Summary
We characterized two sucrose-metabolizing systems – sus and scr – and describe their roles in the physiology and virulence of Streptococcus pneumoniae in murine models of carriage and pneumonia. The sus and scr systems are regulated by LacI family repressors SusR and ScrR respectively. SusR regulates an adjacent ABC transporter (susT1/susT2/susX) and sucrose-6-phosphate (S-6-P) hydrolase (susH). ScrR controls an adjacent PTS transporter (scrT), fructokinase (scrK) and second S-6-P hydrolase (scrH). sus and scr play niche-specific roles in virulence. The susH and sus locus mutants are attenuated in the lung, but dispensable in nasopharyngeal carriage. Conversely, the scrH and scr locus mutants, while dispensable in the lung, are attenuated for nasopharyngeal colonization. The scrH/susH double mutant is more attenuated than scrH in the nasopharynx, indicating SusH can substitute in this niche. Both systems are sucrose-inducible, with ScrH being the major in vitro hydrolase. The scrH/susH mutant does not grow on sucrose indicating that sus and scr are the only sucrose-metabolizing systems in S. pneumoniae. We propose a model describing hierarchical regulation of the scr and sus systems by the putative inducer, S-6-P. The transport and metabolism of sucrose or a related disaccharide thus contributes to S. pneumoniae colonization and disease.
Introduction
Carbon acquisition and metabolism are central to life. The regulation of these processes in bacteria and eukaryotes is complex and necessarily involves timely and appropriate responses to changes in nutrient availability to maintain optimal cellular growth rates (Titgemeyer and Hillen, 2002). Adequate control over processes involved in nutrient acquisition and metabolism are likely to be central to host niche dominance and fitness of microbial pathogens during carriage and infection. The role of carbon metabolism and its relevance in determining in vivo fitness, while intuitively important, has only recently begun to be appreciated (Smith, 2000; Chico-Calero et al., 2002; Munoz-Elias and McKinney, 2006). Recently studied examples include carbon metabolic pathways such as gluconeogenesis and the TCA cycle (Tchawa Yimga et al., 2006), the Entner–Doudoroff pathway (Sweeney et al., 1996a,b; Peekhaus and Conway, 1998), and specific carbon utilization genes (Chang et al., 2004) in the enterics, Escherichia coli and Salmonella sp. In addition to sugars, metabolism of fatty acids has also recently been shown to be required for the virulence and persistence of the lung pathogen, Mycobacterium tuberculosis (McKinney et al., 2000; Munoz-Elias and McKinney, 2005) and has revealed novel and unexpected bacterial enzyme targets for future drug therapies.
Streptococcus pneumoniae is a Gram-positive encapsulated bacterium that is the leading cause of bacterial meningitis, pneumonia and otitis media (McCullers and Tuomanen, 2001; Paton and Giammarinaro, 2001; Hava et al., 2003a; Bogaert et al., 2004). While the study of S. pneumoniae pathogenesis has focused on factors involved in host attachment, damage and immune evasion (Gosink et al., 2000; Hava et al., 2003b; Johnston et al., 2004; Orihuela et al., 2004; Shaper et al., 2004; Barocchi et al., 2006), scant information is available on metabolic pathways and specific carbon sources used during colonization and infection within the host. Signature-tagged mutagenesis screens designed to identify genes with roles in colonization and infection have revealed mutations in several systems involved in carbon metabolism. Many of these insertions localized to carbohydrate uptake and catabolic systems, including both ATPase-Binding Cassette (ABC) transporters and Phosphotransferase Systems (PTS) (Polissi et al., 1998; Lau et al., 2001; Hava and Camilli, 2002) underscoring the potential involvement of these genes in in vivo fitness.
The LacI/GalR family are well-studied transcriptional repressors that typically mediate inducible regulation of operons involved in carbon metabolism (Weickert and Adhya, 1992). One example of this family, CcpA (Catabolite control protein A), first discovered as a repressor of the Bacillus subtilis amylase operon (Henkin et al., 1991), has since been shown to be vital to the co-ordinated expression of a variety of carbon utilization operons, including the processes of carbon catabolite repression and activation in Gram-positive bacteria (Luesink et al., 1998; Stulke and Hillen, 2000; Kim et al., 2002; Viana et al., 2005) including streptococci (Simpson and Russell, 1998; Rogers and Scannapieco, 2001; Asanuma and Hino, 2006). In previous studies, it was established that the S. pneumoniae CcpA orthologue contributes to the ability of this pathogen to colonize the mouse nasopharynx and infect the lung (Iyer et al., 2005) and bloodstream (Iyer et al., 2005). Although the molecular underpinnings of the ccpA defect in colonization and disease are unclear, the attenuation seen for the ccpA strain in vivo highlights the role of carbon source regulation in in vivo fitness in S. pneumoniae. This prompted us to investigate carbohydrate metabolic systems that are specifically induced during infection and explore the contribution of other LacI/GalR members to the regulation of such systems. There are five additional LacI/GalR family members in S. pneumoniae, one of which, RegR, has already been shown to contribute to virulence (Chapuy-Regaud et al., 2003). Two genes encoding other members of this family are proximal to and likely to regulate two putative sucrose transport systems. We investigated if the ability to transport and metabolize sucrose contributes to in vivo fitness of this pathogen. Here we show that sucrose uptake and catabolism play important roles in colonization of the murine nasopharynx and an accessory role in the infection of the lung, but is dispensable in a model of bacteraemia. Our data also indicate that the two sucrose uptake systems are differentially expressed such that one is dominant during lung infection while the other is dominant in colonization of the nasopharynx. Thus, the disaccharide sucrose is a major carbon source utilized by S. pneumoniae during colonization and infection of mucosal surfaces.
Results
The sus locus and the putative sucrose phosphate hydrolase, SusH, contribute to in vivo fitness in lung
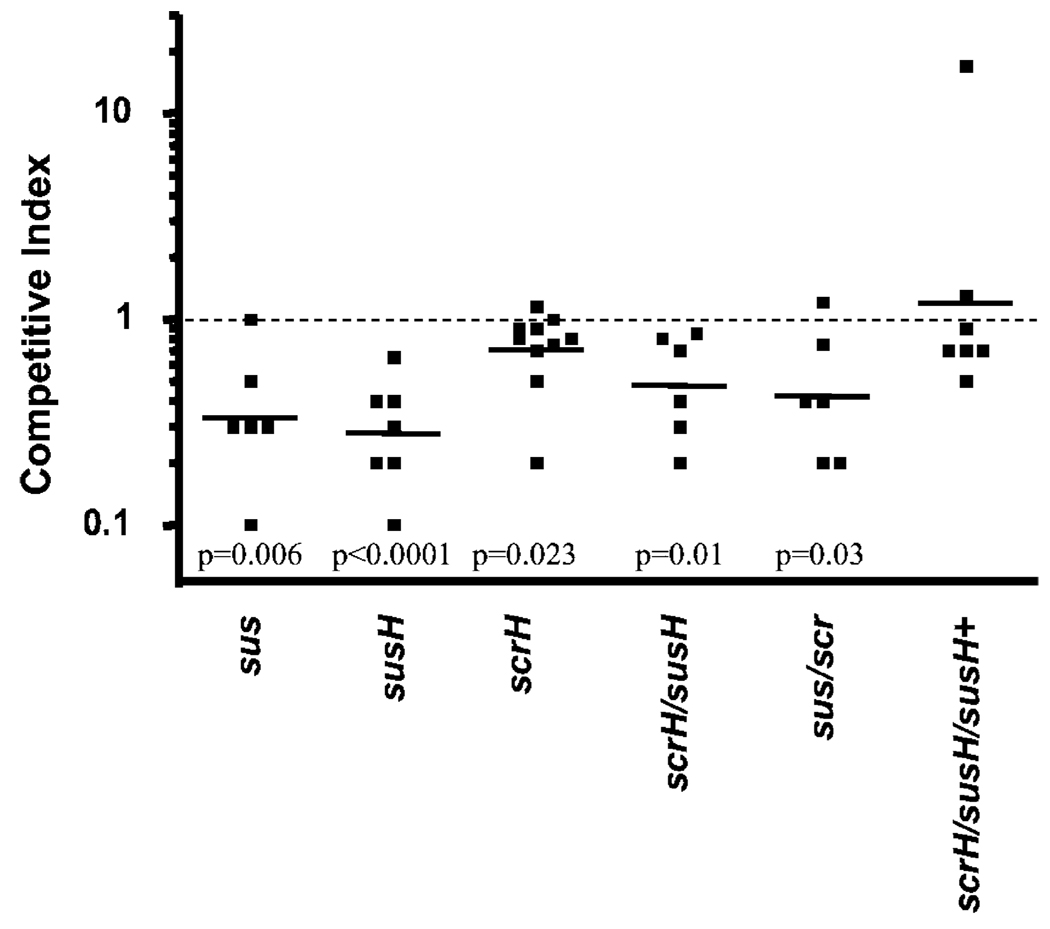
SP1799, herein named SusR for sucrose utilization system repressor, is a LacI family member that lies adjacent to a putative ABC transporter locus, containing the solute-binding lipoprotein SusX, transmembrane components SusT1 and SusT2, and a cytoplasmic sucrose-6-phosphate (S-6-P) hydrolase, SusH. Our prediction that this locus mediates sucrose metabolism was based on our observation that both SusH and its paralogue (ScrH, see below) contain a signature β-fructosidase motif (NDPNG) (Ritsema et al., 2004) which is conserved across bacteria, planta and fungi. To test if this locus contributes to in vivo fitness, we deleted the entire set of genes leaving behind a chloramphenicol (Cm) resistance cassette (cat). The deletion of this locus leads to a threefold decrease in bacterial load in the lung in a competition with wild type (Fig. 1). Consistent with a concerted role for this locus in transport of sucrose, strains either lacking the membrane components, susX/susT1/susT2 (not shown), or just the putative S-6-P hydrolase, susH, are mildly attenuated in the lung (Fig. 1).
