Abstract
Proteases present in oral fluid effectively modulate the structure and function of some salivary proteins and have been implicated in tissue destruction in oral disease. To identify the proteases operating in the oral environment, proteins in pooled whole saliva supernatant were separated by anion-exchange chromatography and individual fractions were analyzed for proteolytic activity by zymography using salivary histatins as the enzyme substrates. Protein bands displaying proteolytic activity were particularly prominent in the 50–75 kDa region. Individual bands were excised, in-gel trypsinized and subjected to LC/ESI-MS/MS. The data obtained were searched against human, oral microbial and protease databases. A total of 13 proteases were identified all of which were of mammalian origin. Proteases detected in multiple fractions with cleavage specificities toward arginine and lysine residues, were lactotransferrin, kallikrein-1, and human airway trypsin-like protease. Unexpectedly, ten protease inhibitors were co-identified suggesting they were associated with the proteases in the same fractions. The inhibitors found most frequently were alpha-2-macroglobulin-like protein 1, alpha-1-antitrypsin, and leukocyte elastase inhibitor. Regulation of oral fluid proteolysis is highly important given that an inbalance in such activities has been correlated to a variety of pathological conditions including oral cancer.
Keywords: Degradation, Enzymatic, Histatin, Oral, Proteolytic
1 Introduction
Human whole saliva (WS) is the fluid which bathes oral hard and soft tissues and its constituents are responsible for the maintenance of a healthy oral condition. WS is derived from different sources that are exocrine and non-exocrine in nature. The exocrine contributions, also called salivary glandular secretions, include the fluids from the parotid, submandibular, and sublingual glands. In addition, minor salivary glands are lined throughout most of the oral cavity, including Von Ebner’s glands in circumvallate papillae of the tongue. The primary secretion flows through a network of ducts toward the oral cavity. Once the glandular secretions emanate into the oral environment, they are mixed with a number of non-exocrine components, which include gingival crevicular fluid, oral microorganisms, desquamated oral epithelial cells, other host and bacteria-derived components and dietary factors, ultimately forming WS.
The proteinaceous elements that are functionally responsible for oral health are expected to be effective in WS which contains a mixture of enzymes. These enzymes, which can be derived from either of the various sources eluded to above, have been described to proteolytically degrade some salivary proteins with high efficiency. Hence, oral fluid proteolysis is an important factor to be taken into account in saliva protein structure–function analyses and in diagnostic exploitations. Bacterial as well as human-derived enzymes have also been implicated in the destruction of oral tissues leading to pathological conditions. Approaches exploring specific oral enzyme activities have shown that hydrolysis of benzoyl-arginine naphthylamide in saliva correlated with the numbers of spirochetes in subgingival plaque samples, and to be an indicator of periodontal disease [1]. The role of collagenases, in particular matrix metalloproteinase-8 (MMP-8), have been well established in tissue destruction associated with periodontitis and implantitis [2, 3]. Additional investigations focusing on the role of oral enzymes in malignant transformations have implicated polymorphism in the gene expression of MMP-13 with a highly aggressive form of oral cancer [4]. Despite the evidence of a relationship between oral proteolytic activities and oral health and disease, the identities of most enzymes operating in the oral cavity are elusive.
One group of salivary proteins that is exceptionally susceptible to WS proteolysis is human histatins [5–9]. Histatins are non-glycosylated small histidine-rich salivary proteins. Structural analysis of fragments derived from histatins and other peptides in WS has provided a first, albeit indirect, insight into the saliva-associated enzymatic cleavage specificities. The actual proteases responsible for the degradation of histatins and other salivary proteins in the oral cavity are essentially unknown. The present study employed a histatin zymography approach in conjunction with LC-ESI-MS/MS to achieve the first global activity-based identification and characterization of proteolytic enzymes in oral fluid.
2 Materials and methods
2.1 WS collection
Stimulated WS was obtained from five orally healthy subjects. Prior to participation, all subjects consented to participation in the study according to approved protocols of the Institutional Review Board at Boston University Medical Center. WS (5 mL) was collected between 10 and 11 a.m. at least 1 h after the last meal. WS was stimulated using 1 g of paraffin wax for mastication and was collected by expectoration (Parafilm, American National Can™, Chicago, IL). Immediately after collection WS was centrifuged at 14000 × g for 20 min at 4°C, and the resultant WS supernatant (WSS) from each of the five subjects was pooled.
2.2 Anion-exchange chromatography
An aliquot of 15 mL of pooled WSS was dialyzed against milliQ water for 16 h. The sample was loaded on a strong anionic-exchange column (MonoQ™ HR16/10; Amersham Biosciences, Piscataway, NJ) connected to a fast protein liquid chromatography (FPLC) (Pharmacia Biotech, Piscataway, NJ). Proteins were eluted at a flow rate of 2 mL/min using buffer A (50 mM NaCl, 50 mM Tris-HCl, pH 8.0) and buffer B (1 M NaCl, 50 mM Tris-HCl, pH 8.0) applying the following gradient scheme: 0–45 min, 0% buffer B; 46–275 min, 0–25% buffer B; 276–300 min, 25–40% buffer B; 301–325 min, 40–100% buffer B; 325–355 min, 100% buffer B; 355–370 min, 100–0% buffer B. The absorbance was monitored at 214 nm and the eluate was collected in 10 mL aliquots. Fractions were dialyzed against water using dialysis tubing with a molecular weight cut-off of 1000 and lyophilized (Virtis, Gardiner, NJ). The dried 10 mL sample was dissolved in 1 mL of milliQ water for further analysis.
2.3 Synthetic histatins
Synthetic histatins 1, 3, and 5 were obtained from commercial sources (Quality Controlled Biochemicals, Hopkinton, MA; American Peptide Company, Sunnyvale, CA). Proteins were dissolved in milliQ water to approximately 10 mg/mL and their precise concentrations were determined by measuring the absorbance at 215 nm using a specific absorption coefficient (ε) of 20 mL mg−1 cm−1.
2.4 Cationic PAGE
WS and WSS sample aliquots (100 µL) to be analyzed by cationic PAGE were dried using a Vacufuge concentrator (Eppendorf Westbury, NY) and resuspended in sample solution containing 0.04% methyl green (Thermo Fisher, Waltham, MA) in 40% sucrose (Sigma, St. Louis, MO). Cationic PAGE was carried out as previously described [10, 11]. Gel polymerization was accelerated by exposing the gels to a light source (60 Watts). Gels were stained with Coomassie Brilliant Blue R-250 (0.1% v/v) in 10% v/v acetic acid and 40% v/v methanol, and destained in the same solution not containing the dye.
2.5 Zymography
WS, WSS and anion-exchange fractions of WSS (100 µL aliquots) were dried and subjected to zymography. Zymogram analysis was performed essentially as described [12] with the modification that synthetic histatin 1, 3, or 5 were used as enzyme substrates and incorporated in the separating gel mixture. Proteins in WS and WSS were separated by electrophoresis. The separating gel mixture contained 0.1% w/v histatin 1, 3, or 5, 8% w/v acrylamide, 0.4% w/v bisacrylamide, 0.1% w/v SDS and 375 mM Tris-HCl, pH 8.8, and the stacking gel contained 4% w/v acrylamide, 0.2% w/v bisacrylamide, 0.1% w/v SDS, and 126 mM Tris-HCl, pH 6.8. Polymerization was initiated upon the addition of 0.05% w/v ammonium persulfate and 0.1% v/v TEMED. Before loading onto the gel, samples to be analyzed (WSS, WS or anion-exchange fractions) were dried by a Vacufuge concentrator (Eppendorf) and mixed with 20 µL zymogram sample buffer containing 25% glycerol, 4% SDS, 0.01% Bromophenol blue, and 62.5 mM Tris-HCl, pH 6.8. The running buffer contained 25 mM Tris-HCl, 192 mM glycine, 0.1% SDS, pH 8.3. Gel electrophoresis was carried out at a constant voltage of 120 V for 1 h at 4°C. After electrophoresis, enzymatic conversion of the gel-incorporated histatin proteins was facilitated by renaturing of the gel for 30 min at room temperature in 2.5% Triton X-100 in milliQ water. Gels were washed in buffer containing 20 mM Tris-HCl and 1 mM CaCl2, pH 7.5, and developed for 16 h at 37°C in the same buffer. Staining of the gel was achieved in 0.1% w/v Coomassie Brilliant Blue R-250 in 10% v/v acetic acid and 40% v/v methanol, and destaining was carried out by boiling the gel in milliQ water for 8 min to visualize the histatin-degrading proteases.
