SUMMARY
Neuroanatomical and functional asymmetries are universal features of the vertebrate CNS, but how asymmetry is generated is unknown. Here we show that zebrafish fgf8 mutants do not elaborate forebrain asymmetries, demonstrated by the failure of the parapineal nucleus to migrate from its initial midline position to the left side of the brain. Local provision of Fgf8 restores the asymmetric migration of parapineal cells, usually to the left, irrespective of the location of the Fgf8 source. This laterality bias is due to left-sided Nodal signaling and when the bias in Nodal signaling is removed, parapineal cells migrate toward the source of Fgf8 protein. This study presents a mechanism for breaking neuroanatomical symmetry through Fgf8-dependent regulation of bistable left- or right-sided migration of the parapineal. The combined action of Fgf and Nodal signals ensures the establishment of neuroanatomical asymmetries with consistent laterality.
INTRODUCTION
Brain asymmetry is conserved among all vertebrates studied and is thought to confer greater efficiency of processing, whereby specialization of one hemisphere leaves the opposite free to perform other tasks (Vallortigara and Rogers, 2005). Compromised brain asymmetries have been linked to several neuropathologies including schizophrenia, autism, and neuronal degenerative diseases (Escalante-Mead et al., 2003; Li et al., 2007; Toth et al., 2004).
The best-described example of a conserved brain asymmetry is displayed in the diencephalic epithalamus of vertebrates (Concha and Wilson, 2001; Concha, 2004; Bianco and Wilson, 2009). In zebrafish embryos, bilaterally positioned parapineal precursors migrate leftward from the dorsal midline, establishing a left-sided nucleus (Concha et al., 2003; Signore et al., 2009). Subsequently, the parapineal promotes the elaboration of left-sided character in habenular neurons, such that the paired habenular nuclei show left-right (L/R) asymmetries in gene expression, neuropil organization, and axonal projections (Aizawa et al., 2005; Bianco et al., 2008; Concha et al., 2003; Gamse et al., 2003, 2005).
The leftward migration of the parapineal nucleus is dependent on left-sided epithalamic Nodal signaling (Concha et al., 2000), which is itself dependent on left-sided Nodal signals from the lateral plate mesoderm (Carl et al., 2007; Inbal et al., 2007; Long et al., 2003). Crucially, in the absence of unilateral Nodal signaling, brain asymmetries develop but are randomized (Concha et al., 2000), with left- or right-sided migration of the parapineal and corresponding habenular asymmetry equally likely outcomes. Therefore, while consistent directional laterality (handedness) relies on Nodal signaling, development of an asymmetric brain per se does not, and must be dependent on other signals. The ability to produce either laterality state suggests that both sides of the brain are equally competent to initiate and reinforce asymmetric development.
In order to elucidate the genetic basis underlying the Nodal-independent breaking of brain symmetry, we screened lines of fish for mutant phenotypes in which the epithalamus appeared symmetric. Here, we describe the phenotype of the fgf8 mutant, acerebellar (Reifers et al., 1998), which shows symmetric development of the epithalamus. We demonstrate that Fgf8, expressed bilaterally in habenular precursor cells, is required for the asymmetric migration of the parapineal nucleus and that in the absence of Nodal signaling, Fgf8 is sufficient to direct the laterality of migration. This study describes a genetic basis for breaking symmetry in the brain, and suggests that mechanisms to generate asymmetry and direct laterality can be uncoupled and probably evolved sequentially.
RESULTS
Fgf8 Is Required to Break Symmetry in the Brain
To elucidate the genetic mechanisms underlying the Nodal-independent breaking of brain symmetry, we screened lines of fish for mutant phenotypes in which the epithalamus appeared symmetric. We observed that in fgf8 mutants (acerebellarti282 [aceti282] or ace; Reifers et al., 1998) and morphants (Draper et al., 2001), parapineal and habenular nuclei develop symmetrically (Figures 1A-1H). Although a discrete parapineal nucleus is not evident in ace embryos (Figures 1A and 1B), the expression of parapineal-specific markers confirms that these cells are specified but fail to migrate from their initial midline position at the rostral limit of the pineal nucleus (Figures 1C-1F). At the stage when the parapineal initiates migration, the habenulae are morphologically evident and contain neuronal precursors/neurons (data not shown). By later stages, markers of habenular asymmetry are, however, reduced and symmetrically expressed in ace mutants (Figures 1G and 1H and data not shown). The loss of asymmetry in habenular markers could in part be due to defective parapineal migration because the parapineal influences lateralized gene expression in the left habenula (Concha et al., 2003; Gamse et al., 2003). However, the bilaterally reduced expression of both asymmetric and symmetric markers (Figures 1I and 1J) suggests that Fgf signaling is required during the development of both left and right habenulae. Altogether, these data indicate that Fgf8 activity is required for the leftward migration of parapineal cells and for the subsequent elaboration of neuroanatomical asymmetries in the epithalamus. We next addressed where and when Fgf8 is required to promote the migration of the parapineal primordium.
Figure 1. The fgf8 Mutant Has a Symmetric Epithalamus.
