Abstract
OBJECTIVE
We tested putative functional single nucleotide polymorphisms (SNPs) in genes which regulate the folate/homocysteine metabolism pathway for their contribution to spina bifida (SB) susceptibility.
STUDY DESIGN
The study consisted of 610 unrelated simplex SB patient families. Genotypes of 46 SNPs located in the coding sequence or promoter region of 11 genes were investigated. Associations between transmission of alleles and SB in the offspring were examined using the reconstruction-combined transmission disequilibrium test.
RESULTS
Significant association of SNP rs5742905 in cystathionine-β-synthase (CBS), rs1643649 in dihydrofolate reductase (DHFR), rs2853533 in thymidylate synthetase (TYMS), and rs3737965 in methylene-tetrahydrofolate-reductase (MTHFR) was found (p= 0.015, 0.041, 0.021, and 0.007 respectively).
CONCLUSION
Transmission disequilibrium of SNP alleles in CBS, DHFR, MTHFR and TYMS confers an increased susceptibility to SB.
Key words/phrases: spina bifida, single nucleotide polymorphism, folate/homocysteine metabolism genes
Introduction
Neural tube defects (NTDs) are one of the most common birth defects in the United States, occurring in 2/10,000 births annually.1 After a significant decline in the rates of spina bifida from 1995–1999, recent data demonstrates from 2003–2005, the rate of spina bifida has remained essentially unchanged.1, 2 Neurulation, the process of neural tube formation, is completed 28 days after conception.3 Disruption of neurulation which occurs during or prior to this time may result in a defect, or failure of closure of the neural tube. A variety of clinical abnormalities can result. The most common defects occur cranially (anencephaly) or caudally (spina bifida). Open NTDs are the most severe with anencephaly being lethal. Individuals affected with spina bifida (SB) who survive are at increased risk of early death and typically require extensive medical and surgical care.4
The etiology of most NTDs are multifactorial, or nonsyndromic, in origin resulting from both genetic and environmental influences. Studies have shown that in pregnancies affected by NTDs, the mothers have lower plasma levels of vitamin B12 and folate than in those not affected.5 Maternal folate status is foremost in the prevention of occurrence and recurrence of many NTDs.6–8 Despite all of the positive results from epidemiologic studies and interventions regarding folate, the mechanisms underlying the relationship of folate status and NTD risk are not fully understood.9, 10
Genetic background has been implicated as an important risk factor for NTDs. In addition, the incidence of NTDs varies geographically and can vary within ethnic and racial groups as well. In the United States Hispanics have one of the highest rates of NTDs. 1, 4, 11 The genetic contribution of folate metabolism to NTDs has been investigated for two decades. The gene encoding the enzyme methylenetetrahydrofolate reductase (MTHFR) which catalyzes the reduction of 5, 10-methylenetetrahydrofolate (5, 10-MTHF) to 5-MTHF, a methyl group donor to convert homocysteine to methionine by methyltransferases, has been well studied. The most commonly studied variants in this gene are the polymorphisms C677T and A1298C. Both of these variants have been associated with increased risk of NTDs in various populations12–15 but the association is not universal.16 The MTHFR 667T variant has been shown to be more common in Hispanic individuals.4, 17 The enzyme activity of MTHFR with the 677T variant only produces approximately 50% of the wildtype 677C variant.18 This association of the MTHFR polymorphism with NTDs has led to inquires into other genes involved in folate transport and metabolism including thymidylate synthase (TYMS), dihydrofolate reductase (DHFR), and methylenetetrahydrofolate dehydrogenase (MTHFD1) genes that have also demonstrated varying associations to spina bifida.19–22
Research into the genetic contribution of complex diseases that result from concomitant effects of environment, behavior, and genetic factors has been stimulated by the immense amount of data generated by the Human Genome Project. Research has revealed that while 99% of human DNA sequences are the same, variation also exists. There are different types of variation in DNA with the simplest and most frequent being single nucleotide polymorphisms (SNPs). SNPs are variations in the DNA sequence that occur when a single nucleotide (A, T, G or C) in the genome can be replaced by another nucleotide, with each nucleotide representing an allele of the SNP. In the vast majority of cases SNPs do not cause disease but such variants are stronger candidates than other genetic markers for locating disease susceptibility loci. SNPs occur approximately 1 in every 1000 base pairs of a sequence, with each individual having their own unique heritable pattern. SNPs may be linked, or inherited together as a consequence of their physical proximity on a single chromosome and may then be transmitted in groups, or linkage blocks, in a non-random fashion. In contrast, linkage disequilibrium (LD) is the nonrandom occurrence of specific SNP allele patterns at different loci. The distinction between the two is that linkage is within a family association, while linkage disequilibrium is a population association. By comparing unique SNP patterns between two groups of individuals having different phenotypes (e.g. eye or hair color), geneticists can pinpoint the SNP patterns and genes associating with the phenotypes (i.e. genetic association). The aim in our study was to analyze the SNP patterns of parents and affected children with SB to determine how often these unique patterns were transmitted to affected offspring. The transmission disequilibrium test (TDT) measures association and linkage in families comparing the frequency of transmission of SNPs from parents to affected offspring. If there is no association with the disease the two alleles of a SNP have an equal chance of being transmitted, however if one allele of the SNP increases the risk of disease, it will be transmitted to the affected offspring more often than expected by chance alone.
