Abstract
Structural magnetic resonance imaging data from 308 twins, 64 singleton siblings of twins, and 228 singletons were analyzed using structural equation modeling and selected multivariate methods to identify genetically mediated intracortical associations. Principal components analyses (PCA) of the genetic correlation matrix indicated a single factor accounting for over 60% of the genetic variability in cortical thickness. When covaried for mean global cortical thickness, PCA, cluster analyses, and graph models identified genetically mediated fronto-parietal and occipital networks. Graph theoretical models suggest that the observed genetically mediated relationships follow small world architectural rules. These findings are largely concordant with other multivariate studies of brain structure and function, the twin literature, and current understanding on the role of genes in cortical neurodevelopment.
Keywords: child development, genetics, neuroanatomy, small world
Introduction
Neuroimaging studies of twins have just started to discern the relative effects of genes and environment on brain anatomy. Heritabilities (i.e., the proportion of phenotypic variance due to genetic sources) differ by age and region, but for volumes of total brain or large brain subdivisions such as frontal, parietal, and temporal lobes reported values are usually around 0.80 (Baare et al. 2001; Geschwind et al. 2002; Wallace et al. 2006); for smaller structures the highest heritability measures are approximately 0.50 (Wright et al. 2002), but the accuracy of these estimates is somewhat limited by small samples. A previous study of regional heritability of cortical thickness in a population of 600 healthy children and adolescents found values ranging to approximately 0.60 (Lenroot et al. 2007).
Multivariate analyses provide a means to address the degree to which the heritabilities of different structures are influenced by the same genetic factors. Shared genetic factors may reflect overlapping ontogeny, neural function, or postnatal development. A greater understanding of these shared genetic factors may facilitate gene discovery by suggesting novel endophenotypic constructs and provide insights into the nature of individual differences in neuroanatomy. To date, there have been very few genetically informative studies to investigate brain structure using multivariate tools, and none in large typical populations at high levels of spatial resolution.
In this study, we apply multivariate techniques to explore the role of genes in cortical patterning. Structural equation modeling was used to estimate the genetic contributions to individual differences in cortical thickness of 54 cerebral parcellations in a population of 600 pediatric subjects in an extended twin design. Gyral levels of cortical parcellation were chosen for the present study as perhaps being closer to functionally discrete anatomical subunits than lobar level analyses but less prohibitive in terms of computational demands and difficulty of interpretation than individual voxels. We then applied 3 distinct multivariate techniques (clustering, principal components analysis [PCA], and graph theory) to determine if phenotypic variance in different cortical regions is driven by common genetic factors.
Methods
Participants
Participants were recruited as part of an ongoing longitudinal study of pediatric brain development at the Child Psychiatry branch of the National Institutes of Mental Health (NIMH). Recruitment was performed via local and national advertisements and participants were screened via an initial telephone interview, parent, and teacher rating versions of the Child Behavior Checklist (Achenbach and Ruffle 2000), and physical and neurological assessment. General exclusion criteria included psychiatric diagnosis in the subject or a 1st degree relative and head injury or other conditions that might have affected gross brain development. Exclusion criteria related to pregnancy and birth events specifically included gestational age of <30 weeks; very low birth weight (<3 lbs 4 oz.), any known exposure to psychotropic medications during pregnancy, and significant perinatal complications.
Twin zygosity was determined by DNA analysis of buccal cheek swabs using 9–21 unlinked short tandem repeat loci for a minimum certainty of 99%, by BRT Laboratories, Inc. (Baltimore, MD). Twins were included in the analysis only if quantifiable magnetic resonance imaging (MRI) scans free from motion or other artifact were obtained on both twins at the same age. Written assent from the child and written consent from a parent were obtained for all participants. The study protocol was approved by the institutional review board of the NIMH.
The resultant sample consisted of 600 children (mean age 11.1, SD 3.4, range 5.4–18.7), including 214 monozygotic (MZ) and 94 dizygotic (DZ) twins, 64 singleton siblings of twins (1–2 per family), 116 members of entirely singleton families (2–5 members per family), and 112 unrelated singletons. The distribution of subjects and basic demographic information are given in Table 1.
Table 1.
Demographic characteristics of the sample
| MZ twins | DZ twins | Sibs of twins | Singletons | Total sample | |
| N (% of total) | 214 (35.7) | 94 (15.7) | 64 (10.7) | 228 (38.0) | 600 |
| Age | |||||
| Mean (SD): | 11.03 (3.16) | 11.20 (3.80) | 11.62 (3.53) | 10.92 (3.48) | 11.08 (3.43) |
| Range: | 5.37–18.72 | 5.55–19.34 | 4.99–19.11 | 5.16–18.88 | 4.99–19.34 |
| Sex (% of subsample) | |||||
| Male | 117 (55) | 53 (56) | 31 (48) | 132 (58) | 332 (55) |
| Female | 97 (45) | 41 (44) | 33 (52) | 96 (42) | 268 (45) |
| Race (% of subsample) | |||||
| Caucasian | 202 (94) | 92 (98) | 63 (98) | 172 (75) | 529 (88) |
| African-American | 6 (3) | 0 (0) | 0 (0) | 36 (16) | 42 (7) |
| Asian-American | 2 (<1) | 0 (0) | 0 (0) | 8 (4) | 10 (2) |
| Hispanic | 4 (2) | 2 (2) | 1 (2) | 11 (5) | 18 (3) |
| Unspecified | 0 (0) | 0 (0) | 0 (0) | 1 (<1) | 1 (<1) |
| Handedness (% of subsample) | |||||
| Right | 189 (88) | 78 (83) | 56 (88) | 199 (87) | 513 (86) |
| Mixed | 19 (9) | 11 (12) | 7 (11) | 21 (9) | 36 (6) |
| Left | 18 (8) | 9 (10) | 10 (16) | 32 (14) | 37 (6) |
| Undetermined | 6 (3) | 2 (2) | 4 (6) | 2 (<1) | 14 (2) |
| SES | |||||
| Mean (SD) | 43.49 (18.40) | 42.92 (13.87) | 40.11 (16.89) | 40.70 (20.57) | 41.99 (18.49) |
| Range | 20–89 | 20–70 | 20–77 | 20–95 | 20–95 |
Image Acquisition
All subjects were scanned on the same GE 1.5-Tesla Signa scanner using the same 3-dimensional spoiled gradient recalled echo in the steady state imaging protocol (axial slice thickness = 1.5 mm, time to echo = 5 ms, repetition time = 24 ms, flip angle = 45°, acquisition matrix = 192 × 256, number of excitations = 1, and field of view = 24 cm). A clinical neuroradiologist evaluated all scans and no gross abnormalities were reported.
Image Processing
The native MRI scans 1st were registered into standardized stereotaxic space using a linear transformation (Collins et al. 1994) and subsequently corrected for nonuniformity artifacts (Sled et al. 1998). The registered and corrected volumes were segmented into white matter, gray matter, cerebrospinal fluid, and background using a neural net classifier (Zijdenbos et al. 2002). The white and gray matter surfaces are then fitted using deformable models resulting in 2 surfaces with 81920 polygons each (MacDonald et al. 2000). The white and gray matter surfaces are resampled into native space and computed tomography (CT) was then computed in native space. Each subject's cortical thickness map was then blurred using a 30-mm surface based blurring kernel, which respects anatomical boundaries (Lerch and Evans 2005).
The cortical regions are the same as those described by He et al. (2007). A probabilistic atlas was used to assign cortical points to specific neuroanatomic regions (Collins et al. 1999). Mean CT was calculated for each of 54 cortical subregions (Table 2), which roughly corresponded to cerebral gyri and were based on the sulcal definitions of Ono et al. (1990).
Table 2.
