Abstract
Understanding how DNA is packaged in the mammalian sperm cell has important implications for human infertility as well as for the cell biology. Recent advances in the study of mammalian sperm chromatin structure and function have altered our perception of this highly condensed, inert chromatin. Sperm DNA is packaged very tightly to protect the DNA during the transit that occurs before fertilization. However, this condensation cannot sacrifice chromosomal elements that are essential for the embryo to access the correct sequences of the paternal genome for proper initiation of the embryonic developmental program. The primary levels of the sperm chromatin structure can be divided into three main categories: the large majority of DNA is packaged by protamines, a smaller amount (2–15%) retains histone-bound chromatin and the DNA is attached to the nuclear matrix at roughly 50 kb intervals. Current data suggest that the latter two structural elements are transferred to the paternal pronucleus after fertilization where they have important functional roles. The nuclear matrix organization is essential for DNA replication, and the histone-bound chromatin identifies genes that are important for embryonic development. These data support the emerging view of the sperm genome as providing, in addition to the paternal DNA sequence, a structural framework that includes molecular regulatory factors that are required for proper embryonic development.
Keywords: sperm chromatin, nuclear matrix, histones, protamines
Introduction
The paternal genome in mammalian spermatozoa is condensed in a manner that is specific to the cell type presumably to protect the DNA during the transit from the male to the oocyte prior to fertilization. The existence of this unique chromatin packaging has important consequences for both the development of improved diagnostics for medical infertility and for the study of higher order DNA structures in the field of cell biology. Infertility researchers are interested in understanding sperm chromatin structure in order to determine how to best interpret assays for DNA integrity, which affects the outcome of assisted reproductive technologies (ART) (Agarwal and Said, 2003; Evenson and Jost, 2000; Morris et al., 2002; Sakkas et al., 2002; Tomsu et al., 2002; van der Heijden et al., 2008). Cell biologists view the unique structure of mammalian sperm chromatin as an important model for how DNA is folded by cells into different functional domains (Adenot et al., 1997; Ajduk et al., 2006; Blow et al., 1989; Derijck et al., 2006; Haaf and Ward, 1995; Yamauchi et al., 2009). The different endeavors in these two areas of research are mutually supportive.
This review summarizes the recent advances in the field, and highlights some of the implications of these new findings for infertility and cell biology. Mammalian sperm chromatin can be divided into three major structural domains: (1) the vast majority of sperm DNA is coiled into toroids by protamines (Hud et al., 1995), (2) a much smaller percent remains bound to histones (Adenot et al., 1997; Churikov et al., 2004; Gineitis et al., 2000; Hammoud et al., 2009; Pittoggi et al., 1999), and (3) the DNA is attached to the sperm nuclear matrix at MARs (matrix attachment regions) at medium intervals of roughly 50 kb throughout the genome (Martins et al., 2004; Nadel et al., 1995). Recent data suggest that these domains are related to different functions. This review first discusses each of these structural domains, then concludes with implications of these findings for infertility and cell biology research. While many questions have yet to be resolved with respect to even the most basic structure of sperm chromatin, recent findings suggest that some important principles about the relationship of sperm structure and function can now be drawn.
New developments
Protamine-bound sperm chromatin
The vast majority of mammalian sperm chromatin is compacted into toroids that contain roughly 50 kb of DNA (Hud et al., 1993; Hud et al., 1995; Brewer et al., 2003) (Fig. 1A). Protamines have large tracts of positively charged arginine residues that neutralize the negative phosphodiester backbone of the DNA (Balhorn, 1982). This neutralization mimics that of divalent cations that can also cause DNA to form similar toroids with similar amounts of DNA (Hud and Vilfan, 2005). This condensation is so complete that most of the DNA is hidden within the toroid (Vilfan et al., 2004). This component of the sperm DNA exists in semi-crystalline state and is resistant to nuclease digestion (Sotolongo et al., 2003). Mammalian protamines also contain several cysteines that are thought to confer an increased stability on sperm chromatin by intermolecular disulfide cross-links. Sperm DNA cannot be decondensed in vitro without reducing reagents (Perreault and Zirkin, 1982; Ohsumi et al., 1988; Balhorn et al., 1991), and the disulfide cross-links increase as the sperm cells transit the epididymis after they exit the testis.
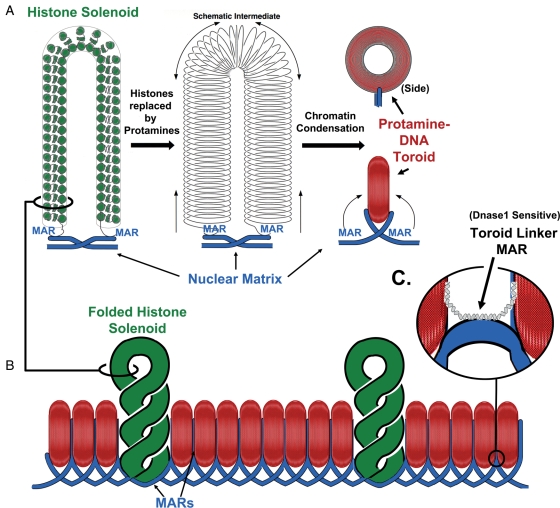
Figure 1.