2.6 In-gel trypsinization of zymogram bands
After zymography, the bands displaying enzymatic activities were excised and cut into 1 mm3 pieces using a new razor blade for each band. The gel pieces were destained in washing buffer (50% ACN in 100 mM ammonium bicarbonate) for 16 h, followed by removal of ACN, dehydration of the gel pieces in 100% ACN, and drying in a vacuum concentrator (Speedvac, Millipore). A 50 µL aliquot of digestion solution containing 12.5 µg/mL modified sequencing-grade trypsin (Promega, Madison, WI) in 50 mM ammonium bicarbonate (pH 8.0), was added followed by incubation at 37°C for 16 h. Peptides were recovered from the supernatant after washing the gel pieces with solution containing 50% ACN and 5% acetic acid. The extracts were dried in a speed-vac and the samples were then stored at 4°C until analysis.
2.7 LC-ESI-MS/MS
The dried, in-gel zymogram digests were reconstituted in 5–10 µL of solvent A containing 2.5% ACN and 0.1% formic acid. Samples were applied to a nano-scale RP-HPLC capillary column (75 µm inner diameter × ~10 cm length) which was created in-house by packing 5 µm C18 spherical silica beads (Micron Bioresource, Auburn, CA) into a fused-silica capillary with a flame-drawn tip. After equilibrating the column, each sample (3 µL) was loaded via a Famos auto sampler (LC Packings, San Francisco CA) onto the column. Peptides were eluted with a linear gradient of increasing buffer B (97.5% ACN, 0.1% formic acid) from 0 to 100% over a 15 min time interval followed by ESI (LTQ linear ion-trap mass spectrometer; ThermoFisher, San Jose, CA) yielding tandem mass spectra of specific fragment ions for each peptide.
2.8 Mass spectrometric data analysis
The MS/MS data were searched against three main databases: (i) the human protein database (Swiss-Prot and TrEMBL, Swiss Institute of Bioinformatics, Geneva, Switzerland, http://expasy.org/sprot), (ii) a database containing all proteases known to date across the plant and animal kingdom ([13] MEROPS, http://merops.sanger.ac.uk/), and (iii) an oral microbial database consisting of all fully and partly sequenced oral microbial organisms to date (http://www.HOMD.org). In addition, the data were searched against databases of each of the microbial species individually to avoid the assignment of two peptides from one protein homologue to different microbial species. Searches were performed essentially as described [14] using Bioworks software (version 3.3.1) with the following parameters selected: (i) fully tryptic fragment; (ii) Delta CN≥0.1; (iii) peptide probability≤0.5, XCorr score≥2.0 and 3.5 for z = 2 and 3, respectively. All spectra were manually examined for the presence of at least three consecutive b- and y-ions. Proteins of which at least two different bona fide peptides were identified were considered a confident match. Searches of the MS/MS data against the human database with sequences in reversed order yielded 0.25% false positive identifications by protein among which there were no proteases.
3 Results
3.1 Comparison of protease activities in WS and WSS
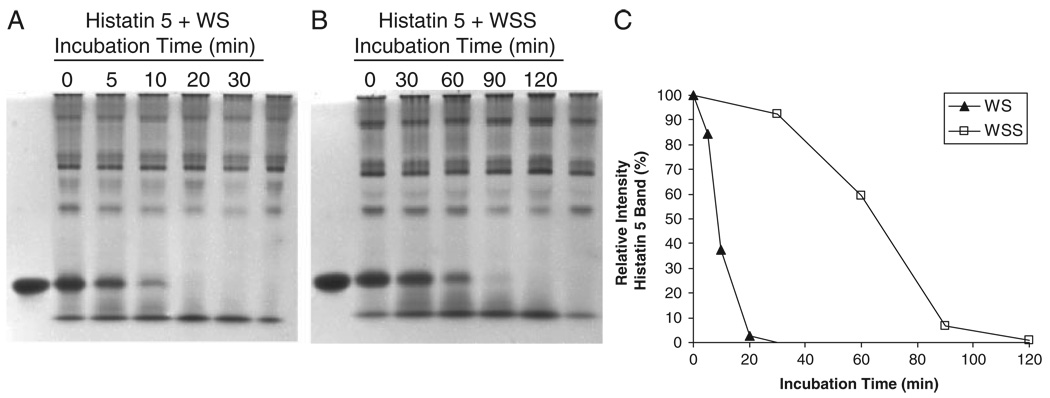
To determine the distribution of WS enzymatic activities in the particulate and soluble fraction of WS, histatin 5 was added to WS and WSS and its degradation was followed as a function of time (Fig. 1). Natural histatin 5 levels in WS and WSS (right lanes) were negligible as compared to the added amount of histatin 5 (left lanes). Histatin disappearance was quantitated by densitometric analysis of the histatin band at each of the sampling time points (Fig. 1C). From the densitometric plots, it could be established that the half-life of the disappearance of histatin 5 was 8.7 min in WS and 65.3 min in WSS. From these data, it can be estimated that the degradation rates in WS are approximately 7.5 times faster than in WSS, and that 88.2% of the proteolytic activity in WS is associated with the non-soluble components.
Figure 1.
Comparison of protease activities in WSS and WS. Synthetic histatin 5 (100 µg) was added to 1 mL of WS (A) or WSS (B) and incubated at 37°C. Aliquots of 100 µL were removed after the indicated time intervals, boiled, dried, and analyzed by cationic-PAGE. Left lanes in (A) and (B): 10 µg histatin 5; right lanes in (A) and (B): 100 µL WS and WSS, respectively. (C) densitometric analysis of histatin 5 as a function of incubation time in WS and WSS. The intensity of the histatin 5 band immediately upon addition to WS or WSS (t = 0) was set to 100%.