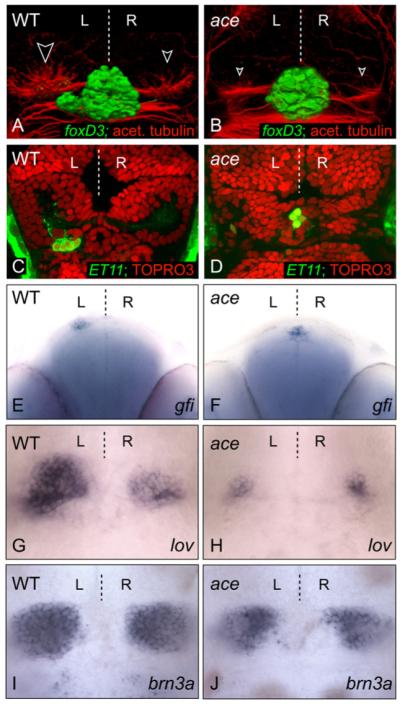
(A-D) Dorsal views of confocal images of the epithalamus in wild-type and ace 3 dpf embryos, with anterior to the top. (A and B) 3D reconstructions of pineal/parapineal nuclei and axons [green, Tg(foxD3:GFP)] and neuropil of the habenular nuclei (red, anti-acetylated tubulin; white-edged arrowheads). (C and D) The parapineal-specific marker Tg(ET11:GFP) is present (green), but expressing cells remain at the midline in the ace embryo compared with those of the wild-type embryo at 3 dpf. Brain morphology is visualized using the nuclear marker TOPRO3 (red). A single z-slice is presented for each example. (E and F) Frontal view of parapineal-specific gfi expression in wild-type and ace 3 dpf embryos, with dorsal to the top. Parapineal cells are at the midline in ace (F). (G-J) Dorsal views of habenular lov (G and H) and brn3a (I and J) expression in wild-type and ace embryos at 4 dpf; expression of both markers is reduced in left and right habenulae.
Fgf8 Is Expressed Adjacent to Migrating Parapineal Cells and Is Required during the Period of Migration
fgf8 is expressed bilaterally in the epithalamus from stages prior to the leftward migration of parapineal cells (22 somites [ss]; Figure S1A available online) and persists until at least 3 days postfertilization (dpf) (Figure S1C). High-resolution analysis revealed fgf8 expression to be subtly asymmetric, such that at 22 ss, expression on the right is usually slightly higher than on the left. (Figure S1A, Table S1 available online). At 24 hr postfertilization (hpf), in addition to overtly symmetric bilateral expression domains, fgf8 is expressed in a small group of cells immediately rostral to the parapineal nucleus, which, prior to migration, is evident as a coherent cluster of cells at the dorsal midline (Figures 2A and 2B). By 28 hpf, when the parapineal initiates migration, most embryos have higher levels of fgf8 expression on the left (Figure 2D, Table S1). Later expression is restricted to the anterior and medial part of the habenulae (Figures 2E, S1B, and S1C). To better understand which cells are able to respond to Fgf8 signals, we analyzed the expression of the four known Fgf receptors (FgfRs) in this region. Although all four FgfRs are widely expressed in the brain (data not shown), fgfr4 (Thisse et al., 1995) shows elevated levels of expression in parapineal cells (Figure 2F) as does etv5 (Figure S1D), an Ets family gene likely to be a target of the Fgf pathway (Roussigne and Blader, 2006). These results suggest that parapineal cells are able to respond to Fgf signals during their unilateral migration. To determine when Fgf signaling is required for parapineal cells to migrate, we abrogated Fgf signaling in a temporally controlled manner, using the SU5402 drug (Mohammadi et al., 1997).
Figure 2. Temporally Controlled Abrogation of Fgf Signaling Identifies a Critical Window for Fgf-Dependent Parapineal Migration.
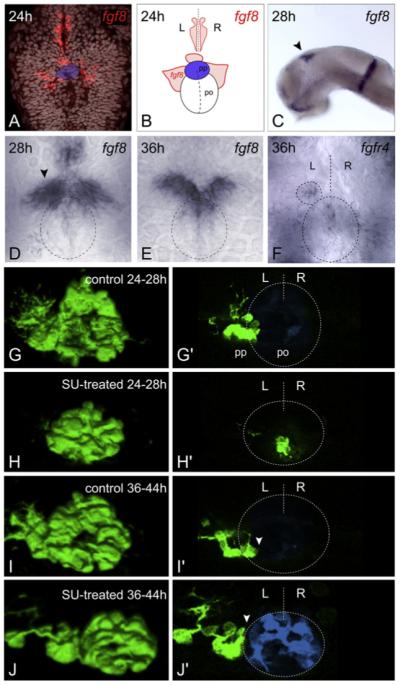
(A and C-E) Expression of fgf8 in the epithalamus at 24 hpf ([A], dorsal view, TOPRO3 nuclear marker in gray, parapineal primordium highlighted in blue), 28 hpf ([C], lateral, black arrowhead; [D], dorsal, black arrowhead denotes stronger left-sided expression), and 36 hpf ([E], dorsal). Dashed ellipse indicates position of pineal nucleus (D and E). (B) Schematic depicting pineal organ (po), parapineal nucleus (pp, blue), fgf8 expression domains (red), and midline (dashed line), as visualized in (A). (F) fgfR4 expression at 36 hpf (dorsal). Dashed lines indicate position of pineal nucleus (large ellipse), parapineal nucleus (small ellipse), and midline (straight line). (G-J) 3D reconstructions and (G′-J′) single z-slices of dorsal views of the epithalamus in control- (G, G′, I, and I′) and SU5402- (H, H′, J, and J′) treated Tg(foxD3:GFP) 4 dpf embryos, with anterior to the top. Pineal cells have been pseudocolored in blue in single z-slices and dashed lines indicate position of pineal nucleus and midline (G′-J′). SU5402 treatment at 24–28 hpf completely abolished the initial leftward migration of the parapineal to lateral and dorsal positions (H, and H′). Treatment at 36–44 hpf abrogated later translocation of the parapineal to ventral and medial locations relative to the pineal nucleus, and parapineal cells remain at dorso-lateral positions adjacent to the pineal ([J and J′], white arrowheads). L, left; R, right.