Three major structural elements of sperm chromatin.
(A) During spermiogenesis, histones are replaced by protamines, condensing the DNA into tightly packaged toroids. Each protamine toroid is a loop domain (part A of this figure is modified from a similar figure published in Ward, 1993). (B) Protamine toroids may be organized by stacking side to side. Recent evidence suggests that some large tracts of DNA retain histone (green solenoid). These may be entire loop domains that are not condensed by protamines. (C) The DNA strands that link the protamine toroids are nuclease sensitive, and may be bound to histones, as well. MAR, matrix attachment region
The protection that protamine binding provides to sperm chromatin was demonstrated unintentionally by an experiment in which mouse sperm were briefly sonicated before microinjection into oocytes (Kuretake et al., 1996). The sonication was used to separate sperm heads from the tails, and the heads were then injected into ooctyes. The fertilized oocytes developed into live born pups, indicating that the sonication did not significantly damage the sperm DNA. Even the mild sonication that was used in these experiments would be enough to introduce a few DNA breaks in somatic cell chromatin. Such treatment of somatic cells causes so many breaks in the histone-bound chromatin that the cell does not survive. This experiment supports the hypothesis that the evolutionary pressure that resulted in this unique type of condensation was the protection of the paternal genome during fertilization.
It is important to note that protamines are found only in mature spermatozoa, not in any other cell type. The other two types of structural domains of sperm chromatin, histone-bound chromatin and MARs, are both found in somatic cells and can be presumed to be residual from the sperm progenitor cells from which spermatozoa are produced (this concept is detailed below). Protamine toroids are unique to the mature sperm cell. Thus, this most common structural domain offers an important boundary for understanding sperm chromatin.
Another important aspect of this, largest structural domain of sperm chromatin is that its major function is almost certainly only for fertilization, and not for embryonic development. Protamine binding also silences gene expression during spermiogenesis (Martins et al., 2004; Carrell et al., 2007; Rathke et al., 2007), but its role during fertilization and beyond is probably protective. Three separate lines of evidence support this contention. First, protamines are completely replaced in the first 2–4 h after fertilization by histones so that the paternal chromatin has the same accessible chromatin as all other somatic cells (Kopecny and Pavlok, 1975; van der Heijden et al., 2005; Ajduk et al., 2006). Second, as mentioned above, sperm chromatin is resistant to much greater mechanical disruption than somatic cells, supporting the protamines’ role in DNA protection. Finally, when round spermatids, the first haploid cell type resulting from spermiogenesis, were injected into mouse oocytes, pups developed normally (Ogura et al., 1994). Since round spermatids do not have any protamines, one can conclude that this level of sperm chromatin structure is not required for proper embryogenesis. As we shall see, this is not true for the other two levels of sperm chromatin structure. The structural organization of both histone-bound chromatin and sperm MARs are probably transmitted to the newly formed paternal pronucleus after fertilization and evidence suggests that both are required for proper embryogenesis.
Finally, the major unanswered question of protamine-bound chromatin structure concerns the secondary organization. Mudrak et al. (2009) have provided evidence that the protamine toroids are stacked side to side like a package of lifesavers (Fig. 1B). This model is the most efficient form in which the protamine toroids could be condensed into a highly protective chromatin. Variations of this theme are certainly possible, for example the protamine ‘lifesaver’ chromatin might be compacted so that two adjacent lines of toroids are aligned together with alternating toroids on the same chromatin being packaged in one line (Mudrak et al., 2009). This is an interesting area of sperm chromatin research that still needs investigation. However, from a functional standpoint, the condensation of this DNA into the crystalline-like toroids already infers the most important functional characteristics of protamine-bound sperm DNA—that it is unlikely to be active until after decondensation in the oocyte, and that it serves a largely protective function during fertilization.
Histone-bound sperm chromatin
Depending on the species and the experiment used to quantify it, between 2 and 15% of mammalian sperm chromatin is bound to histones, rather than protamines (Bench et al., 1996; Adenot et al., 1997; Pittoggi et al., 1999; Gineitis et al., 2000; Churikov et al., 2004; Hammoud et al., 2009). Three important questions regarding sperm histones have recently been addressed.
The first question was whether sperm histones are associated with specific sequences within the sperm chromatin, or positioned randomly within the chromatin fiber as the result of incomplete protamine deposition. This idea was first approached in 1987 (Gatewood et al., 1987), but has been more completely defined by two other laboratories. One group initially focused on the protamine gene locus of human sperm that spans 28 kb flanked by two MAR regions (Wykes and Krawetz, 2003). This entire region seems to be preferentially associated with histones in human spermatozoa, but does contain some protamine-associated DNA, as well. The same group recently surveyed the entire human genome, and concluded that histones were interspersed throughout the genome, primarily at gene promoters (Arpanahi et al., 2009). A separate group performed a similar study and concluded that entire gene families that were important for early development were preferentially associated with histones in human spermatozoa (Hammoud et al., 2009). The work from both groups indicates that histones are non-randomly distributed in the sperm genome, and are associated with specific genes.