3.2 Comparison of protease profiles in WS and WSS
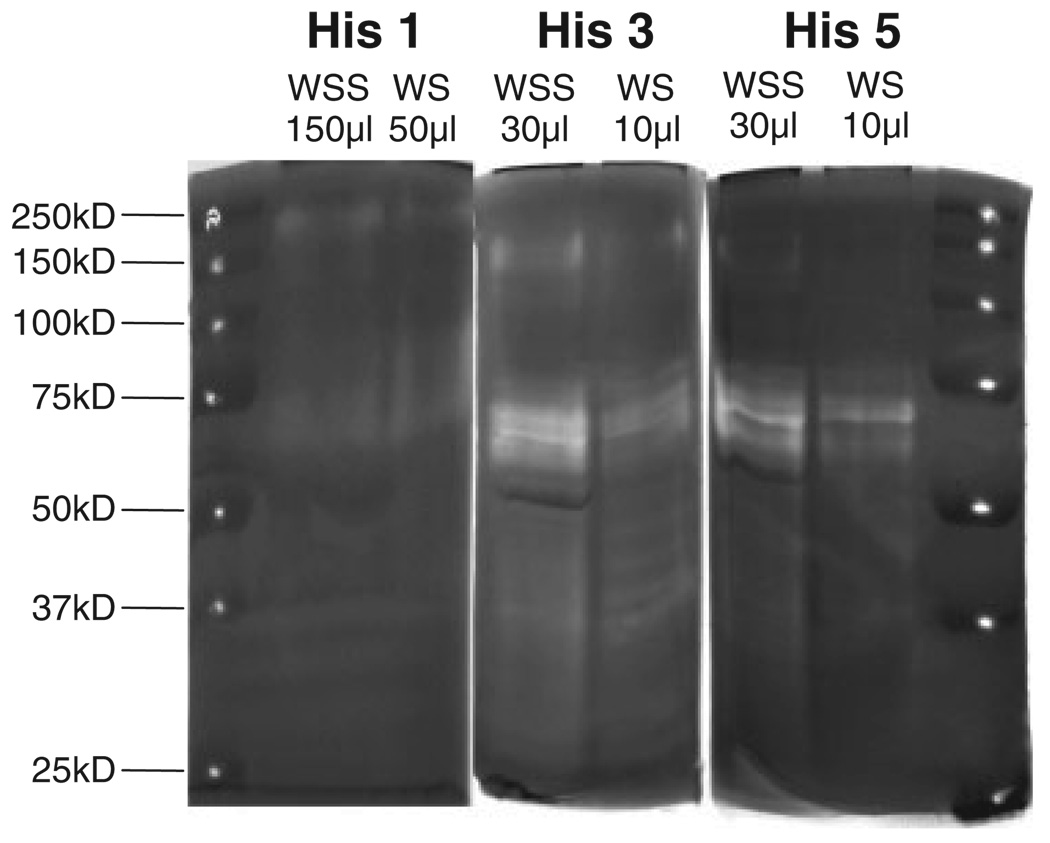
To gain insight into the approximate molecular weights of the histatin-degrading proteases, zymography was carried out with histatin 1, 3, or 5 copolymerized in the separating gel (Fig. 2). Different volumes of WS and WSS were loaded on the gel to compensate for the higher overall proteolytic activity of WS as compared with WSS enabling a comparison of the protease banding patterns. WS and WSS generally displayed similar protease activity patterns toward all three histatins, although higher activities were observed toward histatin 3 and 5 as compared with histatin 1 (Fig. 2). This corroborates with earlier reports that histatin 1 is more resistant to proteolytic breakdown in saliva than histatins 3 and 5 [8, 15]. Multiple protease bands were noted in the 50–75 kDa molecular weight region. Based on the qualitative similarities in the zymogram patterns of WS and WSS, WSS was further utilized for the isolation of the proteolytic enzymes.
Figure 2.
Histatin zymography of WSS and WS. WSS and WS samples were analyzed on zymogram gels with incorporated histatin 1 (left panel), histatin 3 (middle panel) or histatin 5 (right panel). The volumes loaded of each of the saliva sample are indicated. The position of the molecular weight standards (pierced) are depicted in the far left and right lanes.
3.3 Fractionation of WSS by FPLC anion-exchange chromatography
To reduce the complexity in the WSS sample, WSS proteins were dialyzed and fractionated by anion-exchange chromatography (monoQ). The choice for anion-exchange resin material was based on a preliminary experiment in which WSS was incubated with DEAE resin material (anionexchange) followed by batch-wise elution with increasing concentrations of NaCl. It was noticed that very little activity was recovered in the void volume, indicating that the enzymes targeting histatins were predominantly negatively charged. Proteins were eluted from the monoQ column using a gradient from 50 to 100 mM NaCl in 50 mM Tris-HCl (pH 8) (data not shown). The bound (anionic) proteins eluted primarily as three major and two minor peaks. Nevertheless, all 65 eluting fractions were collected for protease activity evaluation.
3.4 Protease activity in anion-exchange fractions
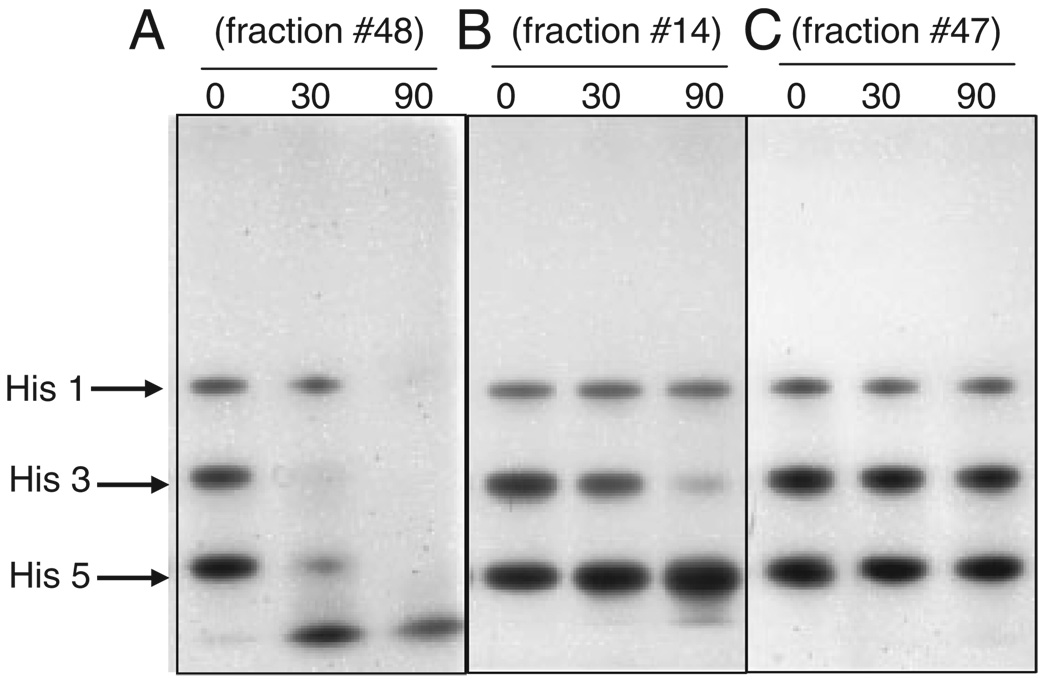
All 65 FPLC fractions were desalted and concentrated tenfold. Protease activity in each fraction was assessed by adding a mixture of histatins 1, 3, and 5 and monitoring their degradation over a 90 min time interval by cationic PAGE (Fig. 3). The selected 90 min time interval stems from results presented in Fig. 1 and allowed for a relative comparison in activities between fractions. Based on the degradation results obtained, the fractions could be divided into three different groups. A first group comprising fractions 20, 21, 25, 26, 31, 36, 48, and 49, displayed a generalized activity toward all three histatins, noted by the disappearance of all three proteins over time and the concomitant appearance of smaller molecular weight fragments (Fig. 3A). Proteases in the second group, which included fractions 6, 9, 10, and 14 to 18, appeared to selectively target histatin 3, given the relative stability of the histatin 1 and histatin 5 bands (Fig. 3B). The remainder of the fractions exhibited no or negligible proteolytic activity toward any of the histatins (Fig. 3C). The thirteen most active fractions, eight from the first group and five from the second group, were chosen for further analysis by histatin zymography.
Figure 3.
Assessment of histatin-degradating activity in FPLC fractions from WSS. A mixture of synthetic histatin 1, 3, and 5 (5 µg each) was added to 161 µL of individual FPLC fractions of WSS proteins. After 0, 30, and 90 min of incubation, 50 µL aliquots were boiled, dried, and analyzed by cationic PAGE. (A) example of a fraction displaying activity toward all histatins, (B) example of a fraction displaying activity toward histatin 3 only. (C) example of a fraction devoid of proteolytic activity toward any of the histatins.