Blocking Fgf signaling in the period immediately preceding migration disrupted parapineal migration, with most cells remaining at the midline (Figures 2G-2H′). Parapineal nuclei were often disaggregated such that a few cells were scattered to the left or right of the midline, but these never migrated laterally as a cohesive group and remained ventral to the pineal nucleus. This largely phenocopies the parapineal migration defect of ace mutants, although the extent of disaggregation of the parapineal primordium was more severe following SU5402 treatment (data not shown). In addition, blocking signaling during the migratory phase led to arrest of parapineal migration, suggesting a continuous requirement for Fgf activity (Figures 2I-2J′). However, if SU5402 was applied to embryos earlier in development and was then washed out before migration started, parapineal migration proceeded as normal and epithalamic asymmetry was undisturbed (data not shown). These data support the idea that parapineal cells require Fgf signaling to initiate and maintain their migration.
Locally Applied Exogenous Fgf8 Rescues Leftward Parapineal Migration
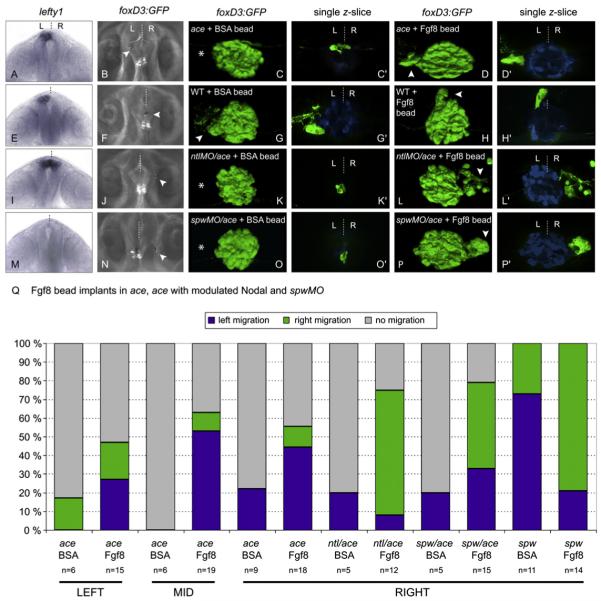
To test the hypothesis that it is the local activity of Fgf8 that is required for the parapineal primordium to move leftward from its initially symmetric location, we provided ace embryos with a focal source of exogenous Fgf8 protein. This was achieved by implantation of Fgf8-loaded microbeads rostral to the pineal complex at 26 ss-24 hpf. Beads were implanted either at, or to the left or right of, the midline (Figures 3B, 3F, 3J, and 3N; Table S2).
Figure 3. Local Provision of Fgf8 Restores Parapineal Migration in ace Embryos and Directs Laterality of Migration in the Absence of a Nodal Signaling Bias.
(A, E, I, and M) Frontal views of lefty1 expression (as a marker of Nodal signaling) in ace (A), wild-type (E), ntlMO/ace (I), and spawMO/ace (M) embryos at 20 hpf, with dorsal to the top. (B, F, J, and N) dorsal views of live brains showing Fgf8-soaked beads implanted rostrally and to the left (B), right (J and N), and midline (F) of the pineal complex visualized by Tg(foxD3:GFP) expression, in ace (B), wild-type (F), ntlMO/ace (J), and spawMO/ace (N) embryos. Anterior is to the top. (C, C′, D, D′, G, G′, H, H′, K, K′, L, L′, O, O′, P, and P′) 3D reconstructions and single z-slices of dorsal views of the epithalamus in ace (C-D′), wild-type (G-H′), ntlMO/ace 1(K-L′), and spawMO/ace (O-P′) Tg(foxD3:GFP) embryos at 3 dpf implanted with BSA- (C, C′, G, G′, K, K′, O, and O′) or Fgf8- (D, D′, H, H′, L, L′, P, and P′) loaded beads. Pineal/parapineal complex (green) is visualized as before. Pineal cells have been pseudocolored in blue in single z-slices. Anterior is to the top. (Q) Graph representing proportions of embryos with right (green), left (blue), or static (gray) parapineal nuclei after epithalamic implantation of BSA- and Fgf8-loaded beads in ace embryos, ace embryos with modulated Nodal, and spawMO embryos. Results are grouped according to position (left, middle, or right) of bead implantation.
Exogenous Fgf8 efficiently restored lateralized parapineal migration in ace mutants such that by 3 dpf, 56% of ace embryos with an implanted Fgf8 bead showed a migrated parapineal nucleus (Figures 3D and 3D′; **p = 0.0015 for ace + Fgf8 where 29/52 migrated, versus ace + BSA where 3/18 migrated; see Statistics in Experimental Procedures), whereas BSA-soaked beads had no effect on parapineal migration (Figures 3C and 3C′; Table S2). In contrast, habenular development was not obviously restored (Figures S2A-S2D), suggesting that the migration defect in ace mutants is not due to the absence of a suitable substrate for navigation on the habenular nuclei. This result also suggests that persistent Fgf8 is required in both left and right habenulae and that although the parapineal influences habenular development (Concha et al., 2003; Gamse et al., 2003), it cannot compensate for a loss of Fgf8. Surprisingly, in these experiments, 76% of the migrated parapineal nuclei were positioned on the left, irrespective of the location at which the bead was placed (Figure 3Q). This suggests that additional signals influence the direction of migration once movement has been initiated by Fgf8. An obvious candidate is the Nodal signaling pathway because it is known to determine the directional laterality of epithalamic asymmetries (Concha et al., 2000). Therefore, we next assessed whether left-sided Nodal signaling was intact in ace mutants.
Analysis of Nodal pathway gene expression in ace mutants revealed that the pathway is still activated unilaterally on the left side of the epithalamus (pitx2, 90% left epithalamic expression, n = 21; Figures S3C and S3D) and body axis (southpaw [spaw], 97% left lateral plate mesoderm expression, n = 73; Figures S3A and S3B) in the majority of ace embryos. Thus, the leftward migration bias of ace mutant parapineal nuclei provided with exogenous Fgf8 is potentially due to left-sided Nodal signaling.
