Abstract
The premise of today’s drug development is that the mechanism of a disease is highly dependent upon underlying signaling and cellular pathways. Such pathways are often composed of complexes of physically interacting genes, proteins, or biochemical activities coordinated by metabolic intermediates, ions, and other small solutes and are investigated with molecular biology approaches in genomics, proteomics, and metabonomics. Nevertheless, the recent declines in the pharmaceutical industry’s revenues indicate such approaches alone may not be adequate in creating successful new drugs. Our observation is that combining methods of genomics, proteomics, and metabonomics with techniques of bioimaging will systematically provide powerful means to decode or better understand molecular interactions and pathways that lead to disease and potentially generate new insights and indications for drug targets. The former methods provide the profiles of genes, proteins, and metabolites, whereas the latter techniques generate objective, quantitative phenotypes correlating to the molecular profiles and interactions. In this paper, we describe pathway reconstruction and target validation based on the proposed systems biologic approach and show selected application examples for pathway analysis and drug screening.
Keywords: iomarker discovery, drug discovery, high-content screen, high-throughput screen, RNAi, system bioinformatics and bioimaging, target validation
I. INTRODUCTION
Instead of studying one gene or one protein at a time, biologists today believe that many genes or proteins interact and that the deciphering and modeling of interaction among them would better assist our understanding of the underlying disease mechanism [1]. Systems biology attempts to identify individual components and molecules by studying the vast, complex networks that biological molecules create to regulate and control life. The analysis and modeling of such complex biological networks beyond the capabilities of existing computational techniques are needed for this new scientific endeavor. In this paper, we discuss two major classes of computational techniques used in system biology: bioinformatics and bioimaging informatics. Bioinformatics in system biology applies to the biomarker discovery and reconstruction of regulatory signals in biological networks. Bioimaging informatics, on the other hand, focuses on the analysis and modeling of biological image data generated for secondary screening, target validation, and drug discovery for lead selection.
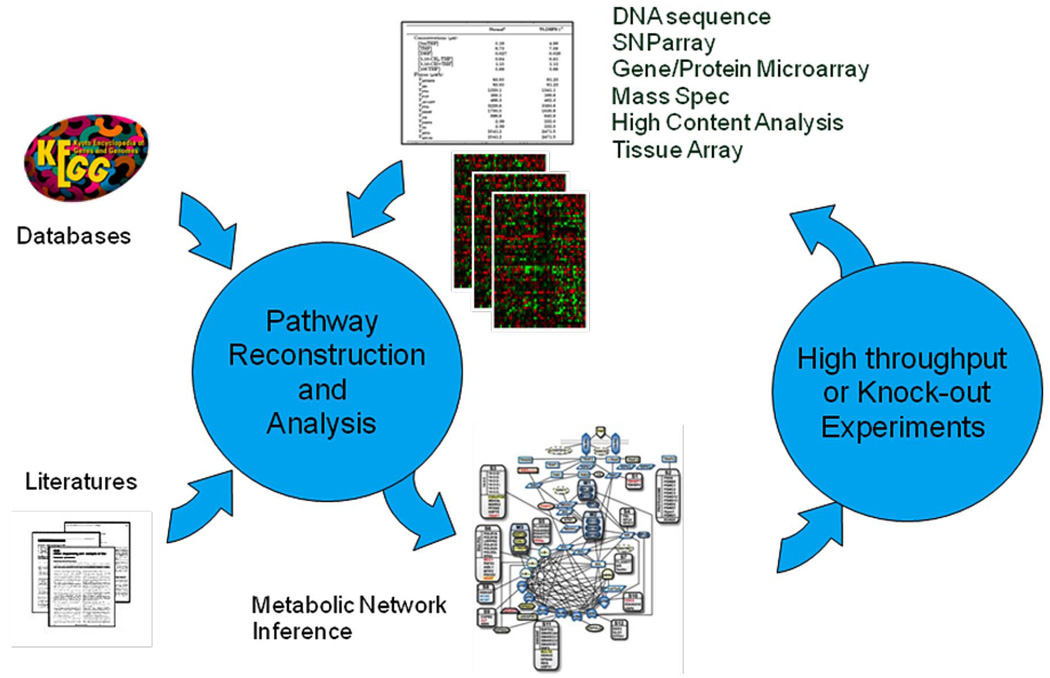
In addition, we emphasize the integration of the two classes of computational techniques in studying disease mechanisms systematically. Fig. 1 summarizes our proposed approach. Starting out from the biological hypothesis of a disease, we ask what the mechanism(s) is behind the disease. The recent advent of high-throughput devices such as gene microarray, single nucleotide polymorphism (SNP) array, and mass spectrometry allow fast screening of a large number of candidates to generate hits of biomarkers. After we identify the candidate biomarkers, the next step is to validate them via techniques such as polymerase chain reaction, RNA interference (RNAi), protein array, and immunohistochemistry and tissue array. Then, we investigate how these biomarkers interact with each other to infect the disease, in the context of signaling pathways or metabolic networks. We reconstruct the signaling pathways by combining and analyzing information available in the public domain, including, but not limited to, DNA sequence data, protein sequence data, information from the literature, pathways recorded in public databases, gene expression data, mass spectrum data, and metabolic profiles. However, these networks often have many false positives or false negatives due to noise and incomplete data.
Fig. 1.
Inferring signal pathway using systematic approach.
In this paper, for the biomarker discovery, we first briefly review discovery from expression data and then focus on biomarker selection from mass spectrometry (MS) data. For the signaling network reconstruction, on the other hand, we will review the network inference from metabolites and gene regulatory networks to protein interactions.
Once we have candidate biomarkers and signaling networks, we can perform target screening to validate biomarkers, drug lead selection, and disease diagnosis and prognosis. This is known as secondary screening. One emerging secondary screening modality is high-content screening (HCS), which involves the automation of light microscopy with multiwell plates, combined with the availability of fluorescent probes that are attached to specific subcellular components, such as chromosomes and microtubules, for visualization of cellular phenotypes and activities, such as mitosis, using epifluorescence microscopy techniques. RNAi screening is another tool to study gene function and target validation, in which double-stranded RNAs (dsRNAs) are applied to a live cell to interfere with a specific mRNA, thereby disrupting expression of a certain gene [2]. Such capability of silencing each individual gene in the whole genome has emerged as arguably one of the best functional genomics tools available to date, providing direct links between genes and biological processes of interest, such as genetic signaling pathways, cell division, cell migration, and disease mechanisms. RNAi differs from other gene-silencing phenomena in that gene silencing caused by RNAi can spread from cell to cell and can generate heritable phenotypes in the first-generation offspring. This paper presents the application of automated RNAi image screening to validate the targets at the cellular biology level. Two examples will be also provided to show how to refine signal pathways: one is the whole genome screening for Rho pathway study and the other is the tuberous sclerosis complex (TSC) pathway study with a gene knockout strategy.
In addition, three types of markers can be used to characterize the spatial features of cells: subcellular structures, location of signaling proteins, and indicators of physiological states. Cellular image analysis methods are being developed to quantify the properties of these structures. Size, shape, intensity, and texture moments are among the common descriptors that can be generated using computational algorithms [3], the Cellprofiler [4], Cellomics [5], GE InCELL 1000, and Q3DM [6]. A few more labs [7]–[9] are also focusing on cellular image segmentation and tracking but do not directly study disease pathways and target validation.
The rest of this paper is organized as follows. Section II reviews the biomarker selection and the signaling network reconstruction, with the focus on the biomarker selection using proteomics techniques. The gene signaling pathway study using genome-wide RNAi screening is presented in Section III, while the analysis of regulatory pathways for dendritic spine morphology is discussed in Section IV. We conclude in Section V.
II. BIOMARKER DISCOVERY AND SIGNAL NETWORK INFERENCE
A. Biomarker Discovery From Gene Microarray
Gene microarrays make it possible to measure simultaneously the expression levels for thousands of genes in a high throughput fashion. By comparing gene expressions in normal and diseased cells, microarrays identify diseased genes and targets for therapeutic drugs. The huge amount of data provided by gene microarray measurement is explored to investigate gene functions and their interdependence, hopefully to provide answers to questions like what type of disease affects a cell or which genes have strong influence on this disease.
Since thousands of genes and only tens of samples exist in microarray data, feature reduction or gene selection is the necessary step for cancer classification. There are mainly two approaches: one is feature reduction and the other is the feature subset selection. For example, transformation methods such as principal component analysis have been applied in cancer classification. Gene selection is an important step in cancer classification and the discovery of gene pathways. Many gene selection methods have been proposed, e.g., the support vector machine method [10], the genetic algorithm [11], the perceptron method [12], Bayesian variable selection [13], [14], and the voting technique [15]. We have also developed new methods in our labs over the years, including the mutual information-based gene and feature-selection method [16] and logistic regression model-based feature-selection method [17]. To cope with the small sample size, the bootstrap technique [18] is employed to obtain more accurate estimation of the mutual information. Although gene selection using entropy and Kulback–Leibler divergence is discussed in Xing et al. [19], that work is based on estimating the distribution of many genes, which is not feasible for classification in this case because the sample size is very small. A linear probit regression classifier was proposed in Albert et al. [20], which is very effective in cancer classification [13]. The strongest genes and features selected by the mutual information criterion actually show a strong nonlinear property because this selection method is a nonlinear procedure [16]. As an application in breast cancer in Fig. 2, we studied 22 breast tumor samples composed of seven BRCA1, eight BRCA2, and seven sporadic. We found gene 1 (tumor protein p53-binding protein, gene 2 (interleukin enhancer binding factor 2, 45kD), and gene keratin 8 are the most important genes [16]. Keratin 8 is a member of the cytokeratin family of genes. Cytokeratins are often used to identify breast cancer metastases by immunohistochemistry.
Fig. 2.
Breast cancer: BRCA1 and BRCA2.
B. Biomarker Discovery From Proteomics
Mass spectrometry (MS) is increasingly used to detect disease-related biomarkers from samples of tissue, serum, or plasma for early diagnosis, prognosis, and the monitoring of disease progression and treatment response. It can be used to differentiate patient samples from one another, such as diseased from normal, or identify patients who are most likely to benefit from particular treatments.
Here we discuss two popular MS techniques. The first technique involves surface-enhanced laser desorption ionization—time of flight (SELDI-TOF) [21], which uses stainless steel- or aluminum-based supports, or chips, engineered with chemical or biological bait surfaces. These varied chemical and biochemical surfaces allow differential capture of proteins based on the intrinsic properties of the proteins themselves. The captured proteins on the chip surface are purified by washing the surface and crystallized with small molecules whose function is to absorb laser energy and transfer it to proteins. Energized protein molecules fly away from the surface into a time-of-flight tube where the time for the molecules to fly through the tube is a function of the molecular weight and charge of the protein.
The second technique, matrix-assisted laser desorption ionization—time of flight (MALDI-TOF) [22], uses a chemical matrix that is premixed with the biological tissues, and when it dries, the mixture will crystallize. The metal plate containing the crystallized sample is then placed into a cavacum chamber where the crystal is bombarded with laser pulses, causing the matrix to vaporize and the protein and peptides within the tissue to ionize. The matrix molecules absorb energy from the laser and transfer it to the proteins, causing them to desorb and ionize and generating a cloud of ionized protein molecules. The electric field accelerates the ionized proteins into a flight tube, where they drift until they strike a detector that records the TOF. Knowing the length of the tube and the applied voltage, researchers can use a quadratic transformation to derive the approximate mass-to-charge ratio of the protein from the observed TOF. The spectral data that result from this experiment consist of the sequentially recorded numbers of ions (the intensities) arriving at the detector coupled with the corresponding mass-to-charge ratio (m/z) values. Peaks in the intensity plot represent proteins that are present in the sample. To identify the proteins, getting the peaks from the background is needed. The background includes noise that results from the application of the matrix and limitation in the detector, and makes the quantification of the spectra difficult. Another artifact affecting the spectra is the baseline, which affects both the peak detection algorithms and sample-to-sample comparisons.
Fig. 3 illustrates a pipeline of biomarker discovery and diagnosis. The computational issues consist of low-level processing, biomarker selection, sample classification, and prediction, as well as protein/peptide identification. Despite their increasing role in biomarker discovery, only a few published papers have dealt with the preprocessing of the MS data. Most researchers get the peaks from the raw data with the software provided by the equipment vendors. For the SELDI data, each peak may correspond to a real protein or peptide. In Yasui et al. [23], the peaks are obtained by examining whether the intensity at the point is the highest among its nearest N points neighborhood set. If it is the highest, it then is taken as a peak. To calibrate the protein m/z measurements across samples, a shift window of size 0.2% of the m/z is defined. The detection of peaks will not be accurate due to noisy background; thus we first provide a statement and a formulation of the low-level processing.
Fig. 3.
A pipeline of biomarker discovery and diagnosis.