3.5 Zymography of FPLC fractions
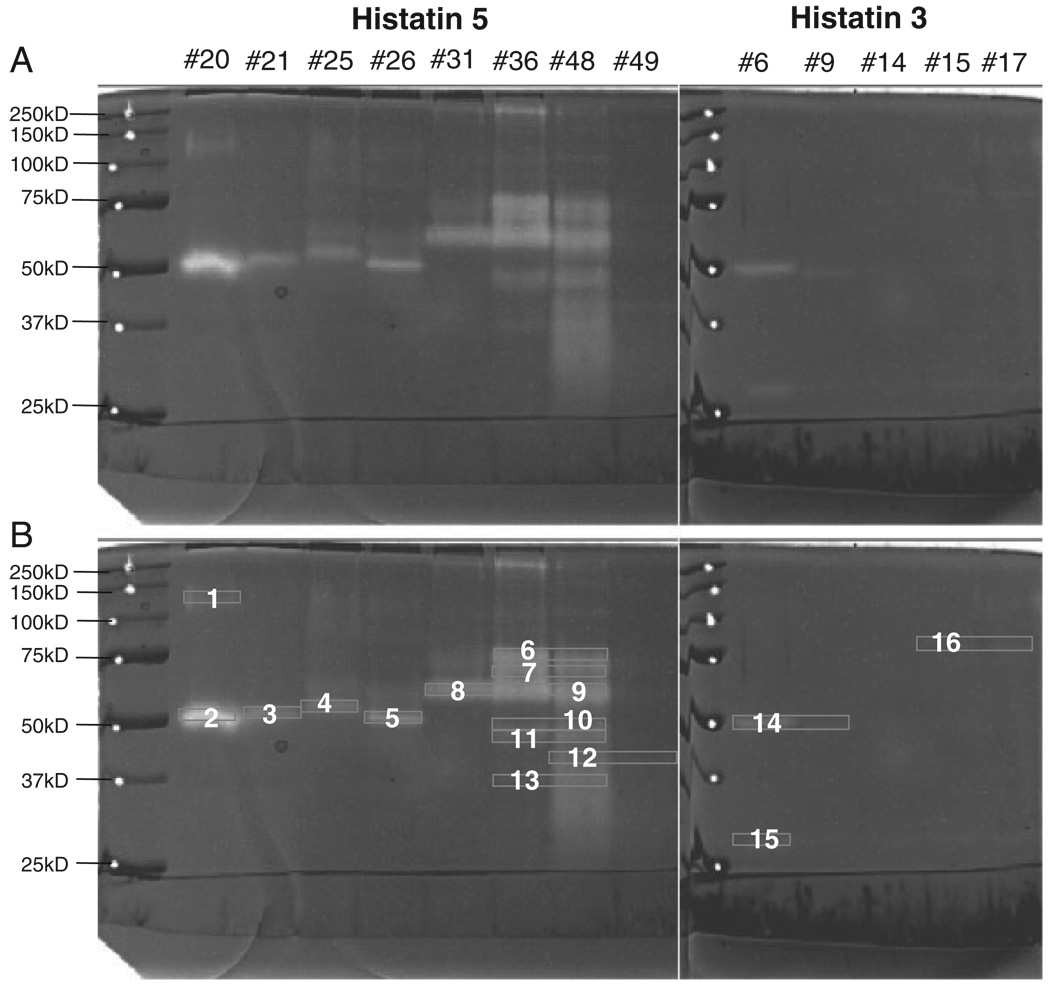
The eight WSS fractions with activity toward all histatins (20, 21, 25, 26, 31, 36, 48, and 49) were subjected to histatin 5 zymography, and the five fractions that appeared selectively active toward histatin 3 (6, 9, 14, 15, and 17) were analyzed by histatin 3 zymography (Fig. 4A). As intended, the complexity of the protease patterns in most fractions were strongly reduced as compared with the patterns observed in unfractionated WSS. Most of the protease bands resided in the 50–75 kDa region, as seen in WSS. The major protein bands exhibiting proteolytic activity were excised and numbered (Fig. 4B). Only in the case of fraction 14, the zymogram bands were very weak. However, bands with electrophoretic mobilities comparable to those in fraction 14 but with higher intensity were observed in other fractions and excised. In some cases, protein bands from different fractions showing similar electrophoretic mobilities were pooled. In total, 16 protease-containing gel pieces were further processed.
Figure 4.
Zymography of active WSS fractions. Proteins in fractions 20, 21, 25, 26, 31, 36, 48, and 49 were analyzed by histatin 5 zymography (left panel) and proteins in fractions 6, 9, 14, 15, and 17 were subjected to histatin 3 zymography (right panel). Left lanes, molecular weight standard. Sixteen protein bands displaying proteolytic activity were excised from the gel for subsequent trypsinization and processing by LC-ESI-MS/MS.
3.6 Mass spectrometric identification of proteases and protease inhibitors in WSS
Proteins in the 16 protein bands were in-gel trypsinized and the peptides extracted and sequenced by LC-ESI-MS/MS. Unexpectedly, searches against the oral microbial databases yielded only four reliable protein identifications among which there was no protease. In a control experiment, a digest of P. gingivalis culture supernatant searched against its respective database yielded the identification of multiple enzymes including the potent proteolytic enzymes gingipain R and gingipain K (data not shown). The results of the searches against the human database are summarized in the Supporting Information Table, which summarizes all proteins identified, and in Table 1 containing the proteases and inhibitors only. Interestingly, in most bands, proteases as well as inhibitors were identified. Some proteases such as kallikrein 1 appeared in multiple zymogram bands. Likewise, some inhibitors such as alpha-1-antitrypsin, were detected in more than one band. Additional characteristics of the proteases and inhibitors identified are listed in Table 2 and Table 3, respectively. Of the 13 proteases found, six belonged to the family of serine proteases (lactotransferrin, myeloblastin, transmembrane protease serine 11D, dipeptidylpeptidase 2, furin and kallikrein-1). The remaining seven proteases included three cysteine proteases, three metallo proteases and one aspartyl protease. Also listed in Table 2 are the preferred cleavage site specificities of each of the enzymes. The serine proteases lactotransferrin, transmembrane protease serine 11D, furin and kallikrein-1 exhibit preferential cleavage specificity after an R or K residue. Of the ten protease inhibitors identified, alpha-2-macroglobulin-like protein 1 (α2ML1), alpha-1-antitrypsin, and leukocyte elastase inhibitor were found most frequently, each displaying inhibitory activities toward various classes of enzymes (Table 3). No additional proteases/inhibitors were identified when the MS/MS data were searched against the designated MEROPS database.
Table 1.