Exogenous Fgf8 Can Specify the Direction of Parapineal Migration in the Absence of Biased Nodal Signaling
To assess whether exogenous Fgf8 can instruct laterality in ace embryos that lack biased Nodal signaling, we generated conditions of symmetric Nodal signaling. Embryos with bilateral and absent Nodal signaling were obtained using notail (ntl) and spaw morpholinos (Concha et al., 2000; Long et al., 2003), respectively (Figures 3I and 3M). In spaw and ntl morphants, parapineal migration occurs normally but with randomized directionality (Concha et al., 2000; Gamse et al., 2005).
To determine whether Fgf8 signaling is likely to be activated in the absence of epithalamic Nodal signaling, we assessed fgf8 expression in spaw morphants at 22 ss and 28 hpf. At both stages, expression levels were similar to those of wild-type, but the subtle L/R differences were usually abolished such that fgf8 was expressed symmetrically across the midline (Table S1). We saw the same loss of asymmetry in late zygotic oep (LZoep) mutant embryos at 28 hpf (Figures S3E and S3F), which also lack epithalamic Nodal signals (Yan et al., 1999). This indicates that Nodal signaling is not required for fgf8 expression but is responsible for the subtle L/R asymmetries in fgf8 expression observed in wild-type embryos.
In embryos where Nodal signaling is either bilaterally symmetric or absent, we find that exogenous Fgf8 is sufficient to direct migration of the parapineal primordium. In ntlMO/ace embryos in which Fgf8-loaded beads were transplanted rostral and to the right of the parapineal, 8 of 9 parapineal nuclei that migrated (9/12 showed migration) did so toward the position of the bead (Figures 3I-3L′ and 3Q). Similarly, in spawMO/ace embryos, rescued parapineal migration was usually toward the bead (n = 7/10 from 15 cases; Figures 3M-3P′ and 3Q). The most parsimonious explanation of these results is that Fgf8 can break initial symmetry by inducing parapineal migration and, in addition, has the potential to influence the laterality of the asymmetry by acting as an attractive signal for migrating cells. To further test this hypothesis, we performed related experiments to assess whether exogenous Fgf8 can influence migration in wild-type embryos, where parapineal cells are able to follow their normal migratory pathways.
In support of the idea that Fgf8 can direct migrating parapineal cells, we found that an exogenous source of Fgf8 rostral to the parapineal primordium of wild-type embryos could, in some cases, direct parapineal cells away from their normal leftward trajectory toward the bead (*p = 0.0111 for wild-type + Fgf8 where 7/33 migrated ectopically, versus wild-type + BSA where 0/31 migrated ectopically; Figures 3E-3H′). Furthermore, exogenous Fgf8 can bias the direction of parapineal migration in spawMO embryos in which the influence of the Nodal pathway is removed but endogenous Fgf8 signaling remains intact. We implanted Fgf8 beads in the right side of the epithalamus of spawMO embryos at 20–22 ss, a stage we considered early enough to ensure that endogenous Fgf8 signaling had not yet committed parapineal cells to migration to either the left or the right. Almost all embryos implanted with a right-sided Fgf8 bead had a right-sided parapineal nucleus (*p = 0.0172 for Fgf8 bead where 11/14 had a right parapineal, versus BSA bead where 3/11 had a right parapineal; Figure 3Q; Table S2). Together, these results strongly support the idea that Fgf8 signaling is indispensable for the initial symmetry break in the epithalamus, and that it can additionally influence the direction of asymmetries if there is no bias conferred by Nodal signaling.
DISCUSSION
Although recent studies have elucidated the signals required for consistent lateralization of the epithalamus in zebrafish (Bianco et al., 2008; Carl et al., 2007; Concha et al., 2000, 2003; Gamse et al., 2003; Inbal et al., 2007; Long et al., 2003), nothing was known about the mechanisms involved in generating asymmetry. The observation that each side of the epithalamus is competent to produce either a “left character” or “right character” laterality state led us to speculate that any signaling pathways required for breaking symmetry in the brain could potentially be activated in a bilateral manner.
We have demonstrated that Fgf8, expressed bilaterally in habenular precursors, is required for the asymmetric migration of parapineal cells. Accordingly, the fgf8 mutant ace and embryos in which Fgf signaling is blocked pharmacologically never initiate parapineal migration and the epithalamus remains symmetric. We were able to effectively rescue parapineal migration in ace mutants by the provision of exogenous Fgf8, and additionally, in ace mutants with no L/R bias in Nodal signaling, exogenous Fgf8 was able to direct the laterality of parapineal migration.
These results suggest Fgf8 signals could be chemotactic for parapineal cells, and/or that exogenous Fgf8 could establish a permissive “pathway,” allowing motility of parapineal cells within areas close to the Fgf8 source. Although motility and directionality are difficult processes to separate in vivo, some of our results lend support to the hypothesis that Fgf8 is chemotactic to parapineal cells. First, in wild-type embryos where endogenous Fgf8 and Nodal signals are intact, an exogenous source of Fgf8 is able to direct parapineal cells away from their usual migratory trajectory. Second, in embryos with no epithalamic Nodal signaling, increasing the levels of Fgf8 on one side positively influences the laterality of migration. Neither result would be expected if Fgf8 acts solely to establish a permissive pathway for parapineal migration. Fgf signaling has been repeatedly implicated in chemotactic migration of many cell types during development, via mechanisms including induction of cytoneme-like filopodia and competition for lead cell position based on levels of FgfR activity (Ghabrial and Krasnow, 2006; Sato and Kornberg, 2002). A future goal will be to analyze the cellular response of the parapineal cells to the reception of Fgf signals.