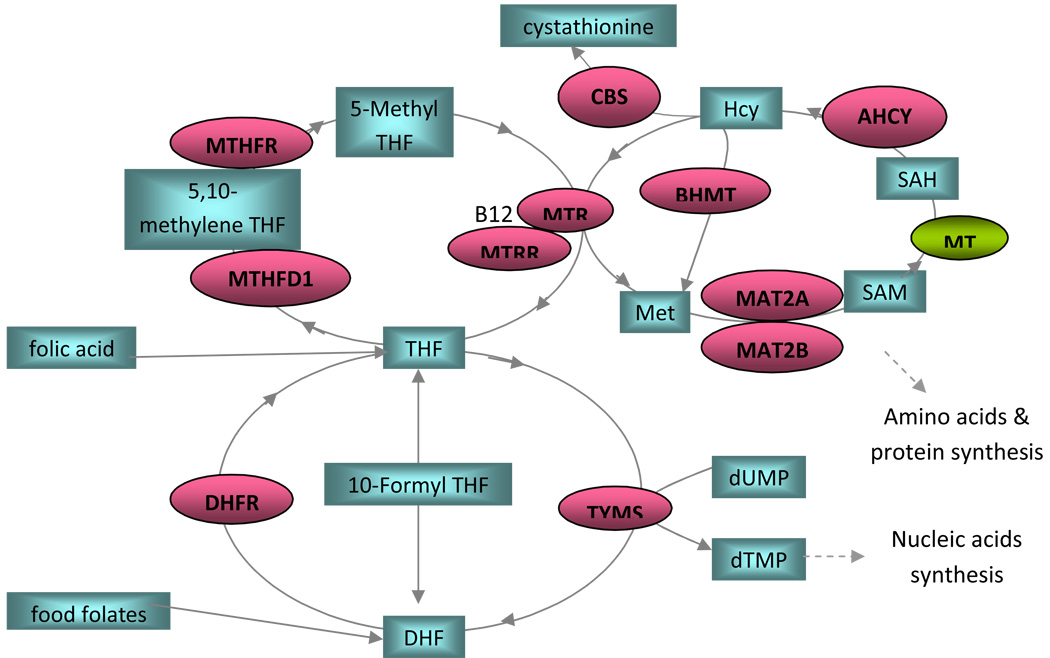
The function and interaction of each of the 11 folate metabolism pathway genes are depicted in Figure 1. In addition to the genes directly coding for enzymes in the folate metabolism pathway shown in Figure 1, there are several other categories of genes involved more indirectly in folate metabolism. While cell proliferation and differentiation are important during neural tube folding, cell death/apoptosis also plays an important role during fusion of the neural roof plate and the surface ectoderm. Delay of the separation between the neural roof plate and surface ectoderm will delay the infiltration of mesenchymal cells between the two tissues layers required for neural arch formation. The MTHFR substrate, 5, 10-N-methylene-THF, donates a methyl group for the synthesis of deoxyribonucleotide dTMP catalyzed by TYMS with dihydrofolate as a byproduct. Interestingly, dihydrofolate and its precursors (dihydropterines) from other pathways are known to be toxic to animals.23, 24 Active cell proliferation during neural tube formation requires a sufficient supply of deoxyribonucleotides and produces a substantial amount of dihydrofolate through TYMS activity. This observation highlighted DHFR’s activity in regulating dihydrofolate concentration within proliferating cells during neurogenesis and initiating apoptosis when suppressed.
Figure 1. Folate Pathway in Humans.
Substrates are shown in rectangular boxes. DHF: dihydrofolate; Hcy: homocysteine; MET: methionine; SAM: S-Adenosyl-methionine; SAH: S-Adenosylhomocysteine; THF: tetrahydrofolate
Enzymes are shown in ellipses. ACHY: S-adenosylhomocysteine hydrolase; BHMT: betaine-homocysteine methyltransferase; CBS: cystathionine-β-synthase; DHFR: dihydrofolate reductase; MAT2A: methionine adenosyltransferase II alpha; MAT2B: methionine adenosyltransferase II beta; MTHFD1: methylenetetrahydrofolate dehydrogenase; MTHFR: methylenetetrahydrofolate reductase; MTR: methyltetrahydrofolate-homocysteine methyltransferase; MTRR: MTR reductase; TYMS: thymidylate synthase; MT: methyl-transferases
Our objective was to test putative functional SNPs of 11 candidate genes known to regulate the folate/homocysteine metabolism pathway for their contribution to spina bifida susceptibility. The 11 candidate genes include methylene-tetrahydrofolate reductase (MTHFR), S-adenosylhomocysteine hydrolase (AHCY), betaine homocysteine methyltransferase (BHMT), cystathionine beta-synthase (CBS), dihydrofolate-reductase (DHFR), methionine adenosyltransferase 2A and 2B enzyme complex (MAT2A, MAT2B), methylenetetrahydrofolate dehydrogenase 1 (MTHFD1), 5-methyltetrahydrofolate homocysteine methyltransferase (MTR), 5-methyltetrahydrofolate-homocysteine methyltransferase reductase (MTRR), and thymidylate synthase (TYMS).
Materials and Methods
Study Population
In this study, patients affected with SB and their parents were enrolled from 1996 to 2006. Our study sample consisted of 610 cases comprised of either triads (affected individual and both parents) or duos (affected individual and one parent) recruited from Houston, Texas; Los Angeles, California; and Toronto, Canada. The study sample included 226 Caucasians, 334 Hispanics of Mexican descent, and 50 of other ethnic background. (Table 1) Level of the defect was determined by reviewing medical records and in some cases by review of X-rays. Sociodemographic, pregnancy history, maternal health history, and maternal exposures were obtained from the mothers of the affected cases. Vitamin supplementation was not elicited in the initial study but a survey to ascertain this information is currently ongoing. The project was approved by the institutional review board of The University of Texas Health Science Center at Houston.
Table 1.
Study Population Characteristics
| Characteristic | Trios | Duos | Total |
|---|---|---|---|
| Race | |||
| Caucasian | 142 | 84 | 226 |
| Mexican American | 168 | 166 | 334 |
| African American | 4 | 14 | 18 |
| Asian American | 2 | 2 | 4 |
| Other | 7 | 5 | 12 |
| Unknown | 6 | 10 | 16 |
| Gender | |||
| Male | 143 | 115 | 258 |
| Female | 150 | 123 | 273 |
| Unknown | 36 | 43 | 79 |
Total of 610 families, 329 trios, 281 duos are genotyped
SNP selection
We searched PubMed and public databases (http://www.ncbi.nlm.nih.gov/SNP, http://genome.cse.ucsc.edu, and http://www.genecards.org/index.shtml) for SNPs either shown to affect biological functions or predicted to affect biologic functions (transcription, splicing, translation, protein). A list of SNPs was submitted to Applied Biosystems Inc. (ABI, Foster City, California) for in silico analyses to exclude incompatible SNPs in the SNPlex multiplex reaction. Probes for a total of 48 compatible SNPs were made by ABI for SNPlex genotyping. (Table 2) Sample preparation
Table 2.
