Abstract
Isochorismate synthase (ICS) converts chorismate to isochorismate for the biosynthesis of phylloquinone, an essential cofactor for photosynthetic electron transport. ICS is also required for salicylic acid (SA) synthesis during Arabidopsis defense. In several other species, including Populus, SA is derived primarily from the phenylpropanoid pathway. We therefore sought to investigate ICS regulation in Populus to learn the extent of ICS involvement in SA synthesis and defense. Arabidopsis harbors duplicated AtICS genes that differ in their exon-intron structure, basal expression, and stress inducibility. In contrast, we found a single ICS gene in Populus and six other sequenced plant genomes, pointing to the AtICS duplication as a lineage-specific event. The Populus ICS encodes a functional plastidic enzyme, and was not responsive to stresses that stimulated phenylpropanoid accumulation. Populus ICS underwent extensive alternative splicing that was rare for the duplicated AtICSs. Sequencing of 184 RT-PCR Populus clones revealed 37 alternative splice variants, with normal transcripts representing ≈50% of the population. When expressed in Arabidopsis, Populus ICS again underwent alternative splicing, but did not produce normal transcripts to complement AtICS1 function. The splice-site sequences of Populus ICS are unusual, suggesting a causal link between junction sequence, alternative splicing, and ICS function. We propose that gene duplication and alternative splicing of ICS evolved independently in Arabidopsis and Populus in accordance with their distinct defense strategies. AtICS1 represents a divergent isoform for inducible SA synthesis during defense. Populus ICS primarily functions in phylloquinone biosynthesis, a process that can be sustained at low ICS transcript levels.
Keywords: defense, phenylpropanoid, splice site sequence, phylloquinone, salicylic acid
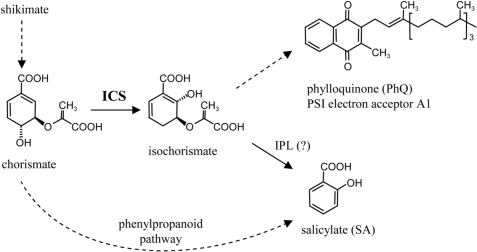
In plants and microorganisms, isochorismate synthase (ICS; EC 5.4.99.6) converts chorismate, a shikimate pathway intermediate, into isochorismate, which then can be channeled toward primary or secondary metabolism (1, 2). Most microorganisms contain two distinct ICS genes: menF is involved in biosynthesis of menaquinones (vitamin K2) for anaerobic electron transport (3), and entC is associated with aerobic metabolism for the production of iron chelators, such as 2,3-dihydroxybenzoic acid (2,3-DHBA), salicylic acid (SA), and complex siderophores, e.g., enterobactin and pyochelin (1, 4). ICS has also been associated with two distinct functions in plants (Fig. 1): biosynthesis of phylloquinone (PhQ, vitamin K1) for photosynthetic electron transport (2, 5) and of SA during stress response (6).
Fig. 1.
Schematic representation of ICS in plant metabolism. Question mark denotes the step catalyzed by isochorismate pyruvate lyase (IPL), an enzyme necessary for ICS-mediated SA biosynthesis in bacteria, but not yet identified in plants.
Arabidopsis contains two highly similar ICS genes, AtICS1 (At1g74710) and AtICS2 (At1g18870), derived from a segmental genome duplication (7). The two genes are differentially regulated, as only AtICS1 is stress inducible for SA-mediated defense (6, 8). Under normal growth conditions, the two isoforms exhibit functional redundancy, with AtICS1 and AtICS2 playing a major and minor role, respectively, in both SA and PhQ biosynthesis (8). In Nicotiana benthamiana, virus-induced gene silencing of a single ICS diminished accumulation of PhQ and UV-induced SA (9), suggesting multiple functions for the single gene. In addition to PhQ and SA, ICS function has been linked to biosynthesis of anthraquinones, indole alkaloids, and other phenolics (10–12). ICS may participate in biosynthesis of salicylate-containing phenolic glycosides (PGs) that are characteristic of the Salicaceae (e.g., Populus and Salix species), but SA involvement in PG synthesis has not been confirmed (13).
The broad suite of pathways supported by ICS, and the functional pleiotropy of ICS isoforms in plants, point to transcriptional regulation as the primary mechanism for modulating ICS function. Because ICS regulates the utilization of chorismate for both photosynthesis and defense, ICS may control cross-talk between primary and secondary metabolism. Coordination of such cross-talk differs between Arabidopsis and species such as Populus that constitutively maintain large and highly variable reserves of nonstructural phenylpropanoid metabolites (e.g., PGs and flavonoid-derived condensed tannins) in their leaves, shoots, and roots (14, 15). Here, we report alternative splicing and gene duplication as two contrasting mechanisms for modulating higher-plant ICS function. Alternative splicing is prevalent for the single-copy ICS genes in Populus and other species, but is rare for the duplicated Arabidopsis ICS genes. The Arabidopsis paralogs are structurally and transcriptionally distinct, consistent with functional divergence following lineage-specific duplication. Alternative splicing of Populus ICS also occurs in the foreign Arabidopsis host, but fails to produce functional transcripts, providing a genetic basis for the splice site sequence evolution. Differential ICS regulation may have evolved in accordance with the chemical defense strategies in Populus and Arabidopsis.
Results
Cloning and Characterization of Populus ICS.