Low-Level Processing
Low-level processing is a key in the MS data analysis. Assume we have n spectra in study, and each takes on the same equally spaced grid of length T of TOF tj with j = 1, 2, … , T. Let xi(tj) denote the observed log spectral intensity for spectra i at TOF tj, while si(tj) consists of a sum of possibly overlapping peaks, each corresponding to a biological molecule. According to the work in Morris et al. [24], the following model is applied to modeling the mass spectra data:
| (1) |
where bi(tj) is the baseline function, Ni is the normalization factor, and noise term eij follows a Gaussian distribution with eij ~ N(0, σ2(tj)). The following steps are necessary to perform.
Calibration maps the observed TOFs tj, j = 1, 2, … , T, to a set of inferred (m/z) ratios xj, j = 1, 2, … , T.
Filtering removes the random noise using adaptive wavelet transform.
Baseline subtraction removes the baseline artifact by computing a monotone local minimum curve on the denoised signal.
Normalization corrects for systematic differences in the total amount of protein desorbed from the sample plate by dividing the total ion current defined to be the mean intensity of the denoised and baseline corrected spectrum. After steps 1)–4), we obtain the estimated signal of si(t).
Peak detection and quantification: First find all local maxima and the associated peak endpoints and then calculate the signal-to-noise ratio (SNR) at each local maximum; the local maxima with SNR being bigger than a threshold.
Peak matching across the samples is necessary to decide which peaks in different samples correspond to the same biomolecule. After those steps, normalization and batch effect removal are performed between sample replicates in different chips and bioprocessors.
MALDI data have a higher resolution than SELDI data. Each true peak corresponds to a cluster of isotopic peaks. But for the baseline correction and denoising, most methods to handle SELDI data can be applied to process MALDI data. The undecimated discrete wavelet transform is applied to do the denoising first in Morris et al. [24] using the model in (1). A monotone decreasing function is assumed for the baseline and then the baseline is removed. For this method, a constant threshold for the wavelet transform is applied. But in practice, the noise does not follow the identical distribution. So this can cause under-smoothing for the high-level noise and oversmoothing of the low-level noise area. The baseline is not always monotone decreasing, and the assumption is dependent on the data sets under investigation. Another paper based on wavelet transform is by Du et al. [25]. This one uses continuous wavelet transform. This method can generate high false negative rates due to the constant sliding window size. A few other approaches are also developed for baseline correction in the MALDI data analysis. Yu and colleagues [26] describe a method to detect the peaks in MALDI data in which the median values within the local neighborhood are computed first and baseline was obtained with the cubic interpolation. Then, the Gaussian filter is applied to smooth the baseline corrected data. A Gabor quadrature filter is applied to extract the envelope signal and get the monoisotopic peak for the isotopic peaks. Finally, the local maximum search is applied to get the peaks. For the Gabor filter to get the monoisotopic peak, it may smooth the peaks. In brief, peak identification is still an open issue.
Biomarker Discovery
Technically, biomarker discovery is intrinsically linked to the variable selection methodology in the area of machine learning and pattern recognition. The mathematical definition of a variable selection problem is to select a subset {y1, y2, … ,yd} given a feature set {x1,x2, … ,xD} to optimize a predefined fitness function J(y1, y2, … ,yd), where d < D.. In the classification or prediction problem, the cost function is usually selected as the accuracy of the predictor. The exhausted search method requires going through all of the possible combinations, which could achieve the exponential computation complexity O(Dd). Therefore, the full search is not feasible for most real applications because of its extremely expensive computation cost. Roughly speaking, there are two categories of variable selection techniques: filter and wrapper [27]. The filter-based techniques use the data statistic characteristics as the criteria to find a subset of features, which can keep most class-relevance and reduce variable redundancy. The wrapper techniques use the accuracy of the predictor as the criteria and then apply some optimization techniques to obtain the global or local optima of the criteria function. Genetic algorithm (GA), originally proposed by Holland, is a conventional wrapper-based method, which mimics the evolutionary process of survival of the fittest. However, the standard GA cannot output the satisfied panel of biomarker selection results in our work because the random search ability severely degenerates due to the high-dimensional biomarker candidate set and the dramatically decreased dimensionality of the target subset. Therefore, we proposed to use the recursively local floating search technique to enhance the individuals for each generation of evolution. Linear discriminant analysis is selected as the fitness function because of its efficient computation [28].
As an example in our Major Adverse Cardiac Events (MACE) proteomic project [28] on 120 patients, the original biomarker set is relatively big for all of the fractions and protein chips. Most traditional pattern classification methods are well established for large data set learning. Here we use two famous small-set and binary response-based classification techniques, partial least squares logistic regression (PLS-LR), and support vector machine classifier, as the classifier for our MACE prediction study. The characteristics of PLS-LR and SVC match well with our MACE prediction problem because SELDI-MS data obtained are binary response biomarkers indicating the MACE group and control group as well as the rather small sample set.
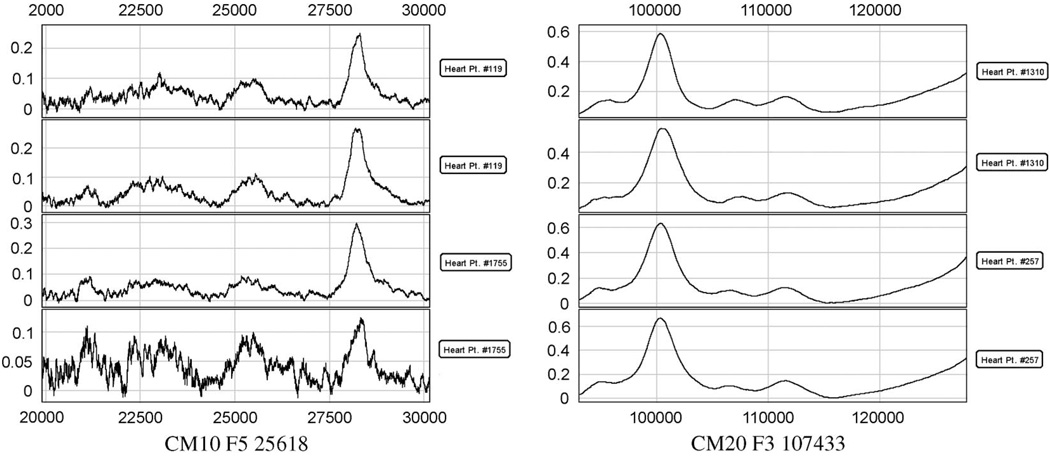
In order to validate the performance of the improved GA (IGA) algorithm, we conducted the biomarker selection experiments on the MACE data set [28], which is described in the Introduction. We used the biomarkers generated from fragment 3 and 4 with the SELDI chips IMAC and CM10. There are a total of 421 biomarkers, including the myeloperoxidase (MPO) value. The objective is to select five biomarkers for MACE prediction. In Fig. 4, we display two samples with 421 biomarkers. One is from the control group and the other is from the MACE group. For the feature-selection procedure, we use the linear discriminant function as the fitness function. The following experiments compare the five selected biomarkers by IGA with the MPO value, five selected biomarkers by T-test, five selected biomarkers by standard GA, and five selected biomarkers by standard sequential forward floating search. Table 1 lists the experimental results. The same parameters for standard GA and improved GA are set: the population size is 60, the maximum generation size is 20, the crossover rate is 0.5, and the mutation rate is 0.3. The experimental results show that the IGA improves the prediction accuracy by 20% compared to MPO and achieves more than 75%. Fig. 5 shows two examples of peaks selected using this approach. Currently, we are studying the protein identification based on the selected candidate biomarkers.
Fig. 4.
Two samples with 421 biomarkers. One is from control group and the other is from MACE group.
Table 1.
The MACE Prediction Accuracy Comparison Study
| Approach | MPO Value |
T- test | Standard GA | SFFS | Improved GA |
|---|---|---|---|---|---|
| Accuracy | 55.25% | 62.23% | 69.05% | 71.92% | 75.16% |
Fig. 5.
Selected peaks in the MACE project.
C. Signaling Network Inference
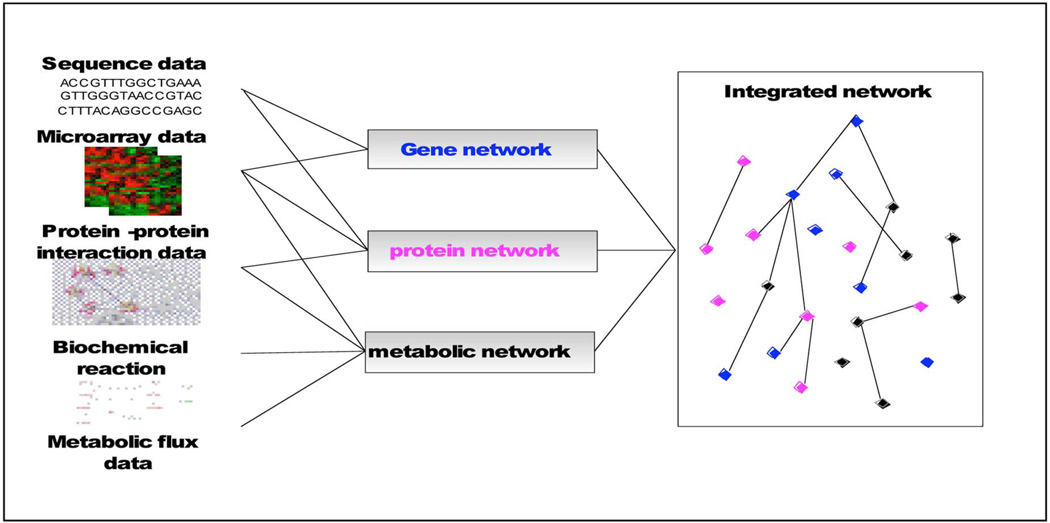
Network modeling plays a central role in systems biology, which attempts to mathematically describe the interactions among the components identified by reductive approaches [29], [30]. There are three levels of networks; gene regulatory networks, protein interaction networks, and metabolic networks. Making use of high-throughput microarray data and DNA sequence data, we can reconstruct gene regulatory networks. The transcription factors are protein molecules that directly interact with genomic DNA to mediate transcription of messenger RNAs. Protein networks can be reconstructed from the literature, public databases, and proteomics data such as mass spectrometry. Metabolic networks can be reconstructed from the literature, public databases, metabolic profiles, and reaction databases, whereas transcriptomics, metabolomics, proteomics, fluxomics and other high-throughput techniques can provide suitable quantitative data for the reconstruction and validation of network models; see Fig. 6. Recently, the literature has reported research progress made in the modeling of integrative networks [31]–[33]. For example, Yeang et al. [34] proposed a joint factor graph model of gene regulation and metabolic reactions, inferring the interactions between metabolites and transcription factors to fit perturbation data. The study of Segal et al. [35] discussed a unified probabilistic algorithm for identifying a “pathway” from gene expression and protein interaction data. Yamanishi et al. [36] presented a supervised kernel-based method to infer enzyme networks from the combination of gene expression, localization data, phylogenetic profile, and chemical compatibility information. Although currently only coarse network reconstruction and modeling are being reported, the integration of heterogenous data at multiple biological levels will undoubtedly open up more vistas to advance systems biology studies.
Fig. 6.
Signaling network inference.
III. SIGNALING NETWORK STUDY VIA HIGH-CONTENT SCREENING
Biological Investigation Using High-Content Screening
Cellular networks can be regarded as three-dimensional (3-D) maps depicting pathways from which higher cellular functions emerge. The dynamics of molecular interactions within these reaction cascades can be assessed in a living cell by the application of fluorescence microscopy, which allows one to correlate such phenomena as cell cycle progression, cell migration and motility, secretion, volume control and regulation of growth, and morphogenesis and cell death. Within fluorescence microscopy, the development of genetically encoded variants of green-fluorescent proteins (GFPs) as tags for proteins and indicators of small solutes has revolutionized our insights into biochemistry at the microscopic level, with the advantage of preserving the cell’s biochemical-connectivity context, compartmentalization, and spatial organization. This development parallels recent progress in genomics, proteomics, and metabolonomics, through which functional attributes are assigned to genes and gene products by alignment to well-characterized sequences or by comparison to models. However, presently, only a small percentage of newly identified products can be categorized in this manner, and further progress depends on the collection of huge amounts of experimental data from functional and microscopic assays.
The analysis of genetically expressed GFP-based fluorochromes is destined to follow the dynamic trafficking and clustering of gene products and the study of spatial-temporal distribution patterns of small solutes in living cells kept under normal physiological conditions. HCS using automated fluorescence microscopy and multiwell plates adds a new dimension to these studies by fast screening hits and functional effectors. Starting from those effectors, we can study their interactions from a systems viewpoint such as that of signaling networks. In this section, we will describe how to study gene function using HCS genome-wide RNAi screening. In Section IV, we will discuss how to validate regulatory pathways through gene knockout via 3-D two-photon microscopy neuron imaging.
