Abstract
Sphingosine 1-phosphate lyase (SPL) is responsible for the irreversible catabolism of sphingosine 1-phosphate, which signals through five membrane receptors to mediate cell stress responses, angiogenesis, and lymphocyte trafficking. The standard assay for SPL activity utilizes a radioactive dihydrosphingosine 1-phosphate substrate and is expensive and cumbersome. In this study, we describe an SPL assay that employs an ω-labeled BODIPY–sphingosine 1-phosphate substrate, allowing fluorescent product detection by HPLC and incorporating advantages of the BODIPY fluorophore. The major aldehyde product is confirmed by reaction with 2,4-dinitrophenylhydrazine. The SPL-catalyzed reaction is linear over a 30 min time period and yields a Km of 35 µM for BODIPY–sphingosine 1-phosphate.
Keywords: Sphingosine 1-phosphate lyase, Sphingosine 1-phosphate, BODIPY, Fluorescent, Sphingolipid, NBD, Assay
Sphingosine 1-phosphate (S1P) lyase (SPL) is a highly conserved enzyme responsible for irreversible catabolism of phosphorylated sphingoid bases generated through the degradation and recycling of membrane sphingolipids [1]. The reaction requires pyridoxal 5′-phosphate as the cofactor and results in the cleavage of the phosphorylated sphingoid base at the C2–C3 carbon–carbon bond, yielding a long-chain aldehyde and ethanolamine phosphate products. The major phosphorylated sphingoid base in humans is S1P, which is synthesized in most cell types and circulates in blood and lymph at high concentrations [2]. S1P signals through a family of five known G protein-coupled membrane receptors that regulate cell survival, migration, and complex processes such as vascular maturation and lymphocyte trafficking [3–5]. SPL activity in tissues and endothelium are required to regulate circulating S1P levels and maintain the chemical gradient that facilitates S1P-mediated lymphocyte egress from peripheral lymphoid organs and thymus [6,7]. Inhibition of SPL through genetic or pharmacological approaches has been shown to block lymphocyte trafficking, indicating that SPL may be a useful target for immune modulation [8].
The SPL gene and gene product are highly conserved throughout evolution and are essential in vertebrates and invertebrates, although its deletion in plants did not lead to deleterious effects [9,10]. Studies in model organisms have demonstrated that SPL expression is necessary for normal development, survival, and tissue homeostasis. The requirement for SPL in mammals is primarily attributed to its regulation of S1P. However, in Leishmania major, a null mutation of SPL blocks infectivity of the parasite due to ethanolamine depletion, demonstrating that product regulation may be critical for some physiological processes [11].
The standard SPL assay employs a tritiated dihydrosphingosine 1-phosphate substrate labeled at C4 and C5 [12]. The assay is radioactive, time consuming, and the substrate is commercially available from only a single source. We previously developed a fluorescent assay employing an ω-labeled NBD–S1P substrate [13]. Although this assay is convenient and involves less exposure to hazardous materials than 4,5-[3H]-dihydro-S1P, the 7-nitrobenz-2-oxa-1,3-diazole (NBD) fluorophore has the disadvantages of being photolabile and polar [14]. The fluorophore consisting of a dipyrrometheneboron difluoride chelate (4,4-difluoro-4-bora-3a,4a-diaza-s-indacene), which is referred to under the trademark BODIPY, possesses several distinctive and useful characteristics, including photochemical and chemical stability, efficient partitioning into membranes, and high molar absorption coefficient and fluorescence quantum yield, which represent significant improvements over the NBD fluorophore [15]. In this study, we have developed a BODIPY-labeled SPL substrate and demonstrated that it can be used effectively to measure SPL activity in biological samples.
Materials and methods
Synthesis of BODIPY–sphingosine
This compound was synthesized by an olefin cross-metathesis reaction between a BODIPY analog bearing a 1-decenyl group at C-8 and an N-protected allylic alcohol derived from (S)-Garner aldehyde, followed by deprotection using boron trifluoride etherate in the presence of molecular sieves [16–18]. The product was purified by column chromatography and characterized by 1H, 13C, and 19F nuclear magnetic resonance spectroscopy and high-resolution mass spectrometry; HRMSm/z: calculated for C26H41BF2N3O2 (MH+), 476.3254; found, 476.3259.
