxI or EGFP-dmCplx. Scale bars: 2 µm. (B) Representative traces of evoked EPSCs. The arrows represent stimulations; artifacts and action potentials are blanked. (C) Representative traces of mEPSCs. (D to G) Summary data for Cplx-TKO and CplxI- or dmCplx-rescued Cplx-TKO neurons. (D) Amplitude of EPSC evoked by single action potentials. Cplx-TKO and CplxI-rescued neurons all showed evoked EPSCs, whereas 22 out of 39 dmCplx-rescued neurons showed no detectable EPSCs. (E) Readily releasable pool vesicle number (transient synaptic charge induced by hypertonic sucrose solution divided by the mEPSC charge). (F) Vesicular release probability (Pvr) of evoked release. (G) mEPSC frequency. Cplx-TKO and CplxI-rescued neurons all showed mEPSCs, whereas 11 out of 38 dmCplx-rescued neurons showed no detectable mEPSCs. Data are expressed as mean ± SEM. *, P < 0.01; **, P < 0.001; ***, P < 0.0001 compared to Cplx-TKO neurons. The numbers of neurons analyzed are shown above the bars.
To study the effect of dmCplx in neurotransmitter release, we used lentiviruses to express wildtype (WT) dmCplx and CplxI at similar levels in cultured hippocampal neurons (Supplemental results and Figure S2). Autaptic Complexin I/II/III triple knockout (Cplx-TKO) neurons were infected with lentiviruses expressing CplxI or dmCplx together with EGFP, or with lentiviruses only expressing EGFP. In response to an action potential, the excitatory postsynaptic current (EPSC) amplitude of Cplx-TKO neurons is severely reduced, and this defect is fully rescued by the expression of CplxI (Figures 1B and 1D; (Xue et al., 2007)). Strikingly, expression of dmCplx in Cplx-TKO neurons almost abolishes the remaining evoked release (Figures 1B and 1D), showing that CplxI and dmCplx affect synaptic transmission in opposite ways in vivo. To directly assess the Ca2+-triggered release efficacy, we determined the vesicular release probability (Pvr), the fraction of the readily releasable vesicles released by one action potential. We measured readily releasable vesicle pool (RRP) by hypertonic sucrose solution (Rosenmund and Stevens, 1996) and Pvr was calculated as the ratio of evoked EPSC charge and RRP charge. Expression of CplxI fully rescues the decreased Pvr of Cplx-TKO neurons (Figure 1F; (Xue et al., 2007)), whereas dmCplx reduces the Pvr by more than 98% compared to Cplx-TKO neurons (Figure 1F). The numbers of fusion competent vesicles in RRP are not significantly different among the three groups (Figure 1E, P = 0.8). Furthermore, expression of CplxI rescues the decreased miniature EPSC (mEPSC) frequency of Cplx-TKO neurons in contrast to dmCplx that suppresses the mEPSC frequency by more than 96% in Cplx-TKO neurons (Figures 1C and 1G). The inhibitory effect of dmCplx is not restricted to hippocampal glutamatergic autapses, as it also inhibits release in striatal GABAergic autaptic neurons (Figure S3) and in hippocampal mass-cultured neurons (Figure S4). In summary, these data show that CplxI facilitate both evoked and spontaneous release, and that dmCplx practically abolishes evoked and spontaneous release in mouse neurons without affecting the number of fusion competent vesicles. Thus, these results are in agreement with the proposed functions for murine and Drosophila Complexins based on genetic loss-of-function studies (Huntwork and Littleton, 2007; Xue et al., 2008b).
Overexpression of murine CplxI and Drosophila Cplx in WT mouse neurons
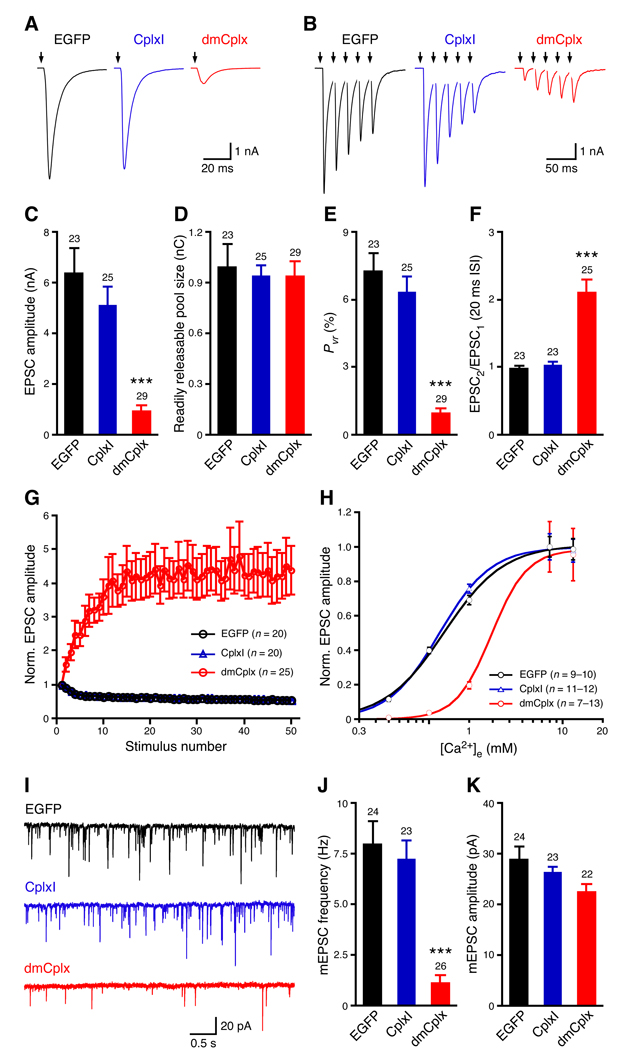
Expression of dmCplx in Cplx-TKO hippocampal neurons blocks both evoked and spontaneous release, whereas CplxI rescues the reduced evoked and spontaneous release of Cplx-TKO neurons (Figure 1). Previous studies showed that more than 10-fold overexpression of CplxI by Semliki Forest viruses in WT hippocampal neurons does not alter neurotransmitter release, indicating that CplxI does not act as a fusion clamp (Xue et al., 2007). If dmCplx functions as a fusion clamp, one would predict that overexpression of dmCplx in WT neurons should inhibit release. To test this hypothesis, we compared WT neurons overexpressing EGFP with those overexpressing dmCplx or CplxI together with EGFP (Figure 2). Consistent with previous results, lentiviral overexpression of CplxI in WT neurons does not significantly affect either evoked or spontaneous release (Figures 2A to 2K, P > 0.05 for all tested parameters). In contrast, overexpression of dmCplx at the similar levels as CplxI (Supplemental results and Figure S2) strongly suppresses EPSC amplitude (Figures 2A and 2C) without affecting the RRP size (Figure 2D). Consequently, the Pvr of evoked release is drastically reduced in WT neurons overexpressing dmCplx (Figure 2E). Consistent with the reduction in Pvr, overexpression of dmCplx alters the short-term synaptic plasticity of WT neurons, from mild depression to strong facilitation during repetitive high-frequency stimulations (Figures 2B, 2F, and 2G). These results indicate that WT neurons overexpressing dmCplx are less sensitive to Ca2+. To test this, we measured evoked EPSC amplitudes as a function of the external Ca2+ concentrations and fitted the data with a standard Hill equation to obtain the dissociation constant (Kd) as a measure of the apparent Ca2+-sensitivity of release. WT neurons overexpressing dmCplx show more than 2-fold increase in Kd compared to WT neurons, indicating that overexpression of dmCplx decreases the apparent Ca2+-sensitivity of release (Figure 2H). Finally, we examined the spontaneous release and found that overexpression of dmCplx markedly reduces the mEPSC frequency (Figures 2I and 2J), but barely affects mEPSC amplitude (Figure 2K). Overexpression of dmCplx does not completely suppress evoked and spontaneous release in WT neurons, presumably because the endogenous murine Complexins compete with dmCplx for the SNARE complex binding and still participate in release. Altogether, these data are consistent with the rescue experiments and show that dmCplx mainly acts as a fusion inhibitor.
Figure 2.