Fig. 1.
The S-6-P hydrolase, susH, and the sus operon contribute to survival of S. pneumoniae in the murine lung. Loss of the S-6-P hydrolase, susH, or loss of the sus operon, sus, causes attenuation in the pneumonia model of infection in competition with wild-type cells. Loss of the second S-6-P hydrolase, scrH, causes only mild attenuation, with no additive attenuation in the double S-6-P hydrolase null, susH/scrH, or the double operon mutant, sus/scr, in competitions with wild type. Provision of a single copy of the susH gene in trans in the double S-6-P hydrolase null complements the in vivo defect.
The SusH paralogue, SP1724, herein named ScrH for sucrose hydrolase, lies divergent from another LacI family member, SP1725 (ScrR), which is part of a putative sucrose-specific PTS. This putative operon contains the PTS components SP1722 (ScrT) and fructokinase, SP1724 (ScrK). We hypothesized that the sus and scr systems might be partially redundant for sucrose uptake during infection and we tested this by comparing the in vivo fitness of each single- and double-hydrolase null, susH/scrH. While the scrH mutant is only slightly attenuated in the mouse lung (Fig. 1), there is no further attenuation for the double-hydrolase mutant, compared with the susH mutant alone (Fig. 1). A similar level of attenuation was observed for a strain missing both sucrose operons, sus/scr (Fig. 1). Provision of susH in trans completely rescues the scrH/susH strain defect in the lung, showing that the loss of the ABC transporter-associated sucrose hydrolase, SusH, contributes to fitness in the lung. Both S-6-P hydrolases are dispensable for survival in a murine model of bacteraemia (data not shown).
The S-6-P hydrolase ScrH contributes to colonization of the murine nasopharynx
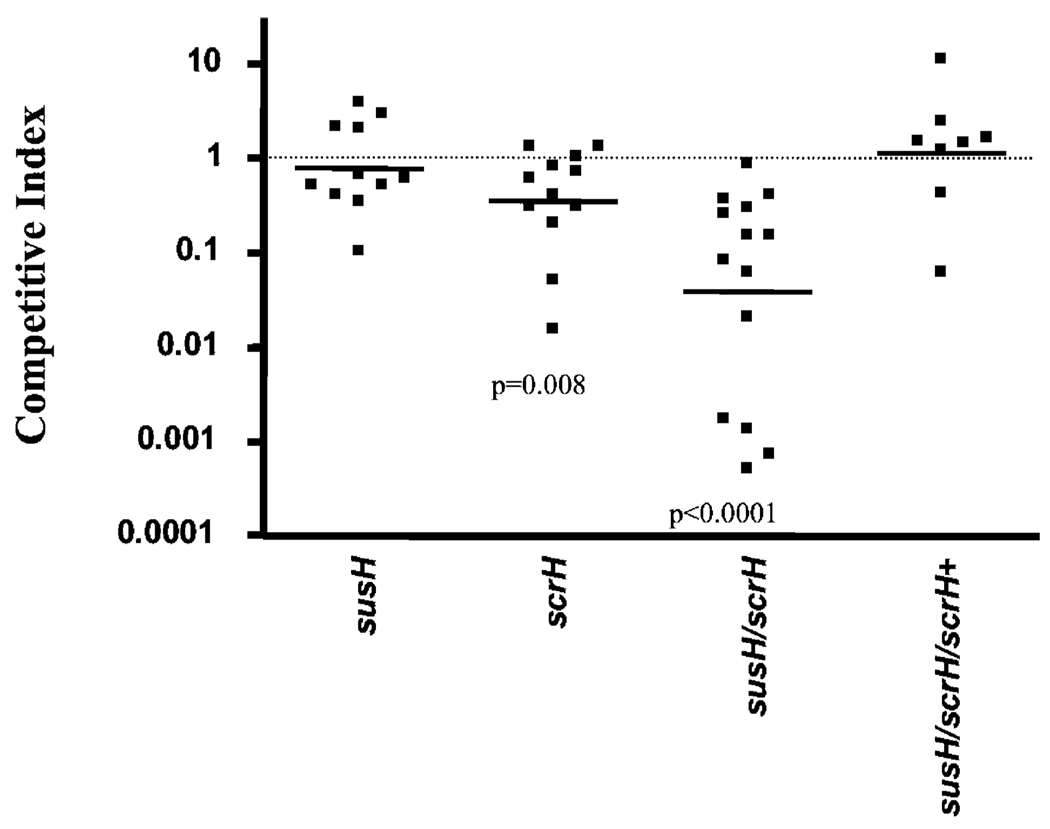
Colonization of the nasopharynx is an important step in the normal life cycle of S. pneumoniae and is thought to precede infection of other sites in diseased patients (Kadioglu et al., 2002; Kwon et al., 2004; Ogunniyi et al., 2007). To assess the contribution of sucrose metabolism to nasopharyngeal colonization, we performed competition assays on the two single- and double-hydrolase mutants. Our results are converse to those in the mouse lung (above) in that the susH strain remains virulent while the scrH null is out-competed threefold in the nasopharynx (Fig. 2). The double mutant is, however, out-competed ~30-fold in the nasopharynx, indicating that SusH is able to substitute at least partially for ScrH function (Fig. 2). Additionally, provision of scrH in trans along with 360 bp of native upstream sequence which should contain its promoter (referred to as NscrH; see Experimental procedures) completely rescued the colonization defect of the susH/scrH mutant (Fig. 2) showing that the S-6-P hydrolase, ScrH, contributes to fitness in the murine nasopharynx. Our data show that the PTS-associated S-6-P hydrolase, scrH, is the main hydrolase required for colonization fitness in the nasopharynx whereas the ABC-associated S-6-P hydrolase is dominant in the mouse lung infection model. Moreover, our results indicate that sucrose (or a related disaccharide) contributes to in vivo fitness for S. pneumoniae in both murine mucosal sites.
Fig. 2.
The S-6-P hydrolase, scrH, contributes to colonization of the murine nasopharynx. Loss of the S-6-P hydrolase, scrH, causes attenuation in the nasopharyngeal colonization model of infection. In contrast, although the susH mutant is not attenuated, the double S-6-P hydrolase mutant susH/scrH is significantly more attenuated than scrH (P = 0.02) indicating that SusH might be active in the scrH mutant. Provision of scrH in trans completely rescues the colonization defect in the susH/scrH mutant.