A. Gene Signaling Pathway Study From Genome-Wide RNAi Screening for Rho GTPase
We reviewed a number of general issues facing microscopy imaging informatics in the paper by Li et al. [37]: normalization, clustering of compounds and cellular phenotypes, representation of compounds, phenotype classification, cell phase identification in cell cycle studies, particle spatial statistical distributions using K functions, and the distribution of the dwelling time of cell phases in time-lapse microscopy images. In this section, we discuss additional challenging factors, including novel phenotype discovery, gene function clustering, gene scoring, and metabolic networks, particularly relating to HCS.
Using images acquired by automated microscopy, biologists visualize phenotypic changes resulting from reverse-functional analysis by the treatment of Drosophila cells in culture with gene-specific dsRNAs [2]. In a small-scale study by manual analysis [38], biologists were able to observe a wide range of phenotypes with affected cytoskeletal organization and cell shape. Nonetheless, without the aid of computerized image analysis tools, it is intractable to characterize morphological phenotypes of a large population of cells quantitatively and to identify genes as well as their dynamic relationships required for distinct cell morphologies on a genome-wide scale.
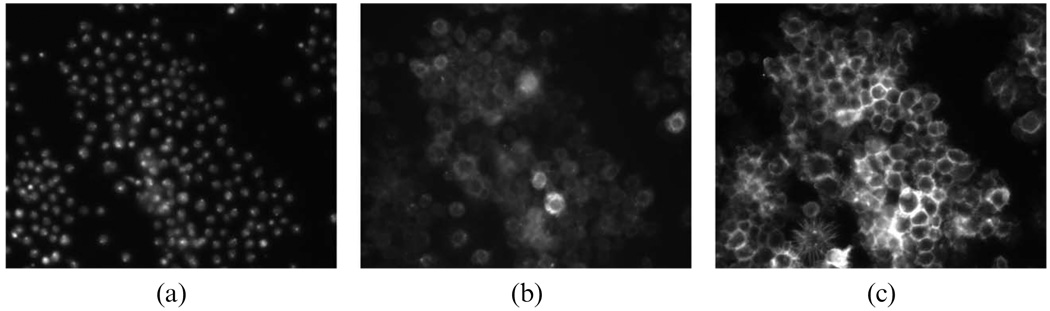
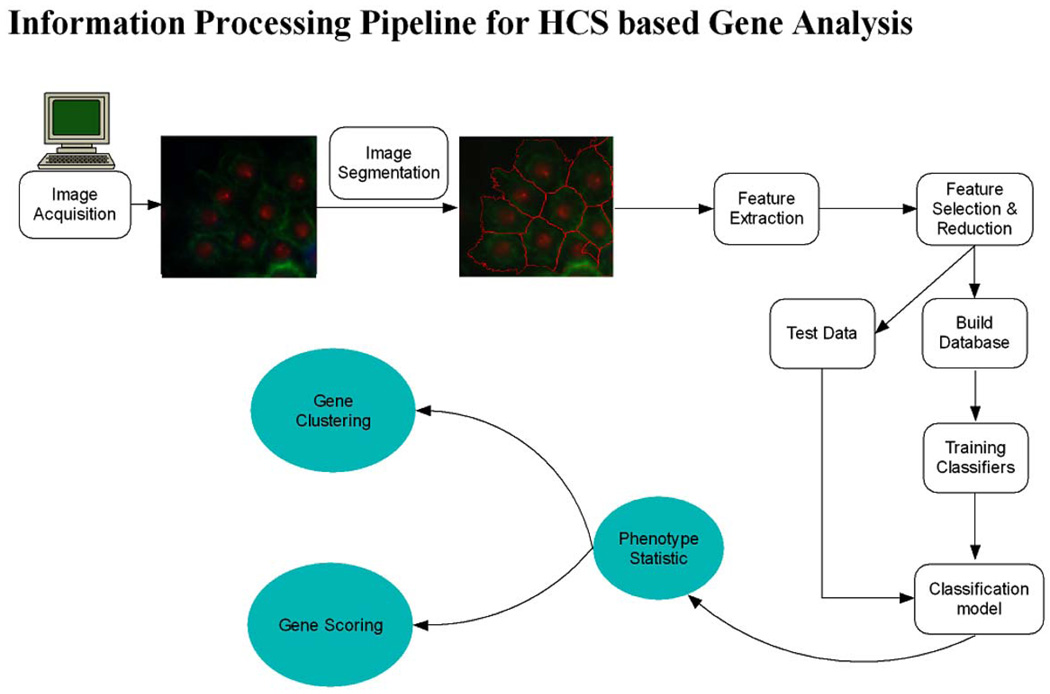
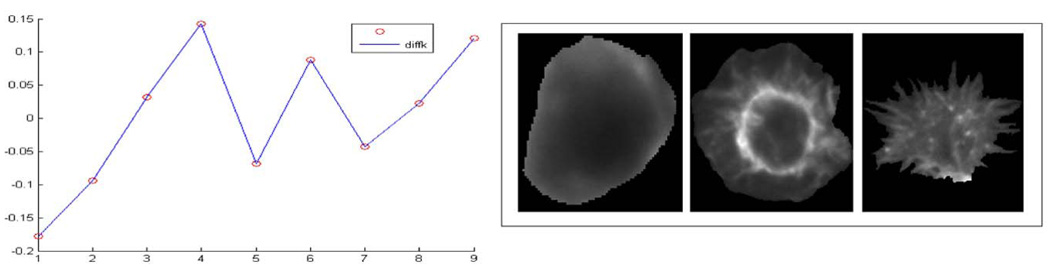
We applied computerized morphological analysis to automate genome-wide RNAi high-content screening for novel effectors of Rho family GTPases in Drosophila cells. About 21 000 dsRNAs specific to predict Drosophila genes were robotically arrayed in 384-well plates. The Drosophila cells were plated and took up dsRNA from culture media. After incubation with the dsRNA, expression of Rac1V12, RhoAV14 or Cdc42V12 is induced, cells are fixed, stained, and imaged by automated microscopy. Each screen will generate ~400 000 images, or millions if including replicates. Biologists of the project have developed a cell-based assay for Rho GTPase activity using the Drosophila Kc167 embryonic cell line. Three-channel images are obtained by labeling F-actin, GFP-Rac, and DNA. Fig. 7 shows an example of RNAi cell images of one well acquired with three channels for phenotypes of (a) DNA, (b) Actin, and (c) Rac. It is difficult to segment the cells from (b) or (c). The two phenotypes can be found from this figure too; S-spiky and R-ruffling, named after the cell shape. The issue is how to identify the three phenotypes automatically for each image in the large population of cells screened. To this, we proposed an information-processing pipeline as shown in Fig. 8. The pipeline consists of image segmentation, phenotype discovery, feature reduction, phenotype classification, gene functional annotation, and gene scoring. The next sections describe these components in more detail.
Fig. 7.
Gives an example of RNAi cell images of one well acquired with three channels for phenotypes of (a) DNA, (b) Actin, and (c) Rac.
Fig. 8.
Informatics pipeline for genome-wide RNAi screening.
B. Image Segmentation
Large-scale intensity variations as well as shading and shadowing effects in our images are often caused by uneven illumination over the field of view. A data-driven approach is employed to deal with this problem [39]. The algorithm works by iteratively making better distinction of the background of the image. A cubic B-spline surface is employed to model the background shading. After removing the shading, we adopt morphological transformation to enhance the image’s contrast. The basic idea is to segment the nuclei in the DNA channel and then segment cell cytoplasm based on the seeds obtained from the DNA channel using level-sets. The key issue is how to keep the details for those cells with rapid changes at the edges.
Cytoplasm Segmentation Using a Novel Deformable Model
An active contour deforms until it reaches the boundary of the object to be detected. This is accomplished by constructing and solving a partial differential equation (PDE) that directs the evolution of the contour from its initial position and shape. Let C(q) : [0,1] → ℝ2 be a parameterized planar curve and let I : [0, a] × [0, b] → ℝ+ be an input image in which the task of segmentation is considered. A fairly generic curve evolution can be defined by the following PDE [40]: , where κ is the Euclidean curvature, N⃗ is the unit inward normal of the curve, α and β are scalar fields, and S⃗ is a vector field defined on I. All of these fields may be position- and time-dependent. The first term expands or shrinks the curve along its normal. The second term deforms the curve guided by the vector field, while the third term uses the curvature to make the curve stay smooth. How to incorporate the image features effectively is the primary challenge when one decides to employ a geometric active contour to achieve one’s goal. Both edge and region properties can be utilized to improve the performance. We designed a novel PDE to solve this problem [41]. The equation is formulated as
| (2) |
where c is a constant. The term g(I) = 1/(1 + |∇I|) is to speed up the flow in those regions where the image gradient is low (g ≅ 1) or slow it down where there is a high gradient (g ≅ 0). The term −∇g(I) acts like a doublet, which attracts the curve closer to the edge, since it points towards the valley of g(I), the centerline of the edge [42]. A term h(I) is introduced using fuzzy c-means clustering [41], and λ is a constant in the range (0,1). Term −∇g(I) increases the capture range of the doublet. A constant d is added to control the strength of doublet force. The curvature term [41] is attenuated by a factor e ∈ (0,1). The factor e is necessary because we want to control the effect of curvature term on contour smoothing in order to handle the spiky shape as well as the normal round shape, both of which are common in RNAi images [42].
For the implementation of our method, we adopt the level-set approach [43]. Instead of manipulating the curve C directly, the curve is embedded as the zero level-set of a higher dimensional function, namely, level-set function Ψ : [0,a] × [0, b] → ℝ. The level-set function is then evolved under the specific PDE. Meanwhile, the status of the curve can be obtained by extracting the zero level-set Ψ(x, t) = 0. Equation (3) can be easily transferred to the level set representation [41]:
| (3) |
Every cell owns one level-set function, initialized from the segmented nucleus and evolved independently from others. This scheme works naturally for the case of isolated cells because the curve will stop at the boundary of the cell under investigation without interfering with other cells [41].
Experimentation and Comparison of Existing Software
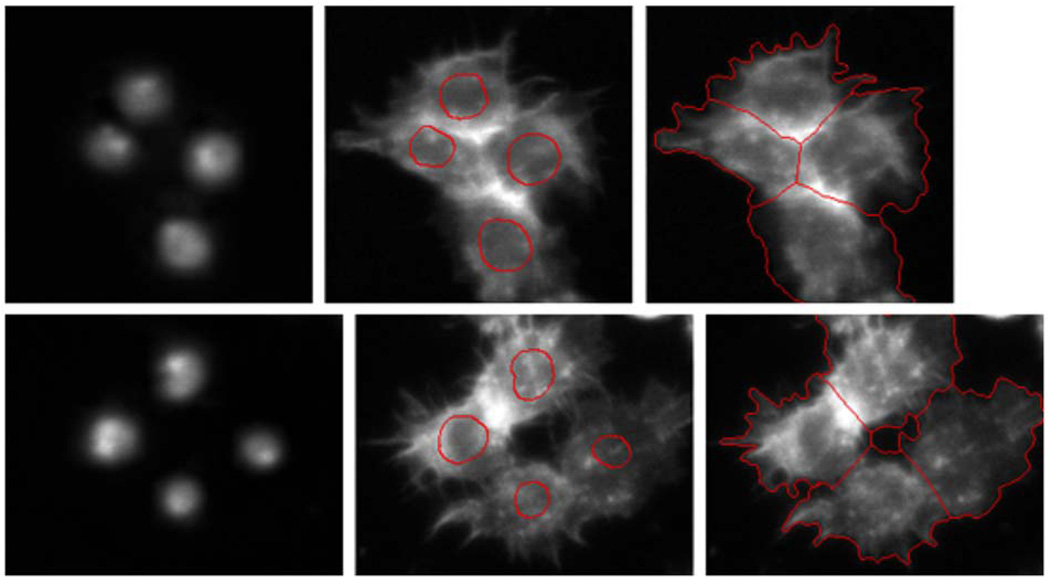
In Fig. 9, we illustrate two segmentation results. The DNA channels are shown in the left panel of Fig. 9. The contours are initialized from nuclei segmentation of the DNA channel shown in the middle panel by the red curve, and they evolve to the final cytoplasm segmentation as shown in the right panel. The relative coincidences of automated and manual segmentation are 98.5%. The comparison of our level-set method with methods of available software packages such as Cellprofiler, Cellomics, GE IN Cell 1000 Developer, and Q3-DM is done based on the image patch, and the segmentation result is shown in Fig. 10. Obviously, Q3DM completely fails, and the boundaries obtained by Cellprofiler, Cellomics, and GE IN Cell 1000 are also not very accurate. The relative coincidences of automated and manual segmentation are 96.4%, 78.1%, 76.9%, 74.3%, and 71.1%, respectively, for the five methods, i.e., our level-sets method, Cellprofiler [4], Cellomics [5], GE IN Cell 1000, and Q3DM [6].
Fig. 9.
The segmentation of spiky shape (a) DNA channels, (b) cells with nuclei segmentation results superimposed, and (c) cytoplasm segmentation.