Cortical ROIs in the present study and abbreviations used in subsequent tables and figures
| Structure name | Left abbreviation | Right abbreviation |
| Superior frontal gyrus | SFG-L | SFG-R |
| Middle frontal gyrus | MFG-L | MFG-R |
| Inferior frontal gyrus | IFG-L | IFG-R |
| Precentral gyrus | PreCG-L | PreCG-R |
| Lateral orbitofrontal gyrus | LFOrbG-L | LFOrbG-R |
| Medial orbitofrontal gyrus | MFOrbG-L | MFOrbG-R |
| Cingulate cortex | Cingulate-L | Cingulate-R |
| Medial frontal gyrus | MedialFG-L | MedialFG-R |
| Superior parietal gyrus | SupParGy-L | SupParGy-R |
| Supramarginal gyrus | SMG-L | SMG-R |
| Angular gyrus | AngularGy-L | AngularGy-R |
| Precuneus | Precuneus-L | Precuneus-R |
| Postcentral gyrus | PostCenGy-L | PostCenGy-R |
| Superior temporal gyrus | STG-L | STG-R |
| Middle temporal gyrus | MTG-L | MTG-R |
| Inferior temporal gyrus | ITG-L | ITG-R |
| Uncus | Uncus-L | Uncus-R |
| Medial occipitotemporal gyrus | MediooccipitemporalGy-L | MediooccipitemporalGy-R |
| Lateral occipitotemporal gyrus | LateraloccipitemporalGy-L | LateraloccipitemporalGy-R |
| Parahippocampal gyrus | ParahippocampalGy-L | ParahippocampalGy-R |
| Occipital pole | OccipitalPole-L | OccipitalPole-R |
| Superior occipital gyrus | SupOccGy-L | SupOccGy-R |
| Middle occipital gyrus | MidOccGy-L | MidOccGy-R |
| Inferior occipital gyrus | InfOccGy-L | InfOccGy-R |
| Cuneus | Cuneus-L | Cuneus-R |
| Lingual gyrus | LingualGy-L | LingualGy-R |
| Insula | Insula-L | Insula-R |
Statistical Analysis
The resultant data were iteratively passed from the statistical programming environment R (Ihaka and Gentleman 1996; R Development Core Team 2005) to Mx (Neale et al. 2002), a matrix-based structural equation modeling package (Neale and Cardon 1992). Univariate variance decomposition was accomplished using an extended twin design of the classical ACE model, which increases the statistical power to detect genetic effects on phenotypes (Posthuma et al. 2000). Models included a simultaneous means regression to adjust for sex, nonlinear age effects, and interactions between age and sex. In these models, the differences in the correlation between MZ twins, DZ twins, and related singletons enabled the parsing of the observed variance in the observed cortical thickness measured into variance of genetic (a2), nongenetic familial (c2), and unique environmental (e2) origin. It is important to note that these statistics represent the proportion of the variance in the population attributable to these factors, rather than the fraction of an individual's phenotype caused by genetic or nongenetic effects. Thus, these measures have the same limitations as other summary statistics (e.g., means).
Optimization was performed using maximum likelihood (ML) (Edwards 1972), which produces unbiased estimates of model parameters. ML also allows for straightforward hypothesis testing, because the removal of parameters of interest from the original model produces nested submodels in which the difference in ML asymptotically follows a χ2 distribution, with degrees of freedom equal to the difference in the number of free parameters (Neale and Cardon 1992). For each neuroanatomic region, we tested for the significance of both genetic and shared environmental effects.
Multivariate Modeling
In order to analyze the pattern of genetic relationships between neuroanatomic structures, we employed a modified version of the multistep multivariate analyses reported by Wright et al. (Wright et al. 2002). First, we constructed extended twin versions of bivariate ACE Cholesky decompositions for each pair of neuroanatomic variables (Fig. 1). In addition to adjusting for age and sex, mean global CT was included as a regressor. Cholesky decomposition factors any symmetric positive definite matrix into a lower triangular matrix postmultiplied by its transpose (Neale and Cardon 1992). This approach places few a priori constraints on the data but allows for the covariance between 2 phenotypes to be decomposed into covariance resulting from either genetic or nongenetic sources. The genetic correlation between any 2 structures was then calculated by standardizing their genetic covariance matrix. The genetic correlation is defined as
where Axy is the genetic covariance between structures x and y, and Ax and Ay represent the heritability of x and y (Falconer and Mackay 1996). The sequential bivariate analyses of 2862 pairwise models populated a 54 × 54 genetic correlation matrix; despite the redundancy, calculations both above and below the diagonal were performed in order to ensure that the optimizer had converged to the proper solution.
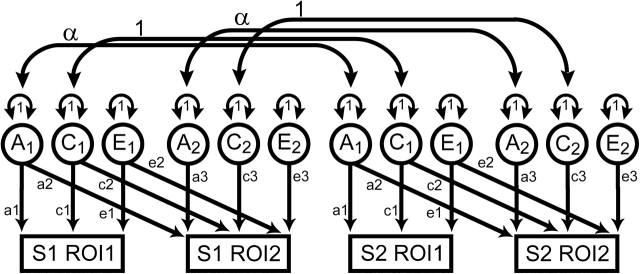
Figure 1.
Example of a path diagram describing the bivariate Cholesky decomposition used to estimate genetic correlations between ROIs. The variance in observed variables (denoted as rectangles) is modeled to be mediated by latent additive genetic (A), shared environmental (C), or unique environmental (E) sources of variance (circles) with latent variances standardized to unity. The model is identified because the correlation between genetic factors (α) is perfect in MZ twins, but one-half between DZ twins and singleton siblings. The expected covariances of this model produce 9 simultaneous equations from which the values of the 9 free parameters (a, c, e) can be estimated. In this example, 2 related family members (S1 and S2) are shown. For families with more than 2 individuals, this model is easily expanded, with families of size k generating (2k)2 informative variance/covariance relationships. Unrelated individuals provide useful information for the estimation of ROI variances as well as the within-person phenotypic covariance.
We then applied 3 multivariate techniques to the subsequent genetic correlation matrix: cluster analysis, PCA, and graph theoretical modeling (details to follow). We used multiple analytic methods for several reasons. First, as the multivariate literature is sparse but employs all of these techniques, using several methods facilitates comparisons with other studies. PCA, for example, is used by the only other extant high-resolution multivariate imaging study on genetically informative data, whereas clustering and graph modeling is commonly employed in the animal literature. Second, all methods have their strengths and weaknesses; in situations in which all methods agree, we will have more confidence that the results are biological rather than statistical artifact. Third, different methods often provide some information not available via other techniques. Graph theoretical models, for example, provide unique information on the connectivity of individual brain regions with other structures, as well as summary statistics for the entire network. In contrast, cluster analysis is uniquely suited for simultaneous inspection of the results and the raw data. The general principles for these 3 multivariate strategies are described below.
Cluster Analysis
The correlation matrix was visualized using the heatmap0.2 function in R (from the gplots package), which also performed hierarchical cluster analysis using Euclidian distances (Warnes et al. 2006). Hierarchical clustering is a form of cluster analysis that requires no a priori specification of the “known” number of clusters present in the data, but rather generates a hierarchy of relationships based on a distance function (Hastie et al. 2001). Heatmap0.2 performs agglomerative clustering, which represents a stepwise “bottom-up” strategy that recursively groups the most related structures until a single cluster remains. In addition to reorganizing the data such that ROIs with similar genetic correlational patterns are spatially proximal in the matrix, a dendrogram also is produced that shows the level of similarity between the ROIs; the shorter the path along the dendrogram between 2 ROIs, the more similar their patterns of genetic correlations. Using this dendrogram, we identified the most prominent high-level clusters in the data.