A single ICS gene (eugene3.00120638) was identified in the sequenced Populus trichocarpa genome (13, 16). The predicted gene contains 16 exons and 15 introns, with intron no. 15 and exon no. 16 both located downstream of the stop codon (Fig. 2). Based on this gene model, the full-length coding sequence was cloned by RT-PCR from leaves of several genotypes, including P. tremuloides and P. fremontii × angustifolia hybrids. The ORF is 1,719 bp and encodes a protein of 572 aa that is highly (97–100%) similar to the JGI model. The predicted amino acid sequence contains a putative chloroplast transit peptide, based on TargetP (17) and Predotar (18) predictions, and a chorismate-binding domain (pfam 00425) conserved in all chorismate-utilizing enzymes (19). Populus ICS shares 64–70% amino acid sequence similarity with the characterized AtICS1, AtICS2, and Catharanthus roseus CrICS (6, 8, 10). We refer to the Populus trichocarpa gene as PtiICS, adopting the three-letter prefix proposed (20). When the context applies to multiple species or genotypes, it is referred to as Populus ICS.
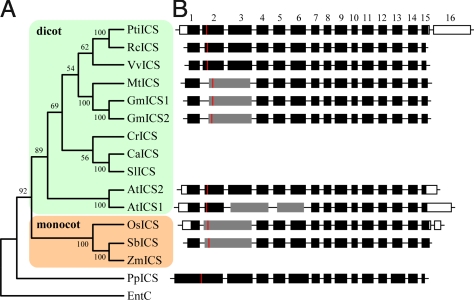
Fig. 2.
Phylogenetic relationship and exon-intron structures of representative plant ICS isoforms. (A) Topology of neighbor-joining tree constructed with predicted amino acid sequences, excluding the putative chloroplast transient peptides. Bootstrap values are indicated near the nodes. (B) Exon-intron structures of ICS genes from eight sequenced plant genomes. Known untranslated regions are shown in white boxes; predicted mature protein start sites are denoted by red lines; and fused exons within the mature proteins are shown in gray. Introns are not drawn to scale. The predicted exons 5 and 12 of PpICS from moss (Physcomitrella patens, JGI e_gw1.24.30.1) are longer than corresponding exons from other species, but with conserved splice sites. Additional sequences are Ricinus communis (EEF52128), Vitis vinifera (XP_002267681), Medicago truncatula (ABE91019), Glycine max (Phytozome Glyma01g25690 [GmICS1] and Glyma03g17420 [GmICS2]), Catharanthus roseus (Q9ZPC0), Capsicum annuum (AAW66457), Solanum lycopersicum (ABJ98719), Oryza sativa (EEC84436), Sorghum bicolor (JGI Sb02g022920), Zea mays (ACG29750), and E. coli entC (AAA16100).
Phylogenetic analysis was conducted using predicted ICS amino acid sequences in the public databases, and included orthologs from other sequenced plant genomes, e.g., rice (Oryza sativa) (21), grapevine (Vitis vinifera) (22), Medicago truncatula, sorghum (Sorghum bicolor) (23), soybean (Glycine max), castor bean (Ricinus communis), and moss (Physcomitrella patens) (24). Higher-plant ICSs form two distinct branches represented by dicot and monocot isoforms (Fig. 2A). As in Populus, the genomes of rice, grapevine, Medicago, sorghum, and castor bean all contain a single ICS. Arabidopsis and soybean both contain two highly similar ICS isoforms. The two AtICSs cluster separately from the rest of the dicot isoforms. The moss genome contains a single PpICS that is phylogenetically distinct from the higher-plant ICSs. It should be noted that PpICS is located within the PHYLLO locus that comprises a tetramodular fusion of four ancestral individual eubacterial genes (menF, menD, menC, and menH) essential for phylloquinone biosynthesis (5). PpICS is therefore allelic to the menF module of PHYLLO in moss, as was found in green algae (5). However, the menF module of PHYLLO in all higher-plants analyzed, including Arabidopsis and Populus, encodes a truncated ICS, lacking the C-terminal chorismate-binding domain (5). The data suggest that the ancestral plant genome contained a single ICS gene of eubacterial origin (menF). The data also support the finding (5) that menF fission (gene duplication followed by differential or partial degeneration) (25) gave rise to a single functional ICS (along with a dysfunctional menF module) during evolution of flowering plants. The present investigation revealed additional ICS gene duplications that occurred independently in the Arabidopsis and soybean lineages, after the divergence of eurosids I and II.
Gene structure analysis of the 11 ICS genes from nine sequenced genomes showed that the exon-intron junction sites within the mature ICS protein coding regions (denoted at the 5′-end by a red line in Fig. 2B) are conserved among PtiICS, RcICS, VvICS, AtICS2, and PpICS. However, AtICS1 contains two fused exons corresponding to PtiICS exons 3–4 and 5–6. The data suggest that AtICS2 is the ancestral gene, and that evolution of the present-day functionally dominant AtICS1 may represent a lineage-specific event. Rice OsICS and sorghum SbICS also contain a fused exon corresponding to PtiICS exons 2–3, and this pattern is shared by the legumes as well. The moss PpICS contains an intron at junction sites conserved in all other angiosperm ICS genes in this region. Therefore, this exon fusion in rice, sorghum, Medicago, and soybean most likely occurred after the divergence of monocots and dicots. Higher-plant ICS genes also contain an intron within the chloroplast-targeting presequence. All other ICS gene structure variations lie primarily within the presequence and UTR regions.
ICS transcripts were detected primarily, and at low levels, in green tissues of Populus (13). ICS expression did not show stress inducibility, based on microarray data mining from a number of inductive treatments [supporting information (SI) Fig. S1]. Instead, Populus ICS was coexpressed with orthologs of Arabidopsis genes involved in PhQ biosynthesis and PSI function (Fig. S1).
