Abstract
Viruses, viroids and prions are the smallest infectious biological entities that depend on their host for replication. The number of pathogenic viruses is considerably large and their impact in human global health is well documented. Currently, the International Committee on the Taxonomy of Viruses (ICTV) has classified ∼4379 virus species while the National Center for Biotechnology Information Viral Genomes Resource (NCBI-VGR) database has mapped 617 705 proteins to eight large taxonomic groups. Despite these efforts, an automated approach for mapping the ICTV master list and its officially accepted virus naming to the NCBI-VGR’s taxonomical classification is not available. Due to metagenomic sequencing, it is likely that the discovery and naming of new viral species will increase by at least ten fold. Unfortunately, existing viral databases are not adequately prepared to scale, maintain and annotate automatically ultra-high throughput sequences and place this information into specific taxonomic categories. ORION-VIRCAT is a scalable and interoperable object-relational database designed to serve as a resource for the integration and verification of taxonomical classifications generated by the ICTV and NCBI-VGR. The current release (v1.0) of ORION-VIRCAT is implemented in PostgreSQL and it has been extended to ORACLE, MySQL and SyBase. ORION-VIRCAT automatically mapped and joined 617 705 entries from the NCBI-VGR to the viral naming of the ICTV. This detailed analysis revealed that 399 095 entries from the NCBI-VGR can be mapped to the ICTV classification and that one Order, 10 families, 35 genera and 503 species listed in the ICTV disagree with the the NCBI-VGR classification schema. Nevertheless, we were eable to correct several discrepancies mapping 234 000 additional entries.
Database URL: http://www.orionbiosciences.com/research/orion-vircat.html
Introduction
Viruses, viroids and prions are the smallest infectious biological entities that depend on their host for replication. Because many species represent a significant threat to global health and can be used as bioweapons; there has been a considerable effort to gain a better understanding of their host range and the molecular forces shaping their adaption and pathogenesis. Periodically the International Committee on the Taxonomy of Viruses (ICTV) generates a master list which currently recognizes about 4379 virus species divided in nine Orders, 98 assigned Families, 26 unassigned Families, 18 assigned Sub-families, 5 unassigned Sub-families, 459 assigned genera and 57 unassigned genera.
The National Center for Biotechnology Information Viral Genomes Resource (NCBI-VGR) (1,2) is a database that uses the Baltimore nomenclature (3) to map ∼1 million protein records to eight large taxonomic groups (excluding unclassified viruses and unclassified bacteriophages) to one Deltavirus species, 96 species of Retro-transcribing viruses, 129 satellites, 601 dsDNA viruses with no RNA stage, 107 species of dsRNA viruses, 353 species of ssDNA viruses, 123 species of ssRNA negative-strand viruses, 580 species of ssRNA positive-strand viruses with no DNA stage, five unclassified archaeal viruses, 35 unclassified phages and nine unclassified viruses.
The ICTVdb is a viral information repository which uses the DELTA system to generate taxonomical reports in HTML format using the ICTV master list (4,5). The ICTVdb uses an eight position decimal code with up to three digit schema similar to that used for enzyme classes to represent order, family, subfamily, genus, species, subspecies, serotype or subtype, and strain or isolate (4,5). This detailed information is linked to approximately 8000 representative sequences from the NCBI database.
In addition to the NCBI-VGR and ICTVdb, several databases covering specific categories of viruses have been implemented. Most are modeled using relational database management systems (RDBMS) and provide standard interfaces like JDBC and ODBC for data and metadata annotation. Some databases support data curation, genome and proteome comparisons (6–8) and have become specialized sources of information for Bunyavirus (9), Flavivirus (10,11). Herpesvirus (12), Coronavirus (12,13), Influenza (14–16), Hepatitis (17–20), HIV (21–23), vaccines (24), ssRNA viruses (25), virulence factors (26), capsid structures (27), siRNA targets (28) and immunogenesis (17,29,30).