Principal Components Analysis
We then completed a PCA on the genetic correlation matrix and extracted the factors with the 6 highest eigenvalues. PCA is a linear transformation that attempts to reduce the dimensionality of the data structure by identifying uncorrelated factors that account for a disproportionate amount of the total variance within the observed measures (Norman and Streiner 2000; Hastie et al. 2001); in essence PCA rotates the axes of measurement to optimally align with the dominant axes of the observed data, with the constraint that all components lie orthogonal to one another. To facilitate interpretation of the factor structure, varimax rotation was subsequently applied (Kaiser 1958). These components describe the predominant relationships between the observed neuroanatomic regions. As PCA was performed on genetic correlation matrices, the resultant factor loadings can be interpreted as correlations between genetic latent factors and regions of interest (ROIs). Though the sign of an individual factor loading is arbitrary, 2 structures with the same sign for a given factor are expected to have a positive partial correlation due to the common genetic factor, whereas structures of opposite sign will have negative partial correlations.
Graph Theory
As an alternate method to characterize relationships between gyral regions, we constructed simple graph theoretical models using Bioconductor (Carey et al. 2005), a collection of R packages for the analysis of genomic data. Graph theory is a branch of discrete mathematics for the analysis of complex networks, with applications in telecommunications, social networking, bioinformatics, and molecular biology, among others. Recently, there has been increasing interest in using graph theory in systems biological analyses of the most complex network known, that is, the brain, with applications ranging from examining neuronal circuitry to understanding structural and functional connectivity between large neuroanatomic regions (Sporns et al. 2004). Prior phenotypic analyses using graph theory have suggested that the brain may possess “small world” properties (Achard et al. 2006), defined as neural networks with a combination of dense local interconnectivity (i.e., clustering) and short average path lengths (Watts and Strogatz 1998) between regions. It has been proposed that this architecture is ideal for functional specialization and integration within the brain (Sporns et al. 2004).
The 2 fundamental components of these models are nodes, which represent units in a large system (e.g., computers, proteins, neurons, or gyri), and edges, which represent the connections between them. To identify important edges within our data, we selected significant positive correlations at an α = 0.05. Significant edges were identified by comparing the fit of an AE Cholesky decomposition with a submodel in which the path allowing for genetic covariance (a2 in Fig. 1) was removed. From this graph, we calculated statistics that evaluate properties of the network, namely the characteristic path length (L) and the clustering coefficient (C) (Watts and Strogatz 1998). The path length simply refers to the average shortest distance between a node and other nodes in the system; the clustering coefficient of a node is the average number of edges connecting a node's neighbors, relative to the total number possible. Small world networks typically have similar L and higher C statistics relative to random networks (Watts and Strogatz 1998).
Within the present data, values of L and C for the system as a whole were calculated by taking mean values for all nodes in the graph. We compared these calculations with those from 1000 simulated random networks, each with the same number of nodes and edges as the real data. In the simulations, for each of i edges, 2 nodes were identified by sampling from a pool of 54 nodes (with replacement) with uniform probability, with the constraints that an edge could not connect a node to itself, nor could edges be redundant. Visualization of graphs was performed using modifications of functions in the GeneTS package (Schafer and Strimmer 2005).
Absolute Measures
Prior multivariate research has shown a strong global genetic factor ubiquitously influencing brain volumes (Schmitt et al. 2007). Thus, we also were interested in understanding how the strength of genetic correlations between CT ROIs would change without the mean global CT covariate. To address this question, we recalculated the genetic correlation matrix after removing the effects of global CT on mean CT of individual gyri. Subsequently, PCA on this matrix was performed identically to the analysis described above.
Results
Variance Component Analyses
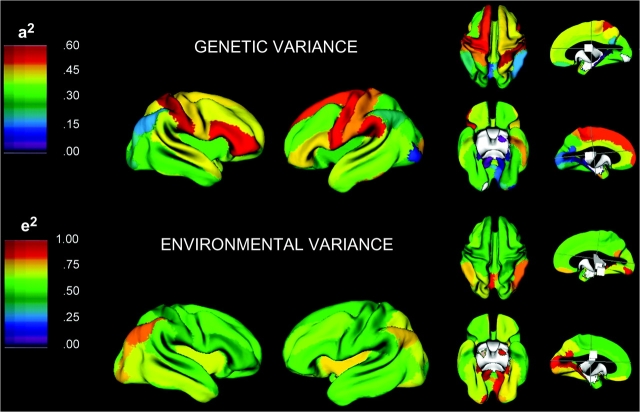
Variance decomposition demonstrated substantial heterogeneity in heritability between cortical regions. Table 3 presents both parameter estimates and tests of the statistical significance of genetic and shared environmental effects to the variation in each region. In general, genetic effects were strongest in frontal lobes, with temporal, parietal, and occipital variance progressively less influenced by genes. The specific regions with the highest heritability included the superior and inferior frontal gyri, the pre- and postcentral gyri, left medial frontal gyrus, left supramarginal gyrus, the left inferior temporal gyrus, and the left occipital pole. Global trends can be seen in Figure 2, which projects point estimates on the brain surface. In contrast to genetic factors, the familial environment appeared to have virtually no role in the observed variability in CT. Although the genetic effects on most of structures were statistically significant at an α of 0.05, no shared environmental factors were significant at this level, and the c2 ML estimate for nearly every structure was zero.