Alternative Splicing of ICS Is Common in Populus.
During RT-PCR cloning and sequencing, several putative splice variants were identified. For validation, the gene was cloned from taxonomically distinct genotypes, including the genome-sequenced P. trichocarpa (section Tacamahaca), P. tremuloides (section Populus), and several P. fremontii × angustifolia cottonwood hybrids (sections Aigeiros and Tacamahaca). Though the RT-PCR amplicon profile differed among genotypes (Fig. S2), sequencing (5–6 clones per genotype) confirmed the occurrence of alternative splicing in all genotypes. We also cloned the 3′-UTR by 3′-RACE from a cottonwood hybrid line (NUL) and found evidence of multiple polyadenylation sites (Fig. S3).
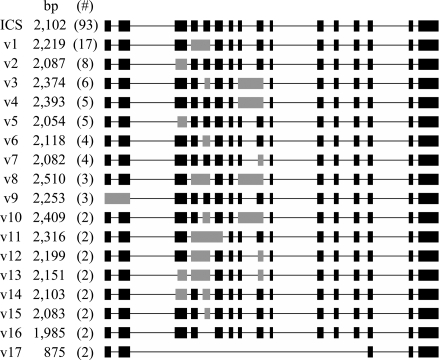
To further assess the extent of ICS alternative splicing in Populus, we sequenced 184 RT-PCR clones from two cottonwood hybrid lines (NUL and 1012). The two hybrid lines exhibit large differences in their foliar PG concentrations (15); however, we were unable to detect a clear genotypic difference in the expression level or alternative splicing frequency of ICS. A total of 38 unique transcripts were identified, ranging from 420 to 2,960 bp (Fig. 3 and Fig. S4). “Normal” transcripts (from the start codon to the 16th exon) were predominant in both genotypes, but represented only 46–55% of the sequenced clones (a combined 93 of 184 clones). Of the 37 alternative splice variants, 17 occurred more than once, representing 71 transcripts in the sequenced population (Fig. 3). Alternative splicing was found to occur at every exon, with the regions spanning exons 4–5 and 8–9 being “hot spots.” Though exon skipping comprises the primary form of alternative splicing in mammals (26), intron retention (56%) and alternative acceptor sites (55%) predominate among the 91 alternative transcripts (128 events total) that we identified. These observations are consistent with other alternative splicing analyses of plant genes (27). A significant portion (32%, or 29 clones) of the alternative transcripts contained multiple alternative splicing events. Nearly all events (63 of 66) were of the intron retention and/or alternative acceptor site types. Exon skipping comprised 7% of alternative splicing events (9 of 128), and 6 of the 9 resulted in loss of multiple (5–13) exons. Alternative splicing via alternative donor sites was less frequent (3 of 91 alternative transcripts) than reported in rice and Arabidopsis (18–22%) (27), and was observed only in transcripts with multiple alternative splicing events.
Fig. 3.
Representative splice variants of Populus ICS. Alternative transcripts (v1–v17) are arranged in order of abundance (frequencies in parentheses). A total of 184 RT-PCR clones were sequenced. Exons affected by alternative splicing are shown in gray. Unique splice variants (v18–v37) are shown in Fig. S4.
A majority of the alternative transcripts (67 of 91) harbor premature stop codons, or are less than 1 kb in length, due to multiexon skipping (four transcripts). These aberrant transcripts may be preferentially targeted for degradation by nonsense-mediated decay, as reported in mammals (28, 29). The predicted proteins from the remaining alternative transcripts contain deletions/insertions/substitutions that impact either the conserved protein domain PRK07054 (seven transcripts) or its upstream region, including several residues that are conserved in all plant ICS proteins (13 transcripts). Therefore, the splice variants are unlikely to produce functional ICS proteins.
BlastN search of GenBank dbEST (release 120508) identified 23 and six entries for AtICS1 and AtICS2, respectively. Only a single AS event was identified—a retention of the last intron in AtICS1. To further investigate the degree of alternative splicing in AtICS1 and AtICS2, we performed RT-PCR cloning and examined the insert size of randomly selected colonies. Consistent with the EST analysis, we found one and two alternative splicing events of 24 AtICS1 (4%) and 23 AtICS2 (9%) recombinant clones, respectively (Fig. S5). This is in sharp contrast with Populus ICS, and suggests that Populus and Arabidopsis ICS genes are subject to divergent regulatory mechanisms in their respective host environments.
Functional Complementation of ICS-Deficient E. coli and Arabidopsis Mutants by PtiICS.
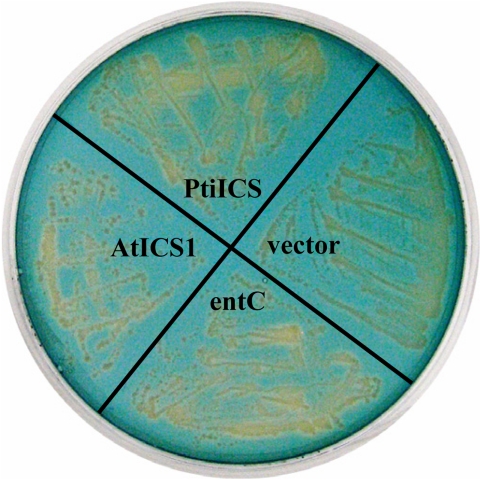
The mature protein-coding region of PtiICS was introduced into the E. coli entC− mutant PBB7 for functional complementation. The PBB7 mutant harbors a truncated copy of the entC, and was compromised in its ability to produce isochorismate-derived enterobactin (3). Conversion of chorismate to isochorismate by a functional ICS would restore siderophore secretion of the mutant, determined by the chrome azurol sulfonate (CAS) assay (30). Both entC and AtICS1 genes were included as positive controls, and as reported (8), both restored siderophore production in the mutant, manifest as orange halos around the colonies (Fig. 4). PtiICS also complemented the mutant in siderophore secretion, providing functional evidence for a role in isochorismate synthesis.
