Abstract
Despite our increasing knowledge on the transcriptional networks connecting abscisic acid (ABA) signalling with the circadian clock, the molecular nodes in which both pathways converge to translate the environmental information into a physiological response are not known. Here, we provide evidence of a feedback mechanism linking the circadian clock with plant responses to drought. A key clock component (TOC1, timing of CAB expression 1) binds to the promoter of the ABA-related gene (ABAR/CHLH/GUN5) and controls its circadian expression. TOC1 is in turn acutely induced by ABA and this induction advances the phase of TOC1 binding and modulates ABAR circadian expression. Moreover, the gated induction of TOC1 by ABA is abolished in ABAR RNAi plants suggesting that the reciprocal regulation between ABAR and TOC1 expression is important for sensitized ABA activity. Genetic studies with TOC1 and ABAR over-expressing and RNAi plants showed defective responses to drought, which support the notion that clock-dependent gating of ABA function is important for cellular homeostasis under dry environments.
Keywords: abscisic acid (ABA), Arabidopsis thaliana, circadian clock, feedback loops, drought
Introduction
Plants, as sessile organisms, have evolved a number of ways to rapidly sense and adjust to the changing environmental conditions. One of the most studied mechanisms involved in these adjustments is the biological or circadian clock, which is able to maintain biological rhythms with a period of approximately 24 h (Más, 2005; McClung, 2008; Harmer, 2009). Circadian clocks were suggested to confer adaptive advantages to organisms (Ouyang et al, 1998; Green et al, 2002; Michael et al, 2003; Dodd et al, 2005) by means of not merely responding to the external signals but also anticipating the predictable environmental variations that occur during the day–night transitions. The presence of an endogenous timing system thereby allows the synchronization of biological activities to occur at specific phase relationships with the environment, permitting the temporal separation of incompatible metabolic events (Más, 2005; Hotta et al, 2007; McClung, 2008; Harmer, 2009).
Intensive research efforts have been devoted in past years to decipher the molecular and biochemical mechanisms underlying circadian clock function. In plants, as in many other organisms, the circadian rhythmicity seems to rely on multiple negative feedback loops at the core the oscillator (Bell-Pedersen et al, 2005; Wijnen and Young, 2006). Molecular-genetic approaches in Arabidopsis thaliana have aided in the identification of clock components and mechanisms of regulation within the circadian oscillator (Más, 2008). The reciprocal regulation between a pair of Myb transcription factors, CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) (Wang and Tobin, 1998) and LATE ELONGATED HYPOCOTYL (Schaffer et al, 1998) with a pseudo-response regulator TIMING OF CAB EXPRESSION 1 (TOC1 or PRR1) (Strayer et al, 2000; Makino et al, 2002) was initially proposed as a core feedback mechanism important for clock function (Alabadí et al, 2001). Computer modelling and experimental validation have subsequently confirmed the existence of additional, interconnected multiple loops that confer robustness and flexibility to the oscillatory activities (Rand et al, 2004; Locke et al, 2006; Zeilinger et al, 2006).
Despite the increasing knowledge of the molecular networks comprising the Arabidopsis circadian oscillator, little is known about the precise biochemical and molecular function of TOC1 within the clock. TOC1 mutant and RNAi plants display a short-period phenotype for clock-controlled gene expression as well as day length-insensitive flowering time (Strayer et al, 2000). The constitutive high expression of TOC1 leads to arrhythmia under constant light conditions in a number of clock outputs (Makino et al, 2002; Más et al, 2003a). TOC1 was also proposed to function between the environmental signals and the circadian and photo-morphogenic outputs (Más et al, 2003a). Further studies have evidenced that a very precise regulation of TOC1 gene and protein rhythmic expression is essential for proper clock function. Different mechanisms contribute to this regulation including changes in chromatin structure (Perales and Más, 2007), transcriptional regulation (Alabadí et al, 2001) and protein degradation by the proteasome pathway (Más et al, 2003b), which together, accurately control the 24 h rhythmic oscillation of TOC1 expression and function.
The use of genome-wide approaches has provided some clues into the global transcriptional networks controlled by the clock. Circadian clock regulation of the Arabidopsis transcriptome is highly extensive although the precise fraction of the genome that is circadian regulated highly varies among the different studies (Harmer et al, 2000; Schaffer et al, 2001; Michael and McClung, 2003; Edwards et al, 2006; Covington and Harmer, 2007; Dodd et al, 2007; Covington et al, 2008; Mizuno and Yamashino, 2008; Michael et al, 2008a, 2008b; Hazen et al, 2009). A recent study has integrated the information from multiple circadian datasets to get a better estimation of the expressed gene fraction that is circadianly regulated (Covington et al, 2008). The use of tilling arrays has also identified the circadian regulation of intergenic regions, introns and natural antisense transcripts, which has extended the pervasiveness of clock function far beyond protein coding genes (Hazen et al, 2009). The functional clustering of clock-regulated genes has also provided insights into metabolic and physiologic pathways that are under circadian control. Indeed, clock-regulated genes are over-represented among several plant hormone and stress-responsive pathways. This is consistent with the diurnal variations of phytohormone abundance (Robertson et al, 2009) and with the gated regulation of plant hormone sensitivity by the clock (Covington and Harmer, 2007; Dodd et al, 2007).