Effects of overexpression of CplxI and dmCplx in WT hippocampal neurons. WT neurons overexpressing CplxI or dmCplx together with EGFP were compared to WT neurons expressing EGFP alone (control neurons). (A and B) Representative traces of basal evoked EPSCs (A) and 5 EPSCs in response to a train of action potentials evoked at 50 Hz (B). The arrows represent stimulations; artifacts and action potentials are blanked. (C) Amplitude of EPSC evoked by single action potentials. (D) Readily releasable pool size determined by the transient synaptic charge induced by hypertonic sucrose solution. (E) Vesicular release probability (Pvr) of evoked release. (F) Paired-pulse ratio at 20 ms inter-stimulus interval (ISI). (G) The amplitudes of EPSCs evoked by a train of stimulations at 10 Hz are normalized to the first EPSC amplitude and plotted against stimulation number. (H) Apparent Ca2+ sensitivity of evoked release. Normalized EPSC amplitudes are plotted as a function of external Ca2+ concentrations ([Ca2+]e) and fitted with the standard Hill equation. EGFP, Kd = 1.30 ± 0.06 mM; CplxI, Kd = 1.20 ± 0.03 mM; dmCplx, Kd = 2.9 ± 0.5 mM. (I) Representative traces of mEPSCs. (J) mEPSC frequency. (K) mEPSC amplitude. Data are expressed as mean ± SEM. ***, P < 0.0001 compared to control neurons. The numbers of neurons analyzed are shown above the bars.
Multiple domains of Drosophila Cplx are required for its inhibitory function
To elucidate the molecular mechanisms underlying the apparently opposite roles of murine and Drosophila Complexins in synaptic exocytosis, we performed a structure-function analysis of Complexins by mutagenesis and chimeric analyses. We used Cplx-TKO neurons expressing EGFP as controls to measure the baseline release. We then rescued Cplx-TKO neurons with different Cplx variants (deletions, point mutations, or chimeric proteins) together with EGFP to assess whether a Cplx variant facilitates or inhibits neurotransmitter release. We used Pvr and mEPSC frequency as two main parameters to examine evoked and spontaneous release efficacy, respectively. Pvr and mEPSC frequency data were normalized to the mean values of the corresponding Cplx-TKO control neurons (red dashed lines and error bars in Figure 3) to allow for comparisons of different Cplx variants across different experiments. Data from all CplxI-or dmCplx-rescued Cplx-TKO neurons are pooled together and shown in Figure 3, as the normalized values do not significantly differ across experiments (P > 0.05). All of the Cplx variants studied in Figure 3 and Figure 4 are properly expressed in neurons and targeted to the synapses (Supplemental results and Figure S5).
Figure 3.
Multiple domains of Drosophila Cplx are required for its inhibitory function in neurotransmitter release. (A, D, and G) Schematic diagrams of domain organization of Cplx variants that correspond to the rescue experiments in (B, C, E, F, H, and I). The residue numbers shown in the domain schemes represent the amino acid numbers of the corresponding WT proteins (Figure S1). CplxI, dmCplx, and CplxIII are represented by black, red and blue, respectively. The grey boxes indicate the central α-helix, and the blue bars indicate the accessory α-helix of CplxI and its homologous sequence of dmCplx. (B, C, E, F, H, and I) Summary data for Cplx-TKO neurons and Cplx-TKO neurons rescued with different Cplx variants. Data are normalized to the mean values of the respective controls (red dashed lines and error bars). Bar graphs show normalized vesicular release probability (Pvr) of evoked release (B, E, and H) and mEPSC frequency (C, F, and I). Data are expressed as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.0001 compared to the corresponding controls (Cplx-TKO). The numbers of neurons analyzed are shown on the bars.
Figure 4.
The N termini of both murine CplxI and Drosophila Cplx facilitate neurotransmitter release. (A) Representative traces of evoked EPSCs. The arrows represent stimulations; artifacts and action potentials are blanked. (B) Representative traces of mEPSCs. (C to F) Summary data for Cplx-TKO neurons and Cplx-TKO neurons rescued with different Cplx variants. (C) Vesicular release probability (Pvr) of evoked release. (D) Paired-pulse ratio at 20 ms inter-stimulus interval (ISI). (E) EPSC amplitude potentiation by elevation of external Ca2+ concentrations. (F) mEPSC frequency. Data are expressed as mean ± SEM. *, P < 0.05; ***; P < 0.0001 compared to Cplx-TKO neurons. The numbers of neurons analyzed are shown above the bars.
The SNARE complex binding mediated by the central α-helix is essential for CplxI to facilitate transmitter release (Xue et al., 2007) and dmCplx has also been shown to bind to the SNARE complex (Schaub et al., 2006). Therefore, we tested whether the SNARE complex binding is also required for dmCplx function. We confirmed that dmCplx binds to the murine SNARE complex in a cosedimentation assay (Figure S6). We mutated two residues, lysine 75 and tyrosine 76 to alanines, whose corresponding residues in CplxI are required for SNARE complex binding (Bracher et al., 2002; Chen et al., 2002; Xue et al., 2007). This mutant dmCplx (K75A Y76A) shows severely diminished binding to the SNARE complex (Figure S6) and fails to block the evoked and spontaneous release in Cplx-TKO neurons (Figures 3A to 3C). These data provide strong evidence that the arrest of both evoked and spontaneous release by dmCplx depends on its specific binding to the SNARE complex. When we replaced the central α-helix of dmCplx with that of CplxI (denoted as dmCplx 1–53-CplxI 48–70-dmCplx 77–143), this substitution does not affect the inhibitory function of dmCplx (Figures 3A to 3C), indicating that SNARE complex binding plays very similar role in dmCplx and CplxI. We therefore examined the functional roles of the sequences that are C-and N-terminal to the central α-helix.
We first deleted the C terminus (residues 84–143) of dmCplx (denoted as dmCplx 1–83). This mutant does not inhibit release (Figures 3D to 3F). When the C terminus of dmCplx is replaced with that of CplxI (denoted as dmCplx 1–76-CplxI 71–134), this chimeric protein shows impaired ability to clamp evoked and spontaneous release, but still partially inhibit mEPSC frequency (Figures 3D to 3F). These results indicate that the C terminus of dmCplx is important for its inhibitory function, and that the C terminus of CplxI is only inhibitory with respect to spontaneous release, but not as strong as that of dmCplx.
The C terminus of dmCplx contains a farnesylation motif (the CAAX box) that is absent from CplxI, but present in murine CplxIII and CplxIV (Figure S1; (Reim et al., 2005)). Farnesylation of the latter two proteins mediates their membrane associations, which leads to the synaptic localization of murine CplxIII and CplxIV in a punctate pattern. A substitution of the cysteine residue in the CAAX box with a serine residue prevents farnesylation of CplxIII and CplxIV, and causes them to distribute diffusely in axonal processes and synapses, similar to CplxI (Reim et al., 2005). To test the role of farnesylation in dmCplx function, we mutated the cysteine residue (Cys140) in the CAAX box to a serine. Similar to dmCplx 1–83, dmCplx C140S loses its ability to inhibit release, indicating that farnesylation is important for the inhibitory role of dmCplx (Figures 3D to 3F). As expected, both dmCplx 1–83 and dmCplx C140S show a diffuse subcellular distribution in axonal processes and synapses (Supplemental results and Figure S5). However, farnesylation is not sufficient to cause inhibition of release by dmCplx because replacement of the dmCplx C terminus with the CplxIII C terminus containing a farnesylation motif (denoted as dmCplx 1–76-CplxIII 81–158) confers a punctate synaptic localization (Figure S5), but does not inhibit release. Instead, this chimeric protein facilitates evoked release (Figures 3D to 3F). Moreover, CplxIII and CplxIV facilitate release in Cplx-TKO neurons (Reim et al., 2005). Taken together, these results indicate that farnesylation is a prerequisite for the inhibitory function of dmCplx, but is not the cause of inhibition. The C terminus of dmCplx is critical for its inhibitory function, which requires both the farnesylation motif and other sequences within residues 84–143.
To assess the role of the sequence that is N-terminal to the central α-helix, we deleted residues 1–53 that contain the N terminus and the accessory α-helix of dmCplx. This mutant (denoted as dmCplx 54–143) can only partially inhibits evoked release, but not spontaneous release (Figures 3G to 3I). When residues 1–53 of dmCplx are replaced with their homologous sequence from CplxI, this chimeric protein (denoted as CplxI 1–47-dmCplx 54–143) partially inhibits evoked and spontaneous release (Figures 3G to 3I). When we removed only the N terminus (residues 1–16) of dmCplx (denoted as dmCplx 17–143), or replaced it with its homologous sequence from CplxI (denoted as CplxI 1–15-dmCplx 17–143), these two mutants are still able to abolish both evoked and spontaneous release, similar to WT dmCplx (Figures 3G to 3I, P > 0.05 compared to dmCplx WT). Altogether, these data indicate that the N terminus (residues 1–16) of dmCplx is not required for its clamping function, whereas the sequence within residues 17–53 including the accessory α-helix is critical for the inhibitory function of dmCplx. It is noteworthy that CplxI 1–47-dmCplx 54–143 inhibits release more strongly than the deletion mutant dmCplx 54–143 (Figures 3H and 3I, P < 0.01). Since the N termini of both CplxI and dmCplx do not inhibit release, this result is consistent with the notion that the accessory -helix (residues 29–47) between the N terminus and the central α-helix of CplxI inhibits release (Xue et al., 2007). Furthermore, CplxI 1–47-dmCplx 54–143 inhibits release less strongly than WT dmCplx (Figures 3H and 3I, P < 0.01), indicating that the accessory α-helix of dmCplx is more inhibitory than that of CplxI.