Fig. 4.
Functional complementation of entC-deficient E. coli mutant PBB7. The mutant strain was transformed with PtiICS, AtICS, entC, or empty vector constructs, and streaked on a CAS blue agar plate. Orange halos around the colonies indicated iron removal from the blue dye as a result of restored siderophore production.
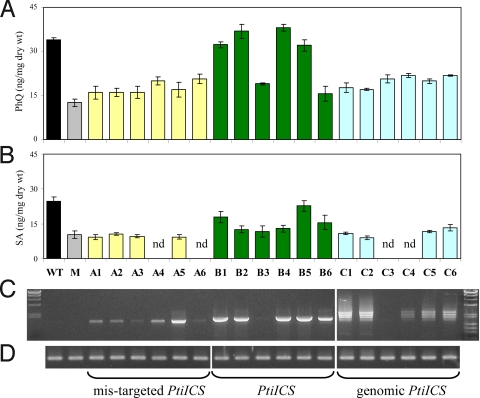
PtiICS was introduced into the Arabidopsis sid2–2 mutant for functional testing in planta. The sid2–2 mutant carries a deletion in AtICS1 (6) and is best characterized by its inability to increase SA synthesis following pathogen, UV, or ozone induction (6, 8, 31). Under normal growth conditions, the mutant also exhibits reduced levels of PhQ and SA (8, 31). We measured the concentration of PhQ and SA in transgenic Arabidopsis harboring the PtiICS cDNA, with or without the putative chloroplast-targeting presequence, under the CaMV 35S promoter. Expression of the full-length PtiICS cDNA in the sid2–2 mutant restored PhQ synthesis, whereas expression of the mis-targeted PtiICS failed to do so (Fig. 5A). Of the six T3 lines homozygous for the full-length PtiICS, only those lines with high levels of expression exhibited fully restored PhQ synthesis (Fig. 5 C and D). The mis-targeted PtiICS was also unable to restore constitutive SA synthesis in the transgenic mutants (Fig. 5B and Fig. S6). Of the four transgenic lines showing full PhQ complementation, only two (B1 and B5) accumulated SA at or near the wild-type concentrations, and the other two (B2 and B4) were more similar to the mutant (Fig. 5B). These findings provide in vivo evidence for PtiICS function in plastidic conversion of chorismate to isochorismate. However, PtiICS may have characteristics that affect product distribution between the downstream pathways leading to PhQ and SA synthesis.
Fig. 5.
Characterization of transgenic Arabidopsis sid2–2 mutant lines harboring various PtiICS constructs. (A) PhQ concentrations, (B) SA concentrations, (C) transcript levels of PtiICS, (D) transcript levels of the housekeeping gene actin. WT, wild type; M, mutant; A1–C6, independent transgenic lines expressing PtiICS cDNA without the plastid-targeting sequence (A1–A6), full-length cDNA (B1–B6), and 6-kb genomic fragment (C1–C6), respectively, under control of the 35S promoter; nd, not determined. Data are means ± SE of 2–6 biological replicates. Molecular weight markers in (C) were Lambda-HindIII (Left) and Lambda-HindIII plus PhiX174-HaeIII (Right).
Alternative Splicing of PtiICS in Transgenic Arabidopsis.
To understand the functional significance of alternative splicing in PtiICS regulation, we introduced two genomic PtiICS constructs into the Arabidopsis Col-0 wild-type and sid2–2 mutant. One construct contained a 6-kb, full-length genomic PtiICS sequence under control of the CaMV 35S promoter, and the other contained an 8-kb genomic fragment, including 1 kb each of the upstream and downstream sequences. RT-PCR analysis of T2 or T3 transgenic plants showed that PtiICS was alternatively spliced in both wild-type and mutant backgrounds, under either promoters used (Fig. 5C and Fig. S2). The alternative splicing patterns, however, differed from those observed in Populus. Constitutively spliced PtiICS transcripts were not detected in any of the transgenic Arabidopsis lines, based on gel electrophoresis of RT-PCR products. Sequencing of 64 RT-PCR clones confirmed the absence of the normal transcript, as well as the occurrence of several alternative splicing events (e.g., alternative donor sites, cryptic exons, and exon skipping) not found in Populus (Fig. S4).
The RT-PCR findings raised the question of whether the mis-spliced PtiICS transcripts could functionally complement the sid2–2 mutant. We measured PhQ and SA concentrations in transgenic lines harboring the 6-kb construct, because both 6-kb and 8-kb constructs gave rise to similar alternative splicing profiles of PtiICS. Transgenic lines expressing mis-spliced PtiICS accumulated both PhQ and SA at levels similar to those found in the sid2–2 mutant (Fig. 5 A and B), indicating that the 6-kb PtiICS gene was not functional in Arabidopsis. The results are consistent with the absence of normal PtiICS transcripts observed in these plants. Taken together, our data suggest that alternative splicing is an integral mechanism of PtiICS regulation and can affect overall ICS activity. Such a mechanism apparently is absent in the duplicated AtICS genes.
Splice-Site Sequence Variation of PtiICS.