Table 3.
ML parameter estimates and P values from hypothesis testing of univariate ACE models
| Variance components (95% CI) | Hypothesis test | |||||
| P valuesa | ||||||
| a2 | c2 | e2 | A | C | A and C | |
| SFG-R | 0.45 (0.20, 0.60) | 0.00 (0.00, 0.15) | 0.55 (0.40, 0.72) | 0.004 | 1.000 | 0.00 |
| SFG-L | 0.51 (0.24, 0.64) | 0.00 (0.00, 0.17) | 0.49 (0.36, 0.65) | 0.002 | 1.000 | 0.00 |
| MFG-R | 0.43 (0.21, 0.59) | 0.00 (0.00, 0.10) | 0.57 (0.41, 0.76) | 0.002 | 1.000 | 0.00 |
| MFG-L | 0.38 (0.05, 0.52) | 0.00 (0.00, 0.21) | 0.62 (0.48, 0.80) | 0.027 | 1.000 | 0.00 |
| IFG-R | 0.52 (0.32, 0.66) | 0.00 (0.00, 0.09) | 0.48 (0.34, 0.67) | 0.000 | 1.000 | 0.00 |
| IFG-L | 0.44 (0.15, 0.58) | 0.00 (0.00, 0.19) | 0.56 (0.42, 0.73) | 0.007 | 1.000 | 0.00 |
| PreCG-R | 0.43 (0.20, 0.58) | 0.00 (0.00, 0.13) | 0.57 (0.42, 0.75) | 0.004 | 1.000 | 0.00 |
| PreCG-L | 0.52 (0.27, 0.65) | 0.00 (0.00, 0.15) | 0.48 (0.35, 0.65) | 0.001 | 1.000 | 0.00 |
| LFOrbG-R | 0.38 (0.12, 0.54) | 0.00 (0.00, 0.15) | 0.62 (0.46, 0.81) | 0.010 | 1.000 | 0.00 |
| LFOrbG-L | 0.34 (0.03, 0.49) | 0.00 (0.00, 0.19) | 0.66 (0.51, 0.84) | 0.037 | 1.000 | 0.00 |
| MForbG-R | 0.22 (0.00, 0.38) | 0.00 (0.00, 0.17) | 0.78 (0.62, 0.96) | 0.099 | 1.000 | 0.05 |
| MForbG-L | 0.27 (0.00, 0.43) | 0.00 (0.00, 0.21) | 0.73 (0.57, 0.91) | 0.087 | 1.000 | 0.01 |
| Cingulate-R | 0.36 (0.16, 0.53) | 0.00 (0.00, 0.09) | 0.64 (0.47, 0.83) | 0.003 | 1.000 | 0.00 |
| Cingulate-L | 0.40 (0.18, 0.56) | 0.00 (0.00, 0.10) | 0.60 (0.44, 0.80) | 0.003 | 1.000 | 0.00 |
| MedialFG-R | 0.38 (0.10, 0.54) | 0.00 (0.00, 0.17) | 0.62 (0.46, 0.80) | 0.016 | 1.000 | 0.00 |
| MedialFG-L | 0.50 (0.27, 0.64) | 0.00 (0.00, 0.13) | 0.50 (0.31, 0.68) | 0.001 | 1.000 | 0.00 |
| SupParGy-R | 0.44 (0.20, 0.59) | 0.00 (0.00, 0.13) | 0.56 (0.41, 0.76) | 0.004 | 1.000 | 0.00 |
| SupParGy-L | 0.30 (0.04, 0.47) | 0.00 (0.00, 0.15) | 0.70 (0.54, 0.89) | 0.032 | 1.000 | 0.01 |
| SMG-R | 0.39 (0.00, 0.53) | 0.00 (0.00, 0.28) | 0.61 (0.47, 0.79) | 0.056 | 1.000 | 0.00 |
| SMG-L | 0.51 (0.22, 0.63) | 0.00 (0.00, 0.21) | 0.49 (0.37, 0.64) | 0.003 | 1.000 | 0.00 |
| AngularGy-R | 0.20 (0.00, 0.39) | 0.00 (0.00, 0.18) | 0.80 (0.61, 0.99) | 0.171 | 1.000 | 0.12 |
| AngularGy-L | 0.24 (0.00, 0.41) | 0.00 (0.00, 0.18) | 0.76 (0.59, 0.95) | 0.113 | 1.000 | 0.04 |
| Precuneus-R | 0.19 (0.00, 0.36) | 0.00 (0.00, 0.21) | 0.81 (0.64, 0.98) | 0.227 | 1.000 | 0.09 |
| Precuneus-L | 0.12 (0.00, 0.28) | 0.00 (0.00, 0.16) | 0.88 (0.72, 1.00) | 0.367 | 1.000 | 0.32 |
| PostCenGy-R | 0.57 (0.36, 0.68) | 0.00 (0.00, 0.13) | 0.43 (0.32, 0.58) | 0.000 | 1.000 | 0.00 |
| PostCenGy-L | 0.48 (0.25, 0.61) | 0.00 (0.00, 0.14) | 0.52 (0.39, 0.68) | 0.001 | 1.000 | 0.00 |
| STG-R | 0.41 (0.13, 0.56) | 0.00 (0.00, 0.17) | 0.59 (0.44, 0.77) | 0.010 | 1.000 | 0.00 |
| STG-L | 0.40 (0.14, 0.55) | 0.00 (0.00, 0.16) | 0.60 (0.45, 0.77) | 0.007 | 1.000 | 0.00 |
| MTG-R | 0.33 (0.00, 0.49) | 0.00 (0.00, 0.21) | 0.67 (0.51, 0.86) | 0.047 | 1.000 | 0.00 |
| MTG-L | 0.39 (0.04, 0.54) | 0.00 (0.00, 0.22) | 0.61 (0.46, 0.80) | 0.031 | 1.000 | 0.00 |
| ITG-R | 0.38 (0.17, 0.53) | 0.00 (0.00, 0.12) | 0.62 (0.47, 0.70) | 0.003 | 1.000 | 0.00 |
| ITG-L | 0.47 (0.18, 0.60) | 0.00 (0.00, 0.20) | 0.53 (0.40, 0.69) | 0.004 | 1.000 | 0.00 |
| Uncus-R | 0.01 (0.00, 0.16) | 0.00 (0.00, 0.09) | 0.99 (0.84, 1.00) | 1.000 | 1.000 | 1.00 |
| Uncus-L | 0.05 (0.00, 0.24) | 0.00 (0.00, 0.10) | 0.95 (0.76, 1.00) | 0.584 | 1.000 | 0.86 |
| MedioOccipitemporalGy-R | 0.31 (0.00, 0.47) | 0.01 (0.00, 0.30) | 0.68 (0.53, 0.87) | 0.196 | 0.938 | 0.00 |
| MedioOccipitemporalGy-L | 0.26 (0.00, 0.42) | 0.00 (0.00, 0.22) | 0.74 (0.59, 0.92) | 0.128 | 1.000 | 0.01 |
| LateralOccipitotrmporalGy-R | 0.33 (0.00, 0.48) | 0.00 (0.00, 0.22) | 0.67 (0.52, 0.85) | 0.052 | 1.000 | 0.00 |
| LateralOccipitotrmporalGy-L | 0.28 (0.00, 0.44) | 0.00 (0.00, 0.20) | 0.72 (0.56, 0.90) | 0.074 | 1.000 | 0.01 |
| ParahippocampalGy-R | 0.06 (0.00, 0.24) | 0.01 (0.00, 0.16) | 0.93 (0.76, 1.00) | 0.808 | 0.929 | 0.61 |
| ParahippocampalGy-L | 0.10 (0.00, 0.33) | 0.06 (0.00, 0.25) | 0.84 (0.67, 0.98) | 0.651 | 0.705 | 0.07 |
| OccipitalPole-R | 0.30 (0.00, 0.50) | 0.05 (0.00, 0.32) | 0.65 (0.50, 0.84) | 0.183 | 0.764 | 0.00 |
| OccipitalPole-L | 0.47 (0.09, 0.60) | 0.00 (0.00, 0.27) | 0.53 (0.40, 0.70) | 0.018 | 1.000 | 0.00 |
| SupOccGy-R | 0.37 (0.01, 0.52) | 0.00 (0.00, 0.24) | 0.63 (0.48, 0.81) | 0.045 | 1.000 | 0.00 |
| SupOccGy-L | 0.31 (0.00, 0.48) | 0.00 (0.00, 0.30) | 0.69 (0.52, 0.91) | 0.216 | 1.000 | 0.01 |
| MidOccGy-R | 0.26 (0.00, 0.43) | 0.00 (0.00, 0.22) | 0.74 (0.57, 0.94) | 0.136 | 1.000 | 0.02 |
| MidOccGy-L | 0.33 (0.04, 0.51) | 0.00 (0.00, 0.16) | 0.67 (0.49, 0.87) | 0.032 | 1.000 | 0.01 |
| InfOccGy-R | 0.23 (0.00, 0.40) | 0.00 (0.00, 0.21) | 0.77 (0.60, 0.96) | 0.193 | 1.000 | 0.04 |
| InfOccGy-L | 0.12 (0.00, 0.49) | 0.21 (0.00, 0.39) | 0.67 (0.50, 0.83) | 0.580 | 0.200 | 0.00 |
| Cuneus-R | 0.35 (0.02, 0.50) | 0.00 (0.00, 0.22) | 0.65 (0.50, 0.83) | 0.039 | 1.000 | 0.00 |
| Cuneus-L | 0.15 (0.00, 0.32) | 0.00 (0.00, 0.19) | 0.85 (0.68, 1.00) | 0.307 | 1.000 | 0.25 |
| LingualGyrus-R | 0.01 (0.00, 0.33) | 0.13 (0.00, 0.25) | 0.86 (0.67, 0.98) | 1.000 | 0.349 | 0.06 |
| LingualGyrus-L | 0.22 (0.00, 0.39) | 0.00 (0.00, 0.24) | 0.78 (0.61, 0.97) | 0.325 | 1.000 | 0.06 |
| Insula-R | 0.30 (0.00, 0.46) | 0.00 (0.00, 0.20) | 0.70 (0.54, 0.88) | 0.059 | 1.000 | 0.01 |
| Insula-L | 0.26 (0.00, 0.43) | 0.00 (0.00, 0.20) | 0.74 (0.57, 0.95) | 0.120 | 1.000 | 0.03 |
P values test the hypotheses of no genetic (A), shared environmental (C), or familial (A and C) effects on phenotypic variance. Statistically significant effects (at an α = 0.05) are shown in boldface.