In summary, our mutagenesis and chimeric analyses indicate that the inhibitory function of dmCplx in Cplx-TKO neurons depends on its binding to the SNARE complex, and requires the C terminus and the accessory α-helix, but not the N terminus. Note that the effects of some Cplx variants on evoked release and spontaneous release are quantitatively different, but the changes are qualitatively similar and are in the same direction compared to the controls (Figure 3).
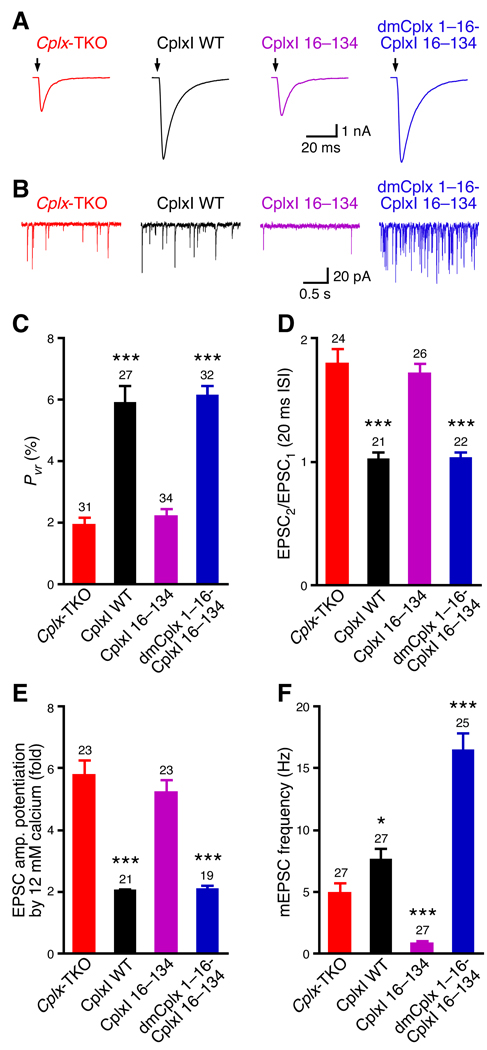
The N terminus of Drosophila Cplx facilitates neurotransmitter release
We previously showed that the N terminus of CplxI are essential to facilitate Ca2+-triggered neurotransmitter release (Xue et al., 2007). The sequence homology between dmCplx and CplxI is relatively high in this region (Figure S1), suggesting that their functions may be similar. We suspected that the N terminus of dmCplx may also facilitate release, but when present in the context of the full-length dmCplx, the facilitatory function of this region is masked by the overwhelming inhibitory function of the rest of the protein. To test this hypothesis, we investigated whether the N terminus of dmCplx can functionally substitute that of CpxI in the rescue experiments with Cplx-TKO neurons. Consistent with previous findings (Xue et al., 2007), deletion of the N terminus (residues 1–15) of CplxI (denoted as CplxI 16–134) results in a strong reduction in evoked release efficacy compared to WT CplxI (Figure 4). Cplx-TKO neurons rescued with CplxI 16–134 show similar Pvr of evoked release (Figures 4A and 4C) and paired-pulse ratio (Figure 4D) to Cplx-TKO neurons (P > 0.3 for both parameters). The apparent Ca2+-sensitivity of release in Cplx-KOs neurons is reduced about 2-fold compared to WT neurons (Reim et al., 2001; Xue et al., 2008b). Due to this right-shift of the Ca2+-dose response curve, when the external standard Ca2+ concentrations (2 mM Ca2+, 4 mM Mg2+) is changed to high Ca2+ concentrations (12 mM Ca2+, 1 mM Mg2+), evoked EPSC amplitude show greater potentiation in Cplx-KOs neurons than in WT neurons (Xue et al., 2007). We therefore measured EPSC amplitude potentiation upon elevation of external Ca2+ concentrations as an indication of apparent Ca2+-sensitivity of release. Cplx-TKO neurons rescued with CplxI 16–134 show larger potentiation than WT CplxI rescue, indicating a lower Ca2+-sensitivity of release (Figure 4E).
To test whether the N terminus of dmCplx can functionally substitute the CpxI N terminus, we generated a chimeric protein dmCplx 1–16-CplxI 16–134, in which the N terminus of CplxI is substituted by the homologous sequence from dmCplx. Remarkably, dmCplx 1–16-CplxI 16–134 behaves like WT CplxI in evoked release, as it completely restores the defects of Cplx-TKO in Pvr, paired-pulse ratio, and Ca2+-sensitivity (Figures 4A, 4C, 4D, and 4E, P > 0.6 for all parameters compared to WT CplxI), indicating that the N termini of both CplxI and dmCplx promote evoked release.
We further examined the effect of the N terminus on spontaneous release. Loss of the first 15 residues in CplxI (CplxI 16–134) strongly suppresses mEPSC frequency to about 17% of Cplx-TKO neurons (Figures 4B and 4F). Together with the finding that CplxI 1–47-dmCplx 54–143 inhibits release more strongly than dmCplx 54–143 (Figures 3H and 3I), this result indicates that the N terminus of CplxI promotes, whereas the accessory α-helix inhibits spontaneous release. This result also implies that the reduced spontaneous release in Cplx-TKO neurons results from the compound effect of losing both facilitatory (the N terminus) and inhibitory (the accessory α-helix) mechanisms associated with distinct domains of Complexins. CplxI 16–134 lacks the facilitatory N terminus but contains the inhibitory accessory α-helix; hence, the reduction of spontaneous release in CplxI 16–134-rescued Cplx-TKO neuron is stronger than in Cplx-TKO neurons.
We then rescued Cplx-TKO neurons with dmCplx 1–16-CplxI 16–134. This chimeric protein not only rescues the reduced mEPSC frequency of Cplx-TKO neurons, but also enhances spontaneous release compared to WT CplxI (Figures 4B and 4F, P < 0.001). Collectively, these data indicate that despite the overall dominant inhibitory function of dmCplx, the N terminus of dmCplx facilitates both evoked and spontaneous release, similar to or better than that of CplxI. The facilitatory function of the dmCplx N terminus is masked by the inhibitory function of other domains in full-length dmCplx, resulting a seeming lack of facilitatory effect of dmCplx in Cplx-TKO neurons (Figures 1, S3 and S4).
Rescue of Drosophila Cplx null mutants with murine and Drosophila Cplx
The rescue experiments in Cplx-TKO mouse neurons show that both murine and Drosophila Complexins facilitate and inhibit neurotransmitter release via distinct domains, but with varying strengths (Figure 3 and Figure 4). To compare murine and Drosophila Complexins in fly synapses, we generated transgenic flies carrying UAS-dmCplx, UAS-CplxI or UAS-CplxIII, and used the GAL4-UAS system (Brand and Perrimon, 1993) to express them in flies (Figure S7). We chose CplxI and CplxIII because they are the representative paralogs of two subfamilies of murine Complexins (Reim et al., 2005). The Drosophila Cplx null mutants (dmCplxSH1) are semilethal (Huntwork and Littleton, 2007), and the escaper adults are sterile and usually die within 2 weeks after eclosion (Figure S8). Pan-neuronal expression of UAS-dmCplx, UAS-CplxI or UAS-CplxIII with the neuronal driver C155-GAL4 (Lin and Goodman, 1994) rescues the lethality and sterility of dmCplx null mutants (Figure S8 and data not shown), indicating that murine and Drosophila Complexins can substitute each other in vivo. Immunostainings show that CplxI and CplxIII are properly targeted to the presynaptic boutons of third instar larval neuromuscular junctions (Figure S9).