Cell lines and preparation of cell extracts
Human SPL and murine sphingosine kinase 1 (Sphk1) were expressed in HEK293 cells. HEK293 cells expressing murine Sphk1 were generated using an adenoviral expression system as described [13]. Infection occurred at nearly 100% efficiency as determined by GFP fluorescence. Fresh whole cell extracts were prepared by tip sonication on ice for 30 s in SK extraction buffer (20 mM Tris–HCl buffer, pH 7.4, 1 mM EDTA, 0.5 mM deoxypyridoxine [19], 15 mM NaF, 20% glycerol, and Roche EDTA-free protease inhibitor cocktail solution). Cells were infected with adenovirus expressing human SPL and a GFP marker (Ad-SPL) at a MOI of 100. The infection efficiency was determined to be >90% by quantifying GFP-positive cells using fluorescent microscopy. Cells were lysed by tip sonication for 30 s on ice in SPL extraction buffer (0.05 M potassium phosphate buffer, pH 7.2, 2 mM EDTA, 0.2 mM pyridoxal 5′-phosphate, 2 mM 2-mercaptoethanol, 11% glycerol, 1 mM PMSF, Roche EDTA-free protease inhibitor cocktail solution).
Phosphorylation of BODIPY–sphingosine
To generate the SK product BODIPY–S1P, 100 nmol of BODIPY–sphingosine was mixed with 1-palmitoyl-lysophosphatidylcholine (LPC) 1:9 (mol/mol) in chloroform, and the solvent was evaporated with a flow of nitrogen. The substrate mixture was resuspended in 250 µl of 20mM MOPS, pH 7.4, 1 mM EDTA, 0.5 mM DOP, 15 mM NaF, and 1 mM 2-mercaptoethanol by tip sonication for 30 s followed by the addition of 500 µl of reaction buffer containing 100 mM MOPS, pH 7.2, 5 mM 2-mercaptoethanol, 15 mM magnesium chloride, and extract from HEK293 (Ad-mSK1) cells containing 100 µg protein. The SK reaction was started by the addition of 250 µl of 10mM ATP and allowed to proceed with shaking for 2–3 h, at 37 °C which afforded an 85–90% of conversion of the substrate BODIPY–sphingosine to product BODIPY–S1P.
Purification of BODIPY–S1P
At the end of the phosphorylation reaction, BODIPY–S1P was purified by solid-phase extraction on a C18-E strata reverse phase column. The column was wetted with 100% methanol and equilibrated with 1–2 ml of 30% methanol in water. The SK reaction mixture was made 30% with methanol and added to the C18-E strata column. The column was washed with 1–2 ml of 30% methanol in water, and BODIPY–S1P was eluted with 4 ml of 100% methanol. The recovery and purity of the BODIPY–S1P eluted from the C18-E strata column were evaluated by HPLC as described below and by TLC using the Skipski running system (chloroform/ methanol/acetic acid/saline 100:50:16:5 (v:v:v:v) and Hard layer, Silica Gel HL, 250 µm particle, 60 Å pore TLC plates (Analtech Inc) [20]. The recovery was 60–70% and the purity was 70–80%. The impurities found were BODIPY–sphingosine and LPC.
Preparation of tissue extracts
Mouse euthanasia and tissue harvest were performed in accordance with an approved Institutional Animal Care and Use Committee protocol. After 1–2 cm of small intestine from SPL wild-type an SPL knockout mice were homogenized at 0 °C by tip sonication in tissue lysis buffer (5 mM MOPS, 1 mM DTT, 1 mM EDTA, 0.25 M sucrose, 10% glycerol with PMSF, and Roche EDTA-free protease inhibitor cocktail solution), the homogenates were subjected to centrifugation at 500g to clarify the extracts. The supernatant was used for SPL activity assays.
SPL assay
The assay was performed using 5 nmol of C18-E strata purified BODIPY–S1P as substrate. The substrate was dispersed in 100 µl of SPL reaction buffer (0.6 mM EDTA, 0.4 mM pyridoxal 5′- phosphate, 3 mM DTT, 70 mM sucrose, 36 mM potassium phosphate buffer, 36 mM NaF) containing 0.08% Triton X-100 by sonication for 10 min. Cell extracts containing 25–50 µg of total protein in a volume of 20 µl were added to start the reaction, which proceeded at 37 °C for 30 min or longer, as indicated. With the exception of assays for Km determination, the final concentration of BODIPY–S1P was kept at 40–50 µM to assure maximum reaction rates. The concentration of Triton X-100 was kept at 1 mM to assure formation of micellar structures. Boiled cell extracts were used as a blank. The reaction was stopped by adding 350 µl of 1% perchloric acid, followed by 2 ml of chloroform/methanol 1:2 (v:v). The phases were separated by addition of 700 µl of 1% perchloric acid and 700 µl of chloroform. The lower organic phase was washed twice with 1% perchloric acid/methanol 8:2 (v:v), dried, resuspended in methanol, and injected onto the HPLC column (4.6 × 75 mm Luna C18-column (Phenomenex, Torrance, CA). BODIPY–sphingosine was used as an internal standard. BODIPY-labeled compounds were separated by HPLC at a flow rate of 1 ml/min. The mobile phase consisted of solvent A (water) and solvent B (methanol/5 mM acetic acid in water/1 M tetra-n-butylammonium dihydrogen phosphate (TBAP), 95:4:1 (v:v:v)). The gradient used was 0–2 min with 30% solvent A/70% solvent B, 2–5 min from 70% to 100% solvent B, and 5–9 min with 100% solvent B. The fluorescent, BODIPY-containing compounds were detected as described above and the peaks were integrated for quantification.