The second, related question was how were these histones distributed—were they interspersed throughout the sperm genome or were they located in discrete regions of the chromatin? The data described above (Arpanahi et al., 2009; Hammoud et al., 2009) suggest that histones are present in two types of distribution—in relatively large tracts of DNA, from 10 to 100 kb, and in smaller tracts of DNA interspersed throughout the genome. This has important implications for the structural organization of sperm chromatin, because if histones were distributed at regular intervals throughout the genome they might be part of a repeating unit of sperm chromatin structure. For example, we had proposed that histone-bound DNA made up the linker regions between each protamine toroid in the chromatin fiber, because these were the regions that were most nuclease sensitive (Sotolongo et al., 2003). The data referenced above, showing that histone-bound segments of DNA are scattered throughout the paternal genome, largely at gene promoter regions (Arpanahi et al., 2009) are consistent with this model.
Finally, a third important question concerning histone-bound DNA in sperm chromatin was whether sperm histones are transmitted to the developing embryo. Shortly after fertilization, the protamines in sperm chromatin are replaced with histones supplied by the oocyte (Kopecny and Pavlok, 1975; van der Heijden et al., 2005; Ajduk et al., 2006), but in those regions where histones are already present in the sperm DNA this may not be necessary. van der Heijden et al. (2006) and van der Heijden et al. (2008) demonstrated that histones with specific modifications in the sperm cell are also present in the paternal pronucleus, suggesting that they were never replaced. The transmission of sperm histones, and the associated chromatin structures, suggest it is possible that the newly fertilized oocytes inherits histone-based chromatin structural organization from the sperm.
The data currently support a model for histone-associated chromatin representing functional genes for both spermiogenesis (possibly representing residual active chromatin that persisted through chromatin condensation) (Martins and Krawetz, 2005; Ostermeier et al., 2005) and for early fertilization (Arpanahi et al., 2009; Hammoud et al., 2009). Moreover, some of these histone-associated sperm chromatin structures may persist during the structural reorganization of the paternal chromatin when the sperm nucleus decondenses to form the paternal pronucleus.
Matrix attachment regions
In both somatic (Vogelstein et al., 1980; Gerdes et al., 1994; Dijkwel and Hamlin, 1995; Linnemann et al., 2009) and sperm nuclei (Ward et al., 1989; Kalandadze et al., 1990; Moss et al., 1993; Choudhary et al., 1995) chromatin is organized into loop domains that are attached every 20–120 kb in length to a proteinaceous structure termed the nuclear matrix (Fig. 1A). This organizes the chromatin into functional loops of DNA that help regulate DNA replication (Vogelstein et al., 1980; Gerdes et al., 1994; Dijkwel and Hamlin, 1995) and gene transcription (Cockerill and Garrard, 1986; Nelson et al., 1986; Choudhary et al., 1995; Ostermeier et al., 2003). This loop domain structure is present throughout the entire sperm chromatin even though the tertiary structure of most of the DNA is very different in spermatozoa (Fig. 1A) (Ward et al., 1989). We have provided evidence to support the hypothesis that each protamine toroid contains a single DNA loop domain (Fig. 1A) (Ward, 1993; Sotolongo et al., 2005). Between each protamine toroid, is a nuclease sensitive segment of chromatin we term the toroid linker, which is also the site of attachment of DNA to the nuclear matrix, or MAR. While it is impossible to predict the size of these linkers with the present data, we would not expect them to be larger than 1000 bp, which is about 2% of the size of the toroid, itself, since 98% of mouse sperm chromatin is associated with protamines (Bench et al., 1996). The nuclease sensitivity suggests that these protamine linker regions are bound by histones (Fig. 1C), and this is consistent with the wide distribution of histones throughout the genome (Arpanahi et al., 2009). Thus, the organization of sperm DNA into protamine toroids and by the nuclear matrix is directly linked.
Several pieces of evidence support a functional role for the sperm nuclear matrix in the function of the paternal genome during early embryogenesis. Spermatozoa with structurally disrupted sperm nuclear matrices do not support embryonic development after ICSI unlike those with intact matrices (Ward et al., 1999). To test the role of the sperm nuclear matrix organization directly, it is experimentally possible to remove the other two types of sperm chromatin organization—protamine condensation and histone-bound nucleosomes—by treatment with high salt and reducing reagent. This treatment leaves only the sperm nuclear matrix with associated loop domains attached and the resulting nuclei are called sperm nuclear halos (Nadel et al., 1995; Kramer and Krawetz, 1996). When sperm halos were injected into oocytes, pronuclei formation was normal and DNA replication proceeded (Shaman et al., 2007). This was true even when up to 50% of the DNA that was not attached to the matrix was removed by restriction endonuclease treatment. DNA, alone, injected into oocytes did not form pronuclei nor did the DNA replicate. However, when the MARs are reversibly cleaved by Topo 2b, the paternal DNA is degraded at the time of the initiation of DNA synthesis 5.5 h after ICSI (Yamauchi et al., 2007a, 2007b). Oocytes injected with intact sperm halos did not develop to the blastocyst stage (unpublished data) suggesting that while the organization of DNA into loop domains by the sperm nuclear matrix is required for DNA replication, it is not sufficient for development.