SNPs Genotyped
| Gene | Putative | |||
|---|---|---|---|---|
| Symbol | Gene Name | SNP ID | Chromosome | Function |
| AHCY | S-adenosylhomocysteine | rs819146 | 20 | Promoter |
| hydrolase | rs819156 | Intron 3 | ||
| rs7271501 | Intron 6 | |||
| rs819133 | Intron 9 | |||
| BHMT | betaine-homocysteine | rs16876512 | 5 | Promoter |
| methyltransferase | rs3733890 | R239Q, ESE | ||
| rs4703772 | L264, ESE | |||
| rs585800 | 3′-UTR, ESE | |||
| CBS | cystathionine-b-synthase | rs1788484 | 21 | Promoter |
| rs5742905 | I278T, ESE | |||
| rs12613 | 3′-UTR, ESE | |||
| rs706209 | 3′-UTR, ESE | |||
| DHFR | dihydrofolate reductase | rs408626 | 5 | Promoter |
| rs1677658 | Promoter | |||
| rs1643649 | Intron | |||
| MAT2A | methionine adenosyltransferase | rs1078004 | 2 | R264, ESE |
| II alpha | rs2043675 | Intron 7 | ||
| MAT2B | methionine adenosyltransferase | rs4869087 | Intron 1 | |
| II beta | rs12515448 | Intron 2 | ||
| rs17061795 | L175, ESE | |||
| rs6898075 | Intron 4 | |||
| MTHFD1 | methylene-tetrahydrofolate | rs8003379 | 14 | Intron 2 |
| dehydrogenase | rs1950902 | K134R | ||
| rs8016556 | Intron 16 | |||
| rs11627387 | Intron 26 | |||
| rs2281603 | Intron 27 | |||
| MTHFR | methylene-tetrahydrofolate | rs3737965 | 1 | Promoter |
| reductase | rs2066470 | P39, ESE | ||
| rs9651118 | Intron 2 | |||
| rs2066462 | S352, ESE | |||
| rs2274976 | R594Q, ESE | |||
| rs4846049 | 3′-UTR | |||
| MTR | methyltetrahydrofolate- | rs1805087 | 1 | D919G, ESE |
| homocysteine | rs2229276 | A1048, ESE | ||
| methyltransferase | rs1131449 | L1192, ESE | ||
| rs11799670 | 3′-UTR, ESE | |||
| MTRR | methyltetrahydrofolate- | rs326118 | 5 | Promoter |
| homocysteine | rs1532268 | S175L | ||
| methyltransferase reductase | rs10064631 | L333V, ESE | ||
| rs162036 | K350R, TRP | |||
| rs2287780 | R415C | |||
| rs16879334 | P450R, ESE | |||
| rs9282787 | 3′-UTR ESE | |||
| rs9282788 | 3′-UTR ESE | |||
| rs9332 | 3′-UTR ESE | |||
| TYMS | thymidylate synthase | rs2853533 | 18 | Intron 1 |
| rs699517 | 3′-UTR ESE | |||
| rs2790 | 3′-UTR ESE |
SNP: single nucleotide polymorphism, ESE: exonic splicing element, UTR: mRNA untranslated region, TRP: triple helix regulatory site, rs: reference SNP
Blood samples and/or saliva samples were obtained from cases and both parents when possible. Genomic DNA from blood lymphocytes was extracted using the Puregene DNA Kit (Gentra Systems Inc, Minneapolis, MN) and DNA in saliva was extracted using the Oragene DNA collection kit (DNA Genotek, Inc., Ottawa, Ontario, Canada). Anonymous control DNA from 92 Hispanic individuals recruited from the Houston area and 92 Caucasian individuals from the Human Variation Panel-Caucasian Panel of 100 (HD100CAU) without a personal or family history of NTDs were used as negative controls and to evaluate the SNPs for Hardy Weinberg Equilibrium (HWE). No significant differences in the proportion of African and European Caucasian (CEU) genetic contributions between negative controls and patient samples were observed as reported in our previous study.17 Quality control of the genotyping was accomplished using known DNA samples from 30 Western European Caucasian families collected and used for building the SNP haplotypes map in humans (HapMap).
DNA Genotyping
SNP genotyping was carried out using the Applied Biosystems Inc. (ABI) SNPlex genotyping platform based on a multiplex oligonucleotide ligation/PCR/probe hybridization assay as described in our previous study.25 Positive control DNA provided by ABI and no DNA controls were included for genotyping quality controls. The positive control DNA contains known genomic DNA to monitor the SNPlex platform for proper performance, while the no DNA control does not contain genomic DNA and will monitor background signals per experiments. Genotype data analyses were performed using the GeneMapper v4.0 software (ABI) and the genotypes called by the software were examined by at least two investigators before exporting and compiling for statistical analysis. Two SNPs were excluded from statistical analysis because they did not meet our criteria for having at least an 85 percent genotype call rate from GeneMapper.
Statistical Analysis
Genotypes of patients and parents were subjected to Mendelian error testing where SNPs with more than 10 families having errors were re-examined. SNPs were not analyzed when more than 10 families demonstrated non-Mendelian errors even after re-examination. In addition, patient families with more than 5 SNPs shown to have non-Mendelian errors were excluded from statistical analyses. Statistical analysis was performed with the model independent reconstruction combined-transmission disequilibrium test (RC-TDT) and standard TDT (S-TDT). RC-TDT is an analytic method for genetic association studies that utilizes affected offspring and is applicable when one of the two parents is missing as the primary data structure.26, 27 RC-TDT estimates the transmitted allele from the parental genotype based on observed genotypes and it is not subject to population stratification. Deviations from the expectation of allele frequencies under random transmission are considered evidence of linkage. All SNPs subjected to RC-TDT were also tested for HWE in NTD negative control samples to evaluate genotype calling errors and equilibrium of allele segregation. SNPs failing the HWE test were not analyzed. LD between SNPs in the same gene was examined using Haploview 4.1 (http://www.broad.mit.edu/haploview/haploview). RC-TDT results of SNP allele(s) with p <0.05 were considered significant. Segregation of alleles for unlinked SNPs is expected to be a random process.
Results
To verify SNPlex genotyping platform accuracy, we compared genotypes we obtained for 45 CEU individuals used in the HapMap project with genotypes published in the SNP database. For all the SNPs successfully genotyped, 100% concordance was observed (data not shown). Duplicates of the positive controls used in separate SNPlex experiments produced reproducible genotypes.
Minor allele frequencies (MAF) were computed for the 46 successfully genotyped SNPs for controls, patients’ and patients’ parents according to their ethnicities and disease status. (Tables 3A and 3B) Frequencies of minor alleles of each SNP between control, patients and patients’ parents appears to be quite similar within same ethnic group. The minor alleles and their frequencies among our Caucasian controls, patients and parents are consistent with those reported by the HapMap CEU. Minor allele and frequencies of the tested SNPs for Mexican American are not available in public databases so we could not carry out comparisons with the data we obtained from the Mexican American controls and patients we studied. Many of the 46 SNPs have minor allele frequency differences from 0.1–0.24 between Caucasian and Mexican Americans. Higher MAF for BHMT rs3733890, rs4703772; MAT2A rs1078004; MTR rs2229276; and MTRR rs326118, rs162036 and rs9332 were seen in Mexican Americans than Caucasian Americans. Lower MAF for BHMT rs585800; DHFR rs1643649; MAT2B rs17061795; MTHFD1 rs8003379; MTHFR rs4846049; and MTRR rs1532268 were seen in Mexican Americans than Caucasian Americans.