Fig. 10.
Performance comparison among commercial software, Cellprofiler, and our level-set method.
In [44], we proposed another interactive model to segment RNAi cell images in which nuclei are first extracted from the DNA channel and used as initialization for segmentation of cells in Actin and Rac channels. Cell boundaries are extracted simultaneously by modeling the interaction between them as well as combining both gradient and region information in the image. Promising experimental results demonstrated that automatic segmentation of high throughput genome-wide multichannel screening can be achieved.
Solve Over-Segmentation Using the Feedback System
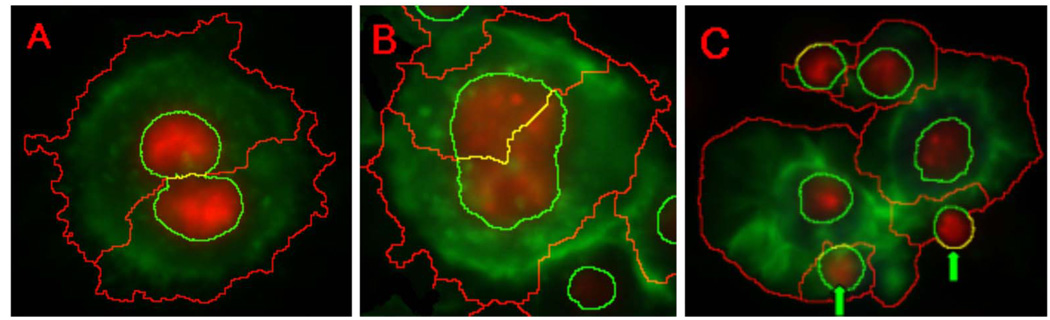
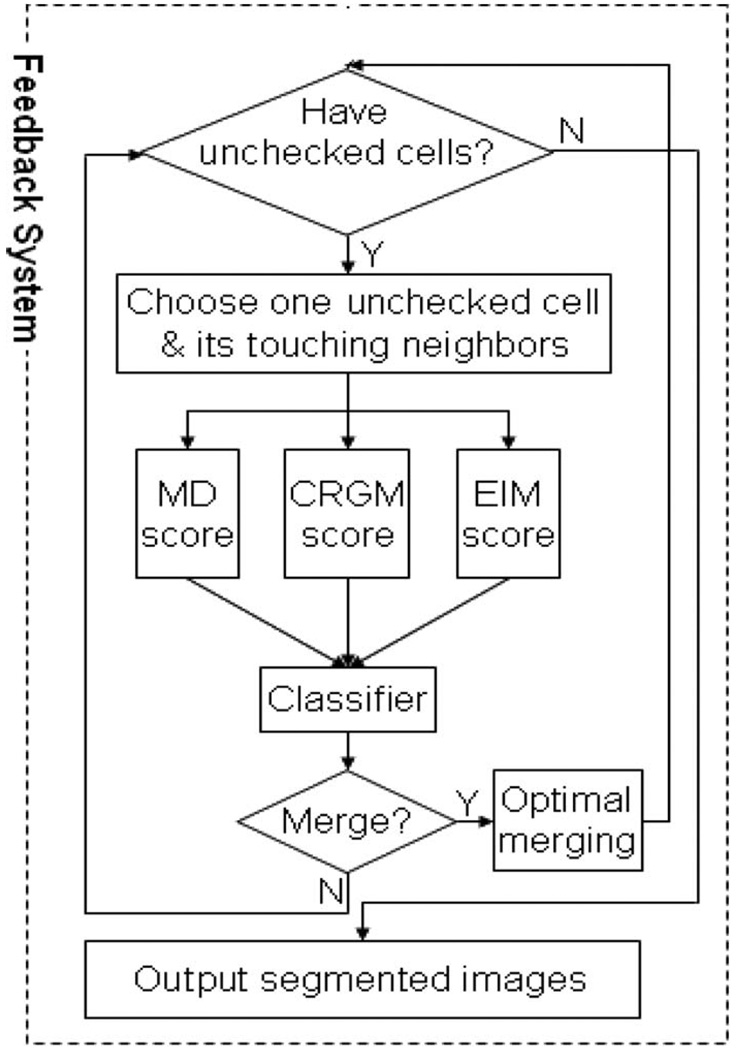
The segmentation methods discussed above are dependent on the segmented nuclei for three-channel based analysis. To visualize nuclei, cells are stained with the ultraviolet-fluorescing DNA-binding molecule 4-in 6-diamidino-2-phenylindole (DAPI). The inherent properties of cells and the DAPI stain lead to oversegmentation. In any given unsynchronized population, cells will be at different stages of the cell cycle, impacting the size, shape, and intensity of the DAPI staining. For example, when cells are in interphase, the DAPI staining is uniform because DNA is dispersed in the entire nucleus. When cells are actively dividing, DNA condenses and separates into two regions within a single cell. The independent DNA segments within one cell result in oversegmentation [Fig. 11(a)]. Additionally, erroneous oversegmentation of nuclei results in oversegmentation of the cell body directly [Fig. 11(b)]. Finally, there are occasionally some nuclei that have little or no cytoplasm (based on staining captured in the F-actin channel) and also result in oversegmentation [Fig. 11(c)]. We developed a fully automated RNAi cell segmentation system that significantly reduces the oversegmentation problem noted in Li et al. [45], and Fig. 12 gives a flowchart of the proposed feedback merging system. The feedback system entails three steps. First, three scoring models capture the specific characteristics of the oversegmented cells. Next, a classifier maps the three scores into a merging decision. Finally, oversegmented cells are detected and merged according to merging decisions.
Fig. 11.
Multiple factors contribute to oversegmentation of cells. Green is F-actin staining, which visualizes the cytoskeleton; red is DAPI staining, which visualizes DNA within the nucleus. (a) An actively dividing cell, which has two DAPI-stained regions, is oversegmented. (b) Oversegmented nucleus results in the oversegmentation of cell directly. (c) Dead nuclei (arrows) result in oversegmentation of cells.
Fig. 12.
An overview flowchart of the entire segmentation system for HCS image analysis.
C. Novel Phenotype Discovery Using Clustering
There is a great deal of interest in automating the discovery of cell-level patterns and in discovering patterns at multiple biological scales via data-mining or machine-learning approaches. Recently, we have been studying three phenotypes that can be distinguished visually. We describe them as normal cell, spiky cell, and ruffling cell. Our preliminary results show a classification accuracy of 76% for the three phenotypes [46]. The low percentage may be due to the presence of other phenotypes, which have different morphological and texture signatures. Moreover, some cells exhibit mixed-phenotype stages, which are also hard to differentiate.
Clustering is a useful method for distinguishing patterns. Two approaches have been used to determine the true number of clusters: one is based on relative criteria and the other based on external and internal criteria. The first approach is to choose the best result from a set of clustering results according to a predefined criterion. The second approach is based on statistical tests and computes both intercluster and intracluster quality to determine the true number of clusters. Recently, the gap statistic [47] has been proposed as a method for estimating the number of clusters. The gap statistic takes the output of any clustering algorithm and compares the change in cluster dispersion to a reference distribution. This method considers the total sum of within-class dissimilarity for a given number of clusters k and data set X, and the clustering solution is Y = Ak(X), where Ak is a clustering algorithm, say, k-means. Assume that Wk corresponds to the squared-error criterion optimized by the k-means algorithm. This method studies the relationship between log(Wk) for different k values and the expectation of log(Wk) for a suitable (null) reference distribution through the definition of the gap. The expected value is estimated by drawing B samples from the reference distribution; hence
| (4) |
where is the total within-cluster scatter for the bth data set drawn from the null reference distribution. Let sk denote the standard deviation of the sampled . Then, the method selects the smallest number of clusters k for which the gap between and log(Wk) is large k̂ = min{k|gapk − gapk+1 − sk+1}. The reference distribution is generated by the reference features from a uniform distribution over a box aligned with the principal components of the data. Fig. 13 shows the discovered three phenotypes N (normal cell), R (ruffling), and S (spiky). Meanwhile, another two patterns are discovered based on the cells that we cannot label as the previous N, R, and S; see the results in Fig. 14.
Fig. 13.
(a) The gap between and log(Wk): the result shows when K = 3, the gap reaches the maximum.
(b) The corresponding three patients discovered by using this method.
Fig. 14.
(a) The gap between and log(Wk): the result shows when K = 2, the gap reaches the maximum.
(b) The corresponding three patients discovered by using this method.
However, when all data sets are merged together, it is hard to identify these patterns. As Wk is assumed to follow spherically distributed clusters, it may contain a structural bias. So we have to do an in-depth investigation of other methods, such as graphic-based cluster validation, and compare these clustering approaches in HCS data.
D. Gene Function Annotation Using Clustering
In a pilot study, Kiger et al. [38] studied genes with known or predicted functions in a cell-cycle progress—specifically, four distinct sets of functionally related genes that regulate the passage from G1 to S phase (cdk4, Cyclin, and the Dp); G2 to M phase (cdc2 and string); the onset of anaphase (fizzy, cdc16, and cdc27); and cyclin-dependent transcription (cdc7 and Cyclin 7). Also, several genes with related phenotypes were identified. Thus, it is possible to use RNAi screening to functionally characterize a large set of genes and, by grouping genes according to morphological criteria, to identify functional modules. Traditionally, scientists clustered gene functions mainly by using cluster analysis, and there are numerous publications describing gene clustering analysis-based gene expression and microarray data. Recently, there have been a number of studies on gene function [48]–[50] based on cellular features, image descriptors, and phenotypes. It has been verified that 16 phenotype classes of the 23 defined phenotype classes are indeed implicated in specific biochemical pathways for genes of known function. It has also been shown that the strength of combined phenotypic and bio-informatics analysis can give considerable predictive information about the function of previously uncharacterized genes.
In our preliminary study, we also investigated clustering analysis of the gene-based Rho protein’s pathway. We extracted 13 features for each gene: the ratio of the numbers of normal, spiky, and ruffling cells (RONN, RONS, and RONR, respectively); the ratio of areas of normal, spiky, and ruffling cells (ROAN, ROAS, and ROAR, respectively); the ratio of perimeters of normal, spiky, and ruffling cells (ROPN, ROPS, and ROP); the number of cells (NOC); the average intensities of cells (AICE); the standard deviation of intensities of cells (SICE); and the average intensities of entire cytoplasm (AICY). We proposed a method of hierarchical clustering with weighted Euclidean distances for the gene clustering. The extracted features were based on the average value of the three sites. The gene data have three prototypes: the negative control (NC) had 48 samples; the screen hits (SH) had 35 samples; and the positive control (PC) had 6 samples. Hence, there were a total of 89 genes in the preliminary study.
The standard hierarchical clustering technique was applied based on similarities in the gene data, and the difference between the gene samples was measured by the Euclidian distance. The visualization was done by dendrograms and heat maps. Fig. 15 shows the gene clustering result. It lists the gene name, gene type (PC, NC, and SH), and gene name in the right side. The clustering result in Fig. 15 is obtained by using hierarchical clustering with weighted features and a weighted Euclidian distance. It can be seen that the clustering result is relatively good, although there are a few genes in the screening where hits are mistakenly clustered into the negative control.
Fig. 15.
Clustering results from RNAi screening project.
E. Screening Hits Selection and Gene Scoring for Effectors Discovery
We created a gene scoring model to quickly process huge volumes of high-content images and to conduct quantitative analysis of the data. Our scoring model has two key components: the gene score regression model, which combines the fuzzy logical concepts, and an EM algorithm, which conducts the rule-model based gene fuzzy membership approximation. The support vector machine technique is applied to derive a single scoring model for each gene class with confidence samples in both the biological and informatics domains. Using fuzzy membership values, a mixture fuzzy model automatically predicted gene scores, which achieved structural and empirical risk optimization. The performance of the automatic gene scoring system was then evaluated by scoring the data with those obtained from manual annotation.
Using the segmentation results and the described phenotype identification method, each individual cell was extracted from the fluorescence image. Fig. 16 displays a snap of the user interference of the genome-wide RNAi cellular image quantitator (G-CELLIQ) system where the phenotype classification is presented. Each cell is labeled by one of the biomarkers normal (N), spiky (S), and ruffling (R). Statistically, we extracted the phenotype distribution properties of the entire fluorescence screen from the labeling results by the following aspects: the ratios of the number of each phenotype; the ratio of their cell areas of each phenotype; and the ratio of the cell perimeters of each phenotype. In the example shown in Fig. 16, we detected 147 normal, 23 spiky, and 8 ruffling cells. The ratio of each phenotype, especially the abnormal phenotypes, spiky and ruffling, was a key factor in evaluating gene function. In order to achieve a stable description, images from three independent sites for each treatment were analyzed; we refer to them as “screened sites.”
Fig. 16.
The user interference of the GCELLIQ system where the phenotype classification is given.