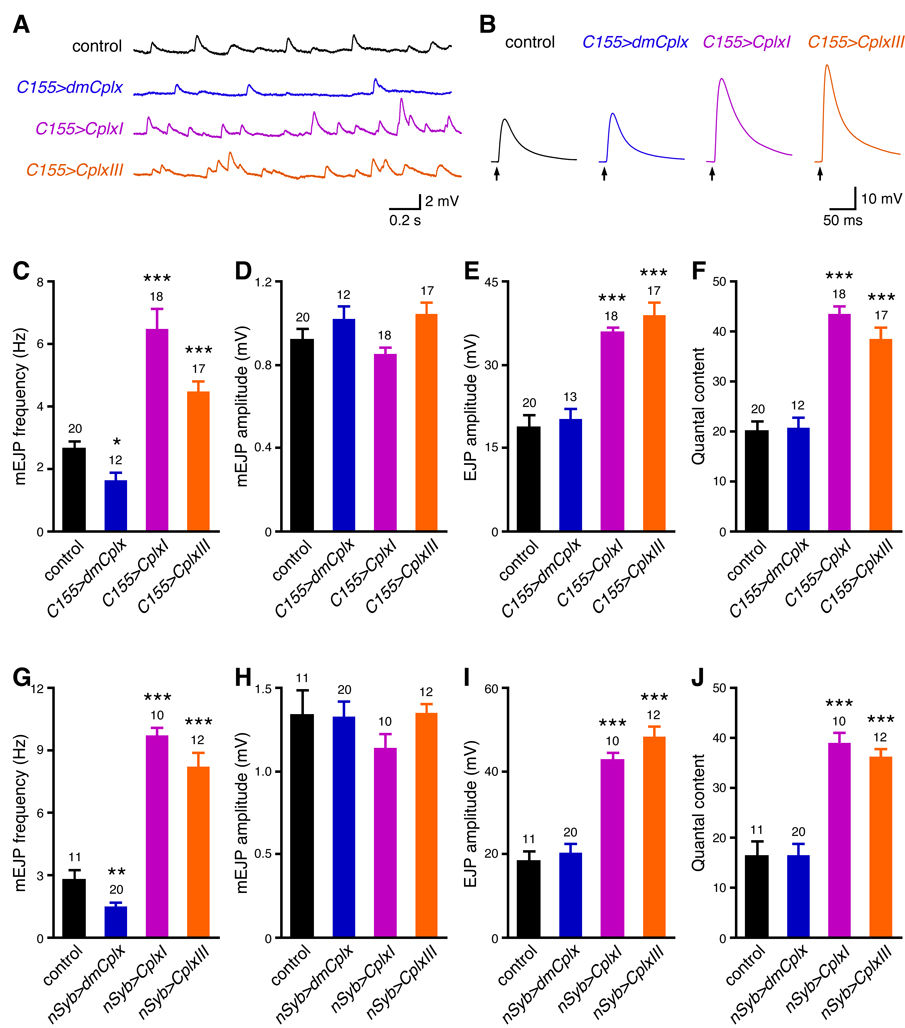
We recorded evoked and miniature excitatory junction potentials (EJPs) of neuromuscular junctions to define the properties of evoked and spontaneous neurotransmitter release in these animals, respectively (Figure 5). Muscle resting membrane potentials and input resistances are not significantly different among all groups (Table S1A, P > 0.05 for both parameters). dmCplx null mutants show reduced evoked release and very high levels of spontaneous release (Huntwork and Littleton, 2007), both of which are completely rescued by neuronal expression of dmCplx (Figures 5A to 5D). Remarkably, neuronal expression of CplxI or CplxIII not only rescues the decreased evoked release in null mutants, but the evoked EJPs are significantly larger than that of control flies or dmCplx-rescued null mutants (Figures 5A and 5C; P < 0.001 for both CplxI rescue and CplxIII rescue compared to control or dmCplx rescue). In contrast, dmCplx null mutants rescued by CplxI or CplxIII still show increased mEJP frequency compared to control flies or dmCplx-rescued null mutants (Figures 5B and 5D; P < 0.001 for both CplxI rescue and CplxIII rescue compared to control or dmCplx rescue). However, both CplxI and CplxIII are able to partially suppress the increased mEJP frequency in dmCplx null mutants (Figure 5D; P < 0.001 for both CplxI rescue and CplxIII rescue compared to null). In fact, this partial suppression by CplxI or CplxIII is underestimated because the quantification of mEJP frequency in null mutants is an underestimate due to the numerous overlaps of mEJPs (Supplemental results and Figure S10). These data show that at fly neuromuscular junctions, murine Cplx and dmCplx play both facilitatory and inhibitory roles in release, but murine Cplx facilitates Ca2+-triggered release more effectively than dmCplx, whereas dmCplx inhibits spontaneous vesicle fusion much more efficiently than murine Cplx.
Figure 5.
Rescue of Drosophila Complexin null mutants with murine Cplx and dmCplx. (A) Representative traces of evoked EJPs. The arrows represent stimulations. (B) Representative traces of mEJPs. (C and D) Summary data for amplitude of EJP evoked by single action potentials (C) and mEJP frequency (D). #, note that the mEJP frequency of null is underestimated due to the overlap of individual mEJPs. Simulation indicates that the true frequency is probably higher than 50 Hz (Supplemental results and Figure S10). Genotypes: control (C155-GAL4/Y), null (C155-GAL4/Y; dmCplxSH1/dmCplxSH1), dmCplx rescue (C155-GAL4/Y; UAS-dmCplx/+; dmCplxSH1/dmCplxSH1), CplxI rescue (C155-GAL4/Y; UAS-CplxI/+; dmCplxSH1/dmCplxSH1), and CplxIII rescue (C155-GAL4/Y; UAS-CplxIII/+; dmCplxSH1/dmCplxSH1). For transgenic rescue experiments, multiple lines show similar results and data are pooled together. Data are expressed as mean ± SEM. **, P < 0.001; ***, P < 0.0001 compared to control. The numbers of neuromuscular junctions analyzed are shown above the bars.
Overexpression of murine and Drosophila Cplx in WT flies
We examined the effects of overexpression of murine and Drosophila Complexins on neurotransmitter release at WT third instar larval neuromuscular junctions. We used two different neuronal driver lines, C155-GAL4 and nSyb-GAL4 (see Experimental Procedures), and the results are similar (Figure 6). Muscle resting membrane potentials and input resistances are not significantly different among all groups (Tables S1B and S1C, P > 0.05 for both parameters). Compared to control, overexpression of dmCplx decreases mEJP frequency (Figures 6A, 6C, and 6G) without affecting mEJP amplitude (Figures 6A, 6D, and 6H), but has no effect on evoked release (Figures 6B, 6E, 6F, 6I, and 6J). Together with the loss-of-function phenotypes, these results indicate that dmCplx has stronger effects on spontaneous release than evoked release at larval neuromuscular junctions. In contrast, overexpression of CplxI or CplxIII markedly increases mEJP frequency (Figures 6A, 6C, and 6G), evoked EJP amplitude (Figures 6B, 6E, and 6I), and the quantal content of evoked release (Figures 6F and 6J). It is likely that CplxI and CplxIII compete with the endogenous dmCplx in WT flies for binding to the SNARE complex, because both murine and Drosophila Complexins bind to the SNARE complex (McMahon et al., 1995; Schaub et al., 2006). This competition leads to a partial replacement of dmCplx by CplxI or CplxIII, thus causing an enhancement of evoked release and a partial loss of inhibition by dmCplx on spontaneous release. Taken together, these data are in line with the rescue experiment results (Figure 5) and support the notion that both murine and Drosophila Complexins facilitate and inhibit release, but with varying strengths.
Figure 6.
Effects of overexpression of dmCplx, CplxI and CplxIII in WT flies. (A to F) C155-GAL4-driven overexpression of dmCplx, CplxI and CplxIII. (A) Representative traces of mEJPs. (B) Representative traces of evoked EJPs. The arrows represent stimulations. (C to F) Summary data of mEJP frequency (C), mEJP amplitude (D), amplitude of EJP evoked by single action potentials (E), and the quantal content of evoked release (F). Genotypes: control (C155-GAL4/Y), C155>dmCplx (C155-GAL4/Y; UAS-dmCplx/+), C155>CplxI (C155-GAL4/Y; UAS-CplxI/+), and C155>CplxIII (C155-GAL4/Y; UAS-CplxIII/+). (G to J) nSyb-GAL4-driven overexpression of dmCplx, CplxI and CplxIII. Bar graphs show the summary data of mEJP frequency (G), mEJP amplitude (H), amplitude of EJP evoked by single action potential (I), and the quantal content of evoked release (J). Genotypes: control (yw/Y; nSyb-GAL4/+), nSyb>dmCplx (yw/Y; UAS-dmCplx/+; nSyb-GAL4/+), nSyb>CplxI (yw/Y; UAS-CplxI/+; nSyb-GAL4/+), and nSyb>CplxIII (yw/Y; UAS-CplxIII/+; nSyb-GAL4/+). Multiple transgenic lines show similar results and data are pooled together. Data are expressed as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.0001 compared to control. The numbers of neuromuscular junctions analyzed are shown above the bars.