ScrH is the major S-6-P hydrolase in vitro
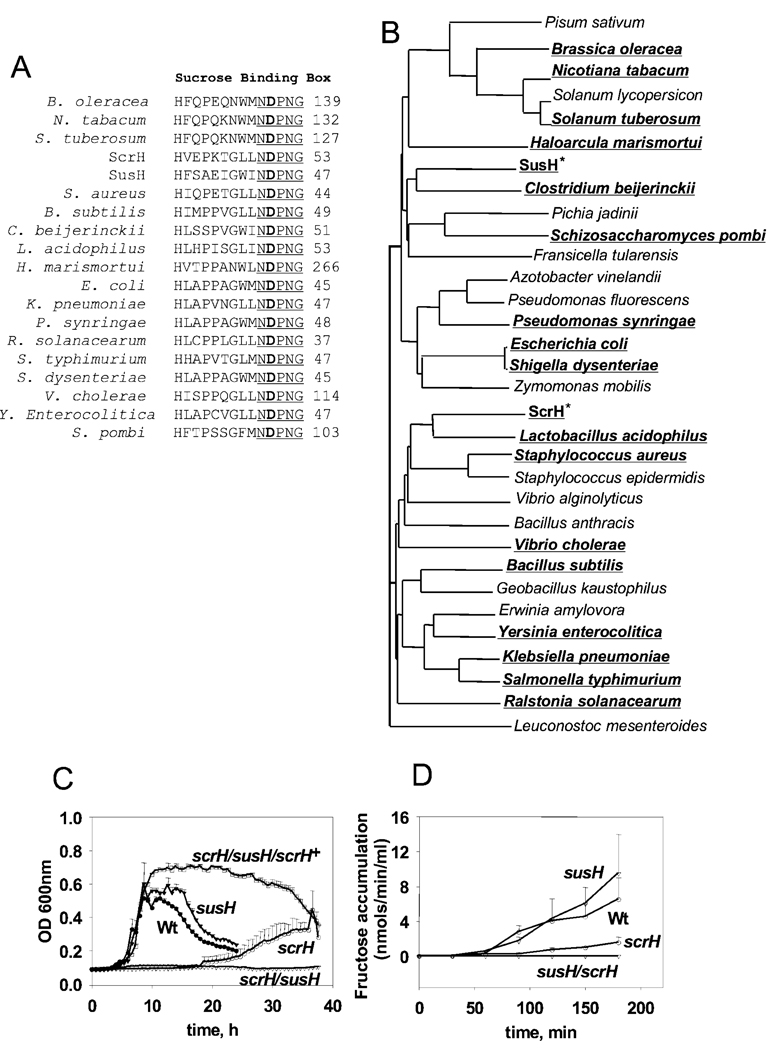
ScrH and SusH belong to the glycosyl hydrolase 32 family that includes the invertases (EC #3.2.1.26) (Henrissat and Bairoch, 1993; Pons et al., 1998), which hydrolyse the glycosidic bond in sucrose to yield glucose and fructose. The two proteins share the penta-amino acid sequence (NDPNG), illustrated in Fig. 3A at their N-termini, which is also called the β-fructosidase motif (Ritsema et al., 2004). This pentapeptide motif in turn is part of a large 14-aminoacid motif shown in Fig. 3A that is called the sucrose-binding box (Ritsema et al., 2006) where the aspartate residue is the catalytic nucleophile required for activity (Reddy and Maley, 1990) and is conserved across a large set of invertases from diverse sources (Ritsema et al., 2006) (Fig. 3B). Looking for invertases in different sources, including plants, fungi and bacteria, reveals that ScrH and SusH cluster into different groups of invertases, with SusH more closely related to invertases from fungi and plants, while ScrH lies within the typical bacterial invertases (Fig. 3B).
Fig. 3.
The S-6-P hydrolase, scrH, is required for growth on sucrose in vitro.
A. Alignment of the conserved sucrose-binding box sequences of SusH and ScrH along with those of some representative invertases from Gram-positive and Gram-negative bacteria, fungi and plants. The conserved β-fructosidase motif is underlined and the conserved aspartate residue is in bold.
B. Phylogenetic tree describing the clustering of SusH and ScrH in relation to S-6-P hydrolases (invertases) from diverse bacterial, fungal and plant sources. SusH is more related to the fungal and plant enzymes while ScrH clusters with typical bacterial invertases. All the members share the sucrose-binding box motif.
C. scrH has a pronounced growth defect on sucrose while susH is dispensable. Loss of both scrH and susH (susH/scrH) abrogates the ability of S. pneumoniae to utilize sucrose as the sole carbon source in vitro. Provision of scrH in trans restores growth of the susH/scrH mutant on sucrose.
D. Wild-type and susH lysates of S. pneumoniae grown on sucrose show ‘sucrolytic’ activity with comparable kinetics of fructose accumulation over time. The scrH lysate shows severely decreased rate of sucrose hydrolysis. There is no detectable sucrose hydrolysis activity in the S-6-P hydrolase double mutant, susH/scrH.
To assess the role of the two hydrolases during growth in vitro, we tested the ability of the single- and double-hydrolase mutants to grow on sucrose as the sole carbon source in a semi-defined minimal medium (SDMM) (Lacks, 1966). While the susH strain grows as well as the wild type (Fig. 3C), the scrH strain shows defective growth on sucrose, indicating that ScrH is the dominant sucrose-splitting enzyme in vitro (Fig. 3C). However, in the absence of scrH, SusH is active as shown by the delayed growth of the scrH mutant but no growth of the double-hydrolase null (Fig. 3C). Provision of either scrH in trans (susH/scrH/scrH+) or susH (data not shown) restores growth of the susH/scrH strain on sucrose, confirming the basis of the sucrose-dependent growth phenotype (Fig. 3C). Unexplicably, the scrH-complemented strain exhibits a higher growth yield than the wild type and shows delayed normal stationary-phase autolysis.
Based on sequence information and our in vitro growth assay results, both genes are likely to be S-6-P hydrolases. To confirm this, and as S-6-P is not commercially available, we assayed the ability of the individual single- and double-hydrolase mutant cell lysates to cleave exogenously added sucrose as reported earlier (Kuramitsu, 1973). This activity was measured by assaying for one of the end-products of the hydrolysis, fructose, using the dinitrosalicylic acid method (Miller, 1959). The susH lysate (Fig. 3D) is able to hydrolyse sucrose to the same extent as the wild type consistent with its limited role during in vitro growth. In contrast, the scrH strain is significantly diminished for sucrose hydrolysis: the remaining residual enzymatic activity in the scrH strain is due to SusH as the double-hydrolase null lacks detectable activity. Therefore, ScrH and SusH and their respective linked transporter systems appear to be the only sucrose metabolic systems that are active in S. pneumoniae TIGR4.
Regulation of the scr and sus loci in S. pneumoniae
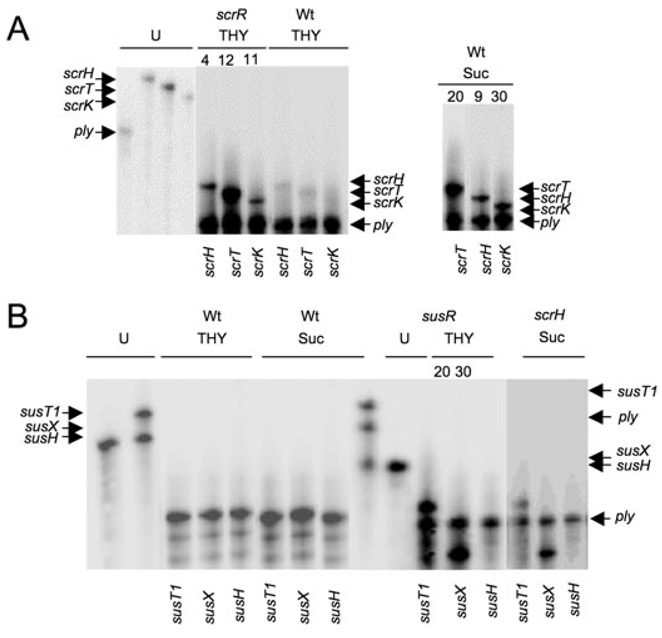
To test the role of the LacI regulators SP1799 (susR) and SP1725 (scrR) in the regulation of the neighbouring sucrose metabolic genes, we measured the levels of expression of the adjoining genes in the respective regulator mutants compared with wild-type cells grown in rich medium and in SDMM with sucrose as the sole carbon source, using RNase protection assays. The transcripts for the sucrose-specific PTS gene, scrT, the sucrose hydrolase, scrH, and the putative fructokinase, scrK, are derepressed 12-, 4- and 11-fold in the scrR strain, respectively, compared with the wild type grown in THY medium (Fig. 4A). Additionally, growth on sucrose induces all three genes 20-, 9- and 30-fold, respectively – levels that are roughly comparable to those in the scrR mutant. Thus, ScrR is a repressor of the divergently transcribed sucrose-inducible PTS transporter operon, as illustrated in the model in Fig. 5A.
Fig. 4.
Ribonuclease protection assays (RPAs) depicting regulation of the scr and sus operons by ScrR and SusR and sucrose. RNA was isolated from wild type, scrR, susR or scrH mutant S. pneumoniae strains after growth in rich medium (THY) or SDMM with sucrose as the sole carbon source (Sucrose). Transcripts were detected in 1–3 µg of total RNA by RPA. Band intensities were quantified and normalized to the intensity of the pneumolysin gene (ply) band. Radioactivity per lane per probe was arbitrarily fixed at 90 000 cpm.
A. ScrR is a repressor of scrH, scrT and scrK (scrR) and growth on sucrose induces scrH, scrT and scrK (sucrose) in wild-type cells. Average intensity relative to the wild type from two independent experiments is reported above each lane. U indicates undigested ply (lane 1), scrH (lane 2), scrT (lane 3) and scrK (lane 4) probes.