Two recent studies have shown Fgf signals to be required for organization and migration of the lateral line primordium (Lecaudey et al., 2008; Nechiporuk and Raible, 2008), a structure that bears some similarity in terms of its organization to the parapineal primordium. In the case of the lateral line primordium, abrogation of Fgf signaling prevents cells from coalescing into the rosette-like structures that constitute nascent neuromasts, and this eventually leads to stalled migration. It is currently not known how Fgf signaling mediates this cohesion and adhesivity, although one possibility is that it may influence epithelialization and consequently the apical junctional complexes that form between polarized epithelial cells (Lecaudey et al., 2008). In ace mutants, we sometimes see disaggregation of a few parapineal cells, but in most cases migration is stalled despite the parapineal forming a coherent and cohesive cluster of cells. Thus in the ace mutants, compromised cohesivity is unlikely to underlie the failure in migration. However, it is intriguing that when Fgf signaling is more severely compromised pharmacologically, we see a greater degree of disaggregation, suggesting that there may be similarities in phenotype between the parapineal and lateral line primordia in conditions where all Fgf signaling is blocked.
We propose that the parapineal acts as a bistable “switch,” whereby once movement is initiated, the midline is an “unstable” location and consequently the parapineal inevitably migrates to more “stable” locations on the left or right, promoting asymmetric development of the adjacent habenula. Constraints inherent in the system ensure that it produces only one outcome (left OR right). For instance, cohesivity coupled with motility of the parapineal nucleus may mean that it can only make one directional choice in response to bilateral signals. During normal development, Nodal signaling biases the laterality choice to the left (Figure 4A). In the absence of Fgf8 signaling, the presence of the left-sided Nodal signal is not sufficient to break anatomical symmetry (Figure 4C). Conversely, in the absence of a Nodal bias, asymmetry still develops dependent on the activity of Fgf8 inducing the migration of parapineal cells (Figure 4B). In such situations laterality is determined by a stochastic mechanism, and we speculate that this could be differences in Fgf8 levels between left and right. In support of this, L/R differences in Fgf activity imposed by exogenous Fgf8 are sufficient to direct the laterality of asymmetries in the absence of unilateral Nodal signaling.
Figure 4. The Role of Fgf8 Signaling in the Generation of Neuroanatomical Asymmetry.
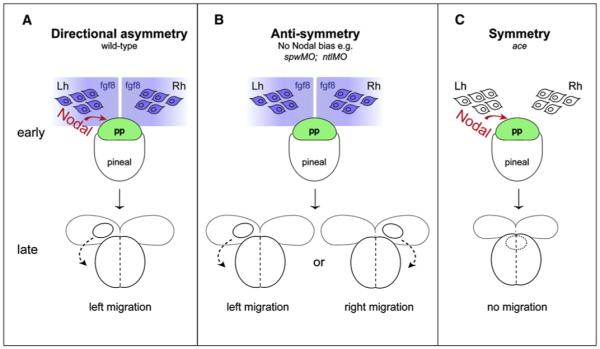
(A) In wild-type embryos: bilateral fgf8 signals from the habenulae (blue) induce migration of the parapineal primordium (green), making the midline an unstable location; Nodal signals (red) ensure a leftward “choice” is made. The parapineal migrates from the “unstable” midline to a “stable” location to the left of the pineal. (B) In embryos lacking Nodal signaling: symmetry is broken by Fgf8 inducing migration; stochastic factors (possibly L/R differences in Fgf8 levels) determine the laterality of migration. Again the parapineal migrates away from the unstable midline location to more stable positions on left or right. (C) In ace mutant embryos: asymmetric Nodal signaling is not sufficient to break anatomical symmetry and the parapineal remains at the midline, unable to migrate in either direction. pp, parapineal; Lh, left habenula; Rh, right habenula; early, 24 hpf; late, 30 hpf.
The relationship between the Fgf and Nodal pathways is not yet fully resolved, but we have found that subtle asymmetries in fgf8 expression are lost in conditions where lateralized Nodal signaling is absent. Could a subtle influence on fgf8 transcription be the primary action of Nodal signaling in biasing parapineal migration to the left? In wild-type embryos, such a mechanism seems reasonable because once parapineal cells initiate migration, the midline may become an unstable location and so a small, transient, and/or localized bias between left and right might be enough to tip the balance in favor of migration in one or other direction. However, such a mechanism cannot easily explain why migration is still usually to the left in ace embryos where the exogenous source of Fgf8 is on the right. This result implies that Nodal can act downstream of the Fgf8 ligand, perhaps facilitating an aspect of migration or morphogenesis that can occur without Nodal but would do so less efficiently.
Our studies support the idea that the evolution of directional asymmetry from a symmetric ancestral structure is likely to proceed in two steps (Palmer, 2004): the first induces asymmetry without a directional bias (antisymmetry) and the second biases this asymmetry in one direction. Loss of the pathway governing the second step should lead to antisymmetry (as is the case with loss of Nodal in the brain). This raises the possibility that the antisymmetry induced by bilateral Fgf8 signaling represents a more ancient mechanism for generating brain asymmetry. Left-sided Nodal signaling regulates visceral asymmetries (Schier, 2003) and so we speculate that the unilateral activation of the Nodal pathway was co-opted from the body axis to provide consistent laterality to brain asymmetry, thereby leading to evolutionary acquisition of a global mechanism for coordinating laterality in the whole embryo. Studies in the worm have demonstrated that an early body axis asymmetry is used to “tip the balance” of a later, bistable asymmetry-generating mechanism in paired neurons (Poole and Hobert, 2006), suggesting that such strategies may be commonly employed.
A mechanism that amplifies stochastic differences between left and right sides of the brain would be sufficient to produce antisymmetry (Cooke, 2004). The mechanism we have described fits this criterion: the cohesive parapineal nucleus is like the prize in a tug-of-war between the habenulae. It is inevitable that it is “pulled” one way or the other, and in doing so breaks the initial symmetry and initiates events that lead to the eventual establishment of lateralized circuitry in the brain.