One of the hormones regulated by the circadian clock is abscisic acid (ABA). This phytohormone is essential in the regulation of many plant growth and development processes as well as in the control of plant responses to stressful environments (Leung and Giraudat, 1998; Finkelstein et al, 2002; Zhu, 2002). The significant number of ABA-related genes that are controlled by the clock has been reported in various studies (Covington and Harmer, 2007; Dodd et al, 2007; Mizuno and Yamashino, 2008). Furthermore, the regulation between ABA and clock signalling pathways is bidirectional as treatment with ABA lengthens the circadian period of gene promoter activity (Hanano et al, 2006). Evidence of a feedback loop between cADPR and the circadian oscillator also suggests a mechanism by which the circadian coordination of ABA-transducing components might be involved in clock-mediated regulation of plant responses to ABA (Dodd et al, 2007). The influence of the clock is not limited to gene expression but is also evidenced in the circadian regulation of physiological processes controlled by ABA. For instance, under water-deficit conditions, ABA induces the closure of the stomatal pore, and this closure was found to be gated by the clock. Indeed, ABA is less effective at closing the stomata in the morning than in the afternoon (Correia et al, 1995), which may ensure that stomata are closed in the heat of the afternoon if water supply is limited (Robertson et al, 2009).
The cellular changes in ABA concentration (Verslues and Zhu, 2007) trigger a downstream network of signalling cascades that ultimately regulate vegetative and reproductive processes and result in enhanced plant tolerance to the environmental stress (Hirayama and Shinozaki, 2007). Essential for ABA signalling is the perception of hormone changes by ABA receptors (Hirayama and Shinozaki, 2007). Among the possible hormone sensors, recent reports have proposed that regulators of ABA-related protein phosphatases 2C may function as ABA receptors within the ABA signalling pathway (Ma et al, 2009; Park et al, 2009). The H subunit of the magnesium-protoporphyrin IX chelatase (ABAR/CHLH/GUN5, At5g13630) was arguably proposed to function as an ABA receptor because of its ABA-binding activity and the ABA-related phenotypes in germination, stomatal closure and responses to water-deficit conditions of ABAR mis-expressing plants (Shen et al, 2006). In broad bean (Vicia faba), ABAR is also involved in ABA-induced stomatal signalling and, equally to its homologous in Arabidopsis, was proposed to specifically bind ABA (Zhang et al, 2002). However, the possible function of ABAR as a receptor was questioned in a study reporting that in barley (Hordeum vulgare L.), the magnesium chelatase large subunit does not bind ABA (Muller and Hansson, 2009). This issue was addressed in a recent study by using a newly developed ABA-affinity chromatography technique, which convincingly showed the specificity of the ABA binding to ABAR (Wu et al, 2009). Furthermore, the authors also assigned ABA-related functions to particular ABAR protein domain (Wu et al, 2009). Altogether, the results in Arabidopsis consign an important role for ABAR within the ABA signalling pathway.
Here, we uncover ABA-related phenotypes for TOC1 mis-expressing plants and confirm a role for ABAR in the ABA signalling pathway. TOC1 is induced by ABA, and this induction is gated by the clock and determines the timing of TOC1 binding to the ABAR promoter. Molecular-genetic studies show the existence of a negative feedback loop in which TOC1 negatively regulates the expression of ABAR, whose activity is in turn necessary for TOC1 activation by ABA. Our studies suggest that proper timing of this feedback loop is important for ABA-mediated changes in gene expression and plant responses to drought conditions.