Discussion
Synaptic exocytosis is exquisitely controlled by a set of facilitatory and inhibitory mechanisms, some of which are often executed by the very same protein (Rizo and Rosenmund, 2008). As a key regulator of the release machinery, Complexins play both facilitatory and inhibitory roles in vesicle fusion through distinct mechanisms (Xue et al., 2007; Yoon et al., 2008). However, the remarkable phenotypic difference between mouse and fly Complexin null animals remained unexplained. In the present work, we compared the functions of murine and Drosophila Complexins in cross-species rescue experiments. Our data establish that murine and Drosophila Complexins share a set of conserved mechanisms in synaptic vesicle fusion.
First, the SNARE complex binding mediated by the central α-helix (residues 48–70 for CplxI and 54–76 for dmCplx) is essential for Complexin function. Mutations that diminish the interaction between the central α-helix and the SNARE complex abolish the functions of both Cplx I (Xue et al., 2007) and dmCplx (Figures 3B, 3C and S6), indicating that the actions of other domains all depend on this high-affinity interaction. The binding of the central α-helix not only can stabilize the assembled SNARE complex (Chen et al., 2002), but perhaps more importantly, can strategically position the accessory α-helix and the N terminus for their actions (Xue et al., 2007).
Second, the accessory α-helix (approximately residues 29–47 for CplxI and 33–53 for dmCplx) located between the N terminus and the central α-helix inhibits vesicle fusion (Figures 3H and 3I). It was proposed that the inhibitory action of the accessory α-helix might arise from its interference with the binding of Synaptobrevin-2 to Syntaxin-1 and SNAP-25 heterodimer and consequently preventing the completely zippering of the SNARE complex (Xue et al., 2007). This model has recently been supported by the findings that Complexins can bind to Syntaxin-1 and SNAP-25 heterodimer in vitro (Guan et al., 2008; Weninger et al., 2008; Yoon et al., 2008) and may form an alternative four-helix bundle with target-SNAREs to inhibit fusion in a reconstituted fusion system (Giraudo et al., 2009).
Third, the N termini (residues 1–16) of both CplxI and dmCplx promote release (Figure 4). It has been speculated that the CplxI N terminus may interact with lipid membranes (Maximov et al., 2009; Xue et al., 2007), but so far, there are no supporting biochemical data. Instead, this facilitatory effect is likely mediated by a direct interaction of the Complexin N terminus with the SNARE complex C terminus. Mutations of methionine 5 and lysine 6 of CplxI disrupt the binding of the CplxI N terminus to the SNARE complex C terminus and abolish the facilitatory activity of the N terminus (M. Xue, T.K. Craig, J. Xu, H.T. Chao, J. Rizo, and C. Rosenmund, unpublished results). Interestingly, methionine 5 is not conserved in dmCplx and an alanine residue is at position 6 (corresponding to residue 5 of CplxI). It is possible that a methionine is not absolutely required for dmCplx and other residues may compensate for the interaction with the SNARE complex C terminus.
Furthermore, at fly neuromuscular junctions, both murine and Drosophila Complexins promote Ca2+-triggered release and suppress spontaneous release, but to very different degrees (Figure 5 and Figure 6). Neuronal expression of murine or Drosophila Complexins rescues the lethality and sterility of Complexin null mutant flies, showing again that murine and Drosophila Complexins share conserved functions (Figure S8).
Therefore, our cross-species rescue experiments show that murine and Drosophila Complexins have both facilitatory and inhibitory functions associated with similar protein domains in synaptic vesicle exocytosis. We propose that the Complexin central α-helix binds to the middle portion of the SNARE complex, stabilizing the SNARE complex and positioning the accessory α-helix and the N terminus (Chen et al., 2002; Xue et al., 2007). The accessory α-helix replaces the C terminus of the Synaptobrevin-2 SNARE motif in the four-helix bundle, preventing the full assembly of the SNARE complex to suppress fusion (Giraudo et al., 2009; Xue et al., 2007). The N terminus directly interacts with the C-terminal portion of the SNARE complex, likely stabilizing this unstable region of the SNARE complex to promotes membrane fusion. However, the relative strengths of these functions are remarkably different between murine and Drosophila Complexins. We propose that the integration of facilitation and inhibition that are associated with distinct domains determines the overall effect of murine and Drosophila Complexins on neurotransmitter release in a given synapse (Figure 7). The overall action of murine and Drosophila Complexins is unlikely to be a linearly additive effect of all facilitatory and inhibitory actions. However, it is clear that the facilitatory function is preponderant in murine Complexins, whereas the inhibitory functions of the accessory α-helix and the C terminus predominate in Drosophila Complexin. Thus, a relative shift in the balance of facilitatory and inhibitory functions results in differential roles of murine and Drosophila Complexins in neurotransmitter release and leads to apparently very different loss-of-function phenotypes in flies and mice. Our results emphasize the functional similarities and differences between murine and Drosophila Complexins, and reconcile previous contradictory hypotheses of Complexin in vivo function (Cai et al., 2008; Huntwork and Littleton, 2007; Reim et al., 2001; Xue et al., 2008b). Moreover, our data illustrate the complexity of Complexin function and strongly support the notion that Complexins play dual roles in vesicle fusion (Xue et al., 2007).
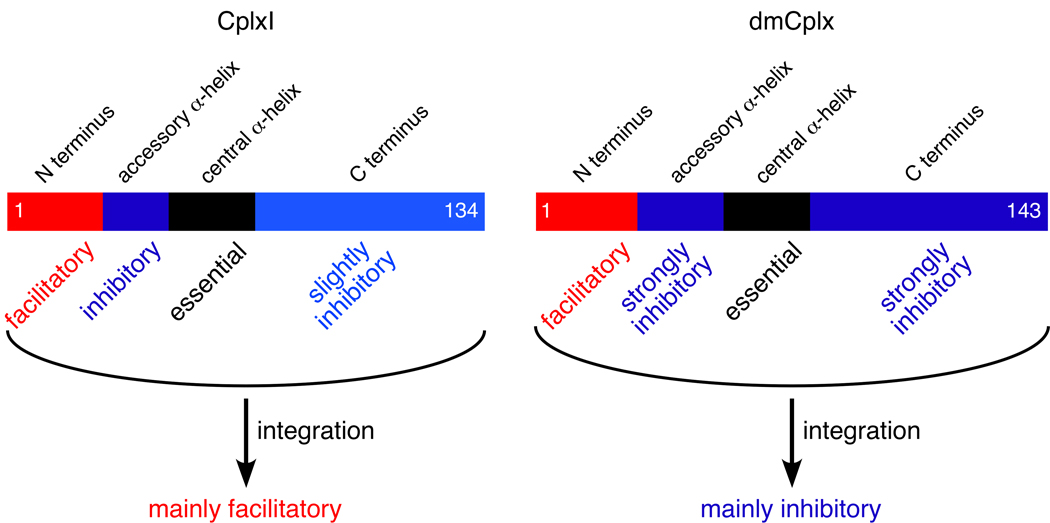
Figure 7.
Distinct functional domains of CplxI and dmCplx. Domains are indicated above the schematic diagrams and the corresponding functions are indicated below for CplxI and dmCplx. The integration of facilitatory and inhibitory functions associated with distinct domains leads to apparently differential effects on neurotransmitter release by CplxI and dmCplx.
Our model is clearly different from the previous models of Complexins based on the fusion clamp hypothesis (Giraudo et al., 2006; Maximov et al., 2009; Schaub et al., 2006; Tang et al., 2006). These models propose that Complexins arrest primed synaptic vesicles at a hemifused and metastable state, which provides the substrate for Ca2+-bound Synaptotagmin-1 to release the clamping function of Complexins, allowing the fast and synchronous fusion. The lack of Complexins and thereby the lack of metastable vesicles for Synaptotagmin-1 action cause excessive spontaneous release and deficient Ca2+-triggered fast release. However, our in vivo results speak against this model because murine Complexins do not completely clamp the excessive spontaneous release in Drosophila Complexin null mutants, yet they actually enhance Ca2+-evoked fast release even better than Drosophila Complexin (Figure 5). This observation indicates that the decreased Ca2+-evoked fast release in Drosophila Complexin null mutants is not functionally coupled to the increased spontaneous release frequency. Could it be that the reduced evoked release in Drosophila Complexin null mutants is due to a partial depletion of readily releasable vesicles by the high frequency spontaneous release? This is unlikely because the vesicle recruitment rate is usually at least 100-fold higher than the spontaneous release rate at resting intracellular Ca2+ level (Neher and Sakaba, 2008) and therefore a 20–30-fold increase in spontaneous release rate should not significantly change the vesicle pool size in Drosophila Complexin null mutants. In addition, murine Complexins-rescued Drosophila Complexin null synapses still exhibit strongly increased spontaneous release, yet the evoked release is even larger than WT synapses, arguing that high frequency spontaneous release in null mutants is unlikely to exhaust vesicles, causing a decreased evoked release.