Based on the phenotype identification results, we used three kinds of statistical phenotype properties to describe the fluorescence images. First, the ratio of different phenotypes was one of the most important parameters. We obtained the statistical result of the numbers of each phenotype as , i = 1, 2, … , K, where i is the index of the screened sites and K is the number of screened sites of the Kc167 cells. We used the average phenotype ratios to represent the gene characteristics. Hence, we transformed the features from to . Secondly, the areas of each segmented cell were calculated and the ratios of each phenotype in the image obtained as . The third statistical screening descriptor was the ratio of the perimeter for each phenotype, which is also based on the sum of the perimeter of all of the cells of a particular phenotype within an image. The statistical ratio of perimeters was . Similarly, the last two types of phenotype descriptions were also calculated using the average value of the K screened sites to achieve reliable and stable results. Finally, we obtained the phenotype statistical property of the HCS as x = {rnum, rarea, rper}.
After the phenotype statistical properties for the HCS were computed, we modeled the relationship between the features’ phenotypes’ properties x and the score y. Once this model was estimated, we predicted the score of the test images. There, we derived the phenotype statistics-based scoring system as F : x = {rnum, rarea, rper} → y, where F is the prediction function and y is the gene score. Fuzzy systems can handle problems with imprecise and incomplete data and can model nonlinear functions of arbitrary complexity. Since the ground-truth score for the training data was obtained manually, the score was only an approximation rather than the exact true value. Moreover, for different gene types, the prediction functions were very distinctive because the manual-score rules vary by gene types. We proposed a single scoring model using a support vector regression model as
| (5) |
where c = 3 because we have three types of data sets: negative control, positive control, and screening hits; P(gi|x, Θ) defines the fuzzy membership function value of gene x belonging to type gi; and fi(x) is modeled by the support vector regression model.
Experimental results: A gene data set was built to score the phenotypes from high-content images. The data set contains three images each for 89 different treatments/ dsRNA, including 54 negative controls, 6 positive controls, and 35 screen hits, which were manually scored, ranging from 0.3 to 5.0, with certain confidence. The fluorescence screening image database of the 89 genes is around 2G data with more than 800 images. From all of the images, 40 221 cells were identified. Following the cellular phenotype classification, the statistics of the phenotype distribution were obtained for each image of every treatment. There were distinctive statistical variations for these fluorescence screenings with a given treatment. We used all of the positive control samples and 80% of the negative controls and screening hits to train the single regression model of each gene class. The remaining 20% of the negative controls and screening hits were used as test samples. The mean square error was used to validate the results, which were calculated as MSE = E(ŷi − yi)2, where ŷi is the predicted score and yi is the target score. The estimated mean square error is 0.021 in the above data set.
IV. ANALYZING AND DISCOVERING PATHWAYS REGULATING DENDRITIC SPINE MORPHOLOGY
In this section, we presented an example of applying computational bioimaging for the discovery of pathways regulating dendritic spine morphology. Section IV-A gives the background about dendritic spines and synapse regulation. Hippocampus slice cultures and RNA-inactivation (image acquisition) are described in Section IV-B. We then present the neuron image quantitator (NeuronIQ) soft-ware1 for dendrite spine segmentation and detection as well as data measurement in Section IV-C, and quantitative data modeling and statistical analysis in Section IV-D.
A. Dendritic Spines and Synapse Regulation
The number of synaptic connections that a neuron makes and the properties of these synapses are regulated by complex and incompletely understood processes. In the mammalian brain, modification of excitatory synapses is often accompanied by morphological changes of specialized cellular compartments called dendritic spines. Each dendritic spine typically contains one synapse and operates as an isolated biochemical compartment that houses the machinery necessary to read out and regulate the activity of that synapse. Synapses are influenced by many factors, including some intrinsic to the neuron, such as its genotype and the ionic channels that it expresses, as well as extrinsic ones, such as circuit firing patterns and the age, health, and sensory experience of the animal [51]–[56]. In addition, synapse function and spine morphology are bidirectionally coupled, such that changes in morphology affect synaptic signaling, and synaptic maturation is accompanied by morphological rearrangements [57], [58]. Since direct measurement of synaptic properties involves electrophysiological analysis of individual cells, spine morphology is often used as an indirect assay for perturbations of synapse function when higher throughput screens are necessary [59], [60]. The limitation in using neuronal spine morphology to identify signaling cascades that regulate synapses or to uncover synaptic defects associated with disease is the enormous effort necessary to perform detailed manual analysis.
Tuberous Sclerosis Complex
TSC is a relatively common inheritable genetic disorder and is characterized by the development of hamartomas in a variety of organs, such as the brain, skin, kidney, lung, and heart [61], [62]. The neurological manifestations of TSC include epilepsy, mental retardation, and autism. Mutations of two tumor suppressor genes TSC1 and TSC2 are equally responsible for familial TSC [63], [64]. TSC1 encodes the protein, hamartin (also known as TSC1), which contains a carboxy-terminal coiled–coiled domain [63], whereas TSC2 encodes tuberin (also known as TSC2), which contains an amino-terminal coiled–coiled domain and a C-terminal Rab guanosine trisphosphatase activation protein (GAP)-like domain [64], [65]. In mice, homozygous inactivation of either TSC1 or TSC2 is embryonic lethal, whereas heterozygous animals are prone to tumors [66]–[68]. It has been well established that components of the insulin pathway are important in cell growth [69], [70]. Members of this pathway include positive regulators: insulin receptor (IR), insulin receptor substrate (IRS), phosphatidylinositol-3-OH kinase (PI3K), phosphoinositide-dependent kinase-1 (PDK-1), Akt, mTOR, S6K, and eukaryote initiation factor 4E (eIF4E), and negative regulator, phosphatases with tensin domain (PTEN). IR, PI3K, Akt, and PTEN, are sitting upstream of the TSC1-TSC2 complex, while mTOR and S6K are sitting downstream of the complex, as shown in Fig. 17 [71].
Fig. 17.
Akt regulates growth by phosphorylating and inhibiting the TSC1-TSC2 complex in drosophila. A schematic representation of genetic epistasis illustrates that stimulation of Akt activity, by activation of IR, PI(3)K, or loss of PTEN, results in TSC2 phosphorylation. This event inactivates TSC1–TSC2 complex. The inactivation results in increased cell growth, mediated by downstream inhibition of mTOR and activation of S6K.
Two-Photon Laser Scanning Microscopy
Two-photon laser scanning microscopy (2PLSM) has become an established technique for the analysis of fluorescence signals from neurons located within brain slices or intact animals (reviewed by Denk and Svoboda [72]). It is particularly well suited for studies of neurons because near-infrared excitation light can be focused deep within scattering tissues, such as the brain, that are opaque to most visible light. It also exploits a nonlinear property of many fluorophores to restrict their excitation to the focal volume, thus avoiding photodamage of out-of-focus structures and permitted prolonged, time-lapse imaging. Neuronal morphology can be determined at high resolution using 2PLSM of cells expressing GFP or of cells filled with synthetic fluorophores. Lastly, synthetic or genetically encoded fluorophores whose fluorescence properties are sensitive to the concentration of specific ions within the cell, or to the activity of specific kinases, have been used to optically monitor biochemical signaling with dendrites and spines.
B. Hippocampal Slice Cultures and RNA-Inactivation
Organotypic hippocampal slice cultures [73] have been prepared from rats and transfected using biolistics [74] with plasmids containing the coding sequence for GFP driven by a human cytomegalovirus promoter and hairpin RNA driven from a U6 promoter. Hairpin sequences that induce RNA-inactivation of the TSC1, TSC2, and PTEN proteins have been produced with loss of protein confirmed by imunofluorescence. In each transfected slice, about five to ten green fluorescent pyramidal cells are found (see Fig. 18). Transfected cells in slice cultures have been maintained for up to 40 days posttransfection.
Fig. 18.
Effects of TSC2 loss on spine morphology and density: Representative images of (a) apical and (b) basal dendrites from (top) GFP or (bottom) αTSC/GFP transfected neurons in HC slice cultures. Field of view is 50/µm across.
Preliminary analysis of the morphology of neurons expressing GFP and a hairpin RNA against TSC2 (αTSC2) has been performed for cells eight to ten days posttransfection and compared to the morphology of cells expressing GFP alone. Three-dimensional image stacks of the neuronal soma and apical and basal dendrites were collected using 2PLSM, which is ideally suited for imaging neuron morphology within scattering tissue such as the brain. As expected from the increased size of cells with knockdown of TSC1/2 pathways seen in other systems, the maximum cross-sectional area of the soma of αTSC2 cells was 105% (n = 4) larger than that of GFP-transfected neurons, with marginally significant p-value.
Each 3-D image stack was analyzed slice by slice. Dendrite length was measured from the two-dimensional (2-D) projection and is an underestimate of the true length in 3-D. The data presented here is from the analysis of 20 neurons, 160 dendritic fields, and 8000–10 000 spines in each condition and represents approximately 35 h of work for a trained technician. If one wants to track richer spine features, such as spine shape, location, and volume, across all image slices to take advantage of the whole 3-D image stack and collect as much quantitative information as possible, the hours of work could increase by an order of magnitude or two more than the manual effort. Developing an automatic segmentation, detection, and extraction toolset will be a critical step towards the realization of large-scale quantitative analysis of microscopic spine image data.
C. NeuronIQ for Dendrite Spine Segmentation and Detection
Optical fluorescence microscopy methods provide powerful tools to study neurons. However, analysis of microscopic images has remained largely manual, extremely time-consuming, and subject to human bias [75]–[79]. Objective, computerized methods are in great need for neuron image analysis. Commercial software like Neurolucida by MicroBrightField,2 Imaris by Bitplane AG,3 and Neuro-Zoom by Neurome4 have different degrees of sophistication in neuron image analysis. These products focus on tracing and extracting features from dendrites and axons but lack capability to analyze and extract spine features. For example, Neurolucida requires the user to first manually mark each branch point on the dendritic tree, then to edit spines to the dendrite. Each subsequent point clicked places the end of a spine, which attaches to the dendritic tree at the nearest branch point. The spine will be attached to the wrong place if the user makes any mistakes in placing the branch points initially. Imaris and NeuroZoom are two products focusing more on displaying neurons but do not have spine detection and analysis functionality. Users must manually mark spines on the image. For example, in NeuroZoom, spines are manually marked and represented by spheres of the same size. Hence it lacks the capability to output features such as the spine size.
Neuron image analysis encompasses dendrite segmentation and spine detection. As neuron images often contain great contrast variations and low contrast details, global thresholding techniques [80] for segmentation will fail. Based on Koh’s approach, Weaver et al. describe a package capable of morphometry on an entire neuron by combining the spine detection algorithms with dendritic tracing algorithms [81]. Although these mentioned algorithms can greatly reduce the human labor by semiautomatic detection, human interference such as a global threshold is still required during the processing. The main difficulty of spine segmentation comes from the shape variation of different spine types as well as dendrite and spine surface irregularities. Several multilevel threshold techniques have been developed to detect the transitions from within an object into the background in 2-D image segmentation. Zheng et al. [82] used three coarsely spaced thresholds and analyzed features such as size growth and central position shift to determine the regions that best represents the target object. Shiffman et al. [83] and Amit [84] used multilevel thresholds and then analyzed the contours pattern to determine the best segmentation point between objects and their backgrounds.
NeuronIQ Ssystem
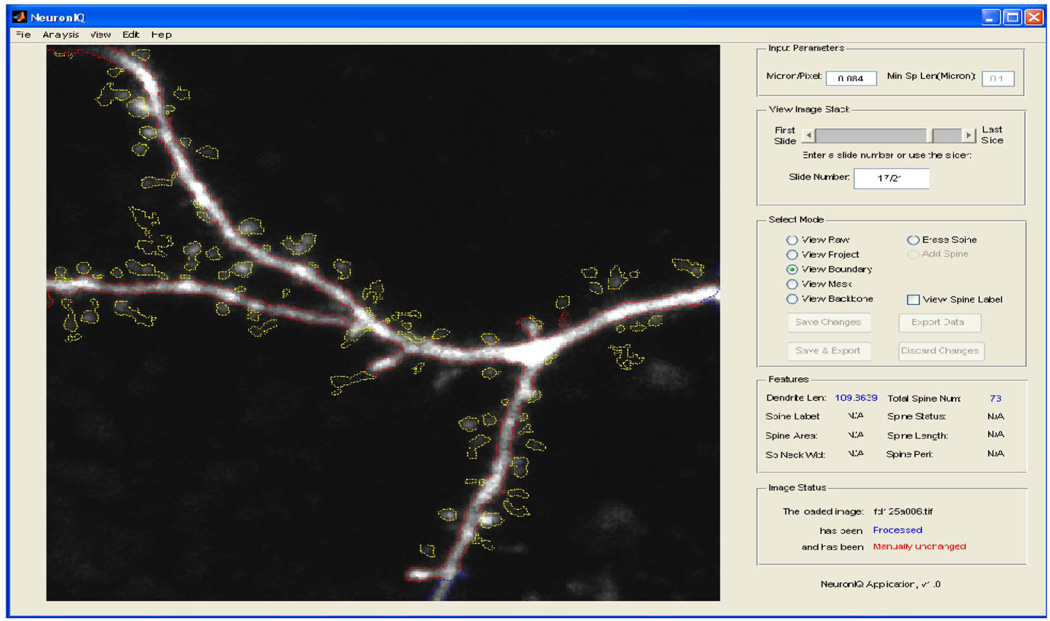
As introduced above, the existing semiautomatic approaches largely reduce user efforts. However, manual interventions, such as setting a global threshold for segmentation, are still needed for these semiautomatic approaches during image processing and thus create certain operator variations. We thus developed a fully automated system to circumvent such problems [85]. The automation includes an adaptive thresholding method, which can yield better segment results than the prevalent global thresholding method. It also introduces an efficient backbone extraction method, an SNR-based, detached spine component detection method, and an attached spine component detection method based on the estimation of local dendrite morphology. Using the Kolmogov–Smirnov test, we find a 99.13% probability that the dendrite length distributions are the same for the automatic and manual processing methods. The spine detection results are also compared with other existing semiautomatic approaches. The comparison results show that our approach has 33% fewer false positives and 77% fewer false negatives on average [85]. Because the proposed detection algorithm requires less user input and performs better than existing algorithms, our approach can quickly and accurately process neuron images without user intervention. The NeuronIQ user interference is shown in Fig. 19.