Results
Genome-wide analysis of TOC1 transcriptional networks
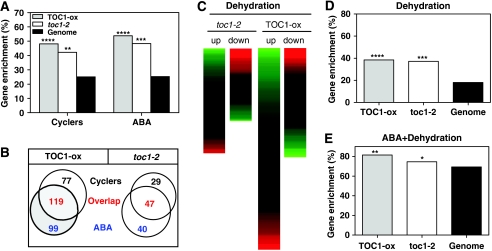
To gain insights into the mechanisms of TOC1 function in the clock, we performed genome-wide transcriptomic analysis of wild-type (WT) and TOC1 over-expressing plants (TOC1 cDNA fused to YFP, herein denominated TOC1-ox) (Más et al, 2003a) synchronized under 12 h light:12 h dark (LD) cycles for 10 days. Statistical analysis showed a total of 245 significantly increased and 160 significantly decreased genes in TOC1-ox compared with WT (Supplementary Table S1). We also analysed transcriptomic data of WT and TOC1 mutant plants (toc1-2) (Strayer et al, 2000) synchronized under LD cycles followed by 2 days under constant light (LL) conditions and found that about 75 genes were downregulated and 105 upregulated in toc1-2 samples (Supplementary Table S2). Using a recently published circadian dataset (Hazen et al, 2009) we found that about 50% (in TOC1-ox, Fisher's exact test P=2.2 × 10−16) and over 40% (in toc1-2, Fisher's exact test P=1.8 × 10−7) of the mis-regulated genes showed circadian fluctuations in mRNA abundance (Figure 1A and Table I). Similar or even higher percentages were obtained when other circadian datasets (Covington et al, 2008) were examined (Table I). Among all of the mis-regulated genes, we were expecting to find a significant proportion of clock-related genes not directly regulated by TOC1. Noticeably, meta-analysis of mis-regulated genes using databases that contain a broad spectrum of transcriptomic profiles (https://www.genevestigator.com/gv/index.jsp) also showed a significant overlap with transcripts involved in ABA signalling pathways (Supplementary Figure S1). Functional clustering using specific ABA-related datasets (Matsui et al, 2008) confirmed the highly significant number of ABA-responsive genes in TOC1-ox (53.82%, Fisher's exact test P=2.2 × 10−16) and in toc1-2 (48.33%, Fisher's exact test P=6.0 × 10−12) (Figure 1A; Table I). Both, upregulated and downregulated set of genes were found to be associated with ABA transcriptional networks (Supplementary Figure S2A). The list of mis-regulated genes included relevant components of the ABA signalling pathway (Finkelstein et al, 2002) such as ABA1, Histone H1-3, RD29B, several late embryogenesis abundant proteins and ABAR/CHLH/GUN5 (Supplementary Tables S3 and S4). We also checked whether clock dysfunction in TOC1 mis-expressing plants was affecting other hormone transcriptional networks. However, functional clustering analysis showed that, with the exception of methyl jasmonate, no other hormone pathways were as significantly affected as the ABA (Supplementary Figure S2B and C). Furthermore, a significant proportion of these ABA-related genes show a circadian oscillation (54.58% in TOC1-ox, Fisher's exact test P=1.36 × 10−7 and 54.02% in toc1-2, Fisher's exact test P=0.6 × 10−4) (Figure 1B and Table I).
Figure 1.
Genome-wide analysis of TOC1 transcriptional networks. (A) Percentages of cyclers and ABA-related genes at the whole-genome level, and in TOC1-ox and toc1-2 transcriptomic datasets (**P-value<10−8; ***P-value<10−11; ****P-value<10−15) (B) Venn diagrams showing the overlap between cyclers and ABA-related genes in TOC1-ox and toc1-2 transcriptomic datasets. (C) Hierarchical clustering of toc1-2 and TOC1-ox mis-regulated genes with transcripts involved in dehydration responses. Percentages of dehydration (D) at the whole-genome level, and in TOC1-ox and toc1-2 transcriptomic datasets, and among these, the ABA-related genes (E) (*P-value<10−3; **P-value<10−6; ***P-value<10−11; ****P-value<10−15). Details of datasets used for analysis are described in Supplementary data.
Table 1.
Meta-analysis of TOC1 mis-regulated genes with publicly available datasets of cycling, ABA- and drought-related genes
| Hormone/ treatment/condition | TOC1-ox | toc1-2 | ||
|---|---|---|---|---|
| Enriched genes (%) | P-value | Enriched genes (%) | P-value | |
| Cyclers (Hazen et al, 2009) | 48.14 | 2.2 × 10−16 | 42.22 | 1.7 × 10−7 |
| Cyclers (Covington et al, 2008) | 55.17 | 2.2 × 10−16 | 59.22 | 5.6 × 10−9 |
| ABA (Matsui et al, 2008) | 53.82 | 2.2 × 10−16 | 48.33 | 6.0 × 10−12 |
| ABA (Matsui et al, 2008) Cyclers (Hazen et al, 2009) | 54.58 | 1.36 × 10−7 | 54.02 | 6.2 × 10−4 |
| Drought (Matsui et al, 2008) | 38.51 | 2.2 × 10−16 | 37.22 | 3.4 × 10−10 |
Our data analysis also showed that about 38% of the mis-regulated genes in TOC1-ox (Fisher's exact test P=2.2 × 10−16) and in toc1-2 (Fisher's exact test P=3.4 × 10−10) were related to plant responses to dehydration (Figure 1D and Table I). These percentages are higher than the 18% of the genes that are associated with drought responses in the genome (Matsui et al, 2008). This is noteworthy because the plant response to changes in water status is one of the many processes controlled by ABA. Consistently, the analysis of common elements in the promoters of the mis-expressed genes also showed a significant over-representation of motifs implicated in molecular responses to dehydration (Supplementary Table S5). Furthermore, two- and three-fold comparisons of circadian, ABA and drought-related genes showed that 81.41% (in TOC1-ox) and a 74.62% (in toc1-2) of the drought-related genes are regulated by ABA (Figure 1E). We also found an important overlapping among ABA and drought genes that were regulated by the circadian clock in both TOC1-ox (Supplementary Figure S2D) and toc1-2 (Supplementary Figure S2E) datasets. Analysis by quantitative PCR (Q-PCR) confirmed the mis-expression of some selected genes in TOC1-ox and toc1-2 mutant plants (Supplementary Figures S3 and S4). When other relevant dehydration-related genes were analysed by Q-PCR, we also found a significant mis-expression, suggesting that the drought-related transcriptional networks altered in TOC1 mis-expressing plants might be even more important than estimated by our microarray data. Altogether, these data led us to further examine the possible link between TOC1 and plant responses to drought mediated by ABA.