Table 3.
A Frequencies of minor alleles for Caucasian American samples
| Caucasian American MAF | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| gene | SNP ID | ||||||||||
| AHCY | rs819133 | G | T | T | 0.16 | 0.09 | 0.19 | 0.14 | 0.13 | ||
| rs7271501 | G | C | C | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | |||
| rs819156 | C | A | A | 0.13 | 0.06 | 0.14 | 0.11 | 0.11 | |||
| rs819146 | G | T | G | 0.13 | 0.07 | 0.14 | 0.11 | 0.11 | |||
| BHMT | rs16876512 | C | T | T | 0.13 | 0.11 | 0.11 | 0.11 | 0.11 | ||
| rs3733890 | G | A | A | 0.30 | 0.26 | 0.30 | 0.28 | 0.26 | |||
| rs4703772 | C | T | T | 0.01 | 0.01 | 0.03 | 0.02 | 0.02 | |||
| rs585800 | A | T | T | 0.26 | 0.28 | 0.26 | 0.27 | 0.26 | |||
| CBS | rs706209 | C | T | T | 0.47 | 0.46 | 0.44 | 0.45 | 0.44 | ||
| rs12613 | G | A | A | 0.13 | 0.12 | 0.13 | 0.13 | 0.12 | |||
| rs5742905 | T | C | C | 0.01 | 0.02 | 0.01 | 0.01 | 0.00 | |||
| rs1788484 | C | T | T | 0.39 | 0.38 | 0.36 | 0.37 | 0.40 | |||
| DHFR | rs1643649 | T | C | C | 0.27 | 0.27 | 0.25 | 0.26 | 0.27 | ||
| rs1677658 | G | T | T | na | na | Na | na | na | |||
| rs408626 | A | G | G | 0.42 | 0.40 | 0.42 | 0.41 | 0.43 | |||
| MAT2A | rs1078004 | C | G | G | 0.48 | 0.50 | 0.49 | 0.50 | 0.45 | ||
| rs2043675 | T | C | C | 0.34 | 0.28 | 0.37 | 0.33 | 0.32 | |||
| MAT2B | rs4869087 | A | C | C | 0.25 | 0.30 | 0.28 | 0.29 | 0.31 | ||
| rs12515448 | A | G | G | 0.48 | 0.50 | 0.49 | 0.50 | 0.48 | |||
| rs17061795 | A | G | G | 0.13 | 0.17 | 0.15 | 0.16 | 0.17 | |||
| rs6898075 | C | T | T | 0.40 | 0.45 | 0.43 | 0.43 | 0.39 | |||
| MTHFD1 | rs8003379 | A | C | C | 0.23 | 0.20 | 0.20 | 0.20 | 0.23 | ||
| rs1950902 | C | T | T | 0.33 | 0.32 | 0.38 | 0.36 | 0.20 | |||
| rs8016556 | T | C | C | 0.32 | 0.31 | 0.35 | 0.33 | 0.35 | |||
| rs11627387 | G | A | A | 0.37 | 0.35 | 0.40 | 0.38 | 0.43 | |||
| rs2281603 | A | G | G | 0.26 | 0.22 | 0.26 | 0.24 | 0.26 | |||
| MTHFR | rs4846049 | G | T | T | 0.29 | 0.28 | 0.32 | 0.31 | 0.35 | ||
| rs2274976 | G | A | A | 0.04 | 0.06 | 0.04 | 0.04 | 0.06 | |||
| rs2066462 | C | T | T | 0.05 | 0.04 | 0.04 | 0.04 | 0.06 | |||
| rs9651118 | T | C | C | 0.21 | 0.23 | 0.21 | 0.21 | 0.23 | |||
| rs2066470 | C | T | T | 0.11 | 0.08 | 0.11 | 0.10 | 0.09 | |||
| rs3737965 | C | T | T | 0.06 | 0.08 | 0.06 | 0.07 | 0.06 | |||
| MTR | rs1805087 | A | G | G | 0.19 | 0.20 | 0.16 | 0.18 | 0.19 | ||
| rs2229276 | A | G | G | 0.47 | 0.46 | 0.47 | 0.47 | 0.39 | |||
| rs1131449 | T | CT | C | 0.39 | 0.38 | 0.41 | 0.40 | 0.44 | |||
| rs11799670 | A | G | G | 0.03 | 0.03 | 0.02 | 0.02 | 0.02 | |||
| MTRR | rs326118 | T | G | G | 0.12 | 0.09 | 0.12 | 0.11 | 0.14 | ||
| rs1532268 | G | A | A | 0.40 | 0.39 | 0.40 | 0.39 | 0.37 | |||
| rs10064631 | G | C | C | na | na | Na | na | na | |||
| rs162036 | A | G | G | 0.12 | 0.10 | 0.12 | 0.11 | 0.14 | |||
| rs2287780 | C | T | T | 0.06 | 0.07 | 0.05 | 0.06 | 0.04 | |||
| rs16879334 | C | G | G | 0.05 | 0.05 | 0.05 | 0.05 | 0.03 | |||
| rs9282787 | T | C | C | 0.24 | 0.21 | 0.24 | 0.23 | 0.18 | |||
| rs9282788 | T | C | C | 0.08 | 0.07 | 0.06 | 0.07 | 0.03 | |||
| rs9332 | C | T | T | 0.13 | 0.11 | 0.13 | 0.13 | 0.14 | |||
| TYMS | rs2853533 | G | C | C | 0.02 | 0.04 | 0.02 | 0.02 | 0.04 | ||
| rs699517 | C | T | T | 0.30 | 0.28 | 0.30 | 0.29 | 0.36 | |||
| rs2790 | A | G | G | 0.19 | 0.16 | 0.21 | 0.19 | 0.24 | |||
| Table 3B Frequencies of minor alleles for Mexican American samples | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Mexican American MAF | |||||||||
| gene | SNP ID | ||||||||
| AHCY | rs819133 | G | T | T | 0.16 | 0.12 | 0.17 | 0.16 | 0.16 |
| rs7271501 | G | C | C | 0.01 | 0.01 | 0.02 | 0.02 | 0.06 | |
| rs819156 | C | A | A | 0.11 | 0.10 | 0.11 | 0.10 | 0.11 | |
| rs819146 | G | T | G | 0.13 | 0.11 | 0.14 | 0.13 | 0.14 | |
| BHMT | rs16876512 | C | T | T | 0.10 | 0.10 | 0.11 | 0.11 | 0.05 |
| rs3733890 | G | A | A | 0.36 | 0.37 | 0.40 | 0.39 | 0.41 | |
| rs4703772 | C | T | T | 0.21 | 0.18 | 0.21 | 0.20 | 0.20 | |