B. SusR is a repressor of susT1 and susX (susR) but growth on sucrose does not induce expression of susT1, susX or susH in wild-type cells. Growth on sucrose induces expression of susT1 and susX in the scrH mutant (scrH). Average intensity relative to the wild type from two independent experiments is reported above each lane, where applicable. Left: U indicates undigested susH (lane 1), susT1 (lane 2 – upper band) and susX (lane 2 – lower band) probes. Right: U indicates undigested susT1 (lane 1 – upper band), ply (lane 1 – middle band), susX (lane 1 – lower band) and susH (lane 2) probes.
Fig. 5.
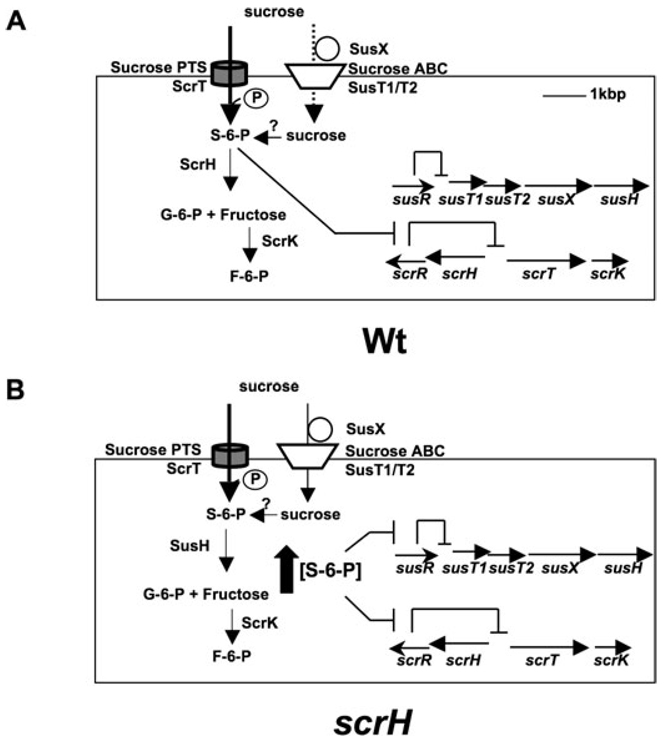
A model describing the hierarchical induction of the sus and scr systems by the putative inducer sucrose-6-phosphate (S-6-P).
A. susR and scrR genes code for the repressors, SusR and ScrR, which normally repress the expression of the downstream genes in the sus and scr operons respectively. S-6-P is formed inside the cell concomitant with entry via ScrT, the sucrose PTS transporter. Once inside, S-6-P derepresses the scr system leading to the expression of the S-6-P hydrolase, ScrH and the fructokinase, ScrK. ScrH activity is the predominant ‘sucrolytic’ activity in wild-type cells under these conditions. ScrH hydrolyses S-6-P to glucose-6-phosphate (G-6-P) and fructose. A small fraction of sucrose could enter the cell via the ABC transporter, SusT1/SusT2 and is converted to S-6-P via the action of an unknown kinase.
B. In the scrH mutant, influx of S-6-P via ScrT causes derepression of the scr operon leading to expression of scrT and scrK. However, the lack of scrH leads to an increase in intracellular concentrations of S-6-P (wide, filled, upward-directed arrow). This condition triggers derepression of the sus operon causing the expression of the ABC system, susT1/susT2, the solute-binding component, susX, and the second S-6-P hydrolase, susH. The sus pathway hydrolase, SusH, hydrolyses S-6-P to G-6-P and fructose.
Analogous results were obtained for the sus locus, where loss of susR leads to the derepression of the ABC transporter gene, susT1, 20-fold and the solute-binding gene, susX, 30-fold (Fig. 4B) compared with wild type grown in rich medium. Although not tested, susT2 is also likely regulated by SusR on the basis of the arrangement of this gene between susT1 and susX in the putative operon. Thus, SusR is a repressor of the adjacent ABC transporter locus (illustrated in Fig. 5A). Although susH appears to be the last gene in the sus operon, we could not detect its derepression in the susR strain suggesting that expression of susH might be at low levels in vitro or that the susH transcript is unstable. Neither repressor (susR and scrR) regulates the expression of the other operon (scr and sus) (data not shown).
In contrast to the scr locus, sucrose did not induce expression of susT1, susX nor susH in wild-type cells grown in SDMM with sucrose (Fig. 4B). We hypothesized that efficient hydrolysis of the putative inducer S-6-P by ScrH in wild-type cells growing in vitro does not allow build-up of inducer concentration to sufficiently high levels to trigger expression from the sus locus. To test this hypothesis, we looked for transcripts of susX, susT1 and susH in scrH mutant cells that were grown in SDMM with sucrose. Sucrose induces expression of the susX and susT1 genes in the scrH mutant (Fig. 4B). This result explains the ability of the scrH mutant to grow on sucrose. We infer that lack of the dominant S-6-P hydrolase, ScrH, allows accumulation of high levels of inducer, S-6-P, in vitro to allow activation of the sus operon (Fig. 5B). It is likely that the scr and sus loci are ‘high’- and ‘low’-affinity sucrose-metabolizing systems, respectively, although this remains to be tested. Again, we were unable to detect susH transcript, possibly due to unstable or low transcript levels. Nevertheless, we were able to detect SusH sucrose hyrolase activity in the scrH mutant when grown in SDMM/sucrose (Fig. 3D), suggesting the gene is expressed.
Discussion
The involvement of carbon metabolic pathways and relevant enzymes in determining in vivo fitness and virulence has been demonstrated for several different pathogens (Munoz-Elias and McKinney, 2005; Barelle et al., 2006; Naderer et al., 2006; Tchawa Yimga et al., 2006). The requirement of these pathways during infection has, in a few cases, led to the identification of specific carbon sources that are utilized in vivo (Chico-Calero et al., 2002; Chang et al., 2004; Exley et al., 2005). In a previous study, we demonstrated the importance of a LacI family carbon regulator, CcpA, in the fitness of S. pneumoniae in the murine lung, bloodstream and nasopharynx (Iyer et al., 2005). Although the mechanistic basis for the ccpA attenuation in vivo is not clear, the importance of this carbon regulator for colonization and disease established a link between carbon metabolism and in vivo fitness and virulence. In this report, we strengthen this link by characterizing two sucrose transport and metabolic systems, sus and scr, which contribute to in vivo fitness. While scr is the dominant sucrose uptake and metabolizing system in vitro, we show that loss of both scrH and susH, encoding the respective S-6-P hydrolases for each system, abrogates the ability to utilize sucrose, indicating that both the sus and scr systems facilitate sucrose uptake and metabolism. In fact, the arrangement of the scr operon, its regulation by scrR and induction by sucrose are similar to the sucrose-metabolizing locus in the oral pathogen, Streptococcus mutans (Sato et al., 1993; Hiratsuka et al., 1998; Wang and Kuramitsu, 2003). Using murine models of carriage and lung infection, we find distinct roles for the two sucrose uptake systems. The scr system is largely dispensable for infection of the mouse lung, but plays a role in colonization of the nasopharynx. In contrast, sus plays a role in fitness in the mouse lung, but is dispensable in the nasopharynx. The distinct roles of the two sucrose utilization systems in two different host sites reveal niche-specific adaptations that we do not fully understand. While sucrose induces both operons and is transported by sus and scr in vitro, the identity of the carbon sources that are transported into S. pneumoniae and that derepress the two operons in vivo is unknown. We hypothesize that both systems are involved in the uptake and metabolism in vivo of sucrose and/or highly similar disaccharides, and we further suggest that the two operons are differentially induced in the lung and the nasopharynx during infection perhaps in response to different disaccharides. We were also able to complement the susH defect in the lung and the scrH defect in the nasopharynx. Provision of either gene in trans was able to restore growth of the double-hydrolase null on sucrose as the sole carbon source, thus confirming the role of ScrH in sucrose metabolism in vitro.
Both the sus and scr systems are inducible by sucrose and are subject to tight regulation by their cognate LacI family regulators, SusR and ScrR (Fig. 5A). Although S-6-P does not readily induce the sus locus in the wild-type strain under the in vitro conditions tested, the derepression of the sus locus in the scrH mutant suggests the need for high intracellular concentrations of S-6-P to counter the repression by SusR (Fig. 5B), a situation that may occur naturally under growth conditions in the lung where sus plays a demonstrable role. However, it remains possible that the natural inducer of the sus system in vivo is not S-6-P, but is instead a related disaccharide found at relevant concentrations in the mouse lung. Both systems appear to be highly specific for the α-1,2 linkage in sucrose as none of the sucrose isomers tested – palatinose (α-1,6), turanose (α-1,3), maltulose (α-1,4) or leucrose (α-1,5) – could be metabolized by wild type or the susH/scrH or sus/scr mutants (Table 1).
Table 1.