These data suggest two roles for the sperm nuclear matrix. First, the proper association of DNA to the nuclear matrix is required for paternal pronuclear DNA replication in the one-cell embryo. Second, the sperm nuclear matrix may act as a checkpoint for sperm DNA integrity after fertilization.
Sperm chromosomes
Although not one of the three structural elements on which this review is focused, a review of sperm chromatin structure must include a brief discussion of the least understood element, the higher order structure of the chromosomes. There are many models for mitotic chromosomes and, while important differences exist, there is general agreement on the basic folding pattern of the DNA (Pienta and Coffey, 1984; Boy de la Tour and Laemmli, 1988). The higher order structure of chromosomes in interphase somatic cells is much less well understood, partially because it is much less uniform. However, it is clear that the DNA is bound to histones in nucleosomes that are in various stages of condensation or openness, depending on their function (Grewal and Elgin, 2007; Kloc and Martienssen, 2008). Mammalian sperm chromosomes fall into a third category in that they are most likely relatively homogenous in structure, but are probably longer and thinner than mitotic chromosomes (Haaf and Ward, 1995). Zalensky et al. have elegantly demonstrated that mammalian sperm chromatin of all species tested are folded into hairpin-like structures with the centromeres positioned near the center of the sperm cell with the telomeres of each chromosome paired and arrayed around the periphery of the sperm nucleus (Zalenskaya et al., 2000; Churikov et al., 2004; Solov'eva et al., 2004). They also demonstrated that individual chromosomes are partitioned into territories that do not overlap (Mudrak et al., 2005; Zalensky and Zalenskaya, 2007). Beyond these facts, models for the higher order structure of sperm chromatin are lacking. Understanding how sperm chromosomes are folded would provide unique insights into the structure of somatic cell chromosomes.
Implications
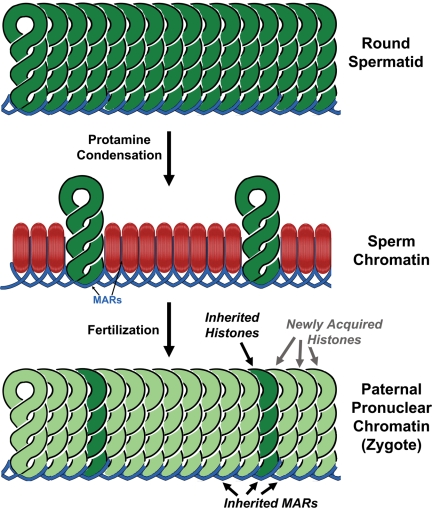
Even though the current status of our knowledge of mammalian sperm chromatin structure lacks many specifics, it does allow us to draw some important conclusions for both medically oriented infertility research and the more basic cell biology questions that employ sperm chromatin as a useful model. Foremost among these is the evidence that supports the conclusion that, of the three types of sperm chromatin structure discussed, two are inherited by the embryo and are probably required for proper development (Fig. 2). This has two important implications. For human infertility research, it suggests that methods to manipulate spermatozoa for ART should be developed that maintain the integrity of the sperm chromatin structure, as well as taking into consideration the integrity of the paternal DNA.
Figure 2.
Inheritance of sperm chromatin structural elements by the embryo. DNA in round spermatids is packaged by histones (top) but during spermiogenesis, most of these are replaced by protamines (middle, red). After fertilization, the protamines are removed, and histones supplied by the oocyte replace them (bottom, light green). However, some histones that were retained in the spermatozoon (middle, dark green) are probably retained in the newly formed paternal pronucleus after fertilization. Sperm nuclear matrix attachment regions (MARs) are probably retained in the paternal pronucleus as well.
The second implication relates to the use of mammalian sperm chromatin as a model for eukaryotic DNA packaging. The complex nature of mammalian sperm chromatin structure allows us to separate functional domains of chromatin, and to understand more about the nature of somatic cell chromatin because of these functional differences. Mammalian sperm chromatin presents an interesting model for eukaryotic chromatin, and for studying the function of the nuclear matrix, in particular, because most of the DNA is condensed into chromatin that is difficult to decondense. Only what is thought to be the most active part of any cell's chromatin, the attachment to the nuclear matrix, required for DNA replication, and important genes for development, is left associated with histones. Thus, the sperm cell has already fractionated the chromatin, naturally, into inactive and active chromatin by condensing most of the DNA with protamines.