Fig. 19.
NeuronIQ user interference.
D. Data Modeling in NeuronIQ for Validation
Three groups of neurons were analyzed: GFP is five neurons expressing GFP alone; αTSC2 is four neurons expressing GFP and a hairpin RNA against TSC2; and αTSC2/rapamycin is five αTSC2 neurons treated with rapamycin. Three-dimensional image stacks of the neuronal soma and apical and basal dendrites were collected from a total of 14 neurons, 160 dendritic fields, and 2719 spines. Morphological features such as number of spines, length of dendrites, density of spines, lengths of spines, and volume of spine in both apical and basal dendrites were analyzed manually. In existing dendrite structure research, spine density was often treated as the most important feature, and the Kolmogorov–Smirnov test (K-S test) [77], [80] was used to discriminate the different phenotypes.
Mutual information is a natural method to measure the dependence between different spine features and neuron phenotypes. Hence mutual information–based feature selection [86] is especially promising among many feature-selection methods. Since the existing mutual information–based feature selection was only suitable for a large sample size [87], we developed a mutual information–based feature-selection method using the bootstrap technique [18] to cope with the small sample size and to obtain more accurate estimation of the mutual information. Our results showed that the proposed mutual information–based spine feature selection could generate satisfying results [88].
Prior preprocessing steps of feature extraction with an appropriate distribution model, and normalization of extracted features into comparable scales [88], were necessary to obtain better performance. After preprocessing of the original data such as feature extraction, scaling, or normalization and missing value estimation, the features were tested and classified into the right phenotype category. In general, a satisfactory normalization yields a good classification result. In our preliminary study, three methods of classifiers—KNN (K= 3), perceptron, and two-layer neural networks were selected to test the extracted features with or without scaling.
Neuron Classification
Using pattern recognition could improve our knowledge in identifying the phenotypes of dendrite structure. Pattern recognition is a major methodology in data mining. Given a set of training patterns from each class, the objective of pattern recognition is to establish decision boundaries in the feature space that separate patterns belonging to different classes [89]. Many methods exist in pattern recognition [89]. We applied K-nearest neighbor K = 3 (KNN), perceptron, and two-layer neural networks (2LNNs) to neuron phenotype identification due to the small sample-size problem. The advantage of KNN is that it is a nonparametric classifier. It is especially effective for small sample problems [86]. The perceptron and neural networks are parameter-based classifiers, where the perceptron is a simple linear repressor with a nonlinear decision and neural networks are complex nonlinear model-based classifiers. Both KNN and perceptron usually work better for small size samples because they have low computational complexity. On the contrary, 2LNNs can deal with complex data models with many parameters but need many samples and causes high computational complexity. A more comprehensive classification method in conjunction with feature selection using mutual information [90] was considered. The motivation for considering mutual information was its capacity to measure a general dependence among random variables.
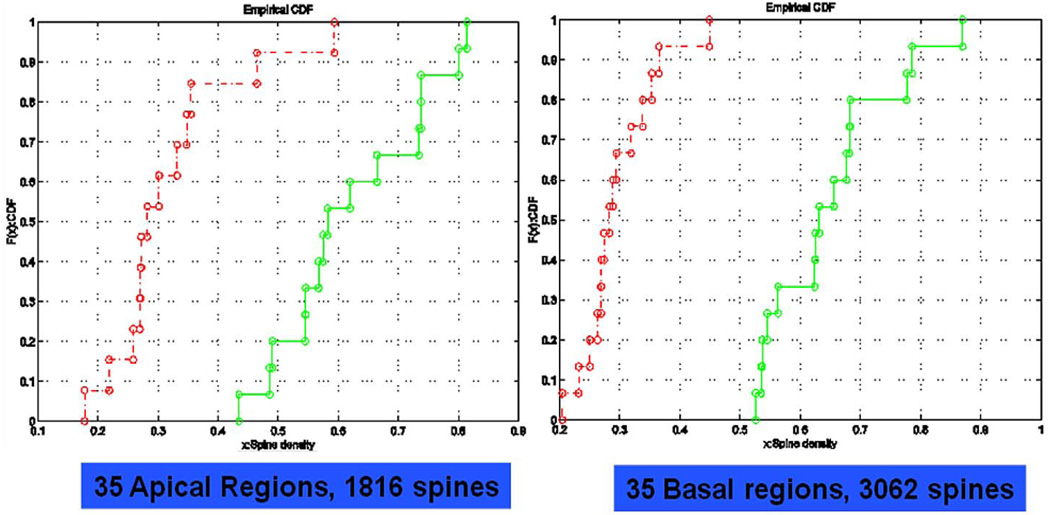
The mutual information of the features for the case GFP control against αTSC2 is listed under Table 2, Case 1. Since we had adopted the normalized mutual information definition, we regarded the features with mutual information as being bigger than one as statistical significance. It is interesting to note that both the mean of spine lengths and the density of spines in basal dendrites had higher mutual information (both > 2.0), and that the density of spines and the mean of spine lengths in apical dendrites had numbers between 1.8 and 1.3. The classification accuracy based on those four features was perfect using the above three classifiers with no errors found. We then performed the K-S test for the two phenotypes [88]. Both apical and basal dendrites revealed significant differences between the cumulative distributions of spine density from GFP control and αTSC2. Table 2, Case 2 is the mutual information of the features for GFP and αTSC2/rapamycin. The spine density in basal dendrites was shown with the highest mutual information (> 2.0). The spine density in apical dendrites was also shown with high mutual information. The mean of the spine lengths in both basal and apical dendrites only had low mutual information (< 1.0). The classification accuracy based on the highest two features, spine density in basal and apical dendrites, was perfect using the above three classifiers. Again, both apical and basal dendrites revealed significant differences between the cumulative distributions of spine density from GFP control and, in this case, αTSC2/ rapamycin [88]. Fig. 20 shows that the cumulative distributed function (cdf) of TSC spine density from the neurons with GFP control (green) or TSC2 RNAi transfection with rapamycin treatment (red).
Table 2.
Mutual Information for Three Cases With Different Features (Case 1: GFP Versus αTSC2; Case 2: Versus αTSC2/rapamycin; Case 3: αTSC2 Versus αTSC2/rapamycin)
| Feature | Case 1 | Case 2 | Case 3 | |||
|---|---|---|---|---|---|---|
| Apical | Basal | Apical | Basal | Apical | Basal | |
| Number of spines | 0.9768 | 1.1609 | 1.6206 | 1.4473 | 1.1712 | 0.7317 |
| Dendrite Length | 1.2216 | 0.9960 | 1.1262 | 0.7492 | 1.1096 | 0.8293 |
| Density of spines | 1.8013 | 2.0024 | 1.9855 | 2.0576 | 0.7898 | 0.8012 |
| Mean of spine length | 1.3158 | 2.0057 | 0.8001 | 0.7896 | 1.3504 | 1.4182 |
| Variance of spine length | 0.8040 | 1.0646 | 0.9731 | 1.0224 | 0.8519 | 0.7640 |
Fig. 20.
CDF of TSC spine density from the neurons with GFP control (green) or TSC2 RNAi transfection with rapamycin treatment (red).
For the αTSC2 and αTSC2/rapamycin, the result is illustrated in Case 3 of Table 2. It is interesting to note that the mean of spine lengths in basal dendrites was the highest mutual information (> 1.4) of all features, and the second highest feature was the mean of spine lengths in apical dendrites (> 1.35). Nevertheless, the spine densities in both apical and basal dendrites had mutual information lower than one. The classification accuracy based on the two features with the highest mutual information, namely, the mean of spine length in the basal and apical dendrites, was perfect using the above three classifiers. Again, we performed the K-S test for the two phenotypes. The cumulative distributions of the spine density overlap in both apical and basal dendrites. Obviously, from this preliminary result, the spine density was no longer the most important feature; instead the spine length was more important.
For the case GFP control versus αTSC2 with rapamycin treatment, only the density of spine in both basal and apical dendrites of αTSC2 with rapamycin treatment displayed a significant difference comparing with GFP control, which suggested rapamycin, an inhibitor of mTOR, reversed the effects of TSC2 inactivation only on the morphological abnormity of length of spine but not density of spine. Moreover, for the case of αTSC2 versus αTSC2 with rapamycin treatment, the significant difference was observed at the length of spine, which further supports the evidence that rapamycin could antagonize the effect of TSC2 loss on the morphological change of the length of spines but not on those of the density of the spine. The mechanism through which rapamycin reversed the effect of TSC2 loss on the morphological abnormity of length of spine but not density was still unclear; and yet, this finding provided a possible new significance of spine lengths and might provide direction in future investigations. Either αTSC2 or αTSC2 with rapamycin treatment could induce morphological change of dendritic spine comparing with GFP control. Since rapamycin could reverse the effect on spine length caused by TSC2 RNA inactivation, the morphological change of spine density was most likely due to a different TSC-coupled mechanism. Furthermore, our results suggested that by using simple normalization methods, the morphological abnormity of density of spine could be detected after TSC2 RNA silencing. However, testing the effect of rapamycin on morphological change induced by TSC2 RNA inactivation needed more sophisticated methods such as pattern-recognition methods, which could deal with more features simultaneously.
V. CONCLUSIONS
Computational systems biology (CSB) is critical in decoding and understanding the biological mechanism of a disease from a system perspective. In this paper, we discussed two classes of computational system biology techniques: bioinformatics and bioimage informatics.
On the bioinformatics part, we first reviewed biomarker discovery using high-throughput screening techniques such as microarray and mass spectrometry. We then introduced signaling network reconstruction from a combination of biological data, such as genomics data, proteomics data, metabolic data, and biochemical reactions, and other data from the published literature. The second part concerned the emerging field of bioimage informatics, currently used for secondary screening for target validation and selection of drug leads. We presented two CSB applications: one was the study of a gene signaling pathway from genome-wide RNAi screening for Rho GTPase, while the other was the analysis and discovery of pathways regulating dendritic spine morphology. Under our paradigm, the signaling pathway would be refined iteratively as shown in Fig. 1 until there is no significant change of the topological structure of the networks.
On the other hand, computational bioimaging is increasingly important in many scientific disciplines, such as drug discovery, cell biology, neurobiology, bioinformatics, and biomedical engineering. Algorithms of cellular segmentation, tracking, and classification are important, but they will have values only when they can be applied to solving real-life problems. The interplay between bioinformatics and computational bioimaging would increase the problem-solving capability of CSB. Our future work will explore this aspect further.
We should point out there are other important areas of computational system biology such as structural and functional informatics, as well as the SNPs for association analyses that identify important genetic variants in populations and genes predisposed to diseases involving complex traits. When scientists are talking about multiscale and multimodality data integration at the systems level, emerging high-throughput biotechnologies, such as SNP array, promoter array, tissue array, and protein array, quickly become common tools for in-vitro studies. On the other hand, clinical imaging informatics and the emerging in-vivo molecular imaging at tissue and organ levels are important tools to assist in candidate biomarker validation, early diagnosis, and therapy monitoring in patients. Although they are beyond the focus of this paper, we expect that eventually the advancement made in computational systems biology would enable us to integrate the nonlinear and heterogeneous information from the molecular level, cellular level, tissue level, and organ level in order to decipher complex mechanisms of diseases and to aid in better therapeutic strategies.
Acknowledgment
The authors would like to express their appreciation of the rewarding collaborations with their biological colleagues, in particular, N. Perrimon and B. Sabatini at Harvard Medical School, as well as H. Wang at the National Institutes of Health. The original image data described in this paper were obtained from the Perrimon and Sabatini laboratories. The image analysis and computational modeling work are the contributions of the authors with the input of other members in their group over the years, notably F. Li, J. Wang, J. Cheng, B. Geng, S. Zhang, and Y. Huang.