Growth of the S-6-P hydrolase mutant (susH/scrH) and the double operon null (sus/scr) on glucose, fructose, sucrose and sucrose isomers.a
| Strain | Glucose | Fructose | Sucrose | Palatinose | Turanose | Maltulose | Leucrose |
|---|---|---|---|---|---|---|---|
| Wild type | + | + | + | − | − | − | − |
| sus/scr | + | + | − | − | − | − | − |
| susH/scrH | + | + | − | − | − | − | − |
Individual sugar (10 mM) in SDMM.
The entire scr locus is conserved in other S. pneumoniae serotype strains that have been sequenced, namely serotypes 2 and 19F. While the sus system genes, susR and susH, are present in these other genomes, exact matches for susT1, susT2 and susX are not found. While SusH is represented within all serotypes (> 97% identity), SusT1, SusT2 and SusX have homologues in serotype 3, 14 and 23F and are absent in serotypes 1, 2 and 19F. However, visual inspection of the susH and susR region in the completely sequenced serotypes, 2 and 19F reveals the existence of a alternative sugar-specific solute-binding/ABC transporter locus in place of susT1, susT2 and susX. The sequence dissimilarity in the ABC transporter membrane components and the solute-binding proteins between the serotypes is interesting. Although speculative, it is possible that the requirement of the transporter system in vivo may subject the locus to immune pressure and select for sequence divergence among serotypes. Examining sequences upstream of the sus operon in all three serotypes revealed a putative cre sequence in the intergenic space between the repressor, susR and SusT1 (serotype 4) or the related ABC transporter gene (serotypes 19F and 2). Cre sequences are known to be regions that are bound by CcpA, which plays an important role in the catabolite repression or activation of neighbouring genes and contributes to regulation of sugar metabolism and virulence in S. pneumoniae. The regulation of the sus locus by CcpA and the potential for interplay between CcpA and SusR in modulating expression of this locus will be interesting as this may address the mechanistic basis of the previously observed attenuation of the ccpA mutant in the lung.
Based on (i) the sucrose-inducible nature of the sus locus in the scrH mutant, (ii) the presence of the sucrose-binding box (NDPNG), in SusH, (iii) the ability of the scrH strain to grow on and hydrolyse sucrose and (iv) the loss of this function in the scrH/susH null, we infer that the solute-binding protein/ABC-transporter/S-6-P hydrolase trio is an alternative system capable of participating in the uptake and metabolism of sucrose. But, how the sus locus is induced in vivo is not clear at present. These results establish, for the first time, a link between a specific carbon uptake and metabolic system in S. pneumoniae and colonization and disease. Specifically, we show that the metabolism of sucrose and/or a related disaccharide is important during colonization of the nasopharynx and might play a minor role in contributing to survival in the lungs. Amino acids and sugars have recently been shown to be essential carbon sources in vivo in both bacterial and protozoan pathogens (Velayudhan et al., 2004; Naderer et al., 2006; Tanzer et al., 2006). In addition, a direct correlation between sucrose and fructose metabolism and in vivo fitness has been described in the phytopathogen, Erwinia amylovora, and the human dental pathogen, Streptococcus mutans, respectively (Bogs and Geider, 2000; Tanzer et al., 2006). The identity of carbon sources that are used by S. pneumoniae during infection and colonization, their roles in contributing to fitness and possible virulence gene induction in different host niches are largely unknown. Our observation that two sucrose transport and metabolizing systems contribute to in vivo fitness of S. pneumoniae in the nasopharynx and to a limited extent, in the lung, represents a significant advance in this direction.
Experimental procedures
Bacterial strains, plasmids and primers for DNA manipulations
Refer to Table 2 for a list of relevant strains and plasmids and the Table S1 for the primers used in this study.
Table 2.
Relevant strains and plasmids.
| Strain/plasmid | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| S. pneumoniae | ||
| AC353 | Spontaneous Smr derivative of TIGR4 | |
| RI2455 | AC353 scrH::cat | This work |
| RI2436 | AC353 susH | This work |
| RI2456 | RI2436 scrH::cat | This work |
| RI2480 | AC353 scr::aad9 | This work |
| RI2479 | AC353 sus::cat | This work |
| RI2481 | RI2479 scr::aad9 | This work |
| RI2587 | RI2456 (SP0474-SP0478::aad9-susH) | This work |
| RI2603 | RI2456 (SP0474-SP0478::aad9-scrH) | This work |
| RI2650 | RI2456 (SP0474-SP0478::aad9-NscrH) | This work |
| RI1932 | AC353 ccpA:cat | Iyer et al. (2005) |
| RI1933 | RI1932 (SP0474-SP0478::aad9-ccpA) | Iyer et al. (2005) |
| RI2678 | RI2456 (SP0474-SP0478::aad9 (tt)-NscrH) | This work |
| E. coli | ||
| AC578 | DH5α λpir(pR412); Apr, Spr | Iyer et al. (2005) |
| AC1000 | DH5α λpir(pAC1000); Apr, Cmr | Iyer et al. (2005) |
| Plasmids | ||
| pAC1000 | S. pneumoniae suicide vector; Cmr | Hava et al. (2003a) |
| pR412 | Has magellan5 minitransposon; Apr, Spr | Martin et al. (2000) |
Construction of the SP1799 (susR) and SP1795 (susH) deletion
The susR (SP1799) and the susH (SP1795) mutant were constructed by deleting the entire 1001 bp and 1300 bp coding sequence respectively. Approximately 800 bp DNA fragments 5′ (F1R1) and 3′ (F2R2) of the susR and susH gene were PCR amplified from AC353 genomic DNA using two primer pair sets, 1799F1–1799R1/1799F2–1799R2 and S6PF2–S6PR2/S6PF1–S6PR1, respectively. Two of the primers in each of the primer pair sets were designed to have a ~20 bp overlap with the 5′ and 3′ sequences of each of the fragments. The two pieces were then fused together using splicing by overlap extension (SOE) (Horton et al., 1990). The susR and the susH DNA fragments were then PCR amplified using the primer pair, XM1799–XH1799 and XB95F1–XH95R2, respectively, that add restriction sites at the ends of the product to facilitate cloning into the multiple cloning site of pAC1000, which was then transformed into AC353 cells using standard transformation techniques (Bricker and Camilli, 1999) and mutants were screened for loss of the susR and susH using PCR and DNA sequencing.
Construction of the scrR, scrH and sus operon deletion
The scrR (SP1725), scrH (SP1724) and sus mutant were constructed by replacing the entire 985 bp, 1450 bp and 6750 bp coding sequence with a CM resistance cassette, cat. The cat cassette, which has its own promoter and conferred CM resistance on E. coli and S. pneumoniae, was PCR amplified from pAC1000 (Hava et al., 2003a) by using primers catF1 and NcatR1 (Iyer et al., 2005). Fragments (650–800 bp) containing the regions 5′ and 3′ of the scrR, scrH gene were PCR amplified from AC353 genomic DNA by using the primer pair sets, 1725F1–1725R1/1725F2–1725R2, 1724F1–24cat/cat24F2–1724R2 and 1799F1–1799R/1799F–S6PR2, respectively. Two of the primers in each of the primer sets were designed to have a 20 bp overlap with the 5′ and 3′ sequences of the cat cassette. The cat cassette was fused to these two PCR products by SOE as before. The final PCR product was introduced into S. pneumoniae TIGR4 by natural transformation as previously described. The double-recombination event was selected for by plating on medium containing 4 µml−1 CM and was confirmed by PCR and DNA sequencing.
Construction of the scr operon deletion
The scr operon deletion was constructed by replacing the entire 5700 bp coding sequence with a spectinomycin (SP) resistance cassette, aad9. The aad9 cassette, which has its own promoter and conferred SP resistance on E. coli and S. pneumoniae, was PCR amplified from pJR412 (Martin et al., 2000) by using primers spcF1 and spcR1. Approximately 800 bp DNA fragments 5′ and 3′ of the sus operon were PCR amplified from AC353 genomic DNA using the primer pair, 2125F1–2125R1 and 2125F2–1725R2. One primer in each of the primer pairs was designed to have a 20 bp overlap with the 5′ and 3′ sequences of the aad9 assette. The aad9 cassette was fused to these two PCR products and the final PCR product was introduced into S. pneumoniae TIGR4 as described earlier. The double-recombination event was selected for by plating on medium containing 200 µg ml−1 SP and was confirmed by PCR and DNA sequencing.
Creation of the double S-6-P hydrolase mutant, susH/scrH
The double mutant was created by transforming the susH strain with genomic DNA from the scrH mutant, using natural transformation as previously described. The recombination event was selected for by plating on medium containing 4 µg ml−1 CM and was confirmed by PCR and DNA sequencing.