ABA-mediated responses to drought are impaired in TOC1 mis-expressing plants
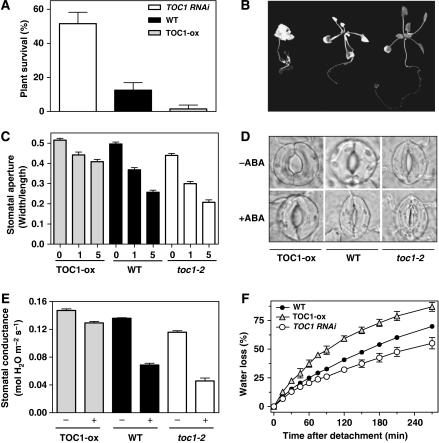
If the changes in gene expression correlate with functional alterations in physiological responses, then plants mis-expressing TOC1 should display ABA-related phenotypes. Data analysis showed that many stress-responsive genes known to be involved in dehydration tolerance (Ingram and Bartels, 1996; Thomashow, 1999) were downregulated in TOC1-ox plants, suggesting that TOC1-ox plants might have reduced tolerance to drought. We therefore analysed responses to drought stress in WT, TOC1-ox, toc1-2 and TOC1 RNAi plants (Más et al, 2003a) by examining percentages of recovery after dehydration using seedlings grown on agar plates (Fujita et al, 2005). Our results showed that most TOC1-ox plants were unable to recover from the drought stress, exhibiting a reduced survival rate (2%) after re-watering (Figure 2A and B). In contrast, approximately 50% of the toc1-2 (not shown) and 52% of TOC1 RNAi plants completely recovered the drought (Figure 2A and B) whereas WT plants showed intermediate phenotypes and about 15% recovered after re-watering (Figure 2A and B). Thus, over-expression of TOC1 significantly reduced the plant tolerance to drought (P-value<0.0001) whereas toc1-2 and TOC1 RNAi plants responded better than WT to water-deficit conditions (P-value<0.0001). The survival rates of TOC1 RNAi plants on dehydration are a long-term response that is unlikely because of altered clock phase in these plants.
Figure 2.
Altered responses to drought conditions of plants mis-expressing TOC1. (A) Plant survival to dehydration stress on agar plates. Data are means±s.e.m. of duplicate experiments with at least 25 plants per genotype. (B) Representative photographs of TOC1-ox (left), WT (middle) and TOC1 RNAi (right) plants of the dehydration experiments. (C) Stomatal aperture in rosette epidermis of TOC1-ox, WT and toc1-2 plants. Stomatal dimensions were measured after incubation for 2 h in a buffer containing 0, 1 or 5 μM ABA. Data are means±s.e.m. of duplicate experiments with at least 100 stomata per genotype and per treatment. (D) Representative images by light microscopy of the stomata guard cells. (E) Stomatal conductance of TOC1-ox, WT and toc1-2 mutant plants. Gas exchange measurements were performed with at least 10 rosettes for genotype. (F) Water-loss rates of detached rosettes from WT, TOC1-ox and toc1-2 plants. Data are means±s.e.m. of triplicate measurements with at least five rosettes for genotype at each time point. Compared with WT, the phenotypic differences were statistically significant with P-values below 0.01 in all cases.
The altered tolerance of TOC1 mis-expressing plants to water stress might be due to deficient stomatal closure under dehydration conditions. Thus, we next examined stomatal guard cell movement that is controlled by both ABA and the circadian clock (Robertson et al, 2009). The experiments were performed at midday, a time when stomata are more responsive to ABA (Robertson et al, 2009). As expected, ABA treatment in WT leaves caused significant stomatal closure (Figure 2C and D). However, this effect was not as pronounced in TOC1-ox whereas stomata in TOC1 RNAi (not shown) and toc1-2 mutant leaves were more effectively closed (Figure 2C and D). Compared with WT, the stomatal response to ABA was significantly altered in TOC1-ox (P-value<0.001), TOC1 RNAi (P-value<0.01) and toc1-2 (P-value<0.01). Consistent with these results, the stomatal conductance of TOC1-ox plants was not as responsive to the treatment with ABA as in WT plants (Figure 2E) whereas toc1-2 mutant plants showed an increased response (Figure 2E). Statistical analysis verified the significance of the differential ABA effects, with P-values<0.001. If the stomatal closure and conductance are impaired in TOC1 over-expressing plants, then the leaf water-loss rates might also be affected. Thus, we next measured the water-loss rates of detached rosettes in TOC1 mis-expressing plants. Our studies showed that 3 h after detachment, the leaves of TOC1 RNAi plants showed a 40% of water loss in contrast to the 54 and 75% of WT and TOC1-ox plants, respectively (Figure 2F). These differences were observed throughout the time course analysis (Figure 2F).