Aldehyde labeling
The aldehyde product from the SPL reaction was identified after reaction with 2,4-dinitrophenylhydrazine (2,4-DNPH), which converts aldehydes to the corresponding 2,4-dinitrophenylhydrazone derivatives. The SPL product mixture was extracted into the organic phase by two-phase extraction as described above in the SPL assay and was dissolved in 20 µl of methanol. The derivatization was conducted by adding 20 µl of a solution of 5 mM 2,4-DNPH in methanol containing 2 M HCl to the methanolic solution of the product. After the reaction was allowed to proceed for 1 h in the dark at room temperature, the reaction mixture was injected onto the HPLC column and the compounds were separated and detected as described above.
Protein determination
Total protein determination was performed by Bradford method [21].
Results and discussion
Synthesis of BODIPY-labeled S1P as a SPL substrate
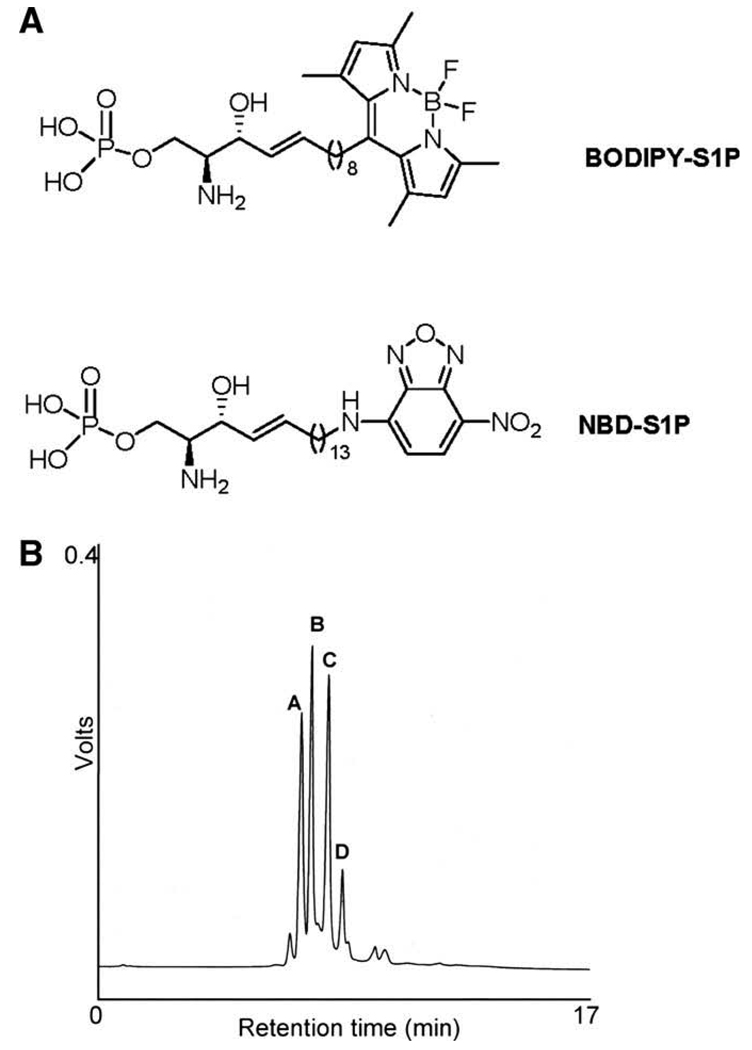
The structure of ω-labeled C13 BODIPY–S1P is shown in Fig. 1. Compared to the ω-NBD-linked S1P analog used in our previous method (see Fig. 1A for structure), the long-chain base linking BODIPY to S1P is shorter by five methylene groups [13]. BODIPY–S1P was prepared by incubating the fluorescently labeled sphingoid base with whole cell extracts from HEK293 cells expressing an adenoviral murine SK construct (Ad-mSK1), as described in Materials and methods. BODIPY–S1P was purified by solid-phase extraction on a C18-E strata column. BODIPY–S1P and a small amount of unreacted BODIPY–sphingosine were detected in the final substrate preparation by HPLC with retention times of 7.0 and 7.3 min, respectively.