Despite the progress, a comprehensive and automated approach for mapping the ICTV master list and its officially accepted virus naming to the NCBI-VGR is not available. This situation does not only limit the development of additional specialized viral databases but makes the cross-validation across them very difficult. As biological databases grow, it is increasingly more difficult to maintain their integrity. In many cases, data entry errors including virus naming and numerical assignment go undetected and errors at the higher levels of taxonomy (e.g. family) are propagated to lower levels (e.g. species) and to external databases. Furthermore, in their current format, available databases cannot scale seamlessly to handle metagenomic sampling. This is particularly relevant because metagenomic datasets will increase the discovery rate and naming of new viral species by at least 10-fold (31).
To address several of the above challenges we report here the implementation of a series of bioinformatic applications and an enterprise database management system to (i) automatically assign each entry of ICTV master list to the NCBI-VGR and determine the level of discrepancy between these two databases. (ii) implement an object-relational genomic catalog storing viral genome information correcting existing discrepacies. Our work empowers virologists to develop specialized databases and it is one of the first steps for the development of a viral ontology.
Methods
Data monitoring, retrieval and integration
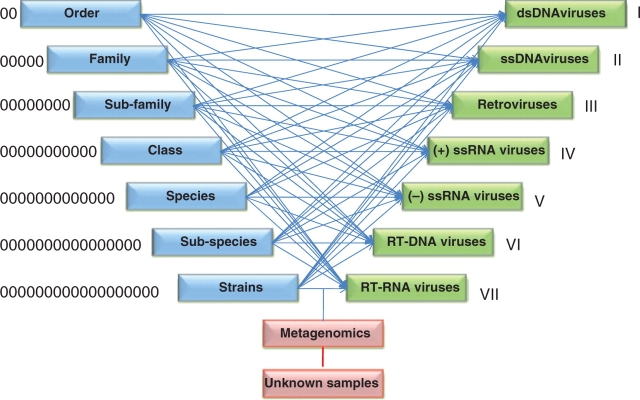
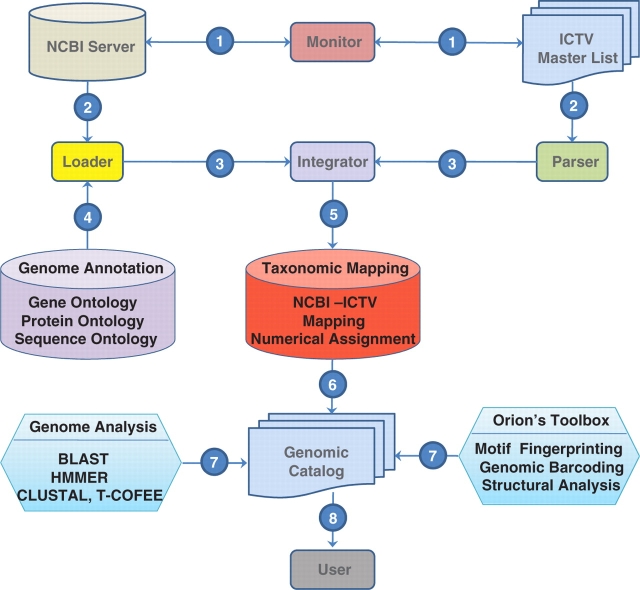
This layer of tools is managed by monitor and adapter modules. The monitor checks periodically the ICTV master list and the NCBI-VGR taxonomical records. In case of change, the monitor module triggers a PERL script named ICTVml_parser.pl which uploads new taxonomical classification and species naming from the ICTV. At the same time, a script, the BioPerlDB class named load_sqdatabase.pl, retrieves and parses new GeneBank records. Once these processes are completed, the NCBI_ICTV_integrator.pl maps the ICTV species naming to the NCBI-VGR taxonID and Baltimore classification schemes (3) (Figure 1). Order, family, sub-family, genus and species naming from the NCBI VGR are flagged and are renamed using the ICTV master list and a virus_synonym table that maintains alternative naming of a virus or strain. When synonyms exist, precedence of the ICTV master list determines the selection of virus names that should be included within a taxonomical category (Figure 2).