This work was supported by the Bioinformatics Program, Harvard Neurodiscovery Center (previously the Harvard Center for Neurodegeneration and Repair), Harvard Medical School, Boston, MA, under a grant and the Bioinformatics Program, The Methodist Hospital Research Institute, Weill Cornell Medical College of Cornell University, New York. The work of S. T. C. Wong was supported by the National Institutes of Health under Research Grants R01 LM008696 and R01 LM009161.
Biographies

Xiaobo Zhou received the B.S. degree from Lanzhou University, Lanzhou, China, in 1988 and the M.S. and Ph.D. degrees from Peking University, Beijing, China, in 1995 and 1998, respectively, both in mathematics.
From 1988 to 1992, he was a Lecturer with the Training Center, 18th Building Company, Chongqing, China. From 1992 to 1998, he was a Research Assistant and Teaching Assistant with the Department of Mathematics, Peking University. From 1999 to 2000, he was a Senior Technical Manager with the 3G Wireless Communication Department, Huawei Technologies Co., Ltd., Beijing. From 1998 to 2004, he was a Postdoctoral Research Fellow with Tsinghua University, Beijing; the University of Missouri-Columbia; Texas A&M University, College Station; and Harvard Medical School, Boston, MA. From 2005 to 2007, he was a faculty with Brigham and Women’s Hospital and Harvard University, Boston, MA. Since 2007, he has been Chief of Bioinformatics and Bioimaging Computing Lab and Associate Professor of Radiology, The Methodist Hospital and Weill Medical College of Cornell University, Houston, TX. His research focus is in developing advanced bioinformatics, bioimage computing tools, and systems biologic approaches for biomarker discovery, pathway analysis, drug target validation, nanomedicine, and clinical diagnosis.

Stephen T. C. Wong received the B.S. (honors) degree in electrical engineering from the University of Western Austrialia and the M.Sc. and Ph.D. degrees in computer science from Lehigh University. He received execution education from Sloan School of Management, Massachusetts Institute of Technology, Cambridge; the Graduate School of Business, Stanford University, Stanford, CA; and the Graduate School of Business, Columbia University, New York. He is a licensed professional engineer (P.E.) since 1991.
He is Professor and Vice Chair of Radiology with The Methodist Hospital Research Institute and Cornell University, Houston, TX. He was Director of the Center for Bioinformatics, Harvard Center of Neurodegeneration and Repair, Boston, MA. He was Director of the Functional and Molecular Imaging Center and an Associate Professor of Radiology, Harvard Medical School and Brigham and Women’s Hospital, Boston. His research theme has been focused on the application of advanced technology to pragmatic biomedical problems and is based on the belief that problems of importance involve the interplay between theory and application. He is an engineer scientist. He has published more than 250 peer-reviewed papers and received seven patents in biomedical informatics. He also served on National Institutes of Health and National Science Foundation scientific review panels. He has broad R&D experience worldwide for two decades with Hewlett-Packard, Bell Labs, ICOT-Japanese 5th Generation Computer Systems project, Philips Electronics, Charles Schwab, and the University of California, San Francisco (UCSF). His earlier research involves pioneering work in optical time-domain reflectometer and optical networks, thinkjet (first inkjet) automation, 1 MB DRAM, and very large-scale integration factory automation before moving into the fields of bioinformatics and medical imaging. He was a key member of the UCSF picture archiving and communication system effort, founded product development departments of Philips Medical Systems, and directed the Web trading development and rearchitecturing of Schwab.com, one of the largest secured e-commerce sites.
Footnotes
Available freely at http://www.mymethodist.com/tmhs/basic-right.do?channelId=-90584&contentId=191152&contentType=GENERIC_CONTENT_TYPE.
To expedite the discovery of new drugs, researchers can analyze the pathways for cellular processes and biological signals, then observe physical characteristics using biological imaging to verify drug effects
REFERENCES
- 1.Kitano H. Systems biology: A brief overview. Science. vol. 295(no 5560):1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 2.Carthew RW. Gene silencing by double-stranded RNA. Curr. Opin. Cell Biol. 2001;vol. 13(no 2):244–248. doi: 10.1016/s0955-0674(00)00204-0. [DOI] [PubMed] [Google Scholar]
- 3.Roques EJ, Murphy RF. Objective evaluation of differences in protein subcellular distribution. Traffic. 2002;vol. 3(no 1):61–65. doi: 10.1034/j.1600-0854.2002.30108.x. [DOI] [PubMed] [Google Scholar]
- 4.Jones TR, Carpenter AE, Golland P. Voronoi-based segmentation of cells on image manifolds; Proc. ICCV Workshop Comput. Vision Biomed. Image Applicat; 2005. pp. 535–543. [Google Scholar]
- 5.Kapur R. High content screening and the CellChip-TM system: Living cells as beacons for drugs and toxins. Eur. Cells Mater. 2001;vol. 2 no. Suppl. 1:7. [Google Scholar]
- 6.Morelock M, Hunter E, Moran T, Heynen S, Laris C, Thieleking M, Akong M, Mike I, Callaway S, DeLeon R, Goodacre A, Zacharias D, Price J. Statistics of assay validation in high throughput cell imaging of nuclear facter kB nuclear translocation. ASSAY Drug Develop. Technol. 2005;vol. 3(no 5):483–499. doi: 10.1089/adt.2005.3.483. [DOI] [PubMed] [Google Scholar]
- 7.Ray N, Acton ST, Ley K. Tracking leukocytes in vivo with shape and size constrained active contours. IEEE Trans. Med. Imag. 2002;vol. 21(no 10):1222–1235. doi: 10.1109/TMI.2002.806291. [DOI] [PubMed] [Google Scholar]
- 8.Zimmer C, Olivo-Marin J-C. Coupled parametric active contours. IEEE Trans. Pattern Anal. Mach. Intell. 2005;vol. 27(no 11):1838–1842. doi: 10.1109/TPAMI.2005.214. [DOI] [PubMed] [Google Scholar]
- 9.Zimmer C, et al. Segmentation and tracking of migrating cells in videomicroscopy with parametric active contours: A tool for cell-based drug testing. IEEE Trans. Med. Imag. 2002;vol. 21(no 10):1212–1221. doi: 10.1109/TMI.2002.806292. [DOI] [PubMed] [Google Scholar]
- 10.Ben-Hur A, Elisseeff A, Guyon I. A stability based method for discovering structure in clustered data; Proc. Pac. Symp. Biocomput; 2002. pp. 6–17. [PubMed] [Google Scholar]
- 11.Li L, et al. Gene selection for sample classification based on gene expression data: Study of sensitivity to choice of parameters of the GA/KNN method. Bioinformatics. 2001;vol. 17(no 12):1131–1142. doi: 10.1093/bioinformatics/17.12.1131. [DOI] [PubMed] [Google Scholar]
- 12.Kim S, et al. Strong feature sets from small samples. J. Comput. Biol. 2002;vol. 9(no 1):127–146. doi: 10.1089/10665270252833226. [DOI] [PubMed] [Google Scholar]
- 13.Lee KE, et al. Gene selection: A Bayesian variable selection approach. Bioinformatics. 2003;vol. 19(no 1):90–97. doi: 10.1093/bioinformatics/19.1.90. [DOI] [PubMed] [Google Scholar]
- 14.Zhou X, Wang X, Dougherty ER. Multi-class cancer classification using multinomial probit regression with Bayesian gene selection. Inst. Elect. Eng. Syst. Biol. 2006;vol. 153(no 2):70–78. doi: 10.1049/ip-syb:20050015. [DOI] [PubMed] [Google Scholar]
- 15.Golub TR, et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science. 1999;vol. 286(no 5439):531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 16.Zhou X, Wang X, Dougherty ER. Nonlinear probit gene classification using mutual-information and wavelet. J. Biol. Syst. 2004;vol. 12(no 3):371–386. [Google Scholar]
- 17.Zhou X, Liu KY, Wong ST. Cancer classification and prediction using logistic regression with Bayesian gene selection. J Biomed. Inf. 2004;vol. 37(no 4):249–259. doi: 10.1016/j.jbi.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 18.Zoubir AM, Boashash B. The bootstrap and its application in signal processing. IEEE Signal Process. Mag. 1998;vol. 15(no 1):56–76. [Google Scholar]
- 19.Xing E, Jordan M, Karp R. Feature selection for high dimensional genomic microarray data. Proc. 8th Int. Conf. Mach. Learn.; Williamstown, MA. 2001. [Google Scholar]
- 20.Albert J, Chib S. Bayesian analysis of binary and polychotomous response data. J. Amer. Stat. Assoc. 1993;vol. 88:669–679. [Google Scholar]
- 21.Petricoin E. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;vol. 359(no 9306):572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 22.Berndt P, Hobohm U, Langen H. Reliable automatic protein identification from matrix-assistant laser desorption/ionization mass spectrometric peptide fingerprints. Electrophoresis. 1999;vol. 20:3521–3526. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3521::AID-ELPS3521>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 23.Yasui Y, et al. An automated peak identification/calibration procedure for high-dimensional protein measures from mass spectrometers. J. Biomed. Biotechnol. 2003;vol. 2003(no 4):242–248. doi: 10.1155/S111072430320927X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris JS, et al. Feature extraction and quantification for mass spectrometry in biomedical applications using the mean spectrum. Bioinformatics. 2005;vol. 21(no 9):1764–1775. doi: 10.1093/bioinformatics/bti254. [DOI] [PubMed] [Google Scholar]
- 25.Du P, Kibbe WA, Lin SM. Improved peak detection in mass spectrum by incorporating continuous wavelet transform-based pattern matching. Bioinformatics. 2006;vol. 22(no 17):2059–2065. doi: 10.1093/bioinformatics/btl355. [DOI] [PubMed] [Google Scholar]
- 26.Yu W, Wu B, Lin N, Stone K, Williams K, Zhao H. Detecting and aligning peaks in mass spectrometry data with applications to MALDI. Comput. Biol. Chem. 2006;vol. 30:27–38. doi: 10.1016/j.compbiolchem.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 27.Kohavi R, John GH. Wrappers for feature subset selection. Artif. Intell. 1997;vol. 97(no 1–2):273–324. [Google Scholar]
- 28.Zhou X, Wang H, Wang J, Hoehn G, Azok J, Brennan ML, Hazen SL, Li K, Wong STC. Biomarker discovery for risk stratification of cardiovascular events using an improved genetic algorithm; Proc. IEEE/NLM International Workshop on Life Science Systems and Applications; 2006. Jul 13–14, pp. 42–44. [Google Scholar]
- 29.Zhou X, et al. A Bayesian connectivity-based approach to constructing probabilistic gene regulatory networks. Bioinformatics. 2004;vol. 20(no 17):2918–2927. doi: 10.1093/bioinformatics/bth318. [DOI] [PubMed] [Google Scholar]
- 30.Sauer U, Heinemann M, Zamboni N. Genetics. Getting closer to the whole picture. Science. 2007;vol. 316(no 5824):550–551. doi: 10.1126/science.1142502. [DOI] [PubMed] [Google Scholar]
- 31.Wang RS, et al. Inferring transcriptional regulatory networks from high-throughput data. Bioinformatics. 2007;vol. 23(no 22):3056–3064. doi: 10.1093/bioinformatics/btm465. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, et al. Inferring gene regulatory networks from multiple microarray datasets. Bioinformatics. 2006;vol. 22(no 19):2413–2420. doi: 10.1093/bioinformatics/btl396. [DOI] [PubMed] [Google Scholar]
- 33.Yeung MK, Tegner J, Collins JJ. Reverse engineering gene networks using singular value decomposition and robust regression. Proc. Nat. Acad. Sci. USA. 2002;vol. 99(no 9):6163–6168. doi: 10.1073/pnas.092576199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeang CH, Vingron M. A joint model of regulatory and metabolic networks. Bioinformatics. 2006;vol. 7:332. doi: 10.1186/1471-2105-7-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Segal E, Wang H, Koller D. Discovering molecular pathways from protein interaction and gene expression data. Bioinformatics. 2003;vol. 19 suppl. 1:i264–i271. doi: 10.1093/bioinformatics/btg1037. [DOI] [PubMed] [Google Scholar]