Figure 2.
Visualization of variance components analysis for 54 measures of cortical thickness. The ML estimates for heritability (a2) and the unique environmental variance (e2) reported in Table 1 are rendered onto the brain surface. Because the estimates for familial variance approach zero for most structures, these are not shown.
Multivariate Relationships
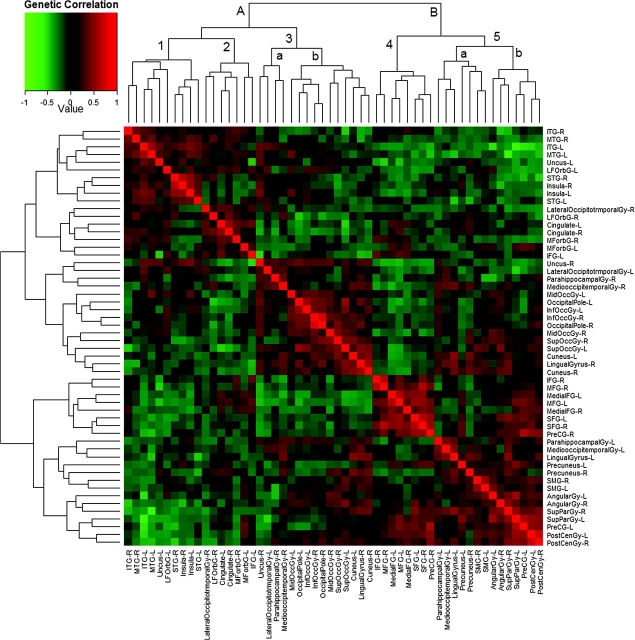
A color map of the genetic correlation matrix is shown in Figure 3. Genetic correlations between the measured cortical structures ranged from −0.67 to 0.76. Left/right gyral homologs were more likely to be positively correlated genetically, as were regions in spatial proximity though this trend was not uniformly observed. The neuroanatomic relationships are apparent in the dendrogram that accompanies the correlation matrix (reproduced on both axes), which displays the results of hierarchical cluster analyses. Two major blocks were identified via cluster analysis: a temporo-occipital cluster and a fronto-parietal cluster. Within these clusters, approximately 5 blocks of related structures emerged: temporal/insular/left lateral orbitofrontal, cingulate/orbitofrontal, occipital/occipitotemporal, frontal (excluding orbitofrontal gyri), and parietal (including precuneus, primary somatosensory cortex bilaterally, and left parahippocampal and lingual gyrus). Positive genetic correlations were largely clustered within these blocks, but there also were several strong correlations between blocks, such as between superior/middle frontal structures and both primary somatosensory cortex and superior parietal lobe. Structures on the inferior of the brain were dispersed between the parietal and occipital blocks and had correlational patterns similar to those of the occipitotemporal structures. Other prominent between-block correlations included between the frontal block and superior parietal lobe, frontal lobe, and cingulate cortices, and between orbitofrontal, cingulate, and middle/inferior temporal structures.
Figure 3.
Heatmap of the genetic correlations between 54 measures of cortical thickness, with results from a hierarchal cluster analysis displayed on the margins. The distance along the dendrogram connecting 2 structures is inversely related to the similarity in their genetic correlational patterns. Two major clusters of structures were identified; a temporo-occipital cluster and a fronto-parietal cluster (lettered A and B, respectively). These clusters in turn divided into roughly 5 subblocks (numbered 1–5); temporal/insular/left lateral orbitofrontal (1), cingulate/orbitofrontal (2), occipital/occipitotemporal (3), frontal (4), and (predominantly) parietal (3). Two of these subblocks had clear, discrete subdivisions within them (lettered a, b).
Principal Components Analysis
The 1st 6 components of the PCA explained over half (58%) of the total genetic variance in all 54 measures, with each factor explaining about 10% of the observed variability. The component loadings are projected onto the brain in Figure 4, which visualizes the most important factors for explaining genetically mediated individual differences in cortical thickness. Like cluster analysis, the genetic factors identified via PCA largely associated regions in spatial proximity. The 1st and most prominent component strongly loaded on frontal and superior parietal structures (1). The 2nd factor loaded predominantly on the lateral surface of the occipital lobe, with high negative loadings in left orbitofrontal gyrus and cingulate gyrus bilaterally (2). The remaining 4 factors explained genetic correlations between ventral occipitotemporal and medial occipital regions (3), gyri of the frontal lobe (excluding orbitofrontal gyrus, 4) temporo-insular and orbititofrontal/cingulate structures (5), and parietal lobe structures and right parahippocampal gyrus (6).
Figure 4.
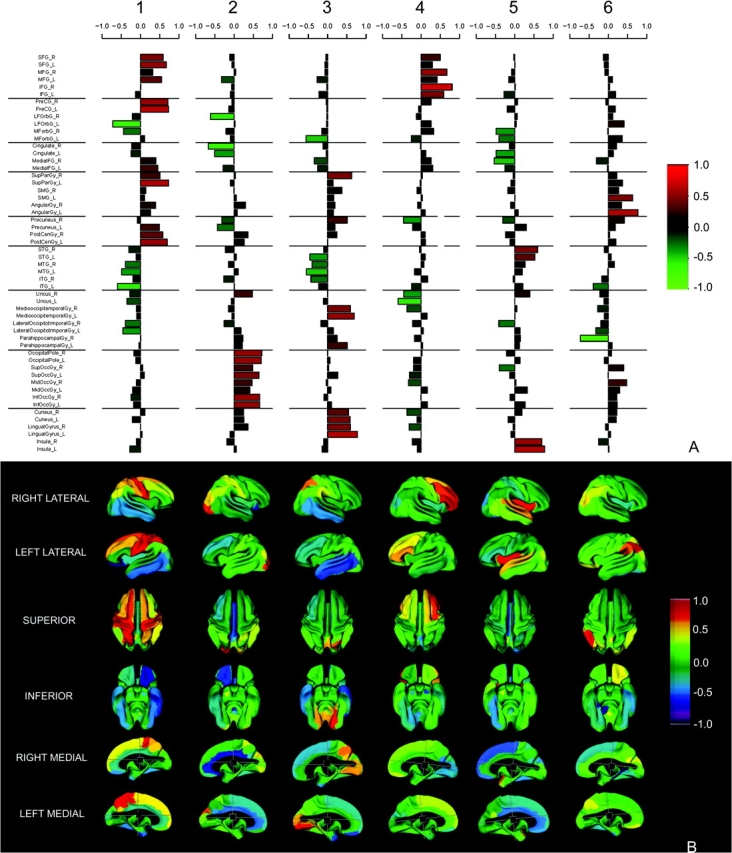
Results of PCA. (A) Barplot of factor loadings on the 54 measured regions, with columns representing the 6 most important components in explaining the total genetic variability in cortical thickness after adjusting for global effects. Because this PCA was performed on a (genetic) correlation matrix, factor loadings can be interpreted as the correlation between an ROI and a (genetic) component. Division lines between ROIs are provided for orientation purposes only. (B) Alternate representation of the same information projected on the brain surface.