Construction of the susH and scrH complementation strains
The susH and scrH complementation strategy involved making transcriptional fusions of susH and scrH with the SP resistance gene, aad9, so that the genes are driven by the aad9 promoter and the construct was recombined into the LacE (SP0474-SP0478) locus (Iyer et al., 2005) to create strains, RI2587 and RI2603. For complementation of scrH in vitro, we also made another construct, where scrH was similarly transcriptionally fused to aad9 (as above) but was placed downstream from its putative regulatory upstream region in an attempt to preserve wild type-like regulation of scrH to create RI2650. This we also refer to as Native scrH (NscrH). To complement scrH in the nasopharynx, we additionally placed a transcriptional terminator at the end of the aad9 gene in the NscrH construct to allow for more controlled expression in vivo, to create strain RI2678. For complementation of the susH mutation, susH, along with its native ribosome binding site, was amplified by PCR from AC353 genomic DNA with primers susHF-susHR. This PCR fragment was used as the template for another PCR amplification step using the primers spsusH and susHR. This PCR adds a 33 bp sequence at the 5′ end that is homologous to the 3′ end of the aad9 (SP resistance cassette) gene. The primers suslac and NLacER were used to PCR amplify the 3′ end of the LacE locus (LacE dn) from AC353 genomic DNA. The suslac primer has a 20 bp sequence at the 5′ end that is homologous to the 3′ end of the susH gene. The suslac-NLacER PCR fragment was joined to the spsusH-susHR fragment using SOE, to create the susH-LacE dn piece. The primers NLacEF and spsusH were used to amplify the region 5′ of the LacE locus (LacE up) along with the aad9 gene from the ccpA+ strain, RI1933, as described previously. The LacE up-aad9 piece was then joined by SOE to the susH-LacE dn fragment to create the final LacE up-aad9-susH-LacE dn construct that was introduced into the susH/scrH strain to create RI2587. The double-recombination event was selected for by plating on medium containing 200 µml−1 SP and was confirmed by PCR and DNA sequencing.
For complementation of the scrH mutation, scrH including its native ribosome binding site, was amplified by PCR from AC353 genomic DNA using primers, scrHF-scrHR. This PCR fragment was used as the template for another PCR amplification step using the primers spscrH and scrHR. This PCR adds a 33 bp sequence at the 5′ end that is homologous to the 3′ end of the aad9 gene. The primers scrlac and NLacER were used to PCR amplify the 3′ end of the LacE locus (LacE dn) from AC353 genomic DNA. The scrlac primer has a 20 bp sequence at the 5′ end that is homologous to the 3′ end of scrH. The scrlac-NLacER PCR fragment was joined to the spscrH-scrHR fragment using SOE, to create the scrH-LacE dn piece. The primers NLacEF and spcR1 were used to amplify the region 5′ of the LacE locus (LacE up) along with the aad9 gene from the ccpA+ strain, RI1933. The LacE up-aad9 fragment was then joined by SOE to the scrH-LacE dn piece to create the final LacE up-aad9-scrH-LacE dn construct that was introduced into the susH/scrH strain to create RI2603. The double-recombination event was selected for by plating on medium containing 200 µml−1 SP and was confirmed by PCR and DNA sequencing.
In another strategy for complementation of the scrH mutation, scrH, along with a putative regulatory region 360 bp upstream including its native ribosome binding site, was amplified by PCR from AC353 genomic DNA. Primers scrupF-scrupR amplified the 360 bp putative regulatory region upstream of scrH to create scrH (upstream). Separately, primers scrHF-scrHR were used to amplify the entire ~1800 bp scrH gene. Using this PCR product as the template, primers upscrHF-scrHR was used to re-amplify scrH, so that primer upscrHF creates a 22 bp addition at the 5′ end of scrH facilitating fusion of this piece with the scrupF-scrupR product using SOE to create scrH (upstream)-scrH. The primers scrlac and NLacER were used to PCR amplify the 3′ end of the LacE locus (LacE dn) from AC353 genomic DNA. The scrlac primer has a 25 bp sequence at the 5′ end that is homologous to the 3′ end of scrH. scrH was then fused to the scrlac-NLacER fragment to create scrH-LacE dn. Subsequently, the scrH (upstream)-scrH fragment was fused to the scrH-LacE dn fragment by SOE to create scrH (upstream)-scrH-LacE dn. In a separate reaction, primers NLacEF and upspcR1 were used to amplify the LacE up-aad9 region from RI1933 (Iyer et al., 2005). The primer upspcR1 creates a 29 bp sequence at the 3′ end of the LacE up-aad9 fragment that is homologous to the 5′ end of scrH (upstream)-scrH-LacE dn. The NLacEF-upspcR1 fragment was then fused to the scrH (upstream)-scrH-LacE dn piece by SOE to create the final ~4200 bp construct, LacE up-aad9-scrH (upstream)-scrH-LacE dn that was introduced into the susH/scrH strain to create RI2650. The double-recombination event was selected for by plating on medium containing 200 µg ml−1 SP and was confirmed by PCR and DNA sequencing.
In order to complement the susH/scrH defect in nasopharyngeal colonization, we placed a transcriptional terminator at the end of the aad9 gene in the NscrH construct. We chose the genomic transcriptional terminator downstream from the SP0479 gene (genomic co-ordinates 459144–459094) for this purpose. We PCR amplified the lacE (up)-aad9 region from RI1933 genomic DNA using primers NLacEF and TTR to create LacEF (up)-TTR. The TTR primer has a 20 bp homology with the 3′ end of the aad9 gene and additionally has a 23 bp overhang that is part of the terminator. Separately, we used primers TTF and scrHR to PCR amplify the scrH (upstream)-scrH fragment from RI2650 genomic DNA to create TTF-scrH (upstream)-scrH. The TTF primer has a 29 bp region of homology with the 5′ end of the scrH (upstream) fragment. The TTF primer also has a 21 bp overhang at the 5′ end that is part of the terminator. We PCR amplified the TTF-scrH (upstream)-scrH fragment using primers TTF1 and scrHR to create TTF1-scrHR. The TTF1 primer shares at its 3′ end 20 bp homology with the 5′ end of TTF and in addition 20 bp with the 5′ end of TTR. This step creates the transcriptional terminator (tt) in the fragment. We then joined the LacEF (up)-TTR fragment to TTF1-scrHR to create LacEF (up)-aad9-tt-scrH (upstream)-scrH using SOE. As described earlier, we fused LacEF (up)-aad9-tt-scrH (upstream)-scrH to LacE (dn) using SOE to create the final ~4.3 kb LacEF (up)-aad9-tt-scrH (upstream)-scrH-LacEF (dn), which was introduced into the susH/scrH strain to create RI2678. The double-recombination event was selected for by plating on medium containing 200 µg ml−1 SP and was confirmed by PCR and DNA sequencing.
Bacterial growth and media
Streptococcus pneumoniae was routinely grown in Todd–Hewitt broth with 1% yeast extract (THY) or on blood agar plates containing 100 µml−1 streptomycin (SM) at 37°C in a 5% CO2 atmosphere. For broth cultures, 5 µl ml−1 Oxyrase (Oxyrase) was added in addition and cultures were typically grown to a final optical density at 600 nm (OD600) of 0.5–0.6. To test growth on sucrose as the sole carbon source and sucrose hydrolysis assays, S. pneumoniae was grown in SDMM without BSA, containing appropriate antibiotics (SM: 100 µg ml−1; CM: 4 µg ml−1; SP: 200 µg ml−1). The concentration of sugar was 10 mM in all cases. There was negligible growth in SDMM in the absence of added sugar.
Animal infections
All animal infections were performed with 8- to 12-week-old male/female Swiss–Webster mice (Taconic Laboratories). Mice were provided with continuous food and water. Bacteria were recovered from plates of tryptic soy agar (TSA) with 5% sheep’s blood, subcultured in THY and grown at 37°C for 4 h to an OD600 ~0.4–0.5. Lung and nasopharyngeal infections in mice were performed as described earlier (Hava and Camilli, 2002; Iyer et al., 2005). Lung infection was carried out for 42 h and nasopharyngeal colonization for 6–7 days, after which the mice were sacrificed and counted for bacteria as described before. Housing of mice and all procedures were performed according to procedures approved by the Tufts University Institution of Animal Care and Use Committee.
RNase protection assays
Total RNA was isolated from 10 ml of S. pneumoniae wild-type, susR and scrR mutant cells grown in THY supplemented with SM (100 µg ml−1) and Oxyrase (5 µl ml−1) or for sucrose induction, from wild-type cells grown in SDMM containing 10 mM sucrose, 30 units/ml catalase and SM (100 µg ml−1). RNA was isolated with a Qiagen RNeasy kit as described earlier. In the case of scrH mutants, cells were grown up in THY as earlier to OD600 ~0.4–0.5. The cells were centrifuged at 4000 r.p.m. for 10 min and the pellet was re-suspended in 10 ml of SDMM containing 10 mM sucrose for 2 h at 37°C and 5% CO2 prior to RNA isolation. Template DNA for the generation of riboprobes was PCR amplified with primer sets, NplyF–rpaply, plyF–rpaply, rp1795–1795R0, rp1796–1796R, rp1798–1798F, rp1724–1725F0, rp1722–1722R and rp1721–1721F.