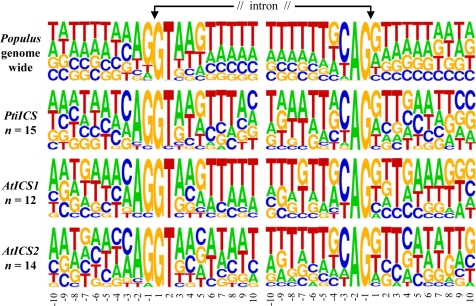
The observation that PtiICS splicing differs between Populus and Arabidopsis suggests an interaction of junction sequences with host splicing machinery. We analyzed the splice-site sequence patterns of PtiICS, AtICS1, and AtICS2, and compared them with the genome-wide consensus sequences compiled from 23,375 multiexon gene models of Populus (see Methods). The junction-site sequence patterns of Populus are highly T-rich (Fig. 6), and very similar to those reported in Arabidopsis and rice (32). However, A and C are more prevalent in the PtiICS splice-site sequences. In comparison, the splice-site intron sequences of AtICS1 and AtICS2 are more conserved, but the exon junction sequences are relatively G-rich. These data support the idea that sequence variation at the junctions sites contributes to the varying degree of alternative splicing observed for Populus and Arabidopsis ICS genes.
Fig. 6.
Splice junction site sequence logos of PtiICS in comparison with AtICS1, AtICS2, and the genome-wide consensus of Populus. A total of 115,273 exon-intron (donor) and 115,271 intron-exon (acceptor) splice sites extracted from 23,375 multiexon genes were used to compute the genome-wide nucleotide frequency at each position.
Discussion
Our finding that the single-copy, weakly expressed ICS gene undergoes extensive alternative splicing in Populus was unexpected, due to the essential role of PhQ in PSI function. Alternative splicing of ICS is common in this genus, affecting ≈50% of the transcripts in the genotypes studied. In contrast, alternative splicing is rare in the duplicated AtICS1 and AtICS2. Because the Populus and Arabidopsis isoforms all catalyze chorismate-to-isochorismate conversion, the differential contribution of alternative splicing to ICS regulation in these two lineages is consistent with an evolutionary impact of alternative splicing on secondary metabolism. A pertinent question is whether alternative splicing of ICS also occurs in other higher-plant lineages. ICS transcripts are poorly represented in public databases, but we were able to find evidence of ICS alternative splicing in rice (Gramene; http://www.gramene.org/) and grapevine (33), where large-scale EST and/or RNA-Seq data are available (Figs. S7 and S8). Alternative splicing may be the “default” mode of regulation for plant ICS genes, and the Arabidopsis homologs appear to have lost this property following lineage-specific duplication. The tendency for duplicated genes to exhibit loss of alternative splicing has been reported in animals (34). In the case of PtiICS, alternative splicing depends in part on the target gene sequence, because the genomic PtiICS also underwent alternative splicing in Arabidopsis. However, the PtiICS splicing pattern differed between the native and foreign hosts, implicating the host splicing machinery as well. PtiICS splicing profiles were similar regardless of whether the Populus or 35S promoter was used, suggesting that certain splicing signals (i.e., cis elements) reside in the introns or junction sites of PtiICS. In accordance with this idea, PtiICS splice-site sequences differ from the genome-wide consensus. A causal link between junction sequence variation, alternative splicing, and transcriptional regulation is therefore supported for plant ICS genes.
Whether alternative splicing of Populus ICS affects plant growth or development is not known, but PhQ is an essential component in photosynthetic PSI electron transfer. In the Arabidopsis sid2–2 mutant, deficiency of the predominant AtICS1 reduced PhQ accumulation by 60–70% with no loss of growth under normal conditions (8). In a separate investigation with a suite of PhQ-deficient mutants (5), PhQ levels at 15–18% of wild type were found to be sufficient for 50–70% PSI activity. This metabolic plasticity of PhQ biosynthesis and PSI electron transfer argues against the idea that alternative splicing of Populus ICS may function in tight regulation of PhQ. Instead, alternative splicing of ICS most likely plays a role that is advantageous in species like Populus but not in Arabidopsis. As reported previously, the ICS pathway is the primary route of SA biosynthesis in Arabidopsis (6), but in other plants, SA can be synthesized via the phenylalanine ammonia-lyase (PAL)-dependent phenylpropanoid pathway (35). For instance, SA is derived from the PAL pathway in ozone-exposed tobacco, and not via ICS as in similarly treated Arabidopsis (36, 37). Arabidopsis accumulates a very low basal level of SA, but has a highly sensitive SA signal perception pathway (38). In contrast, species such as potato, rice, and Populus exhibit constitutively high levels of SA that are 1–3 orders of magnitude higher than those following pathogen attack in Arabidopsis (39–41). In potato and rice, SA originates from the phenylpropanoid pathway and is thought to play a role in constitutive defense (42, 43). Populus species also are capable of accumulating up to 30% of the dry weight in leaves in salicylate-containing PGs as their frontline defense against herbivores (13, 14). Consistent with a phenylpropanoid-based defense, Populus PAL genes exhibit stress inducibility (13, 44), whereas the weakly expressed ICS is not sensitive to the same suite of inductive treatments (Fig. S1). Perhaps alternative splicing plays a regulatory role in species where it is advantageous to disengage ICS from SA biosynthesis, as in Populus. In Arabidopsis, ICS diversification may provide the means for decoupling SA biosynthesis from phenylpropanoid metabolism, thus allowing tighter control of metabolic costs of defense via SA signaling. The degree to which this adaptation is facilitated by reduced alternative splicing of ICS in Arabidopsis remains an open question. Deep sequencing of more plant genomes and associated biochemical analyses will be needed to further substantiate the interrelationship between SA biosynthetic route (phenylpropanoid vs. isochorismate based), ICS transcriptional regulation, and defense strategy used in a broader range of species.