Collectively, our results indicate that proper TOC1 expression is important for stomatal function and regulation of leaf transpiration rates, a notion that is reinforced by the opposite phenotypes of TOC1 over-expressing and mutant plants. In addition to the previously reported and well-characterized clock phenotypes, our study shows an unpredicted role for TOC1 in the control of ABA-mediated plant responses to drought.
TOC1 regulates the diurnal and circadian expression of ABAR
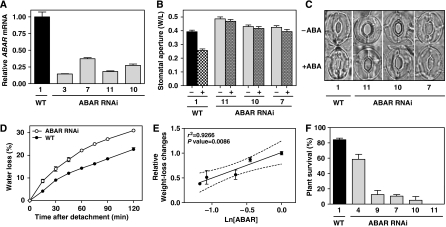
One of the mis-regulated genes in TOC1 microarray datasets was ABAR/CHLH/GUN5 (Shen et al, 2006; Wu et al, 2009). This was of interest because ABAR was initially reported as an ABA receptor and because preliminary studies in the laboratory had linked TOC1 with ABAR (Riechmann JL and Mas P, unpublished results). Although a recent report in barley has questioned the function of ABAR in ABA signalling (Muller and Hansson, 2009), our phenotypic studies using different Arabidopsis ABAR RNAi lines with decreased patterns of ABAR expression (Figure 3A) were consistent with previous reports (Shen et al, 2006; Wu et al, 2009). Indeed, compared with the effect of ABA on WT plants (Figure 3B and C), the stomata in different ABAR RNAi lines were almost insensitive to the treatment with the hormone (Figure 3B and C). Differences were statistically significant in the absence (P-value<0.0001) and in the presence of ABA (P-value<0.0001). Consistent with these and the previously described phenotypes (Shen et al, 2006; Wu et al, 2009), our studies also showed that the water-loss rates of ABAR RNAi plants were significantly higher than in WT (P-value<0.005 at all time points) (Figure 3D). Furthermore, comparisons of ABAR mRNA expression with the severity of water-loss phenotypes in different RNAi lines showed a significant inverse correlation between ABAR mRNA abundance and water-loss rates (Figure 3E). Finally, our studies also showed that the impaired stomatal closure and water-loss rates also correlated with altered responses to dehydration conditions (Figure 3F) with a reduced survival percentage of the different ABAR RNAi plants. Altogether, our results are in concordance with the previous study in Arabidopsis and verify a role of ABAR within the ABA signalling pathway in Arabidopsis.
Figure 3.
Altered responses to drought conditions of plants mis-expressing ABAR. (A) Analysis by Q-PCR of ABAR mRNA expression in WT and several ABAR RNAi lines. (B) Stomatal aperture in rosette epidermis after incubation for 2 h in a buffer containing 0 or 5 μM ABA. Data are means±s.e.m. of duplicate experiments with at least 100 stomata per genotype and per treatment. (C) Representative images by light microscopy of the stomata guard cells. (D) Water-loss rates of detached rosettes from WT and ABAR RNAi plants. Data are means±s.e.m. of triplicate measurements with at least five rosettes for genotype at each time point. (E) Correlation between ABAR mRNA abundance and weight-loss changes of detached rosettes. Weight-loss changes were plotted relative to the WT value. (F) Plant survival to dehydration stress on agar plates. Data are means±s.e.m. of duplicate experiments with at least 25 plants per genotype. Compared with WT, the phenotypic differences were statistically significant with P-values below 0.005 in all cases.