EXPERIMENTAL PROCEDURES
Zebrafish Lines
Embryos were obtained by natural spawning from wild-type (*AB/Tu), aceti282 (Reifers et al., 1998), Tg(foxD3:GFP) (Gilmour et al., 2002), and Tg(ET11:GFP) (Choo et al., 2006) fish. LZoep embryos were generated from MZoep mutants as described (Yan et al., 1999) by injection of oep RNA at the 1-cell stage. All embryos were reared and staged according to standard procedures (Westerfield, 2000). Temperature shifts from 28°C to 25°C at tailbud stage were performed to obtain late somite stage embryos for bead implantation. Lower temperature shifts were never used because these can result in perturbations of laterality (J.C.R., unpublished data). Occasionally 0.002% phenylthiourea was added to fish water from 12 hpf to prevent pigment formation.
Morpholino Antisense Oligonucleotides
spaw morpholino oligonucleotide (spawMO; spaw-MO1; Long et al., 2003) and ntl morpholino oligonucleotide (ntlMO; Feldman and Stemple, 2001) were injected as described. Efficacy of spawMO and ntlMO was confirmed by phenotype and analysis of pitx2 (Bisgrove et al., 1999) expression in the epithalamus (absent in spaw morphants and bilateral in ntl morphants).
Bead Implantation
Fgf8- and BSA-loaded beads were prepared as previously described (Maves et al., 2002) with the exception that 15 μm polystyrene beads (Polysciences) were used. Embryos were encased in 2% agarose and beads were implanted rostral to the pineal complex at 22–24 hpf or 19–20 hpf using a tungsten needle. Embryos were selected on the basis of implantation accuracy with respect to Tg(foxD3:GFP) expression and bead position was recorded. After 1 hr recovery, embryos were released into fish water inoculated with penicillin and streptomycin. Parapineal position was assessed at 2 dpf by live compound imaging and at 3 dpf by confocal microscopy after fixation and immunohistochemistry. Parapineal cells were identified by their ventral and/or lateral location with respect to the pineal nucleus and their stereotypical axonal projections (Concha et al., 2003). Tg(foxD3:GFP)-positive parapineal cells identified in this way corresponded with gfi expression (Dufourcq et al., 2004) in 100% of embryos (n = 6). Efficacy of bead implantation was assessed by analysis of erm expression, an Fgf-target gene (Münchberg et al., 1999), in BSA- and Fgf8-implanted embryos: 0/4 of wild-type and 0/4 of ace embryos implanted with a BSA-loaded bead, and 5/5 of ace and 7/8 of wild-type embryos implanted with an Fgf8-loaded bead showed a ring of erm expression around the bead (Figures S2E-S2H and data not shown). Implantation of Fgf8-loaded beads did not reactivate Nodal signaling in the epithalamus: 0/5 of wild-type embryos implanted with a BSA-loaded bead, and 0/6 of wild-type embryos implanted with an Fgf8-loaded bead at 26 ss, showed pitx2 expression in the epithalamus at 26 hpf (Figures S2I and S2J).
Blocking FgfR Activity
Embryos were treated with the drug SU5402 (Mohammadi et al., 1997) according to standard protocols (Shanmugalingam et al., 2000).
Immunohistochemistry
In situ hybridization and antibody staining were performed as previously described (Macdonald et al., 1994). brn3a (Aizawa et al., 2005), erm (Münchberg et al., 1999), fgf8 (Reifers et al., 1998), fgfR4 (Thisse et al., 1995), gfi (Dufourcq et al., 2004), leftover (kctd12.1) (Gamse et al., 2003), lefty1 (Bisgrove et al., 1999), pitx2 (Bisgrove et al., 1999), and spaw (Long et al., 2003) probes were generated using standard procedures (Macdonald et al., 1994). Embryos were stained using BM Purple (Roche) or BCIP and NBT (Roche) as chromogen. For antibody staining, mouse anti-acetylated tubulin (Sigma, T6793) and rabbit anti-GFP (Torrey Pines Biolabs, TP401) were used at 1:1000 dilutions in blocking buffer (1 × PBS + 0.8% Triton-X + 10% goat serum + 1% DMSO). For nuclear staining, embryos were incubated in 1 × PBS + 0.1% Triton-X + 1% bovine serum albumin containing ToPro (1:1000, Molecular Probes).
Microscopy and Image Manipulation
Fluorescent labeling was imaged by confocal microscopy (Leica SP2) using x25 oil immersion and x40 water-immersion objective lenses. z-stacks were typically acquired at 1–2 μm intervals. 3D projections were generated from the stack of images using Volocity (Improvision) software. Live, bead-implanted transgenic embryos were imaged under x20 water-immersion DIC optics (Axioskop 2 FS microscope, Carl Zeiss). In situ hybridization stainings were photographed using a Jentopix C14 digital camera attached to a Nikon Eclipse E1000 compound microscope. For presentation, image manipulation was performed using Photoshop CS2 (Adobe) software. Parapineal cells have been highlighted in single z-slices in Figures 2 and 3 by pseudocoloring of adjacent pineal cells, which were selected by hand.
Statistics
Statistics were performed using InStat software (Graphpad). Categorical data was analyzed using Fisher’s Exact test, where the p value is two tailed. Confidence is denoted by *p < 0.05 and **p < 0.01.
ACKNOWLEDGMENTS
We thank members of our lab for discussions, reagents, and resources; Daniel Ciantar for confocal help; Carole Wilson and her team for fish care; Vladimir Korzh and Tom Becker for fish lines; Patrick Blader for sharing results and reagents prior to publication; and Adi Inbal, Lila Solnica-Krezel, and other members of the community for reagents and resources. The study was supported by Wellcome Trust; EU and BBSRC grants to S.W.W.; a FEBS fellowship to M.R.; and HHMI, ICM (CHILE), PBCT (Chile), and EU grants to M.L.C.