The data reviewed above are consistent with a model for higher order sperm chromatin structure as depicted in Fig. 1C. As discussed above, at least one group has provided evidence that protamine toroids might be stacked side to side (Mudrak et al., 2009). We have presented evidence that each protamine toroid is one loop domain, with nuclease sensitive regions that link each toroid (Sotolongo et al., 2003). The newer data on sperm histone-bound chromatin suggest that in humans at least a portion of this is present in large, loop-sized tracts of DNA. This suggests that some DNA loop domains may escape protamine condensation altogether (Fig. 1B). The fact that sperm chromatin can survive sonication (Kuretake et al., 1996) suggests that those loop domains that remain bound to histones are also protected. This would be conceivable if each histone-bound loop was packaged between the more stable protamine toroids (Fig. 1B). As discussed above, the data suggest that the structural organization of the histone-bound chromatin and the MARs of the sperm cell are inherited by the paternal pronucleus after fertilization (Fig. 2). Both of these structural elements of sperm chromatin are associated with different functions in the embryo. In the case of MARs, it is clear that the embryo cannot develop past the first cell cycle without proper organization by the nuclear matrix (Shaman et al., 2007), and it is likely that the histone-bound sequences are just as important for later embryonic development (Arpanahi et al., 2009; Hammoud et al., 2009).
Many of the recent advances in this field arose from the fact that the three types of sperm chromatin structure can be biochemically fractionated. Sperm halos that are devoid of both histones and protamines can be prepared by extraction with high salt and reducing reagent (Ward et al., 1989; Kalandadze et al., 1990; Kramer and Krawetz, 1996). The only chromatin structural element these halos retain is the organization of DNA into loop domains at the MARs. Histones can be selectively removed by salt extraction without reducing reagents (Gatewood et al., 1990), leaving behind the protamines and matrix attachment regions intact. This is a particularly intriguing area for future studies as it provides the experimental opportunity to determine the real function of histone retention in mammalian spermatozoa.
These data support an emerging view that sperm nuclei provide much more than half the genetic make-up of the newly fertilized embryo—it presents its DNA in a structural context that is required for the embryo to access the paternal genome in a proper sequence of events.
Funding
This work was supported by the National Institutes of Health (grant number HD28501) and by the Office of Research and Graduate Education at the University of Hawaii at Manoa.
References
- Adenot PG, Mercier Y, Renard JP, Thompson EM. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development. 1997;124:4615–4625. doi: 10.1242/dev.124.22.4615. [DOI] [PubMed] [Google Scholar]
- Agarwal A, Said TM. Role of sperm chromatin abnormalities and DNA damage in male infertility. Hum Reprod Update. 2003;9:331–345. doi: 10.1093/humupd/dmg027. [DOI] [PubMed] [Google Scholar]
- Ajduk A, Yamauchi Y, Ward MA. Sperm chromatin remodeling after intracytoplasmic sperm injection differs from that of in vitro fertilization. Biol Reprod. 2006;75:442–451. doi: 10.1095/biolreprod.106.053223. [DOI] [PubMed] [Google Scholar]
- Arpanahi A, Brinkworth M, Iles D, Krawetz SA, Paradowska A, Platts AE, Saida M, Steger K, Tedder P, Miller D. Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Res. 2009;19:1338–1349. doi: 10.1101/gr.094953.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balhorn R. A model for the structure of chromatin in mammalian sperm. J Cell Biol. 1982;93:298–305. doi: 10.1083/jcb.93.2.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balhorn R, Corzett M, Mazrimas J, Watkins B. Identification of bull protamine disulfides. Biochemistry. 1991;30:175–181. doi: 10.1021/bi00215a026. [DOI] [PubMed] [Google Scholar]
- Bench GS, Friz AM, Corzett MH, Morse DH, Balhorn R. DNA and total protamine masses in individual sperm from fertile mammalian subjects. Cytometry. 1996;23:263–271. doi: 10.1002/(SICI)1097-0320(19960401)23:4<263::AID-CYTO1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Sheehan MA, Watson JV, Laskey RA. Nuclear structure and the control of DNA replication in the Xenopus embryo. J Cell Sci Suppl. 1989;12:183–195. doi: 10.1242/jcs.1989.supplement_12.16. [DOI] [PubMed] [Google Scholar]