| rs585800 | A | T | T | 0.15 | 0.13 | 0.16 | 0.15 | 0.14 | |
| CBS | rs706209 | C | T | T | 0.32 | 0.30 | 0.31 | 0.30 | 0.38 |
| rs12613 | G | A | A | 0.15 | 0.15 | 0.15 | 0.15 | 0.09 | |
| rs5742905 | T | C | C | 0.01 | 0.01 | 0.02 | 0.01 | 0.02 | |
| rs1788484 | C | T | T | 0.31 | 0.29 | 0.32 | 0.31 | 0.31 | |
| DHFR | rs1643649 | T | C | C | 0.20 | 0.18 | 0.21 | 0.20 | 0.12 |
| rs1677658 | G | T | T | na | na | na | na | na | |
| rs408626 | A | G | G | 0.56 | 0.55 | 0.54 | 0.54 | 0.50 | |
| MAT2A | rs1078004 | C | G | G | 0.59 | 0.62 | 0.63 | 0.62 | 0.57 |
| rs2043675 | T | C | C | 0.29 | 0.29 | 0.28 | 0.29 | 0.30 | |
| MAT2B | rs4869087 | A | C | C | 0.28 | 0.24 | 0.28 | 0.27 | 0.34 |
| rs12515448 | A | G | G | 0.47 | 0.43 | 0.46 | 0.45 | 0.42 | |
| rs17061795 | A | G | G | 0.11 | 0.10 | 0.11 | 0.10 | 0.07 | |
| rs6898075 | C | T | T | 0.40 | 0.38 | 0.39 | 0.39 | 0.33 | |
| MTHFD1 | rs8003379 | A | C | C | 0.17 | 0.22 | 0.12 | 0.16 | 0.10 |
| rs1950902 | C | T | T | 0.28 | 0.23 | 0.33 | 0.29 | 0.17 | |
| rs8016556 | T | C | C | 0.28 | 0.26 | 0.25 | 0.26 | 0.28 | |
| rs11627387 | G | A | A | 0.37 | 0.34 | 0.33 | 0.33 | 0.44 | |
| rs2281603 | A | G | G | 0.26 | 0.25 | 0.27 | 0.26 | 0.30 | |
| MTHFR | rs4846049 | G | T | T | 0.20 | 0.23 | 0.19 | 0.21 | 0.14 |
| rs2274976 | G | A | A | 0.03 | 0.04 | 0.05 | 0.05 | 0.04 | |
| rs2066462 | C | T | T | 0.02 | 0.02 | 0.02 | 0.02 | 0.03 | |
| rs9651118 | T | C | C | 0.24 | 0.25 | 0.26 | 0.26 | 0.30 | |
| rs2066470 | C | T | T | 0.11 | 0.10 | 0.11 | 0.11 | 0.06 | |
| rs3737965 | C | T | T | 0.08 | 0.07 | 0.10 | 0.09 | 0.07 | |
| MTR | rs1805087 | A | G | G | 0.23 | 0.19 | 0.24 | 0.22 | 0.16 |
| rs2229276 | A | G | G | 0.47 | 0.46 | 0.47 | 0.47 | 0.49 | |
| rs1131449 | T | CT | C | 0.38 | 0.40 | 0.38 | 0.39 | 0.45 | |
| rs11799670 | A | G | G | 0.10 | 0.10 | 0.11 | 0.10 | 0.09 | |
| MTRR | rs326118 | T | G | G | 0.38 | 0.32 | 0.39 | 0.36 | 0.33 |
| rs1532268 | G | A | A | 0.22 | 0.25 | 0.19 | 0.21 | 0.19 | |
| rs10064631 | G | C | C | na | na | na | na | na | |
| rs162036 | A | G | G | 0.41 | 0.36 | 0.43 | 0.40 | 0.38 | |
| rs2287780 | C | T | T | 0.14 | 0.13 | 0.15 | 0.14 | 0.13 | |
| rs16879334 | C | G | G | 0.12 | 0.12 | 0.14 | 0.13 | 0.11 | |
| rs9282787 | T | C | C | 0.10 | 0.10 | 0.10 | 0.10 | 0.09 | |
| rs9282788 | T | C | C | 0.11 | 0.08 | 0.11 | 0.10 | 0.05 | |
| rs9332 | C | T | T | 0.41 | 0.36 | 0.40 | 0.39 | 0.33 | |
| TYMS | rs2853533 | G | C | C | 0.02 | 0.03 | 0.02 | 0.02 | 0.01 |
| rs699517 | C | T | T | 0.35 | 0.34 | 0.33 | 0.34 | 0.44 | |
| rs2790 | A | G | G | 0.24 | 0.24 | 0.24 | 0.24 | 0.32 | |
MAF = minor allele frequency, na = not available due to low % genotyping efficiency, rs=reference SNP
MAF = minor allele frequency, na = not available due to low % genotyping efficiency, rs=reference SNP
LD analyses on Caucasian samples from HapMap did not suggest the SNPs we tested were linked and segregated together. However, some SNPs we tested were not included in the HapMap study, therefore we performed LD analyses using genotype data from the Caucasian and Hispanic controls in our study using Haploview 4.1. Three LD blocks between the 46 genotyped SNPs were identified with r2 threshold set at 0.8: rs819133, rs819156 and rs819146 in AHCY; rs326118, rs162036 and rs9332 of MTRR and rs2287780 with rs16879334 in MTRR (data not shown). Alleles in the same block are expected to segregate together and show similar significance to disease association. The other SNPs tested are not linked.
The analysis of the entire data set for the 46 successfully genotyped SNPs in the 11 genes found significant preferential transmission of alleles in CBS rs5742905 (T allele, p=0.015), DHFR rs1643649 (T allele, p=0.041), TYMS rs2853533 (G allele, p=0.021), and MTHFR rs3737965 (C allele, p=0.007). (Table 4) All four SNPs in NTD negative controls were in HWE. (Table 3A & 3B) The remaining 42 SNPs did not show significant association by RC-TDT.
Table 4.