A recent fusion clamp model proposes that Complexins control the force transfer from the SNARE complex to the membranes and assist the SNAREs in exerting force on the membranes (Maximov et al., 2009). This model assumes that Complexins are released from the SNARE complex by Synaptotagmin-1 and Ca2+, but it is physically unclear how Complexins can help SNAREs exert force on the membranes if they are dissociated upon Ca2+ influx. In contrast, our model requires Complexins to remain bound to the SNARE complex upon Ca2+ influx and is consistent with the notion that Complexins could function independently from Synaptotagmin-1 (Xue et al., 2007; Yoon et al., 2008).
Drosophila Complexin in Cplx-TKO neurons abolishes both evoked and spontaneous release without altering the number of fusion competent vesicles measured by hypertonic sucrose solution (Figure 1). This effect is intriguing, as very few molecular manipulations specifically block the synaptic vesicle cycle at the final fusion step. Drosophila Complexin does not change the number of primed vesicles, indicating that the initial formation of the SNARE complex is not affected by Drosophila Complexin. The inhibitory effect of Drosophila Complexin requires its binding to the SNARE complex (Figures 3B, 3C, and S6). Hence, we hypothesize that when the Drosophila Complexin central α-helix binds to the partially assembled SNARE complex, the accessory α-helix together with the C terminus prevents the further assembly of the SNARE complex C terminus, thereby arresting vesicles at the primed state. It is currently unknown how mechanistically the C terminus of Drosophila Complexin inhibits release. One possibility is that the C terminus may fold back toward the N-terminal direction and cooperate with the accessory α-helix to inhibit vesicle fusion.
The phenotypic differences between fly and mouse knockouts seem dramatic, but it is worth noting that an increase of just 1.4 kcal/mol in the strength of a protein-protein interaction, which can arise simply from the formation of one hydrogen bond or salt bridge, leads to a 10-fold increase in affinity according to the Boltzmann equation. Hence, subtle changes in the molecular interactions of murine and Drosophila Complexins can suffice to tip the balance between facilitatory and inhibitory strengths. For example, protein sequence alignments show that the lengths and the amino acid compositions of the accessory α-helices differ among different Complexins (Figure S1; (Huntwork and Littleton, 2007; Reim et al., 2005)), which may cause different interactions of the accessory α-helix with Syntaxin-1 and SNAP-25 heterodimer, thus changing its inhibitory strength.
The effects of murine Complexins in murine and fly synapses are not identical, as murine Complexins promote evoked release and inhibit spontaneous release in fly neuromuscular junctions (Figure 5), and promotes both types of release in mouse central synapses (Figure 1; (Strenzke et al., 2009; Xue et al., 2008b)). Likewise, the effects of Drosophila Complexin in murine and fly synapses are not identical either, as it strongly inhibits spontaneous release and mildly promotes evoked release in fly neuromuscular junctions (Figure 5), and strongly inhibits both types of release in mouse synapses (Figures 1, S3, and S4). These observations indicate that in addition to the Complexin intrinsic properties, the molecular differences between species or synapses could differentially affect the facilitatory and inhibitory functions of murine and Drosophila Complexins, thereby tilting the facilitation and inhibition balance and contributing to the phenotypic differences.
Complexins represent a family of proteins that maintain a highly conserved core of sequences and at the same time display great diversity across paralogs and orthologs (Huntwork and Littleton, 2007; Reim et al., 2005). This is likely reflected on their functions, namely conserved facilitatory and inhibitory mechanisms with varying strengths in neurotransmitter release. It will be interesting to test Complexin function in some other model organisms along the phylogenetic tree, such as worm and fish, to determine if and how the balance between facilitatory and inhibitory functions of Complexins have changed during evolution. At different synapses, the strengths of facilitation and inhibition of Complexins may be differentially regulated in a paralog- and ortholog-dependent fashion, thereby regulating release in a synapse-specific manner, and contributing to synaptic diversity and specificity. Furthermore, the ability of Drosophila Complexin to inhibit neurotransmitter release in mammalian neurons potentially provides a powerful tool to manipulate synaptic function to study neural circuits, as one should be able to express Drosophila Complexin to inhibit or even abolish synaptic transmission in a spatially and temporally specific manner.
Experimental Procedures
Lentivirus constructs and production
A modified lentiviral vector (Lois et al., 2002; Xue et al., 2008a) was used, in which a human Synapsin-1 promoter and a ubiquitin C promoter drive the expression of Cplx variants and the reporter, enhanced green fluorescent protein (EGFP), respectively. The same vector without Cplx served as a control construct. For EGFP-tagged fusion proteins, the ubiquitin C promoter and EGFP reporter were removed from the vector. Wildtype (WT) rat CplxI (GenBank accession number: NM_022864), mouse CplxIII (AY264290), and Drosophila Cplx (AY121629) cDNAs were used to generate all Cplx variants by standard recombinant DNA techniques and the cDNAs were subsequently cloned into the lentiviral vector. The resulting constructs were used for both lentiviruses production and transfection of hippocampal neurons. Myc-tagged Cplx variants were generated by fusing a linker (SGGSGGTGG) followed by a c-Myc (EQKLISEEDL) to the C terminus of Cplx variants. EGFP-tagged Cplx variants were generated by fusing EGFP to the N terminus of Cplx variants.
Lentiviruses were produced by co-transfecting HEK 293T cells with the lentiviral vector and two helper vectors, pVSVg and pCMV-delta R8.9 (Lois et al., 2002). Viral supernatants were collected 48–72 hours after transfection and virus particles were concentrated using a centrifugal filter device (Amicon Ultra-15, Millipore). Viruses were titered with wildtype hippocampal mass-cultured neurons. For all the lentiviral expression experiments with cultured neurons, about 2.3 × 106 infectious virus units were used to infect neurons in a 35 mm-diameter well containing 2 ml culture medium within 24 hours after plating neurons.
Mice, neuronal cultures, and transfection
CplxI/II/III-TKO mice were obtained by interbreeding of mice homozygous for the CplxII and CplxIII mutations, and heterozygous for the CplxI mutation as described (Xue et al., 2008b). All procedures to maintain and use these mice were approved by the Institutional Animal Care and Use Committee for Baylor College of Medicine and Affiliates.
Primary neuronal cultures were prepared as described (Xue et al., 2008b). Briefly, hippocampal or striatal neurons were prepared from postnatal day 0 mice and plated at 300 cm−2 density on WT astrocyte microislands for autaptic neuron electrophysiology. Hippocampal neurons were plated at 4,000 cm−2 density on continental WT astrocyte feeder layer for mass-cultured neuron electrophysiology. For Western blotting and immunocytochemistry of protein expression, hippocampal neurons were plated at 10,000 cm−2 and 5,000 cm−2 densities on continental WT astrocyte feeder layer, respectively.
Hippocampal neurons were transfected using the calcium phosphate method as described (Xia et al., 1996) with modification. Briefly, at day in vitro (DIV) 3 or 4, the conditioned neuronal culture medium was removed and saved. Neurons were washed two times with Neurobasal-A medium and incubated in Neurobasal-A medium at 5% CO2 incubator for at least 1 hour. 2 µg DNA was used to prepare calcium phosphate/DNA precipitates for each well (12-well plate). Neurons were incubated with calcium phosphate/DNA precipitates for 10–12 minutes and subsequently washed two times with astrocyte culture medium (DMEM medium supplemented with 10% fetal calf serum). Neurons were then incubated in astrocyte culture medium for 1–2 hours before they were moved back to the conditioned neuronal culture medium.
Drosophila transgenesis and strains
WT rat CplxI, mouse CplxIII, and Drosophila Cplx cDNAs were cloned into the pUAST vector (Brand and Perrimon, 1993) and injected into the yw; Ki, P{Δ2–3}/+ embryos by standard procedures (Rubin and Spradling, 1982) to obtain transgenic lines. C155-GAL4 (Lin and Goodman, 1994) was obtained from the Bloomington Stock Center, nSyb-GAL4 (neuronal Synaptobrevin-GAL4) from Barry Dickson (unpublished, Research Institute of Molecular Pathology, Austria), and dmCplx null mutant (dmCplxSH1, (Huntwork and Littleton, 2007)) from J. Troy Littleton (Massachusetts Institute of Technology, USA). Flies were maintained at 25° C for all experiments. For lifespan tests, adult flies were transferred to fresh food vials every 2 days, and were counted everyday.
Western blotting, immunocytochemistry, and cosedimentation assays
To examine Cplx expressions, proteins were extracted from cultured neurons or fly heads and analyzed by Western blotting. Presynaptic localization of Cplx variants was examined using immunocytochemistry and confocal microscopy. Cosedimentation assays were performed to examine the SNARE complex binding of dmCplx using recombinant glutathione S-transferase (GST)-dmCplx fusion proteins that were generated in HEK293FT cells. These procedures are described in detail in Supplemental Experimental Procedures.