- Boy de la Tour E, Laemmli UK. The metaphase scaffold is helically folded: sister chromatids have predominantly opposite helical handedness. Cell. 1988;55:937–944. doi: 10.1016/0092-8674(88)90239-5. [DOI] [PubMed] [Google Scholar]
- Brewer L, Corzett M, Lau EY, Balhorn R. Dynamics of protamine 1 binding to single DNA molecules. J Biol Chem. 2003;278:42403–42408. doi: 10.1074/jbc.M303610200. [DOI] [PubMed] [Google Scholar]
- Carrell DT, Emery BR, Hammoud S. Altered protamine expression and diminished spermatogenesis: what is the link? Hum Reprod Update. 2007;13:313–327. doi: 10.1093/humupd/dml057. [DOI] [PubMed] [Google Scholar]
- Choudhary SK, Wykes SM, Kramer JA, Mohamed AN, Koppitch F, Nelson JE, Krawetz SA. A haploid expressed gene cluster exists as a single chromatin domain in human sperm. J Biol Chem. 1995;270:8755–8762. doi: 10.1074/jbc.270.15.8755. [DOI] [PubMed] [Google Scholar]
- Churikov D, Siino J, Svetlova M, Zhang K, Gineitis A, Morton Bradbury E, Zalensky A. Novel human testis-specific histone H2B encoded by the interrupted gene on the X chromosome. Genomics. 2004;84:745–756. doi: 10.1016/j.ygeno.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Cockerill PN, Garrard WT. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986;44:273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- Derijck AA, van der Heijden GW, Giele M, Philippens ME, van Bavel CC, de Boer P. GammaH2AX signalling during sperm chromatin remodelling in the mouse zygote. DNA Repair (Amst) 2006;5:959–971. doi: 10.1016/j.dnarep.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Dijkwel PA, Hamlin JL. Origins of replication and the nuclear matrix: the DHFR domain as a paradigm. Int Rev Cytol. 1995;162A:455–484. doi: 10.1016/s0074-7696(08)61236-x. [DOI] [PubMed] [Google Scholar]
- Evenson D, Jost L. Sperm chromatin structure assay is useful for fertility assessment. Methods Cell Sci. 2000;22:169–189. doi: 10.1023/a:1009844109023. [DOI] [PubMed] [Google Scholar]
- Gatewood JM, Cook GR, Balhorn R, Bradbury EM, Schmid CW. Sequence-specific packaging of DNA in human sperm chromatin. Science. 1987;236:962–964. doi: 10.1126/science.3576213. [DOI] [PubMed] [Google Scholar]
- Gatewood JM, Cook GR, Balhorn R, Schmid CW, Bradbury EM. Isolation of four core histones from human sperm chromatin representing a minor subset of somatic histones. J Biol Chem. 1990;265:20662–20666. [PubMed] [Google Scholar]
- Gerdes MG, Carter KC, Moen PT, Jr, Lawrence JB. Dynamic changes in the higher-level chromatin organization of specific sequences revealed by in situ hybridization to nuclear halos. J Cell Biol. 1994;126:289–304. doi: 10.1083/jcb.126.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gineitis AA, Zalenskaya IA, Yau PM, Bradbury EM, Zalensky AO. Human sperm telomere-binding complex involves histone H2B and secures telomere membrane attachment. J Cell Biol. 2000;151:1591–1598. doi: 10.1083/jcb.151.7.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T, Ward DC. Higher order nuclear structure in mammalian sperm revealed by in situ hybridization and extended chromatin fibers. Exp Cell Res. 1995;219:604–611. doi: 10.1006/excr.1995.1270. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hud NV, Vilfan ID. Toroidal DNA condensates: unraveling the fine structure and the role of nucleation in determining size. Annu Rev Biophys Biomol Struct. 2005;34:295–318. doi: 10.1146/annurev.biophys.34.040204.144500. [DOI] [PubMed] [Google Scholar]
- Hud NV, Allen MJ, Downing KH, Lee J, Balhorn R. Identification of the elemental packing unit of DNA in mammalian sperm cells by atomic force microscopy. Biochem Biophys Res Commun. 1993;193:1347–1354. doi: 10.1006/bbrc.1993.1773. [DOI] [PubMed] [Google Scholar]
- Hud NV, Downing KH, Balhorn R. A constant radius of curvature model for the organization of DNA in toroidal condensates. Proc Natl Acad Sci U S A. 1995;92:3581–3585. doi: 10.1073/pnas.92.8.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalandadze AG, Bushara SA, Vassetzky YS, Jr, Razin SV. Characterization of DNA pattern in the site of permanent attachment to the nuclear matrix located in the vicinity of replication origin. Biochem Biophys Res Commun. 1990;168:9–15. doi: 10.1016/0006-291x(90)91667-h. [DOI] [PubMed] [Google Scholar]
- Kloc A, Martienssen R. RNAi, heterochromatin and the cell cycle. Trends Genet. 2008;24:511–517. doi: 10.1016/j.tig.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Kopecny V, Pavlok A. Autoradiographic study of mouse spermatozoan arginine-rich nuclear protein in fertilization. J Exp Zool. 1975;191:85–96. doi: 10.1002/jez.1401910109. [DOI] [PubMed] [Google Scholar]