LC-ESI-MS/MS characterization of proteases and inhibitors
| Band ID | Total no. proteins identified |
Protease | Entry name | Accession no. | No. of peptides | ||
|---|---|---|---|---|---|---|---|
| 1 | 12 | Lactotransferrin | TRFL_HUMAN | P02788 | 7 | ||
| Myeloblastin | PRTN3_HUMAN | P24158 | 2 | ||||
| Alpha-2-macroglobulin-like protein 1 | A2ML1_HUMAN | Q6ZW52 | 41 | ||||
| 2 | 22 | Dipeptidyl-peptidase 1 | CATC_HUMAN | P53634 | 3 | ||
| Lactotransferrin | TRFL_HUMAN | P02788 | 3 | ||||
| Leukotriene A-4 hydrolase | LKHA4_HUMAN | P09960 | 3 | ||||
| Leukocyte elastase inhibitor | ILEU_HUMAN | P30740 | 15 | ||||
| Alpha-1-antitrypsin | A1AT_HUMAN | P01009 | 11 | ||||
| 3 | 14 | Alpha-1-antitrypsin | A1AT_HUMAN | P01009 | 5 | ||
| Leukocyte elastase inhibitor | ILEU_HUMAN | P30740 | 3 | ||||
| 4 | 34 | Lactotransferrin | TRFL_HUMAN | P02788 | 19 | ||
| Dipeptidyl-peptidase 2 | DPP2_HUMAN | Q9UHL4 | 3 | ||||
| Dipeptidyl-peptidase 1 | CATC_HUMAN | P53634 | 2 | ||||
| Alpha-1-antitrypsin | A1AT_HUMAN | P01009 | 25 | ||||
| Leukocyte elastase inhibitor | ILEU_HUMAN | P30740 | 3 | ||||
| Plasma protease C1 inhibitor | IC1_HUMAN | P05155 | 3 | ||||
| Alpha-1-antichymotrypsin | AACT_HUMAN | P01011 | 3 | ||||
| Alpha-2-macroglobulin-like protein 1 | A2ML1_HUMAN | Q6ZW52 | 2 | ||||
| 5 | 27 | Lactotransferrin | TRFL_HUMAN | P02788 | 2 | ||
| Alpha-1-antitrypsin | A1AT_HUMAN | P01009 | 14 | ||||
| Leukocyte elastase inhibitor | ILEU_HUMAN | P30740 | 3 | ||||
| Cystatin-SA | CYTT_HUMAN | P09228 | 2 | ||||
| 6 | 43 | Lactotransferrin | TRFL_HUMAN | P02788 | 17 | ||
| Furin | FURIN_HUMAN | P09958 | 3 | ||||
| Leukocyte elastase inhibitor | ILEU_HUMAN | P30740 | 12 | ||||
| Alpha-1-antitrypsin | A1AT_HUMAN | P01009 | 11 | ||||
| Alpha-1-antichymotrypsin | AACT_HUMAN | P01011 | 7 | ||||
| Angiotensinogen | ANGT_HUMAN | P01019 | 3 | ||||
| Alpha-2-macroglobulin-like protein 1 | A2ML1_HUMAN | Q6ZW52 | 2 | ||||
| Kininogen-1 | KNG1_HUMAN | P01042 | 2 | ||||
| 7 | 37 | Kallikrein-1 | KLK1_HUMAN | P06870 | 3 | ||
| Angiotensinogen | ANGT_HUMAN | P01019 | 5 | ||||
| Neuroserpin | NEUS_HUMAN | Q99574 | 3 | ||||
| Leukocyte elastase inhibitor | ILEU_HUMAN | P30740 | 2 | ||||
| 8 | 30 | Cathepsin D | CATD_HUMAN | P07339 | 8 | ||
| Kallikrein-1 | KLK1_HUMAN | P06870 | 2 | ||||
| Alpha-1-antitrypsin | A1AT_HUMAN | P01009 | 6 | ||||
| Neuroserpin | NEUS_HUMAN | Q99574 | 6 | ||||
| Leukocyte elastase inhibitor | ILEU_HUMAN | P30740 | 2 | ||||
| 9 | 28 | Lactotransferrin | TRFL_HUMAN | P02788 | 16 | ||
| Carboxypeptidase E | CBPE_HUMAN | P16870 | 3 | ||||
| Puromycin-sensitive aminopeptidase | PSA_HUMAN | P55786 | 3 | ||||
| 10 | 11 | Cathepsin L1 | CATL1_HUMAN | P07711 | 5 | ||
| Kallikrein-1 | KLK1_HUMAN | P06870 | 2 | ||||
| 11 | 18 | Puromycin-sensitive aminopeptidase | PSA_HUMAN | P55786 | 3 | ||
| Kallikrein-1 | KLK1_HUMAN | P06870 | 3 | ||||
| Alpha-1-antitrypsin | A1AT_HUMAN | P01009 | 2 | ||||
| 12 | 47 | Puromycin-sensitive aminopeptidase | PSA_HUMAN | P55786 | 6 | ||
| Calpain-1 catalytic subunit | CAN1_HUMAN | P07384 | 5 | ||||
| Alpha-1-antitrypsin | A1AT_HUMAN | P01009 | 3 | ||||
| Plasma protease C1 inhibitor | IC1_HUMAN | P05155 | 3 | ||||
| 13 | 31 | Lactotransferrin | TRFL_HUMAN | P02788 | 17 | ||
| 14 | 18 | Cathepsin D | CATD_HUMAN | P07339 | 3 | ||
| Leukocyte elastase inhibitor | ILEU_HUMAN | P30740 | 3 | ||||
| 15 | 14 | Leukotriene A-4 hydrolase | LKHA4_HUMAN | P09960 | 3 | ||
| Alpha-1-antitrypsin | A1AT_HUMAN | P01009 | 2 | ||||
| 16 | 15 | Cathepsin D | CATD_HUMAN | P07339 | 4 | ||
| Transmembrane protease, serine 11D | TM11D_HUMAN | O60235 | 2 | ||||
| Metalloproteinase inhibitor 1 | TIMP1_HUMAN | P01033 | 2 | ||||
Table 2.
Characteristics of proteases identified by LC-ESI-MS/MS
| Protease | Accession no. | Type | Cleavage site specificitya) | Band ID |
|---|---|---|---|---|
| Lactotransferrin | P02788 | Serine | −/−/−/rk+−/−/−/− | 1, 2, 4, 5, 6, 9, 13 |
| Myeloblastin | P24158 | Serine | −/−/−/vat+−/−/−/− | 1 |
| Transmembrane protease, serine 11D | O60235 | Serine | −/−/−/r+−/−/−/− | 16 |
| Dipeptidyl-peptidase 2 | Q9UHL4 | Serine | −/−/−/Pam+−/−/−/− | 4 |
| Furin | P09958 | Serine | R/k/KR/R+sa/−/−/− | 6 |
| Kallikrein-1 | P06870 | Serine | −/−/−/R+−/−/−/− | 7, 8, 10, 11 |
| Dipeptidyl-peptidase 1 | P53634 | Cysteine | −/esfr+−/−/−/− | 2, 4 |
| Cathepsin L1 | P07711 | Cysteine | −/−/lfy/−+−/−/−/− | 10 |
| Calpain-1 catalytic subunit | P07384 | Cysteine | −/−/l/−+a/−/p/− | 12 |
| Leukotriene A-4 hydrolase | P09960 | Metallo | −/−/−/arpy+−/−/−/− | 2, 15 |
| Carboxypeptidase E | P16870 | Metallo | −/−/−/ap+r | 9 |
| Puromycin-sensitive aminopeptidase | P55786 | Metallo | −/−/−/lar+l/−/−/− | 9, 11, 12 |
| Cathepsin D | P07339 | Aspartic | −/−/−/lf+−/−/−/− | 8, 14, 16 |
Nomenclature of cleavage site specificity used was as reported (http://merops.sanger.ac.uk/). Each cleavage site specificity is illustrated by eight amino acid residues (P4-P3-P2-P1-P1′-P2′-P3′-P4′) around the scissile bond indicated by the symbol +. Amino acids identified with high confidence at the indicated position are capitalized; amino acids identified with lower confidence are indicated in lower case; a dash indicates that the amino acid required at that position has not yet been established.
Table 3.
Characteristics of protease inhibitors identified by LC-ESI-MS/MS
| Protease inhibitor | Accession no. | Target enzymea) | Band ID |
|---|---|---|---|
| Alpha-2-macroglobulin-like protein 1 | Q6ZW52 | Chymotrypsin, subtilisin A, papain, thermolysin | 1, 4, 6 |
| Alpha-1-antitrypsin | P01009 | Elastase-1, elastase-2, cathepsin G, thrombin, plasmin, kallikrein-related peptidase 4 | 2, 3, 4, 5, 6, 8, 11, 12, 15 |
| Leukocyte elastase inhibitor | P30740 | Elastase-2, cathepsin G, myeloblastin | 2, 3, 4, 5, 6, 7, 8, 14 |
| Plasma protease C1 inhibitor | P05155 | Complement component activated C1r and C1s | 4, 12 |
| Alpha-1-antichymotrypsin | P01011 | Kallikrein-related peptidase 14, cathepsin G | 4, 6 |
| Cystatin-SA | P09228 | Cysteine protease | 5 |
| Angiotensinogen | P01019 | Unknown | 6, 7 |
| Kininogen-1 | P01042 | Cysteine proteases (cathepsin L) | 6 |
| Neuroserpin | Q99574 | uPA, tPA, plasmin | 7, 8 |
| Metalloproteinase inhibitor 1 | P01033 | MMPs | 16 |
Based on information available at the MEROPS website (http://merops.sanger.ac.uk/).