Graph Theory
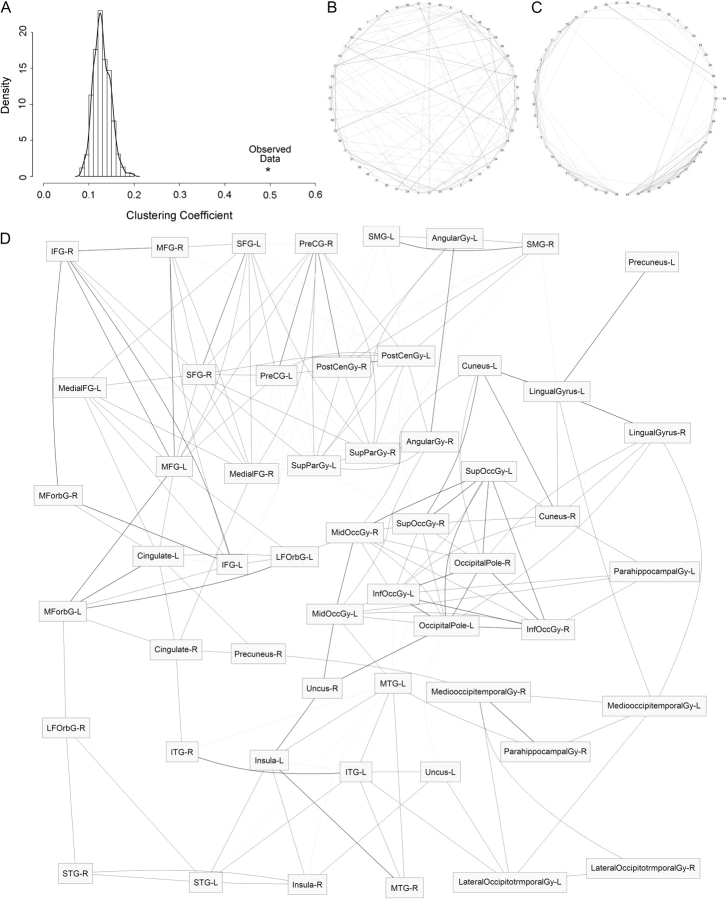
Using statistically significant genetic correlations as a threshold for edge placement, we identified 185 edges connecting the 54 neuroanatomic structures. The degree, clustering coefficient, and average path length for included ROIs are given in the Supplementary Data (Table 1S). The distribution of edges was not uniform; regions with the highest number of edges (i.e., the degree of the node) were the superior and middle frontal gyri, pre- and postcentral gyri, superior parietal gyrus, and the occipital pole. The clustering coefficient and characteristic path length were similarly heterogeneous for different structures, with ranges from 0.17 to 1.0 and 2.3 to 3.7, respectively. The characteristic path length for the entire observed system was 2.8, which was marginally (1.2 times) larger than that in simulation of random networks (mean L = 2.3, SD = 0.03). In contrast, the clustering coefficient for the entire system was 0.50, 4.2 times higher the average value for the simulated networks with randomly assigned edges (mean C = 0.12, SD = 0.03). Figure 5A displays this graphically. The tight local clustering of the data relative to random networks is apparent in Figure 5B,C; in the observed data, most edges from a given node connect to other nodes nearby in the network, with occasional connections to distant nodes. Like PCA and cluster analyses, the graph theoretical approach found strong relationships within fronto-parietal and occipital subnetworks (Fig. 5D).
Figure 5.
Genetically mediated neuroanatomic networks modeled using graph theory. (A) The distribution of the clustering coefficient for 1000 random networks generated using permutation of observed data. The clustering coefficient calculated from the observed data is given as an asterisk. (B) An example of a randomly generated graph, whereas (C) was produced from the observed data. In these graphs, nodes are represented as dots on the periphery of the circle and edges as lines; nodes are placed such that overlap of edges is minimized. (D) An alternative layout of the observed data; darker edges represent stronger correlations. Note the clustering of most frontal and parietal structures (top), occipital structures (middle right), and orbitofrontal, temporal, and insular structures (bottom left).
Absolute Measures
When we repeated the analysis without a global covariate, all structures had strong positive correlations. PCA identified a single genetic factor that could explain over 60% of genetic variability with the 2nd factor explaining more than 10 times less of the total variance (see the Supplementary Data, Fig. 1S). Most structures in the brain showed high loadings on this single factor.
Discussion
Univariate Analyses
The results from univariate analysis found substantial differences in CT heritability throughout the cerebral thickness, with the most heritable structures having about half of their variability explained by additive genetic effects. The most heritable areas were within frontal, somatosensory, supramarginal, or superior temporal regions. In general, gyri in the posterior and inferior cerebrum had lower heritability measures. These findings are largely consistent with the few prior studies at or above the level of resolution of the present study, with the exception that the heritability of cortical thickness appears to be smaller in magnitude when compared with measures of gray matter density (Thompson et al. 2001; Wright et al. 2002). The heritability estimates for cerebral gyri reported here are quite similar to univariate analyses of 40962 cortical points in the same sample (Lenroot et al., 2007). This analysis found significant heritability predominantly in frontal, inferior somatosensory, and left supramarginal regions and low heritability in most posterior and inferior structures, although the statistics reported here blur some fine brain structure that is apparent at higher resolution.
Multivariate Analyses
All 3 multivariate analyses found strong genetically mediated relationships between the frontal lobe and superior parietal cortex, as well as between ROIs in the occipital lobe and other gyri involved in visual processing. The superior fronto-parietal network included regions involved in motoric responses and spatial attention (Mesulam 2000a). Other important components included 2 factors primarily representing structures involved in visual processing (1 including the occipital pole, surrounding occipital gyri, and cingulate cortex, and the 2nd influencing medial and ventral occipital lobe and occipitotemporal gyri), a purely “executive” frontal factor, a “paralimbic” temporo-insular factor, and a component influencing inferior parietal association cortex (Mesulam 1985). Thus, ROIs associated via multivariate analyses tended to have anatomic and/or functional connectivity with each other, which is concordant with prior anatomic studies which show that paralimbic and heteromodal cortical areas have well-developed intraregional connectivity (Mesulam 1985). Cluster analyses, graph models, and PCA all largely reproduced lobar segmentation patterns using the genetic correlations alone, suggesting that genetic variation in cortical thickness may in part be controlled regionally.
There were, however, some important differences between statistical methods; discrepancies between cluster analyses and PCA are at least partially due to the discrete nature of clustering versus the more continuous approach in PCA; in other words, whereas an ROI may belong to only 1 cluster, it can have strong PCA loadings on multiple components. In general, PCA also identified more subtle regional effects than cluster analyses, such as divisions between superior versus inferior parietal lobe or lateral versus medioventral occipital lobe, despite these structures being in close spatial proximity and thought to have related (albeit distinct) functionality. A more subtle examination of the cluster analyses (see Fig. 3) indicates that many subclusters often further subdivide to generate patterns of relatedness that are closer to those observed via PCA (e.g., clusters 5a and 5b). Thus, these multivariate techniques often represent alternative perspectives on the same reality.
Only a few imaging studies have investigated multivariate relationships in genetically informative samples (Pennington et al. 2000; Wright et al. 2002; Schmitt et al. 2007). Of these, by far the most methodologically similar to the present study was that of Wright et al. on 10 MZ and 10 DZ pairs, on which many of our own methods were based. Their analyses found that 2 principal components could account for 24% of the total genetic variance in 92 regions, which they interpreted as representing a fronto-parietal-limbic system and a supraregional network involved in audition. The structures influenced by the 2nd factor primarily were located in temporal lobe, dorsolateral prefrontal and orbitofrontal cortex, insula, and extracortical regions. The factor loadings were quite low for both factors (|loadings| < 0.25). Potential reasons for the discrepancies between the 2 studies are numerous, including 1) differing phenotypes both in measure (volume versus cortical thickness) and parcellation scheme, 2) the bivariate model used by Wright constrained genetic correlations to be positive, whereas the present study allowed for negative values, 3) the former study used twins only, whereas the latter included information from related family members, 4) an adult versus pediatric sample, and 5) a 15-fold difference in sample size with associated differences in power. It is reassuring, however, that the components reported by Wright resemble 2 of the components reported here (components 1 and 5 from Fig. 4).