- Kramer JA, Krawetz SA. Nuclear matrix interactions within the sperm genome. J Biol Chem. 1996;271:11619–11622. doi: 10.1074/jbc.271.20.11619. [DOI] [PubMed] [Google Scholar]
- Kuretake S, Kimura Y, Hoshi K, Yanagimachi R. Fertilization and development of mouse oocytes injected with isolated sperm heads. Biol Reprod. 1996;55:789–795. doi: 10.1095/biolreprod55.4.789. [DOI] [PubMed] [Google Scholar]
- Linnemann AK, Platts AE, Krawetz SA. Differential nuclear scaffold/matrix attachment marks expressed genes. Hum Mol Genet. 2009;18:645–654. doi: 10.1093/hmg/ddn394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins RP, Krawetz SA. RNA in human sperm. Asian J Androl. 2005;7:115–120. doi: 10.1111/j.1745-7262.2005.00048.x. [DOI] [PubMed] [Google Scholar]
- Martins RP, Ostermeier GC, Krawetz SA. Nuclear matrix interactions at the human protamine domain: a working model of potentiation. J Biol Chem. 2004;279:51862–51868. doi: 10.1074/jbc.M409415200. [DOI] [PubMed] [Google Scholar]
- Morris ID, Ilott S, Dixon L, Brison DR. The spectrum of DNA damage in human sperm assessed by single cell gel electrophoresis (Comet assay) and its relationship to fertilization and embryo development. Hum Reprod. 2002;17:990–998. doi: 10.1093/humrep/17.4.990. [DOI] [PubMed] [Google Scholar]
- Moss SB, Burnham BL, Bellve AR. The differential expression of lamin epitopes during mouse spermatogenesis. Mol Reprod Dev. 1993;34:164–174. doi: 10.1002/mrd.1080340208. [DOI] [PubMed] [Google Scholar]
- Mudrak O, Tomilin N, Zalensky A. Chromosome architecture in the decondensing human sperm nucleus. J Cell Sci. 2005;118:4541–4550. doi: 10.1242/jcs.02581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudrak O, Chandra R, Jones E, Godfrey E, Zalensky A. Reorganisation of human sperm nuclear architecture during formation of pronuclei in a model system. Reprod Fertil Dev. 2009;21:665–671. doi: 10.1071/RD08269. [DOI] [PubMed] [Google Scholar]
- Nadel B, de Lara J, Finkernagel SW, Ward WS. Cell-specific organization of the 5S ribosomal RNA gene cluster DNA loop domains in spermatozoa and somatic cells. Biol Reprod. 1995;53:1222–1228. doi: 10.1095/biolreprod53.5.1222. [DOI] [PubMed] [Google Scholar]
- Nelson WG, Pienta KJ, Barrack ER, Coffey DS. The role of the nuclear matrix in the organization and function of DNA. Annu Rev Biophys Biophys Chem. 1986;15:457–475. doi: 10.1146/annurev.bb.15.060186.002325. [DOI] [PubMed] [Google Scholar]
- Ogura A, Matsuda J, Yanagimachi R. Birth of normal young after electrofusion of mouse oocytes with round spermatids. Proc Natl Acad Sci U S A. 1994;91:7460–7462. doi: 10.1073/pnas.91.16.7460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi K, Katagiri C, Yanagimachi R. Human sperm nuclei can transform into condensed chromosomes in Xenopus egg extracts. Gamete Res. 1988;20:1–9. doi: 10.1002/mrd.1120200102. [DOI] [PubMed] [Google Scholar]
- Ostermeier GC, Liu Z, Martins RP, Bharadwaj RR, Ellis J, Draghici S, Krawetz SA. Nuclear matrix association of the human beta-globin locus utilizing a novel approach to quantitative real-time PCR. Nucleic Acids Res. 2003;31:3257–3266. doi: 10.1093/nar/gkg424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermeier GC, Goodrich RJ, Diamond MP, Dix DJ, Krawetz SA. Toward using stable spermatozoal RNAs for prognostic assessment of male factor fertility. Fertil Steril. 2005;83:1687–1694. doi: 10.1016/j.fertnstert.2004.12.046. [DOI] [PubMed] [Google Scholar]
- Perreault SD, Zirkin BR. Sperm nuclear decondensation in mammals: role of sperm-associated proteinase in vivo. J Exp Zool. 1982;224:253–257. doi: 10.1002/jez.1402240215. [DOI] [PubMed] [Google Scholar]
- Pienta KJ, Coffey DS. A structural analysis of the role of the nuclear matrix and DNA loops in the organization of the nucleus and chromosome. J Cell Sci Suppl. 1984;1:123–135. doi: 10.1242/jcs.1984.supplement_1.9. [DOI] [PubMed] [Google Scholar]
- Pittoggi C, Renzi L, Zaccagnini G, Cimini D, Degrassi F, Giordano R, Magnano AR, Lorenzini R, Lavia P, Spadafora C. A fraction of mouse sperm chromatin is organized in nucleosomal hypersensitive domains enriched in retroposon DNA. J Cell Sci. 1999;112:3537–3548. doi: 10.1242/jcs.112.20.3537. [DOI] [PubMed] [Google Scholar]
- Rathke C, Baarends WM, Jayaramaiah-Raja S, Bartkuhn M, Renkawitz R, Renkawitz-Pohl R. Transition from a nucleosome-based to a protamine-based chromatin configuration during spermiogenesis in Drosophila. J Cell Sci. 2007;120:1689–1700. doi: 10.1242/jcs.004663. [DOI] [PubMed] [Google Scholar]