Electrophysiology of cultured neurons
Whole-cell voltage clamp experiments were performed on approximately equal numbers of neurons from every group in parallel on the same day in vitro (9–14 DIV) at room temperature (23–24°C). Neurons were clamped at −70 mV with an Axopatch 200B amplifier (Molecular Devices) under the control of Clampex 10.0 (Molecular Devices). Data were acquired at 10 kHz and low-pass filtered at 5 kHz. The series resistance was compensated about 80% and only cells with series resistances below 10 MΩ were analyzed. The standard extracellular solution contained (mM): NaCl, 140; KCl, 2.4; HEPES, 10; glucose, 10; CaCl2, 2; MgCl2, 4; 300 mOsm; pH 7.4. For Ca2+-sensitivity experiments, the Mg2+ concentration was kept at 1 mM and Ca2+ concentration varied as indicated in extracellular solutions. Hypertonic solution for measuring readily releasable pool (RRP) size was made by adding 500 mM sucrose to the standard extracellular solution. For hippocampal glutamatergic autaptic neurons, the patch pipette solution contained (mM): K-Gluconate, 146; HEPES, 17.8; EGTA, 1; MgCl2, 0.6; ATP-Mg, 4; GTP-Na, 0.3; Phosphocreatine, 12; Phosphocreatine kinase, 50U ml−1; 300 mOsm; pH 7.4. For striatal GABAergic autaptic neurons and hippocampal mass-cultured neurons, the patch pipette solution contained (mM): KCl, 136; HEPES, 17.8; EGTA, 1; MgCl2, 0.6; ATP-Mg, 4; GTP-Na, 0.3; Phosphocreatine, 12; Phosphocreatine kinase, 50U ml−1; 300 mOsm; pH 7.4.
For hippocampal and striatal autaptic neurons, action potential-evoked EPSCs or IPSCs were triggered by a 2 ms somatic depolarization to 0 mV. Neurons were stimulated at 0.2 Hz (for EPSCs) or 0.1 Hz (for IPSCs) in standard extracellular solution to measure basal evoked synaptic responses. RRP size was determined by measuring the charge transfer of the transient synaptic current induced by a pulsed 8 s-long application of hypertonic sucrose solution directly onto the neuron. RRP vesicle numbers in Figure 1 were calculated by the ratio of RRP charges and miniature EPSC (mEPSC) charges. To obtain vesicular release probability (Pvr), the evoked response and the response to the hypertonic sucrose solution were recorded successively from the same neuron. Evoked response was integrated for 1 s (for EPSC) or 2 s (for IPSC) to calculate the charge transfer. Pvr was calculated by the ratio of evoked response charge and RRP size. Short-term plasticity was examined by evoking synaptic responses at 50 or 10 Hz in standard external solution. Paired-pulse ratio was measured by dividing the second EPSC amplitude with the first EPSC amplitude. To examine the apparent Ca2+-sensitivity of release, EPSCs were evoked at 0.2 Hz. Each test measurement at different external Ca2+ concentrations was preceded and followed by a measurement in standard extracellular solution to control for the rundown of synaptic responses. The EPSC amplitude at each Ca2+ concentration was normalized to the amplitude in standard extracellular solution. All data were then normalized to the maximal value. Data were fitted with standard Hill equation: Y = M/(1 + (Kd / X)n); Y, EPSC amplitudes; X, Ca2+ concentrations; M, maximum EPSC amplitude; Kd, dissociation constant; n, Hill coefficient. mEPSCs and mIPSCs were recorded for 1–2 minutes and were blocked by a glutamate receptor antagonist, kynurenic acid (3 mM) or a GABAA receptor antagonist, gabazine (15 µM), respectively. Recordings in the presence of antagonists were used to subtract the false positive events.
For hippocampal mass-cultured neurons, mEPSCs or mIPSCs were recorded from randomly chosen neurons in standard external solution with 0.5 µM tetrodotoxin and 15 µM gabazine or 3 µM NBQX (2,3-Dioxo-6-nitro-1,2,3,4-tetrahydrobenzo[f]quinoxaline-7-sulfonamide), respectively. Recordings in the presence of antagonists were used to subtract the false positive events.
Data were analyzed offline using AxoGraph X (AxoGraph Scientific). To detect mEPSC and mIPSC events, traces were digitally filtered at 1 kHz offline and events were automatically selected with a scaled-template algorithm (Clements and Bekkers, 1997) in AxoGraph X. The template function is a double exponential with a scalable amplitude, a rise time constant of 0.5 ms, a decay time constant of 4 ms (for mEPSC) or 18 ms (for mIPSC), a baseline of 5 ms, and a template length of 10 ms (for mEPSC) or 18 ms (for mIPSC). False positive events were subtracted as described (Xue et al., 2008b). False positive subtraction was not possible to be performed for three dmCplx-rescued Cplx-TKO neurons in Figure 1, and therefore the non-subtracted values were used. Statistic significances were tested using Student’s t-test, the nonparametric Mann-Whitney test, or one-way analysis of variance.
Electrophysiology of Drosophila larval neuromuscular junctions
Male third instar larvae were dissected and bathed in HL-3 solution containing (mM): NaCl, 70; KCl, 5; NaHCO3, 10; HEPES, 5; sucrose, 11.5; trehalose, 5; MgCl2, 20 and CaCl2, 0.7 (for Figure 5) or 0.6 (for Figure 6); pH 7.4. Body wall muscle 6 (segment A3 or A4) was used for intracellular recordings with sharp electrodes filled with 2 M KAc and 1 M KCl (about 80–100 MΩ resistance). Current clamp recordings were performed using an Axoclamp 2B amplifier in bridge mode under the control of AxoGraph X or Clampex 8.2. Data were acquired at 10 kHz and low-pass filtered at 3 kHz. Only the muscles with resting membrane potential below -60 mV were used for experiments. Muscle input resistances were determined by a current injection of −1 nA. Miniature EJPs were recorded for 1–2 minutes. Evoked EJPs were elicited by pulling the cut end of the innervating segmental nerve into a suction electrode and passing a 0.5–0.8 ms depolarizing pulse with a DS2A-Mk. II constant voltage isolated stimulator (Digitimer) to activate both motoneurons. 10–30 evoked EJPs were recorded for each muscle. All experiments were performed at room temperature (20–22°C).
Data were analyzed offline using AxoGraph X. To detect mEJPs, traces were digitally filtered at 1 kHz offline and events were automatically selected with a scaled-template algorithm (Clements and Bekkers, 1997) in AxoGraph X. The template function is a double exponential with a scalable amplitude, a rise time constant of 5 ms, a decay time constant of 25 ms, a baseline of 2 ms, and a template length of 30 ms. Template length was reduced to 12 ms for Drosophila Complexin null mutants and null mutants rescued by CplxI or CplxIII to better detect the high frequency events (for Figure 5). The quantal content of evoked release was calculated by the ratio of evoked EJP amplitude and mEJP amplitude. Evoked EJP amplitude was not corrected for nonlinear summation. For Drosophila Complexin null mutants and null mutants rescued by CplxI or CplxIII, high frequency mEJPs are severely overlapped, which precludes the proper measurement of mEJP amplitude and consequently the calculation of the quantal content of evoked release. Statistic significances were tested using one-way analysis of variance.