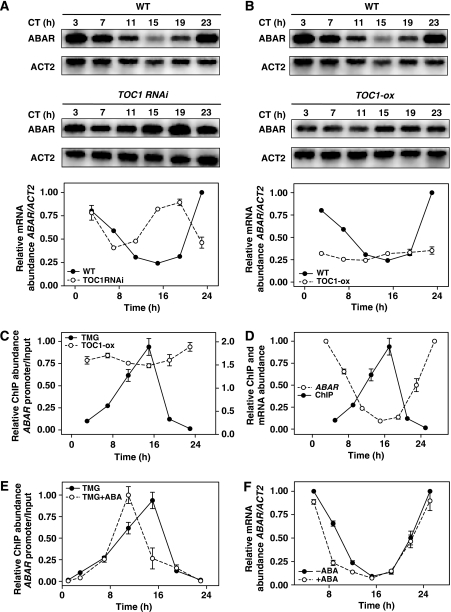
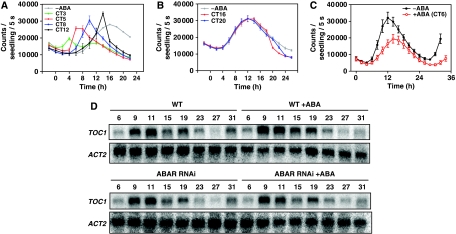
On the basis of the ABA-related phenotypes and the pattern of ABAR expression in TOC1 microarrays, we next explored the possible link between the circadian clock and ABAR expression. To do so, we first performed time course analyses over a diurnal cycle of WT plants grown under short-day (ShD) and long-day (LgD) conditions. Our results showed that ABAR transcript abundance rhythmically oscillated with a peak of expression close to dawn (Supplementary Figure S5A). Similar oscillatory patterns were observed when ABAR expression was examined under LL conditions (Supplementary Figure S5B) indicating that the circadian clock controls the expression of ABAR. As TOC1 is an essential component of the Arabidopsis circadian system, we reasoned that the diurnal and circadian expression of ABAR might be controlled by TOC1. Indeed, our results showed that the waveform of ABAR mRNA was altered in TOC1 RNAi and TOC1-ox plants under LL. The phase of ABAR expression in TOC1 RNAi was advanced, leading to higher mRNA abundance at times when ABAR expression is at its minimum in WT plants (Figure 4A). The waveform of ABAR expression in TOC1 RNAi plants also suggests that ABAR might be complexly regulated in the absence of a functional TOC1. In TOC1-ox plants, ABAR expression was clearly affected, with very reduced abundance throughout the circadian cycle (Figure 4B). Changes in ABAR expression were also observed in TOC1-ox and TOC1 RNAi plants growing under LD cycles (Supplementary Figure S5C and D). The alterations in ABAR expression were also observed when seedlings were treated with ABA (Supplementary Figure S5E and F).
Figure 4.
TOC1 binds to the ABAR promoter and regulates ABAR circadian expression. (A, B) Northern blot analysis of TOC1 RNAi and TOC1-ox plants synchronized under LD cycles followed by 2 days under LL. The waveforms of ABAR expression after mRNA quantification are shown below the blots. CT, circadian time. (C) ChIP assays with TOC1-ox (right axis) and TMG plants (left axis) after TOC1 immuno-precipitation with an antibody to YFP followed by Q-PCR amplification of the ABAR promoter. (D) Comparison of the antiphasic waveforms of ABAR expression by northern blot and TOC1 binding to the ABAR promoter by ChIP assays. (E) Effects of ABA on the waveform of TOC1 binding to the ABAR promoter. (F) Effects of ABA on the ABAR mRNA expression. ChIP abundance was plotted relative to the maximum value.
Together, these results indicate that the circadian clock controls the timing of ABAR expression. Our studies also assign a key function for TOC1 in modulating the phase and amplitude of ABAR diurnal and circadian waveform. The circadian period and/or diurnal phase changes in toc1-2 and TOC1-ox plants might affect the proper expression of ABAR and other ABA/drought-related genes, and this mis-regulation may alter the proper timing of plant responses to drought.
TOC1 is physically associated to the ABAR promoter
As DNA-binding events are crucial in ABA signalling pathways (Busk and Pagès, 1998), we next explored whether regulation of ABAR expression occurred through binding of TOC1 to the ABAR locus. Chromatin immunoprecipitation (ChIP) assays were performed with TOC1-ox and TOC1 minigen plants (TMG, expressing the TOC1 gene under its own promoter) (Más et al, 2003a). The TMG plants more closely resemble the WT conditions (as compared with the arrhythmic TOC1-ox) although the TMG clock runs slower than in WT, with a delayed phase of rhythmic gene expression (Más et al, 2003a). Time course experiments throughout the 24 h cycle showed significant amplification, consistent with the in vivo binding of TOC1 to the ABAR promoter (Figure 4C). Further analysis showed a rhythmic oscillation in the binding, with a peak at a time when ABAR expression is minimal (Figure 4C and D). Our results showed that TOC1 binding is antiphasic to ABAR mRNA expression (Figure 4D) and this is consistent with the inverse correlation between abundance and phenotypes of TOC1 and ABAR. The binding seems to be restricted to the ABAR promoter, and more specifically located around the transcription start site (Supplementary Figure S6). No evident amplification was observed with primers flanking a region in the fourth exon of the ABAR gene (Supplementary Figure S6). Binding ChIP assays with TOC1-ox plants showed significant amplification of the ABAR promoter at all times examined, indicating that TOC1 protein remains bound throughout the cycle (Figure 4C). This constant binding (as opposed to the oscillatory waveform in TMG plants) correlates with a constant repression of ABAR (compare Figure 4B and C) and might account for the ABA-related phenotypes observed in TOC1-ox plants. On the basis of this notion, we expected that treatment with ABA should affect TOC1 binding and/or ABAR expression. To check this possibility, we next performed a time course of ChIP assays in TMG samples treated with the hormone. We also compared by northern blot the pattern of ABAR expression in ABA-treated and untreated plants. Our results showed a transient change in the phase of TOC1 binding to the ABAR promoter in ABA-treated TMG plants (Figure 4E). Concomitantly, we also observed a change in ABAR mRNA waveform, with a significant acute repression (P-value<0.005) around midday, the time window when TOC1 is preferentially bound to the ABAR promoter (Figure 4F). These results indicate a temporal coincidence between the ABA-mediated regulation of ABAR expression and TOC1 binding to the ABAR promoter. The specificity of this association was confirmed by analysing TOC1 binding to other relevant ABA-related loci. Our results showed that with the exception of ABAR, none of the selected loci was amplified (Supplementary Figure S7). Binding was not observed even after treating the seedlings with ABA (Supplementary Figure S8).