4 Discussion
The present work aimed to characterize active enzymes in oral fluid. Before this study, the proteases lactotransferrin, kallikrein-1, dipeptidyl-peptidase and cathepsins [16–22], and the inhibitors alpha-1-antitrypsin, cystatin SA and metalloproteinase inhibitor 1 [23–25] had been detected in saliva and/or gingival fluid by a variety of classical, non-MS-based techniques. In fact, with the exception of alpha-1-antichymotrypsin and kininogen-1, all enzymes and inhibitors listed in Table 1 and Table 2 have been reported recently to be saliva constituents following extensive proteomic analysis of individual salivary secretions [9, 26–30]. In the present investigation, we employed a zymography approach in conjunction with LC-ESI MS/MS. Therefore, the protease identifications made were not only based on mass spectrometric detection (presence), but also on actual activity of the enzyme. Furthermore, different from typical casein or gelatin zymography approaches, histatin zymography facilitated the identification of proteases with activity toward a salivary protein substrate.
4.1 Protease-inhibitor associations
One of the most intriguing and unexpected findings in this exploration was the presence of proteases and inhibitors in multiple bands on the zymogram gel. Evidence for the existence of various isoforms of some proteases have been reported. For instance, kallikrein-1 is variably and extensively post-translationally modified yielding molecular masses of <20, 45, 60, 90 and >200 kDa [31]. Also lactotransferrin has been reported to exist in multiple isoforms [32]. As for most proteases, the majority of the inhibitors appeared in multiple bands. Noticeably, in some cases, the anticipated molecular weight of a protease–inhibitor couple in a particular band on the zymogram did not match the theoretical molecular weights of the identified constituents. For instance, the active component in band 15 migrates with an approximate molecular weight between 25 and 37 kDa, whereas the theoretical molecular weights of its constituents, leukotriene A4 hydrolase and alpha-1-antitrypsin, each exceed this molecular weight. Although fragmentation of the protease and inhibitor could perhaps account for their appearance in the low molecular weight regions, some caution is to be exerted in the interpretation of true protease–inhibitor associations as opposed to coincident identifications.
4.2 Mammalian proteases in WSS
Thirteen proteases from mammalian origin were found upon mass spectrometric processing of the active bands. Three of these proteases, lactotransferrin, kallikrein-1, and human airway trypsin-like protease (HAT) (transmembrane protease, serine 11D) likely play a dominant role in histatin proteolysis, because they are serine proteases exhibiting preferential cleavage specificities C-terminal to arginine (R) and lysine (K) residues, amino acids which are enriched in histatins. Iron-free lactotransferrin cleaves the enzyme substrate Z-YR-AMC, a substrate showing homology to the RGYR template, which is present and targeted in histatin 3 [5]. Because lactotransferrin in human saliva contains very low iron levels (0.0212±0.0081%; [33]), lactotransferrin is predominantly present in the active apo form, which would allow it to function as a protease in oral fluid. Like lactotransferrin, kallikrein-1 is a trypsin-like serine protease with preference for arginine at the P1 position. Its activity has been reported to be inhibited by PMSF, benzamidine, leupeptin, kallistatin, protein C inhibitor, and aprotinin [34], some of which in combination are highly effectively in preventing histatin proteolysis in WSS (Sun et al., unpublished data). Evidence for the likelihood for the involvement of kallikrein-1 in histatin degradation in the oral cavity stems from a recent in vitro study showing that histatin 5 was cleaved by kallikrein-1 C-terminal to R10, K11, and R12 [35]. Another prominent protease identified was transmembrane protease, serine 11D which is also named HAT [36]. HAT protein is expressed in the cells of submucosal serous glands of human bronchi and trachea [36] and it has been detected in human glandular secretion and in WS [26, 29]. The present study unequivocally identifies HAT to be functional in the oral cavity.
4.3 Bacterial proteases in WSS
A body of evidence suggests that a likely source for WS proteases are oral microorganisms [37–39]. Consistent with this, most of the enzymatic activity was found to be associated with the particulate component of WS, of which bacteria are a prominent constituent. Unexpectedly, when the MS/MS data were queried against bacterial databases, a very low number of proteins were identified. Similar low identifications of bacteria-derived proteins in WSS have been reported by other investigators [40]. Frequently, only one peptide was detected from a protein, which did not meet our criteria for protein identification. Given the high probability that oral microbial proteases are at least in part responsible for salivary protein digestion, it is anticipated that further sequencing of oral microbial genomes will enable identifications of bacterial proteases in WS in the near future and expand the numbers of saliva-related protease identifications.
4.4 Protease inhibitors in WSS
Interestingly, we found a relatively large number of protease inhibitors in the zymogram bands. Their identification by multiple peptides suggests that these inhibitors were enriched in the processed zymogram bands. Noticeably, α2ML1, alpha-1-antitrypsin and leukocyte elastase inhibitors, all exhibiting inhibitory activities toward various classes of enzymes, were observed most frequently. The broad-spectrum inhibitory activities of these inhibitors and their association with different proteases and/or isoforms of the same protease could explain their appearance in multiple bands in the zymogram gel. α2ML1 belongs to the αM super family comprising both protease inhibitors and components of the complement system [41]. The ability of αM proteins to inhibit proteases depends on its unique mechanism of inhibition by physical hindrance called the “trap mechanism” [42]. In this mechanism, entrapped proteases lose their ability to cleave high molecular weight substrates but would retain their capacity to cleave smaller molecular weight substrates [43, 44] such as histatin 5. A similar “trap” mechanism of inhibition has been postulated for alpha-1-antitrypsin [45]. Alpha-1-antitrypsin, also named alpha-1 protease inhibitor and alpha-1-antiproteinase, is a serine protease inhibitor belonging to the Serpin family which inhibits a variety of proteases including elastate-1, elastate-2, thrombin, plasmin, cathepsin G, proteinase 3, chymotrypsin A, and kallikrein-related peptidase 4 [46]. Another member of the serpin family, leukocyte elastase inhibitor, was also frequently found in our analyses. It is known to be one of the most efficient protease inhibitors of neutrophil serine proteases including elastase, cathepsin G and proteinase-3 [47–49], and together with α2ML1 and alpha-1-antitrypsin represents an important component of the arsenal of inhibitors in saliva.
Overall, using pure histatins as substrates, the activity-based characterizations have not only enabled the identification of proteases effective in degrading histatins, but also led to the detection of an almost equal number of mammalian-derived protease inhibitors, many with broad-spectrum inhibitory activities. Their identification with high confidence in multiple zymogram bands suggests that they play a crucial role in modulating protease activity in the oral environment. Regulation of such activities is indeed important, given the evidence that uncontrolled proteolytic activities are at the basis of multiple pathological conditions, including oral and duodenal cancer [50–52]. A full understanding of the interplay of these proteases and inhibitors in oral as well as gastro-intestinal physiology would represent a novel area of investigation. Our data suggest that α2ML1, alpha-1 antitrypsin and leukocyte elastase inhibitor may play crucial roles in such regulatory processes.
Supplementary Material
Acknowledgments
The authors thank Ross Tomaino, for carrying out part of the mass spectrometric characterizations at the Taplin Biological Mass Spectrometry Facility at Harvard Medical School Department of Cell Biology. This work was supported by grants DE05672, DE07652, DE18132, DE018448, and DE16699.