RC-TDT Results
| Gene Symbol | dbSNP ID | Allele | RC-TDT p-value |
|---|---|---|---|
| CBS | rs5742905 | T | 0.0156 |
| DHFR | rs1643649 * | T | 0.041 1 |
| MTHFR | rs3737965 | C | 0.0076 |
| TYMS | rs2853533 | G | 0.0213 |
Note: rs11951883 merged with rs1643649. rs=reference SNP
Comment
The study of folate and its association with NTDs is an ongoing endeavor that has led to countless studies of different genes involved in the folate metabolism pathway. We looked for a genetic association of putative functional SNPs between 11 candidate genes and susceptibility to a specific type of NTD, spina bifida. In our study we demonstrated parental transmission disequilibrium in four SNPs, on four different genes not previously reported, which confers an increased susceptibility to SB in their offspring.
The strengths of our study include a patient sample size significantly larger than prior studies and use of the family based RC-TDT method known to be independent of model and ethnicity. Our sample is comprised of approximately 60% Mexican Americans, a group known to have one of the highest incidences of SB. A total of 46 putative functional SNPs on 11 candidate genes in folate/homocysteine metabolism pathways were studied. Forty two of the 46 SNPs examined did not reach significance in our study. There are several possible explanations for this negative finding. First, these 42 SNPs may not have a significant association with SB. Second, the study population size and power may not be sufficient to detect a genetic effect for these 42 SNPs, including the BHMT rs3733890, previously associated with increased risk of spina bifida in a family-based study consisting of 304 Caucasian American families.28 Our study consisted of a total of 610 families; 226 are Caucasian American. We cannot discount that BHMT rs3733890 may be an ethnic specific risk factor that could be shown utilizing a larger sample size study with ethnically matched samples. Consistent with prior reports, our study did not find an association of MTR rs1805087 (p.D919G) and MTRR rs162036 (Lys350Arg) with risk for SB.29
CBS
Many studies have found higher homocysteine in the plasma and amniotic fluid of mothers with NTD affected fetuses suggesting a failure to maintain homocysteine levels may be a contributing factor to NTD susceptibility.30–32 In our patient population the CBS SNP rs5742905, resulting in amino acid change from isoleucine to threonine, is significantly associated to risk of SB. CBS rs5742905 has not been examined previously in relation to SB. The most common CBS mutation studied is the 844ins68 allele, 11 bp downstream of rs5745905, which has shown varied association with NTDs or SB in different ethnic populations.28, 33, 34 Supporting this finding the CBS 844ins68 allele has been associated with lower serum homocysteine in a recent population study of over 10,000 subjects in Norway.35
CBS rs5742905 lle278Thr is associated with pyridoxine-responsive homocystinuria. The CBS 278Thr variant is less stable and has minimal CBS activity.36 Further experiments are needed to determine if 278Thr contributes to risk of SB. Since the frequency of rs5742905 C allele is very low (0.01–0.02), we anticipate to further validate this finding with other linked SNPs in the CBS gene.
DHFR
In addition to converting dihydrofolate to tetrahydrofolate, DHFR also converts food folates into tetrahydrofolate. The DHFR rs1643649 has not been evaluated for its risk association to SB. Previous studies have identified a 19 base pair deletion polymorphism (19bpdel) that has shown to affect expression with conflicting results associated with SB risk in different populations.20, 22, 37, 38 We did not examine the 19bpdel because the SNPlex method is not designed to interrogate polymorphisms involving deletions. More studies will be need to evaluate the role of the 19bpdel to SB risk.
We were able to show increased susceptibility to SB in our study with DHFR rs1643649. The rs1643949 is in intron 3 of DHFR ~10kb downstream from the 19 bpdel. We found the DHFR rs1643649 allele C is predicted to lose pre-mRNA splicing factor binding which may lead to improper splicing. The significance of changes in splicing factor affinity to DHFR pre-mRNA splicing remains to be studied. Alternatively, rs1643649 may be linked to a SB risk allele yet to be identified.
MTHFR
MTHFR has been the major focus of research since the common SNPs C677T (rs1801133) and A1298C (rs1801131) were identified as one of the first genetic links to the NTD risk.12, 14–16, 39 The less frequent 677T variant of MTHFR has half of the enzyme activity of the wildtype enzyme and has been shown to be associated with low serum folate and high homocysteine.35 A novel MTHFR polymorphism conferred increased risk for NTDs in a large Irish NTD cohort and was suggested to be physically linked to C677T.40
MTHFR rs3737965 is located in the promoter sequence and therefore variants may affect transcriptional activity. The T allele variant of rs3737965 is predicted to constitute a progesterone receptor binding motif in contrast to the C allele. We examined rs3737965 and found that it was significantly associated with increased risk transmission in the offspring. We found both C677T and A1298C are not linked with rs3737965 (r2=0.02 & 0.096 respectively). It is predicted the risk mechanism for rs3737965 is not due to simply linking with 677T or 1298C. Further experiments are needed to determine if the rs3737965 variant directly affects MTHFR transcription or is linked with a disease causing allele in our SB patients. Other studies have noted gene-gene interactions in various SNP alleles of MTHFR.28, 34, 41 All of these studies identified risk related to C677T. Two studies failed to show an associated gene-gene interaction but were limited by sample size.33, 42 Our findings demonstrate the importance to evaluate all SNPs throughout a candidate gene that are not linked to known risk alleles.
TYMS
The studies evaluating the 28 bp repeat polymorphism in the 5 prime untranslated region of TYMS association with SB remain inconclusive.43, 19 In our population, TYMS rs2853533 demonstrated an increased risk for SB. The MAF of rs2853533 is low (0.02–0.04); therefore validating this finding with other SNPs linked to rs2853533 is necessary.
In summary, to date no individual polymorphism in any candidate gene has been implicated as a risk factor in all studies. SNPs vary greatly in allele frequency among different geographic regions and ethnicities as we have demonstrated. Given the interconnectedness of the enzymes in the folate-homocysteine pathway we suspect that combinations of genetic variants in these enzymes may underlie NTD susceptibility. Our data contributes to the expanding body of literature.
Acknowledgement
This work was supported by grants from the National Institute of Health (P01 HD35946) and Shriners Hospital for Children (project 8580) to HN and a fellowship grant from the Department of Obstetrics, Gynecology, and Reproductive Sciences at The University of Texas Health Science Center at Houston to CAM. We are grateful to Drs. James Hixson and Lawrence Shimmin for technical support with the SNPlex experiments, to Phong X Tran and Chester Tsai for technical support and data management. We also thank the patients and their families for their participation in this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Presented at The Pregnancy Meeting, 29th Annual Meeting of the Society for Maternal-Fetal Medicine, January 26–31, 2009, San Diego, California.