Figure 1.
Integration process of ORION-VIRCAT. We mapped different ICTV (blue) and NCBI-VGR (green) taxonomies classifications.
Figure 2.
Summary of the implementation of the genomic catalog.
Viral genomic catalog
This object-RDBMS stores metadata and virus genomic sequence information collected by the monitor and adapter modules and join them by the NCBI_ICTV_integrator.pl. ORION-VIRCAT genomic catalog reuses the attributes from BioSQL seqfeatures, annotation, taxon and ontology tables and it is implemented in postgreSQL. In addition, we extended the database schema of BioSQL to include virus morphology description, geographical information, clinical characteristics, isolation location and year, culture passage cycle, and controlled vocabularies. To avoid specific vendor operations we have extended the genomic catalog to ORACLE, mySQL and DB2 and data formats.
Results
The current release (v1.0) of ORION-VIRCAT automatically mapped and joined 617 705 entries from the NCBI-VGR to the viral naming of the ICTV. This detailed analysis revealed that 399 095 entries from the NCBI-VGR can be mapped to the ICTV classification and that one Order, 10 families, 35 genera and 503 species listed in the ICTV disagree with the the NCBI-VGR classification schema (Supplementary Data). Our analysis also found four main types of discrepancies between the ICTV master list and the NCBI-VGR entries. The first level consisted of minor differences in the capitalization between the naming conventions or changes in one letter. For example, the ICTV listed PhiH-like viruses, while the NCBI-VGR listed phiH-like viruses. In a smilar case, the ICTV listed Omicronpapillomavirus while the NCBI-VGR listed Omikronpapillomavirus. The second level of discrepancies included 15 genera remaining unclassified within a particular family in the NCBI-VGR. However, recent updates of the ICTV master list gave these viral groups a genus name. The third level of discrepancy consisted of species belonging to one of four different genera that have been reassigned to a new genus. The fourth level of discrepancy included species listed only in the ICTV master list and classified within a particular taxonomy according to morphological observations but without sequence entries available in the NCBI-VGR.
Discussion
With the advent of genomics several taxonomical classifications have been proposed and have led to the development of several specialized viral databases. However, for the most part, these implementations remain isolated sources of information and lack interoperability and scalability. Here, we report the implementation of ORION-VIRCAT as a progressive step towards the standardization of genomic information about viruses and the development of a scalable system to store viral information at the metagenomic scale. The development of this approach has several implications for the development of viral databases. First, we comprehensively assessed the level of discrepancy between the official naming and taxonomical classification generated by the ICTV master list and the NCBI-VGR. Second, ORION-VIRCAT reconstructed in an object-relational format a genomic catalog mapping all the sequences from NCBI-VGR to the officially accepted naming developed by the ICTV. By using the ICTV we promote the use of officially accepted taxon names developed by the research community and the correct mapping to the information of a particular sequence stored in the NCBI. At the same time, we uncovered genera and species names that need to be revised and updated. Therefore, ORION-VIRCAT promotes nomenclatural clarity through explicit definitions where each taxon has only one accepted name.
By reusing BioPerl and BioSQL, we, in ORION-VIRCAT, adopted widely accepted standards and pseudo-standards that facilitate interoperability with third-party applications. This not only saves considerable time and resources, but allows the implementation of a robust support system for the future development of specialized viral databases. The schema of the genomic catalog is flexible enough to allow addition of new sources of information [e.g. Pathogen Information Markup Languaje (32)]. As a result, ORION-VIRCAT empowers researchers interested in a particular viral taxonomy to download specific sets of information and implement their own databases and extend them with advanced and specific analysis tools. As the ICTV master list generates new names for species, they are added to the table and this way we ensure that every group has the most updated naming convention. Since curators often dedicate much effort to manually annotate group names, we are now developing an annotation tool for data clarification to generate reports to be considered by the ICTV (Table 1).
Table 1.