Abbreviations
- α2ML1
alpha-2-macroglobulin-like protein 1
- FPLC
fast protein liquid chromatography
- HAT
human airway trypsin-like protease
- WS
whole saliva
- WSS
whole saliva supernatant
Footnotes
The authors have declared no conflict of interest.
References
- 1.Bretz WA, Loesche WJ. Characteristics of trypsin-like activity in subgingival plaque samples. J. Dent. Res. 1987;66:1668–1672. doi: 10.1177/00220345870660111301. [DOI] [PubMed] [Google Scholar]
- 2.Chen HY, Cox SW, Eley BM, Mantyla P, et al. Matrix metalloproteinase-8 levels and elastase activities in gingival crevicular fluid from chronic adult periodontitis patients. J. Clin. Periodont. 2000;27:366–369. doi: 10.1034/j.1600-051x.2000.027005366.x. [DOI] [PubMed] [Google Scholar]
- 3.Sorsa T, Tjaderhane L, Salo T. Matrix metalloproteinases (MMPs) in oral diseases. Oral dis. 2004;10:311–318. doi: 10.1111/j.1601-0825.2004.01038.x. [DOI] [PubMed] [Google Scholar]
- 4.Vairaktaris E, Yapijakis C, Nkenke E, Serefoglou ZC, et al. A metalloproteinase-13 polymorphism affecting its gene expression is associated with advanced stages of oral cancer. Anticancer Res. 2007;27:4027–4030. [PubMed] [Google Scholar]
- 5.Castagnola M, Inzitari R, Rossetti DV, Olmi C, et al. A cascade of 24 histatins (histatin 3 fragments) in human saliva. Suggestions for a pre-secretory sequential cleavage pathway. J. Biol. Chem. 2004;279:41436–41443. doi: 10.1074/jbc.M404322200. [DOI] [PubMed] [Google Scholar]
- 6.Helmerhorst EJ, Alagl AS, Siqueira WL, Oppenheim FG. Oral fluid proteolytic effects on histatin 5 structure and function. Arch. Oral. Biol. 2006;51:1061–1070. doi: 10.1016/j.archoralbio.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Messana I, Cabras T, Pisano E, Sanna MT, et al. Trafficking and postsecretory events responsible for the formation of secreted human salivary peptides: a proteomics approach. Mol. Cell. Proteomics. 2008;7:911–926. doi: 10.1074/mcp.M700501-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Payne JB, Iacono VJ, Crawford IT, Lepre BM, et al. Selective effects of histidine-rich polypeptides on the aggregation and viability of Streptococcus mutans and Streptococcus sanguis. Oral Microbiol. Immunol. 1991;6:169–176. doi: 10.1111/j.1399-302x.1991.tb00472.x. [DOI] [PubMed] [Google Scholar]
- 9.Vitorino R, Lobo MJ, Ferrer-Correira AJ, Dubin JR, et al. Identification of human whole saliva protein components using proteomics. Proteomics. 2004;4:1109–1115. doi: 10.1002/pmic.200300638. [DOI] [PubMed] [Google Scholar]
- 10.Baum BJ, Bird JL, Longton RW. Polyacrylamide gel electrophoresis of human salivary histidine-rich-polypeptides. J. Dent. Res. 1977;56:1115–1118. doi: 10.1177/00220345770560091801. [DOI] [PubMed] [Google Scholar]
- 11.Flora B, Gusman H, Helmerhorst EJ, Troxler RF, Oppenheim FG. A new method for the isolation of histatins 1, 3, and 5 from parotid secretion using zinc precipitation. Protein Expr. Purif. 2001;23:198–206. doi: 10.1006/prep.2001.1493. [DOI] [PubMed] [Google Scholar]
- 12.Heussen C, Dowdle EB. Electrophoretic analysis of plasminogen activators in polyacrylamide gels containing sodium dodecyl sulfate and copolymerized substrates. Anal. Biochem. 1980;102:196–202. doi: 10.1016/0003-2697(80)90338-3. [DOI] [PubMed] [Google Scholar]
- 13.Rawlings ND, Morton FR, Kok CY, Kong J, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 2008;36:D320–D325. doi: 10.1093/nar/gkm954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haas W, Faherty BK, Gerber SA, Elias JE, et al. Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Mol. Cell. Proteomics. 2006;5:1326–1337. doi: 10.1074/mcp.M500339-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Baum BJ, Bird JL, Millar DB, Longton RW. Studies on histidine-rich polypeptides from human parotid saliva. Arch. Biochem. Biophys. 1976;177:427–436. doi: 10.1016/0003-9861(76)90455-0. [DOI] [PubMed] [Google Scholar]
- 16.Dipaola C, Mandel ID. Lactoferrin concentration in human parotid saliva as measured by an enzyme-linked immunosorbent assay (ELISA) J. Dent. Res. 1980;59:1463–1465. doi: 10.1177/00220345800590090101. [DOI] [PubMed] [Google Scholar]
- 17.Hofmann W, Junk A, Geiger R. Human tissue kallikrein. II. Isolation and characterization of human salivary kallikrein. Hoppe-Seyler’s Z. Physiol. Chem. 1983;364:425–432. doi: 10.1515/bchm2.1983.364.1.425. [DOI] [PubMed] [Google Scholar]
- 18.Jenzano JW, Courts NF, Timko DA, Lundblad RL. Levels of glandular kallikrein in whole saliva obtained from patients with solid tumors remote from the oral cavity. J. Dent. Res. 1986;65:67–70. doi: 10.1177/00220345860650011201. [DOI] [PubMed] [Google Scholar]
- 19.Jenzano JW, Daniel PA, Kent RT, Leal JL, Koth DL. Evaluation of kallikrein in human parotid and submandibular saliv. Arch. Oral Biol. 1986;31:627–628. doi: 10.1016/0003-9969(86)90088-9. [DOI] [PubMed] [Google Scholar]
- 20.Cox SW, Gazi MI, Eley BM. Dipeptidyl peptidase II-and IV-like activities in gingival tissue and crevicular fluid from human periodontitis lesions. Arch. Oral Biol. 1992;37:167–173. doi: 10.1016/0003-9969(92)90086-n. [DOI] [PubMed] [Google Scholar]
- 21.Kunimatsu K, Yamamoto K, Ichimaru E, Kato Y, Kato I. Cathepsins B, H and L activities in gingival crevicular fluid from chronic adult periodontitis patients and experimental gingivitis subjects. J. Periodont. Res. 1990;25:69–73. doi: 10.1111/j.1600-0765.1990.tb00894.x. [DOI] [PubMed] [Google Scholar]
- 22.Buchmann R, Hasilik A, Nunn ME, Van Dyke TE, Lange DE. PMN responses in chronic periodontal disease: evaluation by gingival crevicular fluid enzymes and elastase-alpha-1-proteinase inhibitor complex. J. Clin. Periodont. 2002;29:563–572. doi: 10.1034/j.1600-051x.2002.290613.x. [DOI] [PubMed] [Google Scholar]
- 23.Michalski JP, McCombs CC, Sheth S, McCarthy M, deShazo R. A modified double antibody sandwich enzyme-linked immunosorbent assay for measurement of alpha-1-antitrypsin in biologic fluids. J. Immunol. Methods. 1985;83:101–112. doi: 10.1016/0022-1759(85)90063-8. [DOI] [PubMed] [Google Scholar]
- 24.Isemura S, Saitoh E, Sanada K. Characterization and amino acid sequence of a new acidic cysteine proteinase inhibitor (cystatin SA) structurally closely related to cystatin S, from human whole saliva. J. Biochem. 1987;102:693–704. doi: 10.1093/oxfordjournals.jbchem.a122107. [DOI] [PubMed] [Google Scholar]
- 25.Hayakawa H, Yamashita K, Ohwaki K, Sawa M, et al. Collagenase activity and tissue inhibitor of metalloproteinases-1 (TIMP-1) content in human whole saliva from clinically healthy and periodontally diseased subjects. J. Periodont. Res. 1994;29:305–308. doi: 10.1111/j.1600-0765.1994.tb01226.x. [DOI] [PubMed] [Google Scholar]