- Sakkas D, Moffatt O, Manicardi GC, Mariethoz E, Tarozzi N, Bizzaro D. Nature of DNA damage in ejaculated human spermatozoa and the possible involvement of apoptosis. Biol Reprod. 2002;66:1061–1067. doi: 10.1095/biolreprod66.4.1061. [DOI] [PubMed] [Google Scholar]
- Shaman JA, Yamauchi Y, Ward WS. The sperm nuclear matrix is required for paternal DNA replication. J Cell Biochem. 2007;102:680–688. doi: 10.1002/jcb.21321. [DOI] [PubMed] [Google Scholar]
- Solov'eva L, Svetlova M, Bodinski D, Zalensky AO. Nature of telomere dimers and chromosome looping in human spermatozoa. Chromosome Res. 2004;12:817–823. doi: 10.1007/s10577-005-5513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotolongo B, Lino E, Ward WS. Ability of hamster spermatozoa to digest their own DNA. Biol Reprod. 2003;69:2029–2035. doi: 10.1095/biolreprod.103.020594. [DOI] [PubMed] [Google Scholar]
- Sotolongo B, Huang TF, Isenberger E, Ward WS. An endogenous nuclease in hamster, mouse and human spermatozoa cleaves DNA into loop-sized fragments. J Androl. 2005;26:272–280. doi: 10.1002/j.1939-4640.2005.tb01095.x. [DOI] [PubMed] [Google Scholar]
- Tomsu M, Sharma V, Miller D. Embryo quality and IVF treatment outcomes may correlate with different sperm comet assay parameters. Hum Reprod. 2002;17:1856–1862. doi: 10.1093/humrep/17.7.1856. [DOI] [PubMed] [Google Scholar]
- van der Heijden GW, Dieker JW, Derijck AA, Muller S, Berden JH, Braat DD, van der Vlag J, de Boer P. Asymmetry in histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mech Dev. 2005;122:1008–1022. doi: 10.1016/j.mod.2005.04.009. [DOI] [PubMed] [Google Scholar]
- van der Heijden GW, Derijck AA, Ramos L, Giele M, van der Vlag J, de Boer P. Transmission of modified nucleosomes from the mouse male germline to the zygote and subsequent remodeling of paternal chromatin. Dev Biol. 2006;298:458–469. doi: 10.1016/j.ydbio.2006.06.051. [DOI] [PubMed] [Google Scholar]
- van der Heijden GW, Ramos L, Baart EB, van den Berg IM, Derijck AA, van der Vlag J, Martini E, de Boer P. Sperm-derived histones contribute to zygotic chromatin in humans. BMC Dev Biol. 2008;8:34. doi: 10.1186/1471-213X-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilfan ID, Conwell CC, Hud NV. Formation of native-like mammalian sperm cell chromatin with folded bull protamine. J Biol Chem. 2004;279:20088–20095. doi: 10.1074/jbc.M312777200. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Pardoll DM, Coffey DS. Supercoiled loops and eucaryotic DNA replicaton. Cell. 1980;22:79–85. doi: 10.1016/0092-8674(80)90156-7. [DOI] [PubMed] [Google Scholar]
- Ward WS. Deoxyribonucleic acid loop domain tertiary structure in mammalian spermatozoa. Biol Reprod. 1993;48:1193–1201. doi: 10.1095/biolreprod48.6.1193. [DOI] [PubMed] [Google Scholar]
- Ward WS, Partin AW, Coffey DS. DNA loop domains in mammalian spermatozoa. Chromosoma. 1989;98:153–159. doi: 10.1007/BF00329678. [DOI] [PubMed] [Google Scholar]
- Ward WS, Kimura Y, Yanagimachi R. An intact sperm nuclear matrix may be necessary for the mouse paternal genome to participate in embryonic development. Biol Reprod. 1999;60:702–706. doi: 10.1095/biolreprod60.3.702. [DOI] [PubMed] [Google Scholar]
- Wykes SM, Krawetz SA. The structural organization of sperm chromatin. J Biol Chem. 2003;278:29471–29477. doi: 10.1074/jbc.M304545200. [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Shaman JA, Boaz SM, Ward WS. Paternal pronuclear DNA degradation is functionally linked to DNA replication in mouse oocytes. Biol Reprod. 2007;a 77:407–415. doi: 10.1095/biolreprod.107.061473. [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Shaman JA, Ward WS. Topoisomerase II mediated breaks in spermatozoa cause the specific degradation of paternal DNA in fertilized oocytes. Biol Reprod. 2007;b 76:666–672. doi: 10.1095/biolreprod.106.057067. [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Ward MA, Ward WS. Asynchronous DNA replication and origin licensing in the mouse one cell embryo. J Cell Biochem. 2009;107:214–223. doi: 10.1002/jcb.22117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalensky A, Zalenskaya I. Organization of chromosomes in spermatozoa: an additional layer of epigenetic information? Biochem Soc Trans. 2007;35:609–611. doi: 10.1042/BST0350609. [DOI] [PubMed] [Google Scholar]
- Zalenskaya IA, Bradbury EM, Zalensky AO. Chromatin structure of telomere domain in human sperm. Biochem Biophys Res Commun. 2000;279:213–218. doi: 10.1006/bbrc.2000.3917. [DOI] [PubMed] [Google Scholar]