Footnotes
SUPPLEMENTAL DATA
The supplemental data for this article include three supplemental Figures and two supplemental Tables and can be found at http://www.neuron.org/supplemental/S0896-6273(08)01052-0.
REFERENCES
- Aizawa H, Bianco IH, Hamaoka T, Miyashita T, Uemura O, Concha ML, Russell C, Wilson SW, Okamoto H. Laterotopic representation of left-right information onto the dorso-ventral axis of a zebrafish midbrain target nucleus. Curr. Biol. 2005;15:238–243. doi: 10.1016/j.cub.2005.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco IH, Carl M, Russell C, Clarke JD, Wilson SW. Brain asymmetry is encoded at the level of axon terminal morphology. Neural Develop. 2008;3:9. doi: 10.1186/1749-8104-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco IH, Wilson SW. The habenular nuclei: a conserved asymmetric relay station in the vertebrate brain. Philos. Trans. R Soc. Lond. B Biol. Sci. 2009 doi: 10.1098/rstb.2008.0213. in press. Published online Decemeber 4, 2008. 10.1098/rstb.2008.0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove BW, Essner JJ, Yost HJ. Regulation of midline development by antagonism of lefty and nodal signaling. Development. 1999;126:3253–3262. doi: 10.1242/dev.126.14.3253. [DOI] [PubMed] [Google Scholar]
- Carl M, Bianco IH, Bajoghli B, Aghaallaei N, Czerny T, Wilson SW. Wnt/Axin1/b-catenin signaling regulates asymmetric nodal activation, elaboration, and concordance of CNS asymmetries. Neuron. 2007;55:393–405. doi: 10.1016/j.neuron.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo BG, Kondrichin I, Parinov S, Emelyanov A, Go W, Toh WC, Korzh V. Zebrafish transgenic Enhancer TRAP line database (ZETRAP) BMC Dev. Biol. 2006;6:5. doi: 10.1186/1471-213X-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha ML. The dorsal diencephalic conduction system of zebrafish as a model of vertebrate brain lateralisation. Neuroreport. 2004;15:1843–1846. doi: 10.1097/00001756-200408260-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha ML, Wilson SW. Asymmetry in the epithalamus of vertebrates. J. Anat. 2001;199:63–84. doi: 10.1046/j.1469-7580.2001.19910063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha ML, Burdine RD, Russell C, Schier AF, Wilson SW. A nodal signalling pathway regulates the laterality of neuroanatomical asymmetries in the zebrafish forebrain. Neuron. 2000;28:399–409. doi: 10.1016/s0896-6273(00)00120-3. [DOI] [PubMed] [Google Scholar]
- Concha ML, Russell C, Regan JC, Tawk M, Sidi S, Gilmour DT, Kapsimali M, Sumoy L, Goldstone K, Amaya E, et al. Local tissue interactions across the dorsal midline of the forebrain establish CNS laterality. Neuron. 2003;39:423–438. doi: 10.1016/s0896-6273(03)00437-9. [DOI] [PubMed] [Google Scholar]
- Cooke J. Developmental mechanism and evolutionary origin of vertebrate left/right asymmetries. Biol. Rev. Camb. Philos. Soc. 2004;79:377–407. doi: 10.1017/s1464793103006298. [DOI] [PubMed] [Google Scholar]
- Draper BW, Morcos PA, Kimmel CB. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: a quantifiable method for gene knockdown. Genesis. 2001;30:154–156. doi: 10.1002/gene.1053. [DOI] [PubMed] [Google Scholar]
- Dufourcq P, Rastegar S, Strähle U, Blader P. Parapineal specific expression of gfi1 in the zebrafish epithalamus. Gene Expr. Patterns. 2004;4:53–57. doi: 10.1016/s1567-133x(03)00148-0. [DOI] [PubMed] [Google Scholar]
- Escalante-Mead PR, Minshew NJ, Sweeney JA. Abnormal brain lateralization in high-functioning autism. J. Autism Dev. Disord. 2003;33:539–543. doi: 10.1023/a:1025887713788. [DOI] [PubMed] [Google Scholar]
- Feldman B, Stemple DL. Morpholino phenocopies of sqt, oep, and ntl mutations. Genesis. 2001;30:175–177. doi: 10.1002/gene.1058. [DOI] [PubMed] [Google Scholar]
- Gamse JT, Thisse C, Thisse B, Halpern ME. The parapineal mediates left-right asymmetry in the zebrafish diencephalon. Development. 2003;130:1059–1068. doi: 10.1242/dev.00270. [DOI] [PubMed] [Google Scholar]
- Gamse JT, Kuan YS, Macurak M, Brösamle C, Thisse B, Thisse C, Halpern ME. Directional asymmetry of the zebrafish epithalamus guides dorsoventral innervation of the midbrain target. Development. 2005;132:4869–4881. doi: 10.1242/dev.02046. [DOI] [PubMed] [Google Scholar]
- Ghabrial AS, Krasnow MA. Social interactions among epithelial cells during tracheal branching morphogenesis. Nature. 2006;441:746–749. doi: 10.1038/nature04829. [DOI] [PubMed] [Google Scholar]
- Gilmour DT, Maischein HM, Nüsslein-Volhard C. Migration and function of a glial subtype in the vertebrate peripheral nervous system. Neuron. 2002;34:577–588. doi: 10.1016/s0896-6273(02)00683-9. [DOI] [PubMed] [Google Scholar]