Sucrose hydrolysis assays
The hydrolysis of sucrose by S. pneumoniae wild-type and mutant lysates was monitored over time by measuring the accumulation of the end-product, fructose, using the dinitro-salicylic acid-phenol (DNS) colorimetric assay as described. Wild-type, susH, scrH and susH/scrH cells were grown up in duplicate in 5 ml of THY (1% yeast extract) to an OD600 ~0.4–0.5. The entire suspension was centrifuged at 4000 r.p.m. for 5 min to pellet the cells. The pellets were washed once in 1 ml of pre-warmed sugar-less SDMM and re-suspended in 1 ml of SDMM. After centrifugation at 4000 r.p.m. for 5 min, the washed pellets were re-suspended in 1 ml of SDMM and inoculated into 10 ml of pre-warmed SDMM with or without 10 mM sucrose and incubated at 37°C and 5% CO2 for 120 min to allow induction of susH or scrH genes. At various time points during the incubation (at ~30–40 min intervals), 1 ml of aliquots were drawn out and centrifuged at 10000 r.p.m. for 1 min and washed once with 500 µl of warm 100 mM K2HPO4 buffer (pH 7.0). After centrifugation, the pellets were re-suspended in 400 µl of warm 100 mM K2HPO4 buffer (pH 7.0). Five microlitres of 10% Triton X-100 was added to the suspension to get a final detergent concentration of 0.1% followed by the addition of 100 µl of 1M sucrose in 100 mM K2HPO4 buffer (pH 7.0) to get a final sucrose concentration of 200 mM. The suspension was mixed by inverting and incubated at 37°C for 70 min. In parallel, we also prepared a similar blank reaction that does not have cells. At the end of the incubation, 300 µl from each reaction was withdrawn and added to a tube containing 300 µl of DNS (Miller, 1959), and incubated at 90°C for 15 min until the development of a red brown colour. The tubes were then transferred to an ice bucket and 100 µl of 40% Rochelle’s salt was added to each reaction and the tubes were left of ice for ~5 min to cool the contents. The absorbance of the reaction was measured at 575 nm against the blank, and the amount of fructose at the end of the reaction was calculated by reading from a separately constructed fructose standard curve.
Supplementary Material
Acknowledgements
We are grateful to members of the Camilli lab for helpful discussions and suggestions during the course of this work. A.C. is an investigator of the Howard Hughes Medical Institute (HHMI). This work was supported by the HHMI.
Footnotes
Supplementary material
This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/j.1365-2958.2007.05878.x
(This link will take you to the article abstract).
Please note: Blackwell Publishing is not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Asanuma N, Hino T. Presence of NAD+specific glyceraldehyde-3-phosphate dehydrogenase and CcpA-dependent transcription of its gene in the ruminal bacterium Streptococcus bovis. FEMS Microbiol Lett. 2006;257:17–23. doi: 10.1111/j.1574-6968.2006.00111.x. [DOI] [PubMed] [Google Scholar]
- Barelle CJ, Priest CL, Maccallum DM, Gow NA, Odds FC, Brown AJ. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell Microbiol. 2006;8:961–971. doi: 10.1111/j.1462-5822.2005.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barocchi MA, Ries J, Zogaj X, Hemsley C, Albiger B, Kanth A, et al. A pneumococcal pilus influences virulence and host inflammatory responses. Proc Natl Acad Sci USA. 2006;103:2857–2862. doi: 10.1073/pnas.0511017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogaert D, De Groot R, Hermans PW. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis. 2004;4:144–154. doi: 10.1016/S1473-3099(04)00938-7. [DOI] [PubMed] [Google Scholar]
- Bogs J, Geider K. Molecular analysis of sucrose metabolism of Erwinia amylovora and influence on bacterial virulence. J Bacteriol. 2000;182:5351–5358. doi: 10.1128/jb.182.19.5351-5358.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricker AL, Camilli A. Transformation of a type 4 encapsulated strain of Streptococcus pneumoniae. FEMS Microbiol Lett. 1999;172:131–135. doi: 10.1111/j.1574-6968.1999.tb13460.x. [DOI] [PubMed] [Google Scholar]
- Chang DE, Smalley DJ, Tucker DL, Leatham MP, Norris WE, Stevenson SJ, et al. Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci USA. 2004;101:7427–7432. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuy-Regaud S, Ogunniyi AD, Diallo N, Huet Y, Desnottes JF, Paton JC, et al. RegR, a global LacI/GalR family regulator, modulates virulence and competence in Streptococcus pneumoniae. Infect Immun. 2003;71:2615–2625. doi: 10.1128/IAI.71.5.2615-2625.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chico-Calero I, Suarez M, Gonzalez-Zorn B, Scortti M, Slaghuis J, Goebel W, Vazquez-Boland JA. Hpt, a bacterial homolog of the microsomal glucose-6-phosphate translocase, mediates rapid intracellular proliferation in Listeria. Proc Natl Acad Sci USA. 2002;99:431–436. doi: 10.1073/pnas.012363899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exley RM, Goodwin L, Mowe E, Shaw J, Smith H, Read RC, Tang CM. Neisseria meningitides lactate permease is required for nasopharyngeal colonization. Infect Immun. 2005;73:5762–5766. doi: 10.1128/IAI.73.9.5762-5766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosink KK, Mann ER, Guglielmo C, Tuomanen EI, Masure HR. Role of novel choline binding proteins in virulence of Streptococcus pneumoniae. Infect Immun. 2000;68:5690–5695. doi: 10.1128/iai.68.10.5690-5695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hava DL, Camilli A. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol Microbiol. 2002;45:1389–1406. [PMC free article] [PubMed] [Google Scholar]
- Hava DL, Hemsley CJ, Camilli A. Transcriptional regulation in the Streptococcus pneumoniae rlrA pathogenicity islet by RlrA. J Bacteriol. 2003a;185:413–421. doi: 10.1128/JB.185.2.413-421.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hava DL, LeMieux J, Camilli A. From nose to lung: the regulation behind Streptococcus pneumoniae virulence factors. Mol Microbiol. 2003b;50:1103–1110. doi: 10.1046/j.1365-2958.2003.03764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkin TM, Grundy FJ, Nicholson WL, Chambliss GH. Catabolite repression of alpha-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacl and galR repressors. Mol Microbiol. 1991;5:575–584. doi: 10.1111/j.1365-2958.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka K, Wang B, Sato Y, Kuramitsu H. Regulation of sucrose-6-phosphate hydrolase activity in Streptococcus mutans: characterization of the scrR gene. Infect Immun. 1998;66:3736–3743. doi: 10.1128/iai.66.8.3736-3743.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton RM, Cai ZL, Ho SN, Pease LR. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- Iyer R, Baliga NS, Camilli A. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J Bacteriol. 2005;187:8340–8349. doi: 10.1128/JB.187.24.8340-8349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JW, Myers LE, Ochs MM, Benjamin WH, Jr, Briles DE, Hollingshead SK. Lipoprotein PsaA in virulence of Streptococcus pneumoniae: surface accessibility and role in protection from superoxide. Infect Immun. 2004;72:5858–5867. doi: 10.1128/IAI.72.10.5858-5867.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadioglu A, Taylor S, Iannelli F, Pozzi G, Mitchell TJ, Andrew PW. Upper and lower respiratory tract infection by Streptococcus pneumoniae is affected by pneumolysin deficiency and differences in capsule type. Infect Immun. 2002;70:2886–2890. doi: 10.1128/IAI.70.6.2886-2890.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Roux A, Sonenshein AL. Direct and indirect roles of CcpA in regulation of Bacillus subtilis Krebs cycle genes. Mol Microbiol. 2002;45:179–190. doi: 10.1046/j.1365-2958.2002.03003.x. [DOI] [PubMed] [Google Scholar]
- Kuramitsu HK. Characterization of invertase activity from cariogenic Streptococcus mutans. J Bacteriol. 1973;115:1003–1010. doi: 10.1128/jb.115.3.1003-1010.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HY, Ogunniyi AD, Choi MH, Pyo SN, Rhee DK, Paton JC. The CIpP protease of Streptococcus pneumoniae modulates virulence gene expression and protects against fatal pneumococcal challenge. Infect Immun. 2004;72:5646–5653. doi: 10.1128/IAI.72.10.5646-5653.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacks S. Integration efficiency and genetic recombination in pneumococcal transformation. Genetics. 1966;53:207–235. doi: 10.1093/genetics/53.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau GW, Haataja S, Lonetto M, Kensit SE, Marra A, Bryant AP, et al. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol Microbiol. 2001;40:555–571. doi: 10.1046/j.1365-2958.2001.02335.x. [DOI] [PubMed] [Google Scholar]