Genera list of discrepancies between ICTV and NCBI
| NCBI-VGR | ICTV | Reference |
|---|---|---|
| Nucleopolyhedrovirus | Alphabaculovirus | PMID: 16648963 |
| Unclassified archaeal viruses | Ampullavirus | PMC: 2566220 |
| Granulovirus | Betabaculovirus | PMID: 16648963 |
| Alphacryptovirus | Betacryptovirus | PMID: 15503213 |
| Unclassified Birnaviridae | Blosnavirus | PMID: 12477876 |
| Unclassified Reoviridae | Cardoreovirus | PMID: 15575876 |
| Unclassified Poxviridae | Cervidpoxvirus | PMC: 1839080 |
| Unclassified Flexiviridae | Citrivirus | PMID: 17362202 |
| Nucleopolyhedrovirus | Deltabaculovirus | PMID: 16648963 |
| Unclassified Lipothrixviridae | Deltalipothrixvirus | PMC: 2224351 |
| Unclassified Reoviridae | Dinovernavirus | PMC: 2409309 |
| Ebola-like viruses | Ebolavirus | PMID: 392795 |
| Unclassified | Elaviroid | PMID: 12743309 |
| Nucleopolyhedrovirus | Gammabaculovirus | PMID: 16648963 |
| Unclassified Entomopoxvirinae | Gammaentomopoxvirus | PMID: 908841 |
| Unclassified Globuloviridae | Globulovirus | PMID: 16682063 |
| Sulfolobus | Guttavirus | PMID: 10873785 |
| Unclassified Drosophila LRT | Hemivirus | PMID: 2410772 |
| Rhadinovirus | Macavirus | PMC: 187546 |
| Unclassified Saccharomyces retrotransposon | Metavirus | PMID: 2159534 |
| Unclassified Reoviridae | Mimoreovirus | PMID: 16603541 |
| Omikronpapillomavirus | Omicronpapillomavirus | PMID: 17554024 |
| Unassigned Herpesviridae | Ostreavirus | PMID: 15604430 |
| Varicellovirus | Percavirus | PMC: 1951306 |
| phiH-like viruses | PhiH-like viruses | |
| Unidentified | Pipapillomavirus | PMID: 17554025 |
| Unclassified Polerovirus | Polemovirus | PMID: 15892965 |
| Unclassified Betaherpesvirinae | Proboscivirus | PMID: 17884307 |
| Unclassified Saccharomyces retrotransposon | Pseudovirus | |
| psiM1-like viruses | PsiM1-like viruses | PMID: 9791169 |
| Unclassified viruses | Raphidovirus | PMID: 16000767 |
| Not listed | Rhizidiovirus | |
| Not listed | Semotivirus | PMID: 3816762 |
| Not listed | Sirevirus | PMID: 16183843 |
Towards a viral ontology
In order to be able to exchange the semantics of information in a database on viruses one first needs to agree on how to explicitly model a virus ontology architecture. Trough the use of ontologies it is possible to develop a mechanism for representing in a formal form the shared descriptions about viruses including taxonomy nomenclature, phylogenetics, molecular and functional biology. We propose starting with the development of a conceptual discussion to define the scope and range of a viral ontology. We believe that the viral ontology should be divided into four parts within two core layers. The first core layer should be a static ontology describing only essential and passive concepts about viruses. The extended layer should describe concepts actively evolving and related to viral naming, taxonomy, phylogenetics, genetics, genomics, biology, host–parasite relations, ecology, morphology and experiments involving viruses. The extended layer should include as a rule, a minimum set of description categories in order to define a species. Representations of the same data by different biologists will likely be different (even when using the same system). Hence, mechanisms for ‘aligning’ different biological schemas or different versions of schemas should be supported.
Since the extended layer is subject to constant changes as biological knowledge on viruses evolves, it is necessary to implement different numerical identifiers for each of the attributes and their concepts. This will allow building a complex concept of cardinality and inheritance for terms while formalizing and verifying their correctness and properties. These behavior constraints can be viewed as temporal logic assertions expressing the evolution of a particular term. At the same time, the extended layer should inherit the ontology terms related to viruses (e.g. Pathogen Transmission Ontology, Diseases Ontology, Phage Ontology, Vaccine Ontology, etc.) from other biomedical ontologies.