Reprints are not requested by the authors.
References
- 1.Centers for Disease Control and Prevention (CDC) Racial/ethnic differences in the birth prevalence of spina bifida – United States, 1999–2005. MMWR Morb Mortal Wkly Rep 2009;57:1409-13. Erratum in MMWR: Morb Mortal Wkly Rep. 2009;58:61. [PubMed] [Google Scholar]
- 2.Martin JA, Hamiliton BE, Sutton PD, et al. Births: final data for 2006. Natl Vital Stat Rep. 2009;57:1–102. [PubMed] [Google Scholar]
- 3.Sadler TW. Embrology of Neural Tube Development. Embryology of neural tube development. Am J Med Genet C Semin Med Genet. 2005;135C:2–8. doi: 10.1002/ajmg.c.30049. [DOI] [PubMed] [Google Scholar]
- 4.Botto LD, Moore CA, Khoury MJ, Erickson JD. Neural-tube defects. N Engl J Med. 1999;341:1509–1519. doi: 10.1056/NEJM199911113412006. [DOI] [PubMed] [Google Scholar]
- 5.Molloy AM, Kirke PN, Troendle JF, et al. Maternal vitamin B12 status and risk of neural tube defects in a population with high neural tube defects prevalence and no folic acid fortification. Pediatrics. 2009;123:917–923. doi: 10.1542/peds.2008-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Czeizel AE, Dudas I. Prevention of the first occurrence of neural-tube defects by periconceptional vitamin supplementation. N Engl J Med. 1992;327:1832–1835. doi: 10.1056/NEJM199212243272602. [DOI] [PubMed] [Google Scholar]
- 7.MRC Vitamin Study Research Group. Prevention of neural tube defects: results of the Medical Research Council Vitamin Study. Lancet. 1991;338:131–137. [PubMed] [Google Scholar]
- 8.Lumley J, Watson L, Watson M, Bower C. Periconceptional supplementation with folate and/or multivitamins for preventing neural tube defects. Cochrane Database of Systematic Reviews. 2001;(Issue 3) doi: 10.1002/14651858.CD001056. [DOI] [PubMed] [Google Scholar]
- 9.Honein MA, Paulozzi LJ, Mathews TJ, Erickson JD, Wong LY. Impact of folic acid fortification of the US food supply on the occurrence of neural tube defects. JAMA. 2001;285:2981–2986. doi: 10.1001/jama.285.23.2981. [DOI] [PubMed] [Google Scholar]
- 10.Berry RJ, Li Z, Erickson JD, et al. Prevention of neural-tube defects with folic acid in China. China-U.S. Collaborative Project for Neural Tube Defect Prevention. N Engl J Med. 1999;341:1485–1490. doi: 10.1056/NEJM199911113412001. Erratum in: N Engl J Med 1999;341:1864. [DOI] [PubMed] [Google Scholar]
- 11.Olney R, Mulinare R. Epidemiology of neural tube defects. Ment Retard Dev Disabil Res Rev. 1998;4:241–246. [Google Scholar]
- 12.van der Put NM, Steegers-Theunissen RP, Frosst P, et al. Mutated methylenetetrahydrofolate reductase as a risk factor for spina bifida. Lancet. 1995;346:1070–1071. doi: 10.1016/s0140-6736(95)91743-8. [DOI] [PubMed] [Google Scholar]
- 13.Volcik KA, Blanton SH, Tyerman GH, Jong ST, Rott EJ, Page TZ, Romaine NK, Northrup H. Methylenetetrahydrofolate reductase and spina bifida: evaluation of level of defect and maternal genotypic risk in Hispanics. Am J Med Genet. 2000;95(1):21–27. [PubMed] [Google Scholar]
- 14.Félix TM, Leistner S, Giugliani R. Metabolic effects and the methylenetetrahydrofolate reductase (MTHFR) polymorphism associated with neural tube defects in southern Brazil. Birth Defects Res A Clinic Mol Tertol. 2004;70:459–463. doi: 10.1002/bdra.20011. [DOI] [PubMed] [Google Scholar]
- 15.Kirke PN, Mills JL, Molloy AM, et al. Impace of the MTHFR C677T polymorphism on risk of neural tube defects: case-control study. BMJ. 2004;328:1535–1536. doi: 10.1136/bmj.38036.646030.EE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Botto LD, Yang Q. 5, 10-methyltetrahydrofolate reductase gene variants and congenital anomalies: a HuGE review. Am J Epidemiol. 2000;151:862–877. doi: 10.1093/oxfordjournals.aje.a010290. [DOI] [PubMed] [Google Scholar]
- 17.Au KS, Tran PX, Tsai CC, et al. Characteristics of a spina bifida population including North American caucasian and Hispanic individuals. Birth Defects Res A Clin Mol Teratol. 2008;82:682–700. doi: 10.1002/bdra.20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin YN, Salavaggione OE, Eckloff BW, Wieben ED, Schaid DJ, Weinshilboum RM. Human methylenetetrahydrofolate reductase pharmacogenomics: gene resequencing and functional genomics. Pharmacogenet Genomics. 2006;16:265–277. doi: 10.1097/01.fpc.0000194423.20393.08. [DOI] [PubMed] [Google Scholar]
- 19.Wilding CS, Relton CL, Sutton MJ, et al. Thymidylate synthase repeat polymorphisms and risk of neural tube defects in a population from northern United Kingdom. Birth Defects Res A Clin Mol Teratol. 2004;70:483–485. doi: 10.1002/bdra.20038. [DOI] [PubMed] [Google Scholar]
- 20.van der Linden IJ, Nguyen U, Heil SG, et al. Variation and expression of dihydrofolate reductase (DHFR) in relation to spina bifida. Mol Genet Metab. 2007;91:98–103. doi: 10.1016/j.ymgme.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 21.van der Linden IJ, Heil SG, Kouwenberg IC, den Heijer M, Blom HJ. The methylenetetrahydrofolate dehydrogenase (MTHFD1) 1958G>A variant is not associated with spina bifida risk in the Dutch population. Clin Genet. 2007;72:599–600. doi: 10.1111/j.1399-0004.2007.00904.x. [DOI] [PubMed] [Google Scholar]
- 22.Johnson WG, Stenroos ES, Spychaia JR, Chatkupt S, Ming SX, Buyske S. New 19 bp deletion polymorphism in intron-1 dihydrofolate reductase (DHFR): a risk factor for spina bifida acting in mothers during pregnancy? Am J Med Genet. 2004;124A:339–345. doi: 10.1002/ajmg.a.20505. [DOI] [PubMed] [Google Scholar]