- Luesink EJ, van Herpen RE, Grossiord BP, Kuipers OP, de Vos WM. Transcriptional activation of the glycolytic las operon and catabolite repression of the gal operon in Lactococcus lactis are mediated by the catabolite control protein CcpA. Mol Microbiol. 1998;30:789–798. doi: 10.1046/j.1365-2958.1998.01111.x. [DOI] [PubMed] [Google Scholar]
- Martin B, Prudhomme M, Alloing G, Granadel C, Claverys JP. Cross-regulation of competence pheromone production and export in the early control of transformation in Streptococcus pneumoniae. Mol Microbiol. 2000;38:867–878. doi: 10.1046/j.1365-2958.2000.02187.x. [DOI] [PubMed] [Google Scholar]
- McCullers JA, Tuomanen EI. Molecular pathogenesis of pneumococcal pneumonia. Front Biosci. 2001;6:D877–D889. doi: 10.2741/mccullers. [DOI] [PubMed] [Google Scholar]
- McKinney JD, Honer zu Bentrup K, Munoz-Elias EJ, Miczak A, Chen B, Chan WT, et al. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 2000;406:735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- Miller GI. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–428. [Google Scholar]
- Munoz-Elias EJ, McKinney JD. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat Med. 2005;11:638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Elias EJ, McKinney JD. Carbon metabolism of intracellular bacteria. Cell Microbiol. 2006;8:10–22. doi: 10.1111/j.1462-5822.2005.00648.x. [DOI] [PubMed] [Google Scholar]
- Naderer T, Ellis MA, Sernee MF, De Souza DP, Curtis J, Handman E, McConville MJ. Virulence of Leishmania major in macrophages and mice requires the gluconeogenic enzyme fructose-1,6-bisphosphatase. Proc Natl Acad Sci USA. 2006;103:5502–5507. doi: 10.1073/pnas.0509196103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogunniyi AD, LeMessurier KS, Graham RM, Watt JM, Briles DE, Stroeher UH, Paton JC. Contributions of pneumolysin, pneumococcal surface protein A (PspA), and PspC to pathogenicity of Streptococcus pneumoniae D39 in a mouse model. Infect Immun. 2007;75:1843–1851. doi: 10.1128/IAI.01384-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orihuela CJ, Gao G, Francis KP, Yu J, Tuomanen EI. Tissue-specific contributions of pneumococcal virulence factors to pathogenesis. J Infect Dis. 2004;190:1661–1669. doi: 10.1086/424596. [DOI] [PubMed] [Google Scholar]
- Paton JC, Giammarinaro P. Genome-based analysis of pneumococcal virulence factors: the quest for novel vaccine antigens and drug targets. Trends Microbiol. 2001;9:515–518. doi: 10.1016/s0966-842x(01)02207-7. [DOI] [PubMed] [Google Scholar]
- Peekhaus N, Conway T. What's for dinner? Entner-Doudoroff metabolism in Escherichia coli. J Bacteriol. 1998;180:3495–3502. doi: 10.1128/jb.180.14.3495-3502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polissi A, Pontiggia A, Feger G, Altieri M, Mottl H, Ferrari L, Simon D. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect Immun. 1998;66:5620–5629. doi: 10.1128/iai.66.12.5620-5629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons T, Olmea O, Chinea G, Beldarrain A, Marquez G, Acosta N, et al. Structural model for family 32 of glycosyl-hydrolase enzymes. Proteins. 1998;33:383–395. doi: 10.1002/(sici)1097-0134(19981115)33:3<383::aid-prot7>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Reddy VA, Maley F. Identification of an active-site residue in yeast invertase by affinity labeling and site-directed mutagenesis. J Biol Chem. 1990;265:10817–10820. [PubMed] [Google Scholar]
- Ritsema T, Verhaar A, Vijin I, Smeekens S. Fructosyltransferase mutants specify a function for the beta-fructosidase motif of the sucrose-binding box in specifying the fructan type synthesized. Plant Mol Biol. 2004;54:853–863. doi: 10.1007/s11103-004-0276-1. [DOI] [PubMed] [Google Scholar]
- Ritsema T, Hernandez L, Verhaar A, Altenbach D, Boller T, Wiemken A, Smeekens S. Developing fructan-synthesizing capability in a plant invertase via mutations in the sucrose-binding box. Plant J. 2006;48:228–237. doi: 10.1111/j.1365-313X.2006.02862.x. [DOI] [PubMed] [Google Scholar]
- Rogers JD, Scannapieco FA. RegG, a CcpA homolog, participates in regulation of amylase-binding protein A gene (abpA) expression in Streptococcus gordonii. J Bacteriol. 2001;183:3521–3525. doi: 10.1128/JB.183.11.3521-3525.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Yamamoto Y, Kizaki H, Kuramitsu HK. Isolation, characterization and sequence analysis of the scrK gene encoding fructokinase of Streptococcus mutans. J Gen Microbiol. 1993;139:921–927. doi: 10.1099/00221287-139-5-921. [DOI] [PubMed] [Google Scholar]
- Shaper M, Hollingshead SK, Benjamin WH, Jr, Briles DE. PspA protects Streptococcus pneumoniae from killing by apolactoferrin, and antibody to PspA enhances killing of pneumococci by apolactoferrin [corrected] Infect Immun. 2004;72:5031–5040. doi: 10.1128/IAI.72.9.5031-5040.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson CL, Russell RR. Identification of a homolog of CcpA catabolite repressor protein in Streptococcus mutans. Infect Immun. 1998;66:2085–2092. doi: 10.1128/iai.66.5.2085-2092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. Questions about the behaviour of bacterial pathogens in vivo. Philos Trans R Soc Lond B Biol Sci. 2000;355:551–564. doi: 10.1098/rstb.2000.0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stulke J, Hillen W. Regulation of carbon catabolism in Bacillus species. Annu Rev Microbiol. 2000;54:849–880. doi: 10.1146/annurev.micro.54.1.849. [DOI] [PubMed] [Google Scholar]
- Sweeney NJ, Klemm P, McCormick BA, Moller-Nielsen E, Utley M, Schembri MA, et al. The Escherichia coli K-12 gntP gene allows E. coli F-18 to occupy a distinct nutritional niche in the streptomycin-treated mouse large intestine. Infect Immun. 1996a;64:3497–3503. doi: 10.1128/iai.64.9.3497-3503.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney NJ, Laux DC, Cohen PS. Escherichia coli F-18 and E. coli K-12 eda mutants do not colonize the streptomycin-treated mouse large intestine. Infect Immun. 1996b;64:3504–3511. doi: 10.1128/iai.64.9.3504-3511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzer JM, Thompson A, Wen ZT, Burne RA. Streptococcus mutans: fructose transport, xylitol resistance, and virulence. J Dent Res. 2006;85:369–373. doi: 10.1177/154405910608500417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchawa Yimga M, Leatham MP, Allen JH, Laux DC, Conway T, Cohen PS. Role of gluconeogenesis and the tricarboxylic acid cycle in the virulence of Salmonella enterica serovar Typhimurium in BALB/c mice. Infect Immun. 2006;74:1130–1140. doi: 10.1128/IAI.74.2.1130-1140.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titgemeyer F, Hillen W. Global control of sugar metabolism: a gram-positive solution. Antonie Van Leeuwenhoek. 2002;82:59–71. [PubMed] [Google Scholar]
- Velayudhan J, Jones MA, Barrow PA, Kelly DJ. l-Serine catabolism via an oxygen-labile l-serine dehydratase is essential for colonization of the avian gut by Campylobacter jejuni. Infect Immun. 2004;72:260–268. doi: 10.1128/IAI.72.1.260-268.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana R, Perez-Martinez G, Deutscher J, Monedero V. The glycolytic genes pfk and pyk from Lactobacillus casei are induced by sugars transported by the phosphoenolpyruvate: sugar phosphotransferase system and repressed by CcpA. Arch Microbiol. 2005;183:385–393. doi: 10.1007/s00203-005-0003-6. [DOI] [PubMed] [Google Scholar]
- Wang B, Kuramitsu HK. Control of enzyme IIscr and sucrose-6-phosphate hydrolase activities in Streptococcus mutans by transcriptional repressor ScrR binding to the cis-active determinants of the scr regulon. J Bacteriol. 2003;185:5791–5799. doi: 10.1128/JB.185.19.5791-5799.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weickert MJ, Adhya S. A family of bacterial regulators homologous to Gal and Lac repressors. J Biol Chem. 1992;267:15869–15874. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.