Fig. 1.
HPLC analysis of BODIPY-labeled SPL products. Structures of BODIPY–S1P and NBD–S1P (A). Chromatogram showing separation of (A) BODIPY–sphingosine, (B) BODIPY–S1P, and (C) and (D) suspected product peaks of the SPL-catalyzed reaction (B).
BODIPY–S1P as a substrate analog of SPL
We have previously shown that ω-linked NBD–S1P is an effective substrate for the SPL reaction [13]. Based on this observation, we reasoned that ω-linked BODIPY–S1P would also be a suitable substrate for the SPL reaction. To test this hypothesis, semi-purified BODIPY–S1P was incubated with cell extracts containing 0.2–0.3 nmol/mg/min SPL activity by virtue of an adenoviral expression construct driving expression of human SPL (Ad-SPL), as described in Materials and methods. The SPL reaction was allowed to proceed for various time periods and stopped by addition of a mixture of perchloric acid, methanol, and chloroform. The reaction products were isolated by chloroform/methanol extraction, dried, and resuspended in methanol. BODIPY-labeled compounds including BODIPY–sphingosine, BODIPY–S1P, and BODIPY-labeled reaction products were then separated by HPLC. The BODIPY–sphingosine and BODIPY–S1P peaks displayed retention times of 7.0 and 7.3 min, respectively. We observed two additional peaks, a major peak at 7.9 min and minor one at 8.4 min, suggesting the formation of SPL products (Fig. 1B).
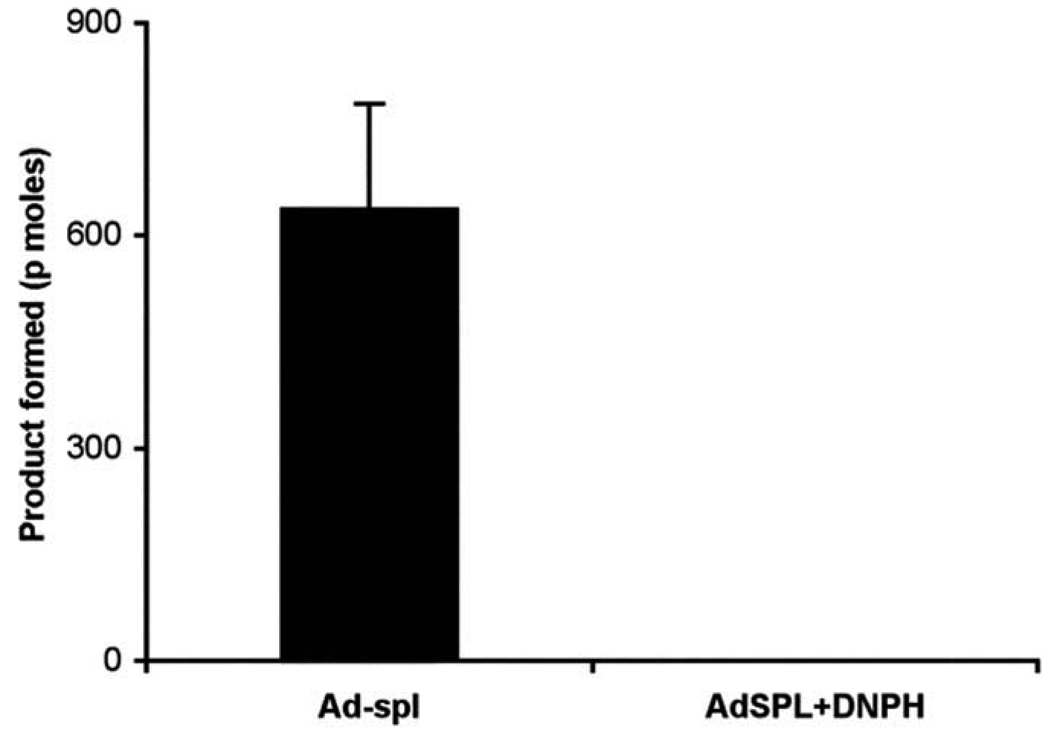
Confirmation of BODIPY-labeled SPL product peak
We have previously shown that the NBD–aldehyde (major) product formed in the SPL reaction can be identified after reaction with 2,4-DNPH, a reagent known to react with carbonyl groups. Therefore, we incubated the BODIPY compounds from the SPL assay with 2,4-DNPH. Following incubation and HPLC separation of the DNPH reaction mixture, the signal from product peak 1, exhibiting a retention time of 7.9 min, was absent (Fig. 2). In contrast, the signals from the later peaks were not affected. Thus, peak 1 was confirmed as the major aldehyde product.
Fig. 2.
Confirmation of BODIPY–aldehyde product by reaction with DNPH. The SPL assay was performed using 25 µg of Ad-SPL total protein extract. The graph shows the SPL product mixture before (Ad-SPL) and after labeling with DNPH (Ad-SPL+DNPH).
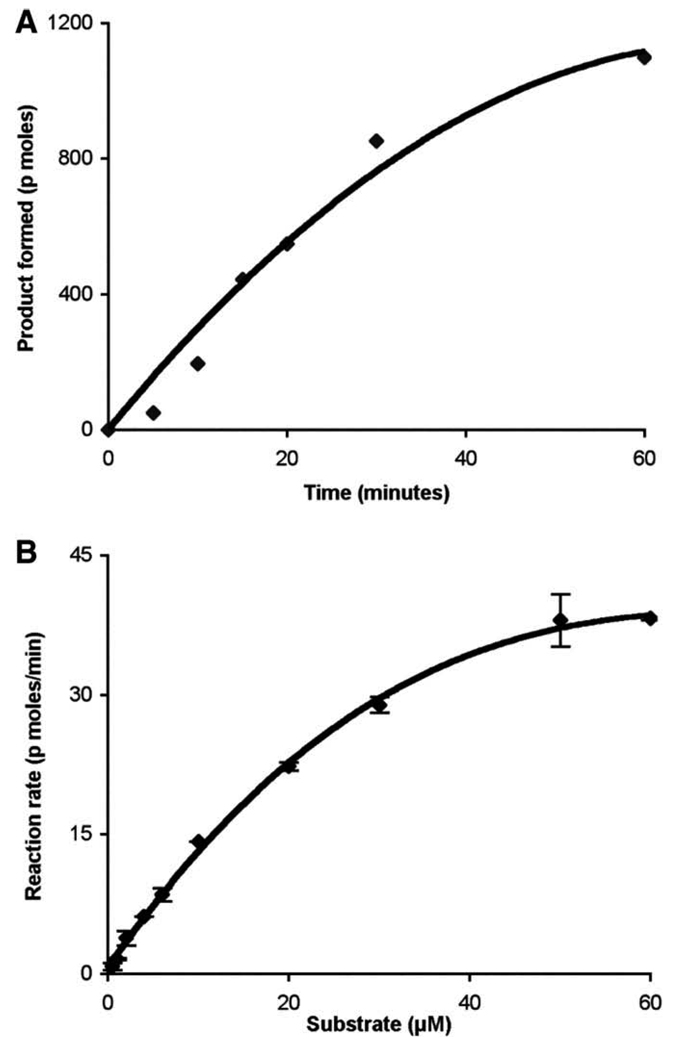
SPL activity as a function of time and substrate concentrations
The amount of product formed with respect to time was linear for up to 30 min (Fig. 3A). To determine the SPL activity at different concentrations of BODIPY–S1P substrate, the concentration of Triton X-100 was maintained at 0.8 mM, which is significantly above the critical micelle concentration of 0.24 mM, whereas the substrate concentration was varied from 1 to 60 µM. The reaction was stopped at 30 min in order to maintain the reaction in the linear range with respect to time. The reaction was linear up to 10 µM substrate, and the Km using BODIPY–S1P was 35 µM (Fig. 3B). This Km is slightly higher than that observed in the standard and NBD-based assays, both of which yielded Km values of 14–20 µM [13,22]. This difference in Km values may be at least partially explained by the different linker lengths in the two model substrates: The BODIPY–S1P substrate used in the current study contains a significantly shorter carbon chain length than the NBD-labeled C18-S1P employed in our previous study, making it less hydrophobic.
Fig. 3.
SPL activity with respect to time and SPL substrate concentration curve using BODIPY–S1P as substrate. Time-dependent formation of SPL product using 25 µg of total protein and 35 µM substrate. The data represent at least three independent experiments (A). Substrate-dependent formation of SPL product using 25 µg of Ad-SPL total protein in 30 min. Each data point represents the mean ± SD of two independent experiments (B).
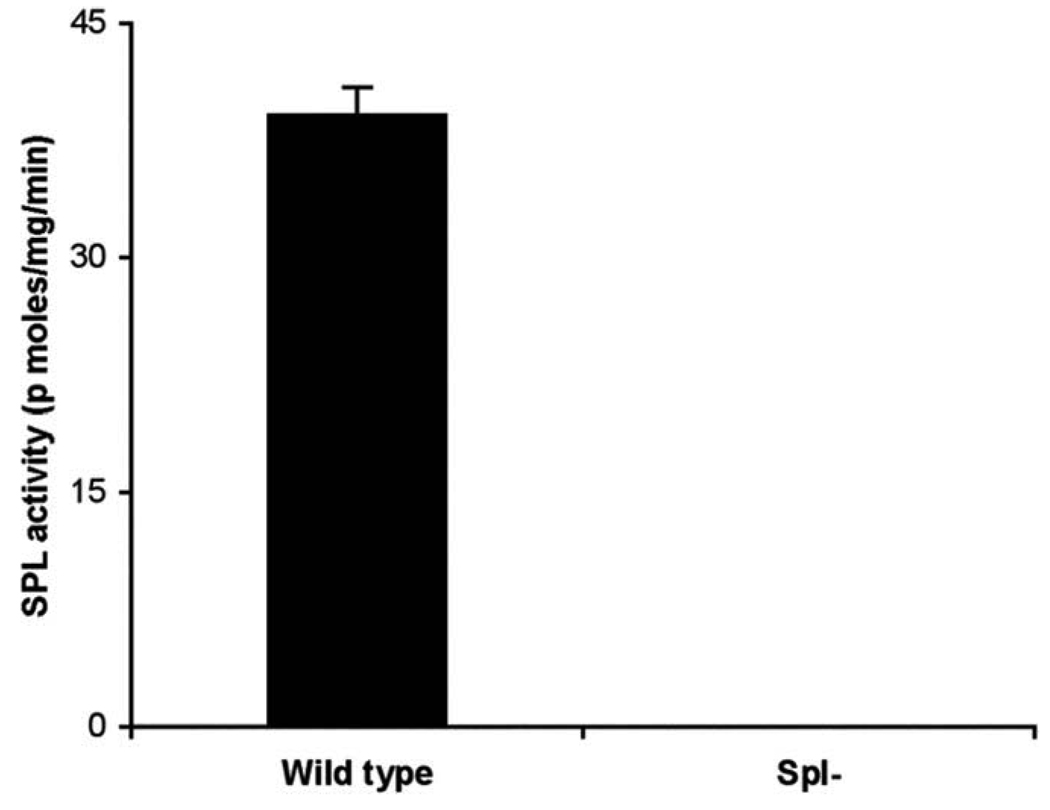
Measurement of SPL activity in genetically modified mouse tissues
To assess the applicability of the BODIPY–S1P fluorescent assay for measurement of SPL activity in mammalian tissues, we measured SPL activity in wild-type mouse small intestine tissues, which normally contain high levels of SPL activity [23], and compared it to that of small intestine tissues from a genetically modified SPL knockout mouse [24]. As shown in Fig. 4, SPL activity in wild-type intestines was approximately 40 nmol product/mg protein/ min, and SPL knockout tissues was not detectable. These results confirm that the BODIPY–S1P SPL assay method is useful for measuring SPL activity in tissues as well as cell extracts.
Fig. 4.
SPL activity in intestinal tissues of SPL knockout mouse and wild-type littermate control. The SPL reaction was carried out for 30 min using 50 µg of wild-type (wild-type) and SPL knockout (Spl-) mouse intestinal extracts. Each data point represents the mean ± SD of at least three independent experiments.
In summary, BODIPY presents a number of advantages as a fluorophore, including chemical and photochemical stability, high fluorescent intensity, insensitivity to the polarity and pH of the environment, and facile integration into membranes. BODIPY has been effectively linked to a variety of lipids including fatty acids, triglycerides, phospholipids, glycolipids, cholesterol, and the S1P analog FTY720 [15,18,25–29]. BODIPY-labeled fluorescent lipids have been used to visualize lipid trafficking in living animals and the subcellular compartments of living cells, to examine membrane microdomains and to quantitate enzyme activity [27,29–32]. In the current study, we have developed a new SPL assay that employs a BODIPY–S1P substrate and is useful for the measurement of SPL activity in cells and tissues. Similar approaches could be used to label a variety of sphingoid base phosphate molecules with BODIPY in order to test enzyme specificity requirements and to measure the activity of SPL homologs using appropriate substrates, i.e., those endogenous to the organism in which the enzyme normally functions.
Acknowledgments
This study was supported by funds from National Institutes of Health Grants GM-66954 and CA-77528 (to J.D.S.) and HL-083187 (R.B.).
Abbreviations
- BODIPY
4,4-difluoro-4-bora-3a,4a-diaza-s-indacene
- DNPH
2,4-dinitrophenylhydrazine
- DOP
deoxypyridoxine
- CLAP
chymotrypsin/leupeptin/antipain/pepstatin A
- GFP
green fluorescence protein
- HPLC
high-performance liquid chromatography
- NBD
7-nitrobenz-2-oxa-1,3-diazole
- SK
sphingosine kinase
- S1P
sphingosine 1-phosphate
- SPL
sphingosine 1-phosphate lyase
- TBAP
tetra-n-butylammonium dihydrogen phosphate
- LPC
1-palmitoyl-lysophosphatidylcholine
References
- 1.Fyrst H, Saba JD. Sphingosine-1-phosphate lyase in development and disease: sphingolipid metabolism takes flight. Biochim. Biophys. Acta. 2008;1781:448–458. doi: 10.1016/j.bbalip.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spiegel S, Milstien S. Functions of the multifaceted family of sphingosine kinases and some close relatives. J. Biol. Chem. 2007;282:2125–2129. doi: 10.1074/jbc.R600028200. [DOI] [PubMed] [Google Scholar]
- 3.Hla T. Signaling and biological actions of sphingosine-1-phosphate. Pharmacol. Res. 2003;47:401–407. doi: 10.1016/s1043-6618(03)00046-x. [DOI] [PubMed] [Google Scholar]
- 4.Rivera J, Proia RL, Olivera A. The alliance of sphingosine-1-phosphate and its receptors in immunity. Nat. Rev. Immunol. 2008;8:753–763. doi: 10.1038/nri2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matloubian M, Lo CG, Cinamon G, Lesneski MJ, Xu Y, Brinkmann V, Allende ML, Proia RL, Cyster JG. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427:355–360. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- 6.Venkataraman K, Lee YM, Michaud J, Thangada S, Ai Y, Bonkovsky HL, Parikh NS, Habrukowich C, Hla T. Vascular endothelium as a contributor of plasma sphingosine-1-phosphate. Circ. Res. 2008;102:669–676. doi: 10.1161/CIRCRESAHA.107.165845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwab S, Pereira J, Matloubian M, Xu Y, Huang Y, Cyster J. Lymphocyte sequestration through S1P lyase inhibition an disruption of S1P gradients. Science. 2005;309:1735–1739. doi: 10.1126/science.1113640. [DOI] [PubMed] [Google Scholar]
- 8.Billich A, Baumruker T. Sphingolipid metabolizing enzymes as novel therapeutic targets. Subcell Biochem. 2008;49:487–522. doi: 10.1007/978-1-4020-8831-5_19. [DOI] [PubMed] [Google Scholar]
- 9.Oskouian B, Saba JD. Death and taxis: what non-mammalian models tell us about sphingosine-1-phosphate. Semin. Cell Dev. Biol. 2004;15:529–540. doi: 10.1016/j.semcdb.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Tsegaye Y, Richardson CG, Bravo JE, Mulcahy BJ, Lynch DV, Markham JE, Jaworski JG, Chen M, Cahoon EB, Dunn TM. Arabidopsis mutants lacking long chain base phosphate lyase are fumonisin-sensitive and accumulate trihydroxy-18:1 long chain base phosphate. J. Biol. Chem. 2007;282:28195–28206. doi: 10.1074/jbc.M705074200. [DOI] [PubMed] [Google Scholar]
- 11.Zhang K, Pompey JM, Hsu FF, Key P, Bandhuvula P, Saba JD, Turk J, Beverley SM. Redirection of sphingolipid metabolism towards de novo synthesis of ethanolamine in Leishmania. EMBO J. 2007;26:1094–1104. doi: 10.1038/sj.emboj.7601565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Veldhoven PP. Sphingosine-1-phosphate lyase. In: Merrill AH Jr., Hannun YA, editors. Sphingolipid Metabolism and Cell Signaling Part A. New York: Academic Press; 2000. pp. 244–254. [Google Scholar]
- 13.Bandhuvula P, Fyrst H, Saba J. A rapid fluorescent assay for sphingosine-1-phosphate lyase enzyme activity. J. Lipid Res. 2007;48:2769–2778. doi: 10.1194/jlr.D700010-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Mazeres S, Schram V, Tocanne JF, Lopez A. 7-Nitrobenz-2-oxa-1,3-diazole-4-yl-labeled phospholipids in lipid membranes: differences in fluorescence behavior. Biophys. J. 1996;71:327–335. doi: 10.1016/S0006-3495(96)79228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loudet A, Burgess K. BODIPY dyes and their derivatives: syntheses and spectroscopic properties. Chem. Rev. 2007;107:4891–4932. doi: 10.1021/cr078381n. [DOI] [PubMed] [Google Scholar]
- 16.Sudhakar N, Kumar A, Prabhakar A, Jagadeesh B, Rao B. The first synthesis of the anhydrophytosphingosine pachastrissamine (jaspine B) from Garner’s aldehyde. Tetrahedron Lett. 2005;46:325–327. [Google Scholar]
- 17.Yamamoto T, Hasegawa H, Hakogi T, Katsumura S. Versatile synthetic method for sphingolipids and functionalized sphingosine derivatives via olefin cross metathesis. Org. Lett. 2006;8:5569–5572. doi: 10.1021/ol062258l. [DOI] [PubMed] [Google Scholar]
- 18.Peters C, Billich A, Ghobrial M, Högenauer K, Ullrich T, Nussbaumer P. Synthesis of borondipyrromethene (BODIPY)-labeled sphingosine derivatives by cross-metathesis reaction. J. Org. Chem. 2007;72:1842–1845. doi: 10.1021/jo062347b. [DOI] [PubMed] [Google Scholar]
- 19.Spyridopoulos I, Mayer P, Shook KS, Axel DI, Viebahn R, Karsch KR. Loss of cyclin A and G1-cell cycle arrest are a prerequisite of ceramide-induced toxicity in human arterial endothelial cells. Cardiovasc. Res. 2001;50:97–107. doi: 10.1016/s0008-6363(01)00196-1. [DOI] [PubMed] [Google Scholar]
- 20.Skipski V, Peterson R, Barclay M. Quantitative analysis of phospholipids by thin-layer chromatography. Biochem. J. 1964;90:374–378. doi: 10.1042/bj0900374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 22.Stoffel W, Bauer E, Stahl J. The metabolism of sphingosine bases in Tetrahymena pyriformis. Sphingosine kinase and sphingosine-1-phosphate lyase. Hoppe Seylers Z. Physiol. Chem. 1974;355:61–74. doi: 10.1515/bchm2.1974.355.1.61. [DOI] [PubMed] [Google Scholar]
- 23.Oskouian B, Sooriyakumaran P, Borowsky A, Crans A, DIllard-Telm L, Tam Y, Bandhuvula P, Saba J. Sphingosine-1-phosphate lyase potentiates apoptosis via p53- and p38-dependent pathways and is downregulated in colon cancer. Proc. Natl. Acad. Sci. USA. 2006;103:17384–17389. doi: 10.1073/pnas.0600050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmahl J, Raymond CS, Soriano P. PDGF signaling specificity is mediated through multiple immediate early genes. Nat. Genet. 2007;39:52–60. doi: 10.1038/ng1922. [DOI] [PubMed] [Google Scholar]
- 25.Li Z, Bittman R. Synthesis and spectral properties of cholesterol- and FTY720-containing boron dipyrromethene dyes. J. Org. Chem. 2007;72:8376–8382. doi: 10.1021/jo701475q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z, Mintzer E, Bittman R. First synthesis of free cholesterol–BODIPY conjugates. J. Org. Chem. 2006;71:1718–1721. doi: 10.1021/jo052029x. [DOI] [PubMed] [Google Scholar]
- 27.Marks DL, Bittman R, Pagano RE. Use of Bodipy-labeled sphingolipid and cholesterol analogs to examine membrane microdomains in cells. Histochem. Cell Biol. 2008;130:819–832. doi: 10.1007/s00418-008-0509-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wood TE, Thompson A. Advances in the chemistry of dipyrrins and their complexes. Chem. Rev. 2007;107:1831–1861. doi: 10.1021/cr050052c. [DOI] [PubMed] [Google Scholar]
- 29.Holtta-Vuori M, Uronen RL, Repakova J, Salonen E, Vattulainen I, Panula P, Li Z, Bittman R, Ikonen E. BODIPY–cholesterol: a new tool to visualize sterol trafficking in living cells and organisms. Traffic. 2008;9:1839–1849. doi: 10.1111/j.1600-0854.2008.00801.x. [DOI] [PubMed] [Google Scholar]
- 30.Kemken D, Mier K, Katus HA, Richardt G, Kurz T. A HPLC-fluorescence detection method for determination of cardiac phospholipase D activity in vitro. Anal. Biochem. 2000;286:277–281. doi: 10.1006/abio.2000.4812. [DOI] [PubMed] [Google Scholar]
- 31.Darroch PI, Dagan A, Granot T, He X, Gatt S, Schuchman EH. A lipid analogue that inhibits sphingomyelin hydrolysis and synthesis, increases ceramide, and leads to cell death. J. Lipid Res. 2005;46:2315–2324. doi: 10.1194/jlr.M500136-JLR200. [DOI] [PubMed] [Google Scholar]
- 32.Burgdorf C, Prey A, Richardt G, Kurz T. A HPLC-fluorescence detection method for determination of phosphatidic acid phosphohydrolase activity: application in human myocardium. Anal. Biochem. 2008;374:291–297. doi: 10.1016/j.ab.2007.10.039. [DOI] [PubMed] [Google Scholar]