- 23.Enzinger C, Wirleitner B, Spottl N, Bock G, Fuchs D, Baier-Bitterlich G. Reduced pteridine derivatives induce apoptosis in PC12 cells. Neurochem Int. 2002;41:71–78. doi: 10.1016/s0197-0186(01)00134-6. [DOI] [PubMed] [Google Scholar]
- 24.Spottl N, Wirleitner B, Bock G, Widner B, Fuchs D, Baier-Bitterlich G. Reduced pteridine derivatives induce apoptosis in human neuronal NT2/HNT cells. Immunobiology. 2000;201:478–491. doi: 10.1016/S0171-2985(00)80100-X. [DOI] [PubMed] [Google Scholar]
- 25.Davidson C, Northrup N, King TM, et al. Genes in glucose metabolism and association with spina bifida. Reprod Sci. 2008;15:51–58. doi: 10.1177/1933719107309590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knapp M. The transmission/disequilibrium test and parental-genotype reconstruction: the reconstruction-combined transmission/disequilibrium test. Am J hum Genet. 1999;64:861–870. doi: 10.1086/302285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knapp M. Reconstructing parental genotypes when testing for linkage in the presence of association. Theor Popul Biol. 2001;60:141–148. doi: 10.1006/tpbi.2001.1540. [DOI] [PubMed] [Google Scholar]
- 28.Boyles AL, Billups AV, Deak KL, et al. Neural tube defects and folate pathway genes: family-based association tests of gene-gene and gene-environment interactions. Environ Health Perspect. 2006;114:1547–1552. doi: 10.1289/ehp.9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Leary VB, Mills JL, Pangilinan F, Kirke PN, Cox C, Conley M, Weiler A, Peng K, Shane B, Scott JM, Parle-McDermott A, Molloy AM, Brody LC. Members of the Birth Defects Research Group. Analysis of methionine synthase reductase polymorphisms for neural tube defects risk association. Mol Genet Metab. 2005;85:220–227. doi: 10.1016/j.ymgme.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 30.Steegers-Theunissen RP, Boers GH, Blom HJ, et al. Neural-tube defects and elevated homocysteine levels in amniotic-fluid. Am J Obstet Gynecol. 1995;172:1436–1441. doi: 10.1016/0002-9378(95)90474-3. [DOI] [PubMed] [Google Scholar]
- 31.Mills JL, McPartlin JM, Kirke PN, et al. Homocysteine metabolism in pregnancies complicated by neural-tube defects. Lancet. 1995;345:149–151. doi: 10.1016/s0140-6736(95)90165-5. [DOI] [PubMed] [Google Scholar]
- 32.Felkner M, Suarez L, Canfield MA, Brender JD, Sun Q. Maternal serum homocysteine and risk for neural tube defects in a Texas-Mexico border population. Birth Defects Res A (Clin Mol Teratol) 2009 Jan 29;85:574–578. doi: 10.1002/bdra.20545. Early view. Epub. [DOI] [PubMed] [Google Scholar]
- 33.Morrison K, Papapetrou C, Hol FA, et al. Susceptibility to spina bifida; an association study of five candidate genes. Ann Hum Genet. 1998;62:379–396. doi: 10.1046/j.1469-1809.1998.6250379.x. [DOI] [PubMed] [Google Scholar]
- 34.de Franchis R, Botto LD, Sebastio G, et al. Spina bifida and folate related genes: a study of gene-gene interactions. Genet Med. 2002;4:126–130. doi: 10.1097/00125817-200205000-00005. [DOI] [PubMed] [Google Scholar]
- 35.Fredriksen A, Meyer K, Ueland PM, Vollset SE, Grotmol T, Schneede J. Large-scale population-based metabolic phenotyping of thirteen genetic polymorphisms related to one-carbon metabolism. Hum Mutat. 2007;28:856–865. doi: 10.1002/humu.20522. [DOI] [PubMed] [Google Scholar]
- 36.Chen X, Wang L, Fazlieva R, Kruger WD. Contrasting behaviors of mutant cystathionine beta-synthase enzymes associated with pyridoxine response. Hum Mutat. 2006;27:474–482. doi: 10.1002/humu.20320. [DOI] [PubMed] [Google Scholar]
- 37.Parle-McDermott A, Pangilinan F, Mills JL, et al. The 19-bp deletion polymorphism in intron-1 of dihydrofolate reductase (DHFR) may decrease rather than increase risk for spina bifida in the Irish population. Am J Med Genet Part A. 2007;143A:1174–1180. doi: 10.1002/ajmg.a.31725. [DOI] [PubMed] [Google Scholar]
- 38.Akar N, Akar E, Egin Y, Deda G, Arsan S, Ekim M. Neural tube defects and 19 bp deletion within intron-1 of dihydrofolate reductase gene. Turk J Med Sci. 2008;38:383–386. [Google Scholar]
- 39.van der Put NMJ, Gabreels F, Stevens EMB, et al. A second common mutation in the methylenetetrahydrofolate reductase gene:an additional risk factor for neural-tube defects? Am J Hum Genet. 1998;62:1044–1051. doi: 10.1086/301825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Leary VB, Mills JL, Parle-McDermott A, et al. Screening for new MTHFR polymorphisms and NTD risk. Am J Med Genet. 2005;138A:99–106. doi: 10.1002/ajmg.a.30846. [DOI] [PubMed] [Google Scholar]
- 41.Relton CL, Wilding CS, Pearce MS, et al. Gene-gene interactions in a folate-related genes and risk of neural tube defects in a UK population. J Med Genet. 2004;41:256–260. doi: 10.1136/jmg.2003.010694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Speer MC, Nye J, McLone D, et al. Possible interaction of genotypes at cystathionine β-synthase and methylenetetrahydrofolate reductase (MTHFR) in neural tube defects. Clin Genet. 1999;56:142–144. doi: 10.1034/j.1399-0004.1999.560208.x. [DOI] [PubMed] [Google Scholar]
- 43.Volcik KA, Shaw GM, Zhu H, Lammer EJ, Laurent C, Finnell RH. Associations between polymorphisms within the thymidylate synthase gene and spina bifida. Birth Defects Res A Clin Mol Teratol. 2003;67:924–928. doi: 10.1002/bdra.10029. [DOI] [PubMed] [Google Scholar]