Although multivariate anatomic studies in humans are, at present, quite sparse, there are several examples of multivariate analyses in the literature on nonhuman primates as well as multivariate functional studies on humans (Bassett and Bullmore 2006). Considering several differences in methodology as well as interspecies neuroanatomical differences, the correlational patterns that we observed are broadly similar to those produced in functional analyses of primate cortical connectivity using the large public database CoCoMac (Collections of Connectivity on the Macaque) (Young 1993; Stephan et al. 2000). By using several different multivariate techniques, including cluster analysis, multidimensional scaling (MDS), and graph theoretical models, Stephen et al. examined functional connectivity via 245 different local cortical applications of strychnine on 3897 tests of activity throughout the cortex. These analyses identified 3 major clusters comprising 1) a “somatomotor” cluster including primary motor area, premotor areas, primary somatosensory cortex, and superior parietal regions (PEp and PEm), 2) a “visual” cluster including primary visual cortex, extrastriate visual cortex, temporo-occipital regions, and medial temporal cortex, and 3) an “orbito-temporal-insular” cortical cluster including frontal operculum, anterior insula, and “polar, medial, and allocortical regions of the temporal lobe.” A separate study by Young (1993) reported similar findings on primate brain structure.
We also observed strong relationships between the frontal lobe and superior parietal cortex, and genetically mediated associations between regions involved in vision as well as between temporo-insular and the orbitofrontal regions. These findings were most evident in PCA, in which somatomotor, visual, and temporal-insular-orbital components were observed. Multivariate functional imaging studies on humans find similar clusters. For example, employing hierarchical cluster analysis and MDS on resting state functional MRI (fMRI) data, Salvador identified approximately 6 regional systems corresponding to the 4 neocortical lobes (with parietal regions and premotor cortex clustered together, as reported here), medial temporal lobe, and the subcortical nuclei (Salvador et al. 2005). The association between parietal and motor cortices, in particular, appears to be consistent across species and methods. Salvador additionally observed strong phenotypic relationships between a given ROI and its contralateral homologues, which also has been observed in anatomic data (He et al. 2007). As we observed strong interhemispheric genetic correlations between these regions, our data suggest that the associations between structural homologues are largely genetically mediated.
Graph theoretical analyses in primates also suggest that the organization of the brain follows small world architectural rules, by which interstructural connectivity is dense within blocks and more sparse between them (Kotter and Sommer 2000; Sporns et al. 2000; Sporns et al. 2002). It has been suggested that this type of architecture is an appealing model of neural function because it allows both for modularization and efficient integration between functional subregions, resulting in enhanced computational power and transmission speeds (Bassett and Bullmore 2006). Despite the fact that structural organization is an area of great interest to the field of systems neuroscience, at present there have been few multivariate structural MRI studies in humans (Crick and Jones 1993; Mesulam 2000b); this fact is somewhat surprising given the rapid increase in the use of multivariate techniques for the analysis of data from functional and diffusion tensor imaging (Ramnani et al. 2004). Though the present study is limited by several factors when compared with other studies (not the least of which are level of resolution, implicit rather than explicit physical distances, and a more simplistic graph theoretical analysis), the dense clustering observable at the gyral level also implies a small world architecture. Several fMRI and DTI studies have reported small world properties in human neural function and white matter orientation (Sporns et al. 2005; Achard et al. 2006), and a recent study by He et al. (2007) found small world architecture with structural measures of cortical thickness and quite similar image processing methodology to that of the present study. The high clustering observed using graph theoretical models on our data also implies small world architecture; however, because our analyses were based on genetic, rather than phenotypic correlations, it appears that genetic factors are involved in the patterning of the human cortex in this manner.
Finally, it is important to keep in mind that these analyses do not account for all of the genetic variation in the cerebral cortex. In particular, the use of a global covariate obscures the presence of a genetic factor that strongly influences the thickness of nearly the entire cerebral cortex. These analyses found that this single factor accounted for over 60% of the total genetic variance. A prior volumetric study by our group showed a similar phenomenon in a multivariate study of cerebrum and 5 other brain structures of diverse ontogenetic origins (cerebellum, basal ganglia, thalamus, corpus callosum, and the lateral ventricles) in a genetically informative sample (Schmitt et al. 2007). Thus, although the present study suggests that some genetic effects on cortical patterning are regionalized, these appear secondary to global effects caused by genetic factors influencing the entire cerebral cortex. As the genes influencing individual differences in brain structure are discovered, it is likely that the genes with the largest effect size will influence the cortex as a whole, with genes of lesser effect acting on specific subregions and local networks.
Supplementary Material
Supplementary material can be found at: http://www.cercor.oxfordjournals.org/
Funding
National Institutes of Health grants (MH-65322, MH-20030, and DA-18673); and the Intramural Program of the NIH, NIMH.
Supplementary Material
Acknowledgments
Conflict of Interest: None declared.
References
- Achard S, Salvador R, Whitcher B, Suckling J, Bullmore ET. A resilient, lowfrequency, small-world brain functional network with highly connected association cortical hubs. J Neurosci. 2006;26:63–72. doi: 10.1523/JNEUROSCI.3874-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achenbach TM, Ruffle TM. The Child Behavior Checklist and related forms for assessing behavioral/emotional problems and competencies. Pediatr Rev. 2000;21:256–271. doi: 10.1542/pir.21-8-265. [DOI] [PubMed] [Google Scholar]
- Baare WF, Hulshoff Pol HE, Boomsma DI, Posthuma D, De Geus EJ, Schnack HG, Van Haren NE, van Oel CJ, Kahn RS. Quantitative genetic modeling of variation in human brain morphology. Cereb Cortex. 2001;11:816–824. doi: 10.1093/cercor/11.9.816. [DOI] [PubMed] [Google Scholar]
- Bassett DS, Bullmore E. Small-world brain networks. Neuroscientist. 2006;12:512–523. doi: 10.1177/1073858406293182. [DOI] [PubMed] [Google Scholar]
- Carey VJ, Gentry J, Whalen E, Gentleman R. Network structures and algorithms in Bioconductor. Bioinformatics. 2005;21:135–136. doi: 10.1093/bioinformatics/bth458. [DOI] [PubMed] [Google Scholar]
- Collins DL, Neelin P, Peters TM, Evans AC. Automatic 3D Intersubject registration of MR volumetric data in standardized Talairach space. J Comput Assist Tomogr. 1994;18:192–205. [PubMed] [Google Scholar]
- Collins DL, Zijdenbos AP, Barre WFC, Evans AC. ANIMAL+INSECT: improved cortical structure segmentation. In: Kuba A, Samal M, Todd-Pokropek A, editors. Information processing in medical imaging: 16th International Conference. Berlin: Springer; 1999. pp. 210–223. [Google Scholar]