- 26.Denny P, Hagen FK, Hardt M, Liao L, et al. The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. J. Proteome Res. 2008;7:1994–2006. doi: 10.1021/pr700764j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu S, Xie Y, Ramachandran P, Ogorzalek Loo RR, et al. Large-scale identification of proteins in human salivary proteome by liquid chromatography/mass spectrometry and two-dimensional gel electrophoresis-mass spectrometry. Proteomics. 2005;5:1714–1728. doi: 10.1002/pmic.200401037. [DOI] [PubMed] [Google Scholar]
- 28.Wilmarth PA, Riviere MA, Rustvold DL, Lauten JD, et al. Two-dimensional liquid chromatography study of the human whole saliva proteome. J. Proteome Res. 2004;3:1017–1023. doi: 10.1021/pr049911o. [DOI] [PubMed] [Google Scholar]
- 29.Xie H, Rhodus NL, Griffin RJ, Carlis JV, Griffin TJ. A catalogue of human saliva proteins identified by free flow electrophoresis-based peptide separation and tandem mass spectrometry. Mol. Cell. Proteomics. 2005;4:1826–1830. doi: 10.1074/mcp.D500008-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Amado FM, Vitorino RM, Domingues PM, Lobo MJ, Duarte JA. Analysis of the human saliva proteome. Expert Rev. Proteomics. 2005;2:521–539. doi: 10.1586/14789450.2.4.521. [DOI] [PubMed] [Google Scholar]
- 31.Jenzano JW, Su HW, Featherstone GL, Lundblad RL. Molecular diversity of tissue kallikrein in human saliva. Agents Actions Suppl. 1992;38:137–144. doi: 10.1007/978-3-0348-7321-5_18. [DOI] [PubMed] [Google Scholar]
- 32.Massucci MT, Giansanti F, Di Nino G, Turacchio M, et al. Proteolytic activity of bovine lactoferrin. Biometals. 2004;17:249–255. doi: 10.1023/b:biom.0000027700.90780.45. [DOI] [PubMed] [Google Scholar]
- 33.Fine DH, Furgang D, Beydouin F. Lactoferrin iron levels are reduced in saliva of patients with localized aggressive periodontitis. J. Periodont. 2002;73:624–630. doi: 10.1902/jop.2002.73.6.624. [DOI] [PubMed] [Google Scholar]
- 34.Barrett AJ, Rawlings ND, Woessner JF. Handbook of Proteolytic Enzymes. London: Academic Press; 2004. p. 1896. [Google Scholar]
- 35.Robinson S, Niles RK, Witkowska HE, Rittenbach KJ, et al. A mass spectrometry-based strategy for detecting and characterizing endogenous proteinase activities in complex biological samples. Proteomics. 2008;8:435–445. doi: 10.1002/pmic.200700680. [DOI] [PubMed] [Google Scholar]
- 36.Yasuoka S, Ohnishi T, Kawano S, Tsuchihashi S, et al. Purification, characterization, and localization of a novel trypsin-like protease found in the human airway. Am. J. Resp. Cell Mol. Biol. 1997;16:300–308. doi: 10.1165/ajrcmb.16.3.9070615. [DOI] [PubMed] [Google Scholar]
- 37.Soder PO. Proteolytic activity in the oral cavity: proteolytic enzymes from human saliva and dental plaque material. J. Dent. Res. 1972;51:389–393. doi: 10.1177/00220345720510022601. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura M, Slots J. Salivary enzymes. Origin and relationship to periodontal disease. J. Periodont. Res. 1983;18:559–569. doi: 10.1111/j.1600-0765.1983.tb00393.x. [DOI] [PubMed] [Google Scholar]
- 39.Helmerhorst EJ, Sun X, Salih E, Oppenheim FG. Identification of Lys-Pro-Gln as a novel cleavage site specificity of saliva-associated proteases. J. Biol. Chem. 2008;283:19957–19966. doi: 10.1074/jbc.M708282200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo T, Rudnick PA, Wang W, Lee CS, et al. Characterization of the human salivary proteome by capillary isoelectric focusing/nanoreversed-phase liquid chromatography coupled with ESI-tandem MS. J. Proteome Res. 2006;5:1469–1478. doi: 10.1021/pr060065m. [DOI] [PubMed] [Google Scholar]
- 41.Galliano MF, Toulza E, Gallinaro H, Jonca N, et al. A novel protease inhibitor of the alpha2-macroglobulin family expressed in the human epidermis. J. Biol. Chem. 2006;281:5780–5789. doi: 10.1074/jbc.M508017200. [DOI] [PubMed] [Google Scholar]
- 42.Feldman SR, Gonias SL, Pizzo SV. Model of alpha 2-macroglobulin structure and function. Proc. Natl. Acad. Sci. USA. 1985;82:5700–5704. doi: 10.1073/pnas.82.17.5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ganrot PO. Alpha-2-anti-trypsin activity and different trypsin substrates. Clin. Chim. Acta. 1966;13:518–521. doi: 10.1016/0009-8981(66)90246-4. [DOI] [PubMed] [Google Scholar]
- 44.Enghild JJ, Salvesen G, Thogersen IB, Pizzo SV. Proteinase binding and inhibition by the monomeric alphamacroglobulin rat alpha 1-inhibitor-3. J. Biol. Chem. 1989;264:11428–11435. [PubMed] [Google Scholar]
- 45.Dobo J, Gettins PG. Alpha1-proteinase inhibitor forms initial non-covalent and final covalent complexes with elastase analogously to other serpin-proteinase pairs, suggesting a common mechanism of inhibition. J. Biol. Chem. 2004;279:9264–9269. doi: 10.1074/jbc.M311731200. [DOI] [PubMed] [Google Scholar]
- 46.Carrell R, editor. Serpins: The Superfamily of Serine Proteinase Inhibitors. Amsterdam: Elsevier Biomedical Press; 1986. pp. 403–419. [Google Scholar]
- 47.Remold-O’Donnell E, Chin J, Alberts M. Sequence and molecular characterization of human monocyte/neutrophil elastase inhibitor. Proc. Natl. Acad. Sci. USA. 1992;89:5635–5639. doi: 10.1073/pnas.89.12.5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benarafa C, Remold-O’Donnell E. The ovalbumin serpins revisited: perspective from the chicken genome of clade B serpin evolution in vertebrates. Proc. Natl. Acad. Sci. USA. 2005;102:11367–11372. doi: 10.1073/pnas.0502934102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cooley J, Takayama TK, Shapiro SD, Schechter NM, Remold-O’Donnell E. The serpin MNEI inhibits elastase-like and chymotrypsin-like serine proteases through efficient reactions at two active sites. Biochemistry. 2001;40:15762–15770. doi: 10.1021/bi0113925. [DOI] [PubMed] [Google Scholar]
- 50.Lopez-Otin C, Matrisian LM. Emerging roles of proteases in tumour suppression. Nat. Rev. 2007;7:800–808. doi: 10.1038/nrc2228. [DOI] [PubMed] [Google Scholar]
- 51.Manzone H, Billings PC, Cummings WN, Feldman R, et al. Levels of proteolytic activities as intermediate marker endpoints in oral carcinogenesis. Cancer Epidemiol. Biomarkers Prev. 1995;4:521–527. [PubMed] [Google Scholar]
- 52.Quesada V, Ordonez GR, Sanchez LM, Puente XS, Lopez-Otin C. The Degradome database: mammalian proteases and diseases of proteolysis. Nucleic Acids Res. 2009;37:D239–D243. doi: 10.1093/nar/gkn570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.