Altogether, our results show that TOC1 binds to the ABAR promoter. This binding is regulated not only by the circadian clock but also by the ABA, which advances the phase of TOC1 binding and acutely represses ABAR expression around midday. On the basis of the phenotypes of ABAR and TOC1 mis-expressing plants, this regulation is most likely important for ABA-mediated plant responses to drought.
TOC1 expression is acutely induced by ABA and this regulation is gated by the clock and requires a functional ABAR
Relevant to our studies was the observation that TOC1 was included in the microarray dataset of genes regulated by ABA (Matsui et al, 2008). To examine this new regulatory mechanism, we monitored bioluminescence rhythms of plants expressing the luciferase gene (LUC) driven by the TOC1 promoter (TOC1:LUC) in the presence of ABA. Our results showed that ABA treatments administered during the subjective day acutely induced TOC1:LUC expression (Figure 5A). The magnitude of induction was not constant but progressively increased throughout the day, reaching maximum values at 5–10 h after the subjective dawn (Figure 5A). Administration of ABA during the subjective night had no clear immediate consequences on TOC1:LUC amplitude or phase (Figure 5B). On subsequent days, the extended time with the hormone led to a decreased amplitude and slight delay in the phase of TOC1∷LUC expression. The time-of-day-dependent regulation of TOC1 expression by ABA indicates that the acute response is gated by the clock and correlates with the timing of TOC1 binding and changes in ABAR expression by ABA.
Figure 5.
TOC1 is acutely induced by ABA and this regulation is gated by the clock and requires a functional ABAR. (A, B) TOC1∷LUC expression in WT plants treated with 25 μM ABA at the indicated circadian times (CT) during the subjective day and night. (C) TOC1∷LUC expression in ABAR RNAi plants treated with 25 μM ABA at the indicated CT during the subjective day. Data are means±s.e.m. of luminescence from 6–12 plants. (D) Northern blot analysis of TOC1 mRNA expression in WT and ABAR RNAi plants. TOC1 expression was compared in the absence or in the presence of ABA.
The transient induction of TOC1 is suggestive of a fine-tuned switch mechanism that regulates the ABA-mediated effects on TOC1 expression. As feedback loops are common mechanisms for precise regulation of gene expression, we explored the possible function of ABAR in TOC1 induction by ABA. We therefore examined the regulation of TOC1:LUC expression in ABAR RNAi plants, lacking a functional expression of the ABAR gene. Our results showed that in the absence of ABA, TOC1∷LUC expression in ABAR RNAi plants was similar to the one observed in WT plants, although we found a slightly advanced phase of TOC1∷LUC expression that was not reproducibly observed in all ABAR RNAi lines (Supplementary Figure S9). After treatment with ABA, the acute induction of TOC1:LUC in WT plants was completely abolished in ABAR RNAi plants (Figure 5C) with a slight delay in the oscillatory phase of TOC1:LUC expression and with bioluminescence signals lower than those of untreated ABAR RNAi samples (Figure 5C). Furthermore, upregulation of TOC1:LUC expression by ABA was not observed at any time during the circadian cycle. The role of ABAR in TOC1 induction was also verified by northern blot analysis. Our results showed that in ABAR RNAi samples, TOC1 mRNA steady state was not upregulated after ABA treatment as opposed to the gated induction observed in WT plants (Figure 5D and E; Supplementary Figure S10). Together, these results indicate that the ABA-mediated acute induction of TOC1 requires the presence of a functional ABAR, which positively regulates the expression of TOC1. Noticeably, this regulation seems to have an important function only in the presence of ABA. In the absence of exogenous hormone, the waveform of TOC1 expression is not clearly affected, with the possible exception of a slightly advanced phase. These results suggest that ABAR is important for regulation of TOC1 expression within the ABA signalling pathway but not within the circadian pathway. Our results also indicate the existence of a regulatory feedback loop involving the reciprocal regulation of ABAR and TOC1.