Alternative splicing of Populus ICS may also have as yet uncharacterized consequences, as PhQ has been implicated in plasma membrane redox regulation and oxidative stress protection (45, 46). Our finding that Populus ICS functionally complemented PhQ synthesis more effectively than SA production in the Arabidopsis sid2–2 mutant also raises the possibility that metabolic channeling differentially directs the downstream uses of isochorismate in the two species. Populus ICS and AtICS1 could exhibit different affinities with other enzyme component(s) in a hypothetical metabolic complex for SA synthesis. Investigating the relationship between alternative splicing and Populus ICS function will require separation of normal transcripts from the pool of complex splice variants. This is presently challenging because fragment sizing using an automated DNA sequencer cannot resolve transcripts larger than ≈1.5 kb (Fig. S9). An RT-PCR cloning and sequencing-based approach, as used in this study, is not feasible for large-scale comparisons. Technical advances will be required to gain further insight into alternative splicing as complex as what we have observed for the Populus ICS. The present investigation reveals that alternative splicing and gene duplication differentially shaped the regulation of ICS in Populus and Arabidopsis, respectively, in accordance with their distinct defense strategies. That ICS exhibits such regulatory complexity may reflect a central metabolic role affecting the shikimate-chorismate pathway, photosynthetic electron transfer, and phenylpropanoid metabolism.
Materials and Methods
Plant Materials and Growth Conditions.
Populus plants were grown in soil in pots and maintained in a greenhouse, as described (15). Arabidopsis plants were grown in Metro-Mix 300 (Sun Gro) and maintained in a growth chamber at 22 °C under a 16-h photoperiod. Leaves were snap-frozen, ground to a fine powder in liquid nitrogen, and stored at −80 °C until use.
RT-PCR Cloning and Sequencing.
Total RNA extraction and first-strand cDNA synthesis were conducted as described (13). RT-PCR was performed using gene-specific primers (Table S1) and cloned into pCRII-TOPO (Invitrogen). The transformants were analyzed by colony PCR and/or sequenced using an Applied Biosystems ABI3730 or a Beckman CEQ8000 sequencer. Small-scale RT-PCR cloning and sequencing (5–6 clones per genotype) was performed for P. trichocarpa (Nisqually-1), P. tremuloides (271), and P. fremontii × angustifolia hybrids (1012, 1979, and NUL). Large-scale sequencing (one 96-well plate per genotype) was conducted for P. fremontii × angustifolia clones 1012 and NUL.
Sequence Analysis.
Sequences obtained from GenBank, JGI, or Phytozome were manually curated for exon predictions, guided by multiple sequence alignment. Phylogenetic analysis was performed using the neighbor-joining method in MEGA 4.1 (47), with Poisson correction, pairwise deletion, and bootstrap test (1,000 replicates). Spliced alignment was performed using Splign (48), and gene structures were displayed using Gene Structure Draw (http://www.compgen.uni-muenster.de/cgi-bin/Tools/StrDraw.pl). Splice-site sequence conservation was analyzed for all 23,375 multiexon gene models contained in the 19 linkage groups of JGI Populus genome v1.1. Genes located in the scaffolds were not included in this analysis. Nucleotide frequency of sequences flanking the splice sites was displayed using the WebLogo program (http://weblogo.berkeley.edu/).
Functional Complementation of E. coli Mutant PBB7.
cDNAs encoding entC and mature PtiICS and AtICS1 were PCR amplified with a 5′-NcoI site (Table S1), cloned into pCRII-TOPO, and sequenced. The 5′-NcoI and 3′-XbaI/3′-EcoRI digested fragments were subcloned into pTV118N (Takara Bio) and transformed into E. coli mutant PBB7 engineered with pRARE2 (Novagene). The CAS assay for siderophore production was conducted as described (30), except that after autoclaving, the CAS-iron solution was mixed 1:10 (vol/vol) with 2% LB molten agar, and then poured into Petri dishes.
Arabidopsis Transformation.
The PtiICS coding region with the plastid-targeting sequence was PCR amplified from a plasmid clone. Two genomic PtiICS fragments were PCR amplified from P. trichocarpa Nisqually-1, one (6 kb) containing the full genomic sequence from start and stop codons, and the other (8 kb) containing 1 kb each of the upstream and downstream sequences. PCR products were cloned into pCRII-TOPO, sequenced, and subcloned into pCambia2300 (8-kb clone) or pCambia1302 behind the CaMV 35S promoter. The standard floral dip protocol was followed for Arabidopsis transformation.
PhQ Analysis.
Concentrations of PhQ were measured by reversed-phase HPLC using freeze-dried leaf samples. The extraction and analytical conditions were carried out as described (49), except that the solid-phase extraction step was omitted due to the low lipid concentrations of the Arabidopsis samples.
SA Analysis.
Total SA levels were determined by GC-MS as described (8) using freeze-dried leaf powder and 80% (vol/vol) aqueous methanol for initial extraction. Derivatized extract was diluted with an equal volume (50 μL) of methylene chloride, and 1 μL of the mixture was analyzed on an Agilent 7890A-5975C GC-MSD system using a DB-5MS column (30 m × 0.25 mm × 0.25 μm with a guard column) and the splitless mode. The inlet temperature was set at 250 °C. The oven temperature was initially held at 80 °C for 5 min, increased to 180 °C at 4 °C/min, and held at 180 °C for 1 min. SA concentrations were determined in SIM mode at m/z 267 by calibration curves developed using an authentic standard (Sigma) and o-anisic acid (m/z 209) as the internal standard.