- Inbal A, Kim SH, Shin J, Solnica-Krezel L. Six3 represses nodal activity to establish early brain asymmetry in zebrafish. Neuron. 2007;55:407–415. doi: 10.1016/j.neuron.2007.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecaudey V, Cakan-Akdogan G, Norton WH, Gilmour D. Dynamic Fgf signaling couples morphogenesis and migration in the zebrafish lateral line primordium. Development. 2008;135:2695–2705. doi: 10.1242/dev.025981. [DOI] [PubMed] [Google Scholar]
- Li X, Branch CA, Ardekani BA, Bertisch H, Hicks C, DeLisi LE. fMRI study of language activation in schizophrenia, schizoaffective disorder and in individuals genetically at high risk. Schizophr. Res. 2007;96:14–24. doi: 10.1016/j.schres.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S, Ahmad N, Rebagliati M. The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development. 2003;130:2303–2316. doi: 10.1242/dev.00436. [DOI] [PubMed] [Google Scholar]
- Macdonald R, Xu Q, Barth KA, Mikkola I, Holder N, Fjose A, Krauss S, Wilson SW. Regulatory gene expression boundaries demarcate sites of neuronal differentiation in the embryonic zebrafish forebrain. Neuron. 1994;13:1039–1053. doi: 10.1016/0896-6273(94)90044-2. [DOI] [PubMed] [Google Scholar]
- Maves L, Jackman W, Kimmel CB. FGF3 and FGF8 mediate a rhombomere 4 signaling activity in the zebrafish hindbrain. Development. 2002;129:3825–3837. doi: 10.1242/dev.129.16.3825. [DOI] [PubMed] [Google Scholar]
- Mohammadi M, McMahon G, Sun L, Tang C, Hirth P, Yeh BK, Hubbard SR, Schlessinger J. Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science. 1997;276:955–960. doi: 10.1126/science.276.5314.955. [DOI] [PubMed] [Google Scholar]
- Münchberg SR, Ober ER, Steinbeisser H. Expression of the Ets transcription factors erm and pea3 in early zebrafish development. Mech. Dev. 1999;88:233–236. doi: 10.1016/s0925-4773(99)00179-3. [DOI] [PubMed] [Google Scholar]
- Nechiporuk A, Raible DW. FGF-dependent mechanosensory organ patterning in zebrafish. Science. 2008;320:1774–1777. doi: 10.1126/science.1156547. [DOI] [PubMed] [Google Scholar]
- Palmer AR. Symmetry Breaking and the Evolution of Development. Science. 2004;306:828–833. doi: 10.1126/science.1103707. [DOI] [PubMed] [Google Scholar]
- Poole RJ, Hobert O. Early embryonic programming of neuronal left/right asymmetry in C.elegans. Curr. Biol. 2006;16:2279–2292. doi: 10.1016/j.cub.2006.09.041. [DOI] [PubMed] [Google Scholar]
- Reifers F, Böhli H, Walsh EC, Crossley PH, Stainier DY, Brand M. Fgf8 is mutated in zebrafish acerebellar mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis. Development. 1998;125:2381–2395. doi: 10.1242/dev.125.13.2381. [DOI] [PubMed] [Google Scholar]
- Roussigne M, Blader P. Divergence in regulation of the PEA3 family of ETS transcription factors. Gene Expr. Patterns. 2006;6:777–782. doi: 10.1016/j.modgep.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Sato M, Kornberg TB. FGF is an essential mitogen and chemoattractant for the air sacs of the Drosophila tracheal system. Dev. Cell. 2002;3:195–207. doi: 10.1016/s1534-5807(02)00202-2. [DOI] [PubMed] [Google Scholar]
- Signore I, Guerrero N, Colombo A, Loosli F, Villalon A, Wittbrodt J, Concha M. Zebrafish and medaka: model organisms for a comparative developmental approach of brain asymmetry. Philos. Trans. R Soc. Lond. B Biol. Sci. 2009 doi: 10.1098/rstb.2008.0260. in press. Published online December 4, 2008. 10.1098/rstb.2008.0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier AF. Nodal signaling in vertebrate development. Annu. Rev. Cell Dev. Biol. 2003;19:589–621. doi: 10.1146/annurev.cellbio.19.041603.094522. [DOI] [PubMed] [Google Scholar]
- Shanmugalingam S, Houart C, Picker A, Reifers F, Macdonald R, Barth A, Griffin K, Brand M, Wilson SW. Ace/Fgf8 is required for forebrain commissure formation and patterning of the telencephalon. Development. 2000;127:2549–2561. doi: 10.1242/dev.127.12.2549. [DOI] [PubMed] [Google Scholar]
- Thisse B, Thisse C, Weston JA. Novel FGF receptor (Z-FGFR4) is dynamically expressed in mesoderm and neurectoderm during early zebrafish embryogenesis. Dev. Dyn. 1995;203:377–391. doi: 10.1002/aja.1002030309. [DOI] [PubMed] [Google Scholar]
- Toth C, Rajput M, Rajput AH. Anomalies of asymmetry of clinical signs in parkinsonism. Mov. Disord. 2004;19:151–157. doi: 10.1002/mds.10685. [DOI] [PubMed] [Google Scholar]
- Vallortigara G, Rogers LJ. Survival with an asymmetrical brain. Behav. Brain Sci. 2005;28:575–633. doi: 10.1017/S0140525X05000105. [DOI] [PubMed] [Google Scholar]
- Westerfield M. A Guide for the Laboratory Use of Zebrafish (Danio rerio) Fourth Edition Univ. of Oregon Press; Eugene: 2000. The Zebrafish Book. [Google Scholar]
- Yan YT, Gritsman K, Ding J, Burdine RD, Corrales JD, Price SM, Talbot WS, Schier AF, Shen MM. Conserved requirement for EGF-CFC genes in vertebrate left-right axis formation. Genes Dev. 1999;13:2527–2537. doi: 10.1101/gad.13.19.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]