Conclusions
With the advent of genomic and metagenomic scale virus genome sampling, using conventional taxonomic criteria based on morphological and developmental properties is considered unpractical. The bioinformatics strategy presented here lends support for future collaborative efforts for a comprehensive, large-scale viral genome analysis system. These systems should allow intelligent software agents and advanced text-mining algorithms to analyze information about viruses and present it in new ways that can not only advance our understanding of viruses, but redefine their classification.
Supplementary data
Supplementary Data are available at Database Online.
Funding
The development of ORION-VIRCAT is partially supported by the Defense Threat Reduction Agency under the contract W81XWH-0720029. Funding for open access charge: Contract W81XWH-0720029.
Conflict of interest statement. None declared.
Acknowledgements
The authors would like to thank Dr Sofi Ibrahim at the US Army Institute for Infectious Diseases (USAMRIID) and Dr Carmenza Spadafora at the Instituto de Investigaciones Científicas y Servicios de Alta Tecnología (INDICASAT) for the helpful discussions and suggestions.
References
- 1.Wheeler D.L., Barrett T., Benson D.A., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bao Y., Federhen S., Leipe D., et al. National center for biotechnology information viral genomes project. J. Virol. 2004;78:7291–7298. doi: 10.1128/JVI.78.14.7291-7298.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baltimore D. Expression of animal virus genomes. Bacteriol. Rev. 1971;35:235–241. doi: 10.1128/br.35.3.235-241.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchen-Osmond C. Further progress in ICTVdB, a universal virus database. Arch. Virol. 1997;142:1734–1739. [PubMed] [Google Scholar]
- 5.Buechen-Osmond C., Dallwitz M. Towards a universal virus database—progress in the ICTVdB. Arch. Virol. 1996;141:392–399. doi: 10.1007/BF01718409. [DOI] [PubMed] [Google Scholar]
- 6.Kulkarni-Kale U., Bhosle S., Manjari G.S., et al. VirGen: a comprehensive viral genome resource. Nucleic Acids Res. 2004;32:D289–D292. doi: 10.1093/nar/gkh098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lefkowitz E.J., Upton C., Changayil S.S., et al. Poxvirus Bioinformatics Resource Center: a comprehensive Poxviridae informational and analytical resource. Nucleic Acids Res. 2005;33:D311–D316. doi: 10.1093/nar/gki110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan Q. Bioinformatics databases and tools in virology research: an overview. In Silico Biol. 2008;8:71–85. [PubMed] [Google Scholar]
- 9.Fourment M., Gibbs M.J. The VirusBanker database uses a Java program to allow flexible searching through Bunyaviridae sequences. BMC Bioinformatics. 2008;9:83. doi: 10.1186/1471-2105-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Misra M., Schein C.H. Flavitrack: an annotated database of flavivirus sequences. Bioinformatics. 2007;23:2645–2647. doi: 10.1093/bioinformatics/btm383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schreiber M.J., Ong S.H., Holland R.C., et al. DengueInfo: a web portal to dengue information resources. Infect. Genet. Evol. 2007;7:540–541. doi: 10.1016/j.meegid.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Alba M.M., Lee D., Pearl F.M., et al. VIDA: a virus database system for the organization of animal virus genome open reading frames. Nucleic Acids Res. 2001;29:133–136. doi: 10.1093/nar/29.