Biological relevance of ABAR and TOC1 interaction
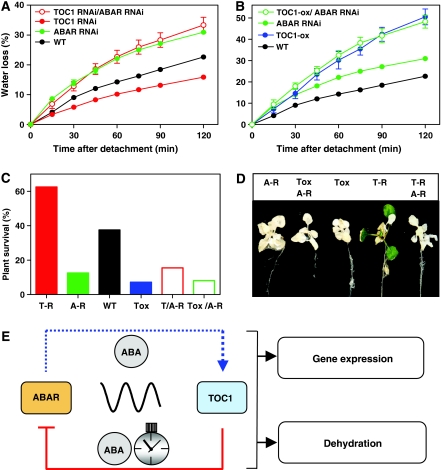
To further investigate whether the reciprocal regulation between TOC1 and ABAR expression is important for ABA signalling, we conducted a genetic study in which the TOC1-ox and the TOC1 RNAi plants were transformed with the ABAR RNAi construct (Shen et al, 2006). Several lines for each genotype were selected and the decreased ABAR expression was verified by Q-PCR (Supplementary Figure S11). Initial studies were focused on water-loss rates, as analysis of different ABAR RNAi transgenic lines showed that the severity of the water-loss phenotypes correlated with the ABAR mRNA abundance (Figure 3). In our genetic studies, we predicted that if the TOC1 RNAi phenotypes were due to increased ABAR expression, then the significantly diminished transpiration rate of TOC1 RNAi plants should be overridden by the opposing phenotype of ABAR RNAi plants. Conversely, if the ABA-related phenotypes of TOC1-ox plants are mediated by the repressed ABAR expression, then the transpiration rates of ABAR RNAi/TOC1-ox plants should be similar to those of plants over-expressing the repressor TOC1. Consistent with these assumptions, our results showed that double TOC1 RNAi/ABAR RNAi plants displayed higher transpiration than WT plants, with similar water-loss rates to those of ABAR RNAi plants (Figure 6A). As predicted, the water-loss phenotype of TOC1 RNAi/ABAR RNAi clearly contrasted to the decreased transpiration of TOC1 RNAi (Figure 6A) indicating that the TOC1 RNAi phenotype is only apparent in the presence of a functional ABAR. Conversely, our results showed that the transpiration rate of TOC1-ox was not significantly affected by the reduced expression of ABAR in TOC1-ox/ABAR RNAi plants. These plants displayed an increased transpiration rate, similar to the phenotype observed for single TOC1-ox (Figure 6B). The ABAR RNAi plants displayed intermediate phenotypes (Figure 6B) as ABAR expression was overall higher in these RNAi lines than in TOC1-ox/ABAR RNAi (Supplementary Figure S11). Similar genetic conclusions were obtained when we performed experiments of dehydration on plates. The high percentages of TOC1 RNAi plant survival after dehydration were completely overridden in the double RNAi plants, suggesting that TOC1 RNAi phenotypes are only evident in the presence of a functional ABAR (Figure 6C and D). Conversely, the reduced survival percentages of TOC1-ox plants were not clearly affected by the reduced ABAR expression in TOC1-ox/ABAR RNAi lines (Figure 6C and D).
Figure 6.
ABAR and TOC1 interaction in plant responses to drought. (A, B) Water-loss percentages of detached rosettes from WT, ABAR RNAi, TOC1 RNAi, double ABAR/TOC1 RNAi, TOC1-ox and TOC1-ox/ABAR RNAi plants. Data are means±s.e.m. of triplicate measurements with at least 10 rosettes for genotype at each time point. (C) Survival percentages of WT, ABAR RNAi (A-R), TOC1 RNAi (T-R), double ABAR/TOC1 RNAi T/A-R, TOC1-ox (Tox) and TOC1-ox/ABAR RNAi (Tox/A-R) plants subjected to dehydration on agar plates. (D) Representative photographs of plants in the dehydration experiments. (E) Schematic representation depicting the reciprocal regulation between TOC1 and ABAR and the implication of ABA and the circadian clock in this regulation.
Altogether, the genetic studies are in agreement with the molecular data and functionally connect the reciprocal regulation of TOC1 and ABAR expression with plant tolerance to drought conditions (Figure 6E). We propose that increased ABA concentrations activate TOC1 expression and this activation is gated by the clock and determines the timing of TOC1 binding to the ABAR promoter and the changes in ABAR expression. Our results indicate that ABAR is required for TOC1 induction by ABA. Proper timing of this feedback loop is important for ABA-mediated changes in gene expression and plant responses to drought conditions.
Discussion
Comparisons of circadian and ABA-responsive transcriptional profiles show that the genome-wide fraction of ABA-related genes controlled by the clock is around 40%, a significant percentage that is in agreement with previous studies (Covington et al, 2008) and with the notion that ABA signalling