Supplementary Material
Acknowledgements
We thank Hongmei Chen and Thea Hellmann for technical assistances; J. Troy Littleton for providing dmCplx antibody and dmCplx null mutant flies; Barry Dickson for nSyb-GAL4 flies before publication; Feng Liu and Kimberley Tolias for sharing transfection protocol and reagents, and Carlos Lois and Ralf Nehring for providing lentiviral vectors. The Brp and Dlg monoclonal antibodies developed by Erich Buchner and Corey Goodman, respectively, were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242. We are grateful to John Clements for assistance with AxoGraph X, Josep Rizo for comments, and Nils Brose for support. This work was supported by Baylor College of Medicine Mental Retardation and Developmental Disabilities Research Center and National Institutes of Health (NS50655 to C.R.). H.J.B. is a HHMI investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bowen ME, Weninger K, Ernst J, Chu S, Brunger AT. Single-molecule studies of synaptotagmin and complexin binding to the SNARE complex. Biophys J. 2005;89:690–702. doi: 10.1529/biophysj.104.054064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracher A, Kadlec J, Betz H, Weissenhorn W. X-ray structure of a neuronal complexin-SNARE complex from squid. J Biol Chem. 2002;277:26517–26523. doi: 10.1074/jbc.M203460200. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brose N. For better or for worse: complexins regulate SNARE function and vesicle fusion. Traffic. 2008;9:1403–1413. doi: 10.1111/j.1600-0854.2008.00758.x. [DOI] [PubMed] [Google Scholar]
- Brunger AT. Structure and function of SNARE and SNARE-interacting proteins. Q Rev Biophys. 2005;38:1–47. doi: 10.1017/S0033583505004051. [DOI] [PubMed] [Google Scholar]
- Cai H, Reim K, Varoqueaux F, Tapechum S, Hill K, Sorensen JB, Brose N, Chow RH. Complexin II plays a positive role in Ca2+-triggered exocytosis by facilitating vesicle priming. Proc Natl Acad Sci U S A. 2008;105:19538–19543. doi: 10.1073/pnas.0810232105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Tomchick DR, Kovrigin E, Arac D, Machius M, Sudhof TC, Rizo J. Three-dimensional structure of the complexin/SNARE complex. Neuron. 2002;33:397–409. doi: 10.1016/s0896-6273(02)00583-4. [DOI] [PubMed] [Google Scholar]
- Clements JD, Bekkers JM. Detection of spontaneous synaptic events with an optimally scaled template. Biophys J. 1997;73:220–229. doi: 10.1016/S0006-3495(97)78062-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudo CG, Eng WS, Melia TJ, Rothman JE. A clamping mechanism involved in SNARE-dependent exocytosis. Science. 2006;313:676–680. doi: 10.1126/science.1129450. [DOI] [PubMed] [Google Scholar]
- Giraudo CG, Garcia-Diaz A, Eng WS, Chen Y, Hendrickson WA, Melia TJ, Rothman JE. Alternative Zippering as an On-Off Switch for SNARE-Mediated Fusion. Science. 2009;323:512–516. doi: 10.1126/science.1166500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan R, Dai H, Rizo J. Binding of the Munc13-1 MUN domain to membrane-anchored SNARE complexes. Biochemistry. 2008;47:1474–1481. doi: 10.1021/bi702345m. [DOI] [PubMed] [Google Scholar]
- Huntwork S, Littleton JT. A complexin fusion clamp regulates spontaneous neurotransmitter release and synaptic growth. Nat Neurosci. 2007;10:1235–1237. doi: 10.1038/nn1980. [DOI] [PubMed] [Google Scholar]
- Ishizuka T, Saisu H, Odani S, Abe T. Synaphin: a protein associated with the docking/fusion complex in presynaptic terminals. Biochem Biophys Res Commun. 1995;213:1107–1114. doi: 10.1006/bbrc.1995.2241. [DOI] [PubMed] [Google Scholar]
- Jahn R, Scheller RH. SNAREs - engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- Li Y, Augustine GJ, Weninger K. Kinetics of complexin binding to the SNARE complex: correcting single molecule FRET measurements for hidden events. Biophys J. 2007;93:2178–2187. doi: 10.1529/biophysj.106.101220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DM, Goodman CS. Ectopic and increased expression of Fasciclin II alters motoneuron growth cone guidance. Neuron. 1994;13:507–523. doi: 10.1016/0896-6273(94)90022-1. [DOI] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Malsam J, Seiler F, Schollmeier Y, Rusu P, Krause JM, Sollner TH. The carboxy-terminal domain of complexin I stimulates liposome fusion. Proc Natl Acad Sci U S A. 2009;106:2001–2006. doi: 10.1073/pnas.0812813106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maximov A, Tang J, Yang X, Pang ZP, Sudhof TC. Complexin Controls the Force Transfer from SNARE Complexes to Membranes in Fusion. Science. 2009;323:516–521. doi: 10.1126/science.1166505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Missler M, Li C, Sudhof TC. Complexins: cytosolic proteins that regulate SNAP receptor function. Cell. 1995;83:111–119. doi: 10.1016/0092-8674(95)90239-2. [DOI] [PubMed] [Google Scholar]
- Neher E, Sakaba T. Multiple roles of calcium ions in the regulation of neurotransmitter release. Neuron. 2008;59:861–872. doi: 10.1016/j.neuron.2008.08.019. [DOI] [PubMed] [Google Scholar]
- Pabst S, Hazzard JW, Antonin W, Sudhof TC, Jahn R, Rizo J, Fasshauer D. Selective interaction of complexin with the neuronal SNARE complex. Determination of the binding regions. J Biol Chem. 2000;275:19808–19818. doi: 10.1074/jbc.M002571200. [DOI] [PubMed] [Google Scholar]
- Pabst S, Margittai M, Vainius D, Langen R, Jahn R, Fasshauer D. Rapid and selective binding to the synaptic SNARE complex suggests a modulatory role of complexins in neuroexocytosis. J Biol Chem. 2002;277:7838–7848. doi: 10.1074/jbc.M109507200. [DOI] [PubMed] [Google Scholar]
- Reim K, Mansour M, Varoqueaux F, McMahon HT, Sudhof TC, Brose N, Rosenmund C. Complexins regulate a late step in Ca2+-dependent neurotransmitter release. Cell. 2001;104:71–81. doi: 10.1016/s0092-8674(01)00192-1. [DOI] [PubMed] [Google Scholar]
- Reim K, Wegmeyer H, Brandstatter JH, Xue M, Rosenmund C, Dresbach T, Hofmann K, Brose N. Structurally and functionally unique complexins at retinal ribbon synapses. J Cell Biol. 2005;169:669–680. doi: 10.1083/jcb.200502115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo J, Rosenmund C. Synaptic vesicle fusion. Nat Struct Mol Biol. 2008;15:665–674. doi: 10.1038/nsmb.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenmund C, Stevens CF. Definition of the readily releasable pool of vesicles at hippocampal synapses. Neuron. 1996;16:1197–1207. doi: 10.1016/s0896-6273(00)80146-4. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Schaub JR, Lu X, Doneske B, Shin YK, McNew JA. Hemifusion arrest by complexin is relieved by Ca(2+)-synaptotagmin I. Nat Struct Mol Biol. 2006;13:748–750. doi: 10.1038/nsmb1124. [DOI] [PubMed] [Google Scholar]
- Strenzke N, Chanda S, Kopp-Scheinpflug C, Khimich D, Reim K, Bulankina AV, Neef A, Wolf F, Brose N, Xu-Friedman MA, Moser T. Complexin-I is required for high-fidelity transmission at the endbulb of held auditory synapse. J Neurosci. 2009;29:7991–8004. doi: 10.1523/JNEUROSCI.0632-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Yamamoto H, Matsuda Z, Ogawa M, Yagyu K, Taniguchi T, Miyata T, Kaba H, Higuchi T, Okutani F, et al. Identification of two highly homologous presynaptic proteins distinctly localized at the dendritic and somatic synapses. FEBS Lett. 1995;368:455–460. doi: 10.1016/0014-5793(95)00713-j. [DOI] [PubMed] [Google Scholar]
- Tang J, Maximov A, Shin OH, Dai H, Rizo J, Sudhof TC. A complexin/synaptotagmin 1 switch controls fast synaptic vesicle exocytosis. Cell. 2006;126:1175–1187. doi: 10.1016/j.cell.2006.08.030. [DOI] [PubMed] [Google Scholar]
- Weninger K, Bowen ME, Choi UB, Chu S, Brunger AT. Accessory Proteins Stabilize the Acceptor Complex for Synaptobrevin, the 1:1 Syntaxin/SNAP-25 Complex. Structure. 2008;16:308–320. doi: 10.1016/j.str.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z, Dudek H, Miranti CK, Greenberg ME. Calcium influx via the NMDA receptor induces immediate early gene transcription by a MAP kinase/ERK-dependent mechanism. J Neurosci. 1996;16:5425–5436. doi: 10.1523/JNEUROSCI.16-17-05425.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M, Ma C, Craig TK, Rosenmund C, Rizo J. The Janus-faced nature of the C(2)B domain is fundamental for synaptotagmin-1 function. Nat Struct Mol Biol. 2008a;15:1160–1168. doi: 10.1038/nsmb.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M, Reim K, Chen X, Chao HT, Deng H, Rizo J, Brose N, Rosenmund C. Distinct domains of complexin I differentially regulate neurotransmitter release. Nat Struct Mol Biol. 2007;14:949–958. doi: 10.1038/nsmb1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M, Stradomska A, Chen H, Brose N, Zhang W, Rosenmund C, Reim K. Complexins facilitate neurotransmitter release at excitatory and inhibitory synapses in mammalian central nervous system. Proc Natl Acad Sci U S A. 2008b;105:7875–7880. doi: 10.1073/pnas.0803012105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon TY, Lu X, Diao J, Lee SM, Ha T, Shin YK. Complexin and Ca2+ stimulate SNARE-mediated membrane fusion. Nat Struct Mol Biol. 2008;15:707–713. doi: 10.1038/nsmb.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.