Supplementary Material
Acknowledgments.
We thank Kate Tay for technical assistance, Frederick Ausubel (Massachusetts General Hospital, Boston) for providing the sid2–2 mutant seed, Eckhard Leistner (Institut fur Pharmazeutische Biologie, Bonn, Germany) for providing the PBB7 mutant, and Toby Bradshaw (University of Washington, Seattle) for providing P. trichocarpa Nisqually-1 cuttings. This work was supported by the U.S. National Science Foundation Plant Genome Program Grants DBI-0421756 and 0836433 (to C.J.T. and S.A.H.) and the U.S. Department of Agriculture, Agricultural Research Service Cooperative Agreement no. 58–1950-7–707 (to X.F. and S.L.B.). Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the authors, and do not necessarily reflect the views of the U.S. National Science Foundation and the U.S. Department of Agriculture.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Populus ICS cDNA sequences, including constitutively and alternatively spliced variants, have been deposited in the GenBank database (accession nos. FJ968815–FJ968854, GQ260071, and GQ260072).
This article contains supporting information online at www.pnas.org/cgi/content/full/0906869106/DCSupplemental.
References
- 1.Walsh CT, Liu J, Rusnak F, Sakaitani M. Molecular studies on enzymes in chorismate metabolism and the enterobactin biosynthetic pathway. Chem Rev. 1990;90:1105–1129. [Google Scholar]
- 2.Poulsen C, Verpoorte R. Roles of chorismate mutase, isochorismate synthase and anthranilate synthase in plants. Phytochemistry. 1991;30:377–386. [Google Scholar]
- 3.Müller R, Dahm C, Schulte G, Leistner E. An isochorismate hydroxymutase isogene in Escherichia coli. FEBS Lett. 1996;378:131–134. doi: 10.1016/0014-5793(95)01436-5. [DOI] [PubMed] [Google Scholar]
- 4.Serino L, et al. Structural genes for salicylate biosynthesis from chorismate in Pseudomonas aeruginosa. Mol Gen Genet. 1995;249:217–228. doi: 10.1007/BF00290369. [DOI] [PubMed] [Google Scholar]
- 5.Gross J, et al. A plant locus essential for phylloquinone (vitamin K-1) biosynthesis originated from a fusion of four eubacterial genes. J Biol Chem. 2006;281:17189–17196. doi: 10.1074/jbc.M601754200. [DOI] [PubMed] [Google Scholar]
- 6.Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414:562–565. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]
- 7.Haas BJ, Delcher AL, Wortman JR, Salzberg SL. DAGchainer: A tool for mining segmental genome duplications and synteny. Bioinformatics. 2004;20:3643–3646. doi: 10.1093/bioinformatics/bth397. [DOI] [PubMed] [Google Scholar]
- 8.Garcion C, et al. Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol. 2008;147:1279–1287. doi: 10.1104/pp.108.119420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Catinot J, Buchala A, Abou-Mansour E, Métraux J-P. Salicylic acid production in response to biotic and abiotic stress depends on isochorismate in Nicotiana benthamiana. FEBS Lett. 2008;582:473–478. doi: 10.1016/j.febslet.2007.12.039. [DOI] [PubMed] [Google Scholar]
- 10.van Tegelen LJP, Moreno PRH, Croes AF, Verpoorte R, Wullems GJ. Purification and cDNA cloning of isochorismate synthase from elicited cell cultures of Catharanthus roseus. Plant Physiol. 1999;119:705–712. doi: 10.1104/pp.119.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Der Heijden R, Jacobs DI, Snoeijer W, Hallard D, Verpoorte R. The Catharanthus alkaloids: Pharmacognosy and biotechnology. Curr Med Chem. 2004;11:607–628. doi: 10.2174/0929867043455846. [DOI] [PubMed] [Google Scholar]
- 12.Han Y-S, Van der Heijden R, Verpoorte R. Biosynthesis of anthraquinones in cell cultures of the Rubiaceae. Plant Cell Tiss Org. 2001;67:201–220. [Google Scholar]
- 13.Tsai CJ, Harding SA, Tschaplinski TJ, Lindroth RL, Yuan Y. Genome-wide analysis of the structural genes regulating defense phenylpropanoid metabolism in Populus. New Phytol. 2006;172:47–62. doi: 10.1111/j.1469-8137.2006.01798.x. [DOI] [PubMed] [Google Scholar]
- 14.Lindroth RL, Hwang SY. Diversity, redundancy and multiplicity in chemical defense systems of aspen. Recent Adv Phytochem. 1996;30:25–56. [Google Scholar]
- 15.Harding SA, et al. Functional genomics analysis of foliar condensed tannin and phenolic glycoside regulation in natural cottonwood hybrids. Tree Physiol. 2005;25:1475–1486. doi: 10.1093/treephys/25.12.1475. [DOI] [PubMed] [Google Scholar]
- 16.Tuskan GA, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 17.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 18.Small I, Peeters N, Legeai F, Lurin C. Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics. 2004;4:1581–1590. doi: 10.1002/pmic.200300776. [DOI] [PubMed] [Google Scholar]
- 19.Dosselaere F, Vanderleyden J. A metabolic node in action: Chorismate-utilizing enzymes in microorganisms. Crit Rev Microbiol. 2001;27:75–131. doi: 10.1080/20014091096710. [DOI] [PubMed] [Google Scholar]