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Y., Lau S.K., Woo P.C., et al. CoVDB: a comprehensive database for comparative analysis of coronavirus genes and genomes. Nucleic Acids Res. 2008;36:D504–D511. doi: 10.1093/nar/gkm754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang S., Zhang J., Liao X., et al. Influenza Virus Database (IVDB): an integrated information resource and analysis platform for influenza virus research. Nucleic Acids Res. 2007;35:D376–D380. doi: 10.1093/nar/gkl779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu G., Rowley T., Garten R., et al. FluGenome: a web tool for genotyping influenza A virus. Nucleic Acids Res. 2007;35:W275–W279. doi: 10.1093/nar/gkm365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Squires B., Macken C., Garcia-Sastre A., et al. BioHealthBase: informatics support in the elucidation of influenza virus host pathogen interactions and virulence. Nucleic Acids Res. 36:D497–D503. doi: 10.1093/nar/gkm905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hraber P.T., Leach R.W., Reilly L.P., et al. Los Alamos hepatitis C virus sequence and human immunology databases: an expanding resource for antiviral research. Antivir. Chem. Chemother. 2007;18:113–123. doi: 10.1177/095632020701800301. [DOI] [PubMed] [Google Scholar]
- 18.Panjaworayan N., Roessner S.K., Firth A.E., et al. HBVRegDB: annotation, comparison, detection and visualization of regulatory elements in hepatitis B virus sequences. Virol. J. 2007;4:136. doi: 10.1186/1743-422X-4-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Combet C., Penin F., Geourjon C., et al. HCVDB: hepatitis C virus sequences database. Appl. Bioinformatics. 2004;3:237–240. doi: 10.2165/00822942-200403040-00005. [DOI] [PubMed] [Google Scholar]
- 20.Kuiken C., Hraber P., Thurmond J., et al. The hepatitis C sequence database in Los Alamos. Nucleic Acids Res. 2008;36:D512–D516. doi: 10.1093/nar/gkm962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan C., Kim J., Chen L., et al. The HIV positive selection mutation database. Nucleic Acids Res. 2007;35:D371–D375. doi: 10.1093/nar/gkl855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Araujo L.V., Soares M.A., Oliveira S.M., et al. DBCollHIV: a database system for collaborative HIV analysis in Brazil. Genet. Mol. Res. 2006;5:203–215. [PubMed] [Google Scholar]
- 23.Doherty R.S., De Oliveira T., Seebregts C., et al. BioAfrica's HIV-1 proteomics resource: combining protein data with bioinformatics tools. Retrovirology. 2005;2:18. doi: 10.1186/1742-4690-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiang Z., Todd T., Ku K.P., et al. VIOLIN: vaccine investigation and online information network. Nucleic Acids Res. 2008;36:D923–D928. doi: 10.1093/nar/gkm1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snyder E.E., Kampanya N., Lu J., et al. PATRIC: the VBI PathoSystems Resource Integration Center. Nucleic Acids Res. 2007;35:D401–D406. doi: 10.1093/nar/gkl858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou C.E., Smith J., Lam M., et al. MvirDB—a microbial database of protein toxins, virulence factors and antibiotic resistance genes for bio-defence applications. Nucleic Acids Res. 35:D391–D394. doi: 10.1093/nar/gkl791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shepherd C.M., Borelli I.A., Lander G., et al. VIPERdb: a relational database for structural virology. Nucleic Acids Res. 2006;34:D386–D389. doi: 10.1093/nar/gkj032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naito Y., Ui-Tei K., Nishikawa T., et al. siVirus: web-based antiviral siRNA design software for highly divergent viral sequences. Nucleic Acids Res. 2006;34:W448–W450. doi: 10.1093/nar/gkl214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lundegaard C., Lamberth K., Harndahl M., et al. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8-11. Nucleic Acids Res. 2008;36:W509–W512. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yusim K., Richardson R., Tao N., et al. Los alamos hepatitis C immunology database. Appl. Bioinformatics. 2005;4:217–225. doi: 10.2165/00822942-200504040-00002. [DOI] [PubMed] [Google Scholar]
- 31.Valdivia-Granda W. The next meta-challenge for Bioinformatics. Bioinformation. 2008;2:358–362. doi: 10.6026/97320630002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He Y., Vines R.R., Wattam A.R., et al. PIML: the Pathogen Information Markup Language. Bioinformatics. 2005;21:116–121. doi: 10.1093/bioinformatics/bth462. [DOI] [PubMed] [Google Scholar]