- 36.Yamanishi Y, Vert JP, Kanehisa M. Supervised enzyme network inference from the integration of genomic data and chemical information. Bioinformatics. 2005;vol. 2 suppl. 1:i468–i477. doi: 10.1093/bioinformatics/bti1012. [DOI] [PubMed] [Google Scholar]
- 37.Zhou X, Wong STC. Informatics challenges of high-throughput cellular and molecular microscopy. IEEE Signal Process. Mag. 2006;vol. 23(no 3):63–72. [Google Scholar]
- 38.Kiger AA, et al. A functional genomic analysis of cell morphology using RNA interference. J. Biol. 2003;vol. 2(no 4):27. doi: 10.1186/1475-4924-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindblad J, et al. Image analysis for automatic segmentation of cytoplasms and classification of Rac1 activation. Cytometry A. 2004;vol. 57(no 1):22–33. doi: 10.1002/cyto.a.10107. [DOI] [PubMed] [Google Scholar]
- 40.Caselles V, Kimmel R, Sapiro G. Geodesic active contours. Int. J. Comput. Vision. 1997;vol. 22(no 1):61–79. [Google Scholar]
- 41.Xiong G, Zhou X, Ji L. Automated segmentation of Drosophila RNAi fluorescence cellular images using deformable models. IEEE Trans. Circuits Syst. I, Reg. papers. 2006;vol. 53(no 11):2415–2424. [Google Scholar]
- 42.Deriche R. Fast algorithms for low-level vision. IEEE Trans. Pattern Anal. Mach. Intell. 1990;vol. 12(no 1):78–87. [Google Scholar]
- 43.Sethian JA. Cambridge Monographs on Applied and Computational Mathematics. vol. 3. Cambridge, U.K.: Cambridge Univ. Press; 1996. Level set methods: evolving interfaces in geometry, fluid mechanics, computer vision, and materials science; p. xviii. [Google Scholar]
- 44.Yan P, Zhou X, Shah M, Wong STC. Automatic segmentation of RNAi fluorescent cellular images with interaction model. IEEE Trans. Inf. Technol. Biomed. doi: 10.1109/TITB.2007.898006. to be published. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li F, et al. An automated feedback system with the hybrid model of scoring and classification for solving over-segmentation problems in RNAi high content screening. J. Microsc. 2007;vol. 226(pt 2):121–132. doi: 10.1111/j.1365-2818.2007.01762.x. [DOI] [PubMed] [Google Scholar]
- 46.Wang J, Zhou X, Bradley PL, Perrimon N, Wong STC. Henotype recognition for high-content RNAi genome-wide screening. J. Mol. Screening. 2007 to be published. [Google Scholar]
- 47.Efron B, et al. Empirical Bayes analysis of a microarray experiment. J. Amer. Statist. Assoc. 2001;vol. 96(no 456):1151–1160. [Google Scholar]
- 48.Sonnichsen B, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;vol. 434(no 7032):462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- 49.Sachse C, et al. High-throughput RNA interference strategies for target discovery and validation by using synthetic short interfering RNAs: Functional genomics investigations of biological pathways. Meth. Enzymol. 2005;vol. 392:242–277. doi: 10.1016/S0076-6879(04)92015-0. [DOI] [PubMed] [Google Scholar]
- 50.Gunsalus KC, et al. Predictive models of molecular machines involved in Caenorhabditis elegans early embryogenesis. Nature. 2005;vol. 436(no 7052):861–865. doi: 10.1038/nature03876. [DOI] [PubMed] [Google Scholar]
- 51.Shors TJ, Chua C, Falduto J. Sex differences and opposite effects of stress on dendritic spine density in the male versus female hippocampus. J. Neurosci. 2001;vol. 21(no 16):6292–6297. doi: 10.1523/JNEUROSCI.21-16-06292.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lolova I. Dendritic changes in the hippocampus of aged rats. Acta Morphol. Hung. 1989;vol. 37(no 1–2):3–10. [PubMed] [Google Scholar]
- 53.Philpot BD, et al. Visual experience and deprivation bidirectionally modify the composition and function of NMDA receptors in visual cortex. Neuron. 2001;vol. 29(no 1):157–169. doi: 10.1016/s0896-6273(01)00187-8. [DOI] [PubMed] [Google Scholar]
- 54.Comery TA, et al. Abnormal dendritic spines in fragile X knockout mice: Maturation and pruning deficits. Proc. Nat. Acad. Sci. USA. 1997;vol. 94(no 10):5401–5404. doi: 10.1073/pnas.94.10.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Passafaro M, et al. Induction of dendritic spines by an extracellular domain of AMPA receptor subunit GluR2. Nature. 2003;vol. 424(no 6949):677–681. doi: 10.1038/nature01781. [DOI] [PubMed] [Google Scholar]
- 56.Sala C, et al. Inhibition of dendritic spine morphogenesis and synaptic transmission by activity-inducible protein Homer1a. J. Neurosci. 2003;vol. 23(no 15):6327–6337. doi: 10.1523/JNEUROSCI.23-15-06327.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Majewska A, Tashiro A, Yuste R. Regulation of spine calcium dynamics by rapid spine motility. J. Neurosci. 2000;vol. 20(no 22):8262–8268. doi: 10.1523/JNEUROSCI.20-22-08262.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dailey ME, Smith SJ. The dynamics of dendritic structure in developing hippocampal slices. J. Neurosci. 1996;vol. 16:2983–2994. doi: 10.1523/JNEUROSCI.16-09-02983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nimchinsky EA, Oberlander AM, Svoboda K. Abnormal development of dendritic spines in FMR1 knock-out mice. J. Neurosci. 2001;vol. 21(no 14):5139–5146. doi: 10.1523/JNEUROSCI.21-14-05139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ji Y, et al. Apolipoprotein E isoform-specific regulation of dendritic spine morphology in apolipoprotein E transgenic mice and Alzheimer’s disease patients. Neuroscience. 2003;vol. 122(no 2):305–315. doi: 10.1016/j.neuroscience.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 61.Young J, Povey S. The genetic basis of tuberous sclerosis. Mol. Med. Today. 1998;vol. 4(no 7):313–319. doi: 10.1016/s1357-4310(98)01245-3. [DOI] [PubMed] [Google Scholar]
- 62.Gomez MR. Phenotypes of the tuberous sclerosis complex with a revision of diagnostic criteria. Ann. New York Acad. Sci. 1991;vol. 615:1–7. doi: 10.1111/j.1749-6632.1991.tb37742.x. [DOI] [PubMed] [Google Scholar]
- 63.van Slegtenhorst M, et al. Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science. 1997;vol. 277(no 5327):805–808. doi: 10.1126/science.277.5327.805. [DOI] [PubMed] [Google Scholar]
- 64.Consortium. Identification and characterization of the tuberous sclerosis gene on chromosome 16: The European Chromosome 16 Tuberous Sclerosis Consortium. Cell. 1993;vol. 75(no 7):1305–1315. doi: 10.1016/0092-8674(93)90618-z. [DOI] [PubMed] [Google Scholar]
- 65.Maheshwar MM, et al. The GAP-related domain of tuberin, the product of the TSC2 gene, is a target for missense mutations in tuberous sclerosis. Human Mol. Genetics. 1997;vol. 6(no 11):1991–1996. doi: 10.1093/hmg/6.11.1991. [DOI] [PubMed] [Google Scholar]
- 66.Onda H, et al. Tsc2(+/−) mice develop tumors in multiple sites that express gelsolin and are influenced by genetic background. J. Clin. Invest. 1999;vol. 104(no 6):687–695. doi: 10.1172/JCI7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Au KS, et al. Germ-line mutational analysis of the TSC2 gene in 90 tuberous-sclerosis patients. Amer. J. Human Genetics. 1998;vol. 62(no 2):286–294. doi: 10.1086/301705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kobayashi T, et al. A germ-line Tsc1 mutation causes tumor development and embryonic lethality that are similar, but not identical to, those caused by Tsc2 mutation in mice. Proc. Nat. Acad. Sci. USA. 2001;vol. 98(no 15):8762–8767. doi: 10.1073/pnas.151033798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kozma SC, Thomas G. Regulation of cell size in growth, development and human disease: PI3K, PKB and S6K. Bioessays. 2002;vol. 24(no 1):65–71. doi: 10.1002/bies.10031. [DOI] [PubMed] [Google Scholar]
- 70.Stocker H, Hafen E. Genetic control of cell size. Curr. Opin. Genetics Dev. 2000;vol. 10(no 5):529–535. doi: 10.1016/s0959-437x(00)00123-4. [DOI] [PubMed] [Google Scholar]
- 71.Potter CJ, Pedraza LG, Xu T. Akt regulates growth by directly phosphorylating Tsc2. Nature Cell. Biol. 2002;vol. 4(no 9):658–665. doi: 10.1038/ncb840. [DOI] [PubMed] [Google Scholar]
- 72.Denk W, Svoboda K. Photon upmanship: Why multiphoton imaging is more than a gimmick. Neuron. 1997;vol. 18:351–357. doi: 10.1016/s0896-6273(00)81237-4. [DOI] [PubMed] [Google Scholar]
- 73.Stoppini L, Buchs PA, Muller DA. A simple method for organotypic cultures of nervous tissue. J. Neurosci. Meth. 1991;vol. 37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- 74.Lo DC, McAllister AK, Katz LC. Neuronal transfection in brain slices using particle-mediated gene transfer. Neuron. 1994;vol. 13:1263–1268. doi: 10.1016/0896-6273(94)90412-x. [DOI] [PubMed] [Google Scholar]
- 75.Elston GN, Tweedale R, Rosa MG. Supragranular pyramidal neurones in the medial posterior parietal cortex of the macaque monkey: Morphological heterogeneity in subdivisions of area 7. Neuroreport. 1999;vol. 10(no 9):1925–1929. doi: 10.1097/00001756-199906230-00024. [DOI] [PubMed] [Google Scholar]
- 76.Jacobs B, et al. Regional dendritic and spine variation in human cerebral cortex: A quantitative golgi study. Cerebr. Cortex. 2001;vol. 11(no 6):558–571. doi: 10.1093/cercor/11.6.558. [DOI] [PubMed] [Google Scholar]
- 77.Nimchinsky EA, Sabatini BL, Svoboda K. Structure and function of dendritic spines. Annu. Rev. Physiol. 2002;vol. 64:313–353. doi: 10.1146/annurev.physiol.64.081501.160008. [DOI] [PubMed] [Google Scholar]
- 78.Matsuzaki M, et al. Structural basis of long-term potentiation in single dendritic spines. Nature. 2004;vol. 429(no 6993):761–766. doi: 10.1038/nature02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zito K, et al. Induction of spine growth and synapse formation by regulation of the spine actin cytoskeleton. Neuron. 2004;vol. 44(no 2):321–334. doi: 10.1016/j.neuron.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 80.Koh IYY, et al. An image analysis algorithm for dendritic spines. Neural Comput. 2002;vol. 14(no 6):1283–1310. doi: 10.1162/089976602753712945. [DOI] [PubMed] [Google Scholar]
- 81.Weaver CM, et al. Automated algorithms for multiscale morphometry of neuronal dendrites. Neural Comput. 2004;vol. 16(no 7):1353–1383. doi: 10.1162/089976604323057425. [DOI] [PubMed] [Google Scholar]
- 82.Zheng B, Chang YH, Gur D. Computerized detection of masses in digitized mammograms using single-image segmentation and a multilayer topographic feature analysis. Acad. Radiol. 1995;vol. 2(no 11):959–966. doi: 10.1016/s1076-6332(05)80696-8. [DOI] [PubMed] [Google Scholar]
- 83.Shiffman S, Rubin GD, Napel S. Medical image segmentation using analysis of isolable-contour maps. IEEE Trans. Med. Imag. 2000;vol. 19(no 11):1064–1074. doi: 10.1109/42.896782. [DOI] [PubMed] [Google Scholar]
- 84.Amit Y. Graphical shape templates for automatic anatomy detection with applications to MRI brain scans. IEEE Trans. Med. Imag. 1997;vol. 16(no 1):28–40. doi: 10.1109/42.552053. [DOI] [PubMed] [Google Scholar]
- 85.Cheng J, Zhou X, Miller E, Witt RM, Zhu JM, Sabatini BL, Wong STC. A novel computational approach for automatic dendrite spines detection in two-photon laser scan microscopy. J. Neurosci. Meth. 2007 September 15;vol. 165(no 1):122–134. doi: 10.1016/j.jneumeth.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhou X, Wang X, Dougherty ER. Nonlinear probit gene classification using mutual information and wavelet-based, feature selection. J. Biol. Syst. 2004;vol. 12(no 3):371–386. [Google Scholar]
- 87.Battiti R. Using mutual information for selecting features in supervised neural-net learning. IEEE Trans. Neural Netw. 1994;vol. 5(no 4):537–550. doi: 10.1109/72.298224. [DOI] [PubMed] [Google Scholar]
- 88.Zhou X, et al. Mutual information-based feature selection in studying perturbation of dendritic structure caused by TSC2 inactivation. Neuroinformatics. 2006;vol. 4(no 1):81–94. doi: 10.1385/NI:4:1:81. [DOI] [PubMed] [Google Scholar]
- 89.Duda RO, Hart PE, Stork DG. Pattern Classification. 2nd ed. New York: Wiley; 2001. [Google Scholar]
- 90.Cover TM, Thomas JA. Elements of Information Theory. New York: Wiley; 1991. [Google Scholar]