- 20.Kumar M, et al. An update on the nomenclature for the cellulose synthase genes in Populus. Trends Plant Sci. 2009;14:248–254. doi: 10.1016/j.tplants.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 21.Yu J, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 22.Jaillon O, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 23.Paterson AH, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 24.Rensing SA, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- 25.Wang W, Yu H, Long M. Duplication-degeneration as a mechanism of gene fission and the origin of new genes in Drosophila species. Nat Genet. 2004;36(5):523–527. doi: 10.1038/ng1338. [DOI] [PubMed] [Google Scholar]
- 26.Leipzig J, Pevzner P, Heber S. The Alternative Splicing Gallery (ASG): Bridging the gap between genome and transcriptome. Nucleic Acids Res. 2004;32:3977–3983. doi: 10.1093/nar/gkh731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barbazuk WB, Fu Y, McGinnis KM. Genome-wide analyses of alternative splicing in plants: Opportunities and challenges. Genome Res. 2008;18:1381–1392. doi: 10.1101/gr.053678.106. [DOI] [PubMed] [Google Scholar]
- 28.Baker KE, Parker R. Nonsense-mediated mRNA decay: Terminating erroneous gene expression. Curr Opin Cell Biol. 2004;16(3):293–299. doi: 10.1016/j.ceb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 29.Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 31.Nawrath C, Metraux J-P. Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell. 1999;11:1393–1404. doi: 10.1105/tpc.11.8.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reddy ASN. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Ann Rev Plant Biol. 2007;58:267–294. doi: 10.1146/annurev.arplant.58.032806.103754. [DOI] [PubMed] [Google Scholar]
- 33.Denoeud F, et al. Annotating genomes with massive-scale RNA sequencing. Genome Biol. 2008;9:R175. doi: 10.1186/gb-2008-9-12-r175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xing Y, Lee C. Alternative splicing and RNA selection pressure—Evolutionary consequences for eukaryotic genomes. Nat Rev Genet. 2006;7:499–509. doi: 10.1038/nrg1896. [DOI] [PubMed] [Google Scholar]
- 35.Pierpoint WS. Salicylic-acid and its derivatives in plants: Medicines, metabolites and messenger molecules. Adv Bot Res. 1994;20:163–235. [Google Scholar]
- 36.Ogawa D, et al. Salicylic acid accumulation under O3 exposure is regulated by ethylene in tobacco plants. Plant Cell Physiol. 2005;46:1062–1072. doi: 10.1093/pcp/pci118. [DOI] [PubMed] [Google Scholar]
- 37.Pasqualini S, et al. Ozone and nitric oxide induce cGMP-dependent and -independent transcription of defence genes in tobacco. New Phytol. 2009;181:860–870. doi: 10.1111/j.1469-8137.2008.02711.x. [DOI] [PubMed] [Google Scholar]
- 38.Dong XN. SA, JA, ethylene, and disease resistance in plants. Curr Opin Plant Biol. 1998;1:316–323. doi: 10.1016/1369-5266(88)80053-0. [DOI] [PubMed] [Google Scholar]
- 39.Koch JR, et al. Ozone sensitivity in hybrid poplar correlates with insensitivity to both salicylic acid and jasmonic acid: The role of programmed cell death in lesion formation. Plant Physiol. 2000;123:487–496. doi: 10.1104/pp.123.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morse AM, et al. Salicylate and catechol levels are maintained in nahG transgenic poplar. Phytochemistry. 2007;68:2043–2052. doi: 10.1016/j.phytochem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 41.Raskin I, Skubatz H, Tang W, Meeuse BD. Salicylic acid levels in thermogenic and non-thermogenic plants. Ann Bot. 1990;66:369–373. [Google Scholar]
- 42.Coquoz JL, Buchala AJ, Meuwly P, Metraux JP. Arachidonic acid induces local but not systemic synthesis of salicylic acid and confers systemic resistance in potato plants to Phytophthora infestans and Alternaria solani. Phytopathology. 1995;85:1219–1224. [Google Scholar]
- 43.Silverman P, et al. Salicylic acid in rice: Biosynthesis, conjugation, and possible role. Plant Physiol. 1995;108:633–639. doi: 10.1104/pp.108.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kao YY, Harding SA, Tsai CJ. Differential expression of two distinct phenylalanine ammonia-lyase genes in condensed tannin-accumulating and lignifying cells of quaking aspen. Plant Physiol. 2002;130:796–807. doi: 10.1104/pp.006262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lohmann A, et al. Deficiency in phylloquinone (vitamin K1) methylation affects prenyl quinone distribution, photosystem I abundance and anthocyanin accumulation in the arabidopsis AtmenG mutant. J Biol Chem. 2006;281:40461–40472. doi: 10.1074/jbc.M609412200. [DOI] [PubMed] [Google Scholar]
- 46.Lochner K, Doring O, Bottger M. Phylloquinone, what can we learn from plants? BioFactors. 2003;18:73–78. doi: 10.1002/biof.5520180209. [DOI] [PubMed] [Google Scholar]
- 47.Kumar S, Nei M, Dudley J, Tamura K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kapustin Y, Souvorov A, Tatusova T, Lipman D. Splign: Algorithms for computing spliced alignments with identification of paralogs. Biol Direct. 2008;3:20. doi: 10.1186/1745-6150-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu X, et al. 9-Cis retinoic acid reduces 1α,25-dihydroxycholecalciferol-Induced renal calcification by altering vitamin K-dependent γ-carboxylation of matrix γ-carboxyglutamic acid protein in A/J male mice. J Nutr. 2008;138:2337–2341. doi: 10.3945/jn.108.093724. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.