- Crick F, Jones E. Backwardness of human neuroanatomy. Nature. 1993;361:109–110. doi: 10.1038/361109a0. [DOI] [PubMed] [Google Scholar]
- Edwards AWF. Likelihood. London: Cambridge; 1972. [Google Scholar]
- Falconer DS, Mackay TFC. Introduction to quantitative genetics. Essex (UK): Longman; 1996. [Google Scholar]
- Geschwind DH, Miller BL, DeCarli C, Carmelli D. Heritability of lobar brain volumes in twins supports genetic models of cerebral laterality and handedness. Proc Natl Acad Sci U S A. 2002;99:3176–3181. doi: 10.1073/pnas.052494999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Friedman S. The elements of statistical learning; data mining, inference, and prediction. New York: Springer; 2001. [Google Scholar]
- He Y, Chen ZJ, Evans AC. Small-world anatomical networks in the human brain revealed by cortical thickness from MRI. Cereb Cortex. 2007 doi: 10.1093/cercor/bhl149. Advance Access published January 4, 2007. [DOI] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. R: a language for data analysis and graphics. J Comput Graph Statist. 1996:299–314. [Google Scholar]
- Kaiser HF. The varimax criterion for analytic rotation in factor analysis. Psychometrika. 1958;23:187–200. [Google Scholar]
- Kotter R, Sommer FT. Global relationship between anatomical connectivity and activity propagation in the cerebral cortex. Philos Trans R Soc Lond B Biol Sci. 2000;355:127–134. doi: 10.1098/rstb.2000.0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenroot RK, Schmitt JE, Ordaz SJ, Wallace GL, Neale MC, Lerch JP, Kendler KS, Evans AC, Giedd JN. Differences in genetic and environmental influences on the human cerebral cortex associated with development during childhood and adolescence. Hum Brain Mapp. 2008 doi: 10.1002/hbm.20494. Forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch JP, Evans AC. Cortical thickness analysis examined through power analysis and a population simulation. Neuroimage. 2005;24:163–173. doi: 10.1016/j.neuroimage.2004.07.045. [DOI] [PubMed] [Google Scholar]
- MacDonald D, Kabani N, Avis D, Evans AC. Automated 3-D extraction of inner and outer surfaces of cerebral cortex from MRI. Neuroimage. 2000;12:340–356. doi: 10.1006/nimg.1999.0534. [DOI] [PubMed] [Google Scholar]
- Mesulam M-M. Patterns in behavioral neuroanatomy: association areas, the limbic system, and hemispheric specialization. In: Mesulam M-M, editor. Principles of behavioral neurology. Philadelphia (PA): F.A. Davis Company; 1985. [Google Scholar]
- Mesulam M-M. Behavioral neuroanatomy: large-scale networks, association cortex, frontal syndromes, the limbic system and hemisphereic specializations. In: Mesulam M-M, editor. Principles of behavioral and cognitive neurology. New York: Oxford University Press; 2000a. [Google Scholar]
- Mesulam M-M. Brain, mind, and the evolution of connectivity. Brain Cogn. 2000b;42:4–6. doi: 10.1006/brcg.1999.1145. [DOI] [PubMed] [Google Scholar]
- Neale MC, Boker SM, Xie G, Maes HH. 5th ed. Richmond (VA): Department of Psychiatry, Medical College of Virginia, Virginia Commonwealth University; 2002. Mx: statistical modeling. [Google Scholar]
- Neale MC, Cardon LR. Methodology for genetic studies of twins and families. Dordrecht (The Netherlands): Kluwer Academic; 1992. [Google Scholar]
- Norman GR, Streiner DL. Hamilton (ON): B.C. Decker, Inc; 2000. Biostatistics: the bare essentials. [Google Scholar]
- Ono M, Kubik S, Abernathey CD. Atlas of the cerebral sulci. New York: Thieme; 1990. [Google Scholar]
- Pennington BF, Filipek PA, Lefly D, Chhabildas N, Kennedy DN, Simon JH, Filley CM, Galaburda A, DeFries JC. A twin MRI study of size variations in human brain. J Cogn Neurosci. 2000;12:223–232. doi: 10.1162/089892900561850. [DOI] [PubMed] [Google Scholar]
- Posthuma D, De Geus EJ, Neale MC, Hulshoff Pol HE, Baare WEC, Kahn RS, Boomsma D. Multivariate genetic analysis of brain structure in an extended twin design. Behav Genet. 2000;30:311–319. doi: 10.1023/a:1026501501434. [DOI] [PubMed] [Google Scholar]
- R Development Core Team . Vienna (Austria): R Foundation for Statistical Computing; 2005. R: a language and environment for statistical computing. [Google Scholar]
- Ramnani N, Behrens TEJ, Penny W, Matthews PM. New approaches for exploring anatomical and functional connectivity in the human brain. Biol Psychiatry. 2004;56:613–619. doi: 10.1016/j.biopsych.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Salvador R, Suckling J, Coleman MR, Pickard JD, Menon D, Bullmore E. Neurophysiological architecture of functional magnetic resonance images of human brain. Cereb Cortex. 2005;15:1332–1342. doi: 10.1093/cercor/bhi016. [DOI] [PubMed] [Google Scholar]
- Schafer J, Strimmer K. An empirical Bayes approach to inferring large-scale gene association networks. Bioinformatics. 2005;21:754–764. doi: 10.1093/bioinformatics/bti062. [DOI] [PubMed] [Google Scholar]
- Schmitt JE, Wallace GW, Rosenthal MA, Molloy EA, Ordaz S, Lenroot R, Clasen LS, Blumenthal J, Kendler KS, Neale MC, et al. A multivariate analysis of neuroanatomic relationships in a genetically informative pediatric sample. Neuroimage. 2007;35:70–82. doi: 10.1016/j.neuroimage.2006.04.232. [DOI] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Sporns O, Chialvo DR, Kaiser M, Hilgetag CC. Organization, development and function of complex brain networks. Trends Cogn Sci. 2004;8:418–425. doi: 10.1016/j.tics.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Edelman GM. Theoretical neuroanatomy: relating anatomical and functional connectivity in graphs and cortical connection matrices. Cereb Cortex. 2000;10:127–141. doi: 10.1093/cercor/10.2.127. [DOI] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Edelman GM. Theoretical neuroanatomy and the connectivity of the cerebral cortex. Behav Brain Res. 2002;135:69–74. doi: 10.1016/s0166-4328(02)00157-2. [DOI] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Kotter R. The human connectome: a structural description of the human brain. PLoS Comput Biol. 2005;1:32–42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan KE, Hilgetag CC, Burns GA, O'Neill MA, Young MP, Kotter R. Computational analysis of functional connectivity between areas of primate cerebral cortex. Philos Trans R Soc Lond B Biol Sci. 2000;355:111–126. doi: 10.1098/rstb.2000.0552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Cannon TD, Narr KL, van Erp T, Poutanen VP, Huttunen M, Lonnqvist J, Standertskjold-Nordenstam CG, Kaprio J, et al. Genetic influences on brain structure. Nat Neurosci. 2001;4:1253–1258. doi: 10.1038/nn758. [DOI] [PubMed] [Google Scholar]
- Wallace GW, Schmitt JE, Lenroot R, Viding E, Ordaz S, Rosenthal MA, Molloy EA, Clasen LS, Kendler KS, Neale MC, et al. A pediatric study of brain morphometry. J Child Psychol Psychiatry. 2006;47:987–993. doi: 10.1111/j.1469-7610.2006.01676.x. [DOI] [PubMed] [Google Scholar]
- Warnes GR, Bolker B, Lumley T. R package version 2.3.1. 2006. gplots: various R tools for plotting data. http://cran.r-project.org/src/contrib/Descriptions/gplots.html. [Google Scholar]
- Watts DJ, Strogatz SH. Collective dynamics of ‘small-world’ networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- Wright IC, Sham P, Murray RM, Weinberger DR, Bullmore ET. Genetic contributions to regional variability in human brain structure: methods and preliminary results. Neuroimage. 2002;17:256–271. doi: 10.1006/nimg.2002.1163. [DOI] [PubMed] [Google Scholar]
- Young MP. The organization of neural systems in the primate cerebral cortex. Proc Biol Sci. 1993;252:13–18. doi: 10.1098/rspb.1993.0040. [DOI] [PubMed] [Google Scholar]
- Zijdenbos AP, Forghani R, Evans AC. Automatic “pipeline” analysis of 3-D MRI data for clinical trials: application to multiple sclerosis. IEEE Trans Med Imaging. 2002;21:1280–1291. doi: 10.1109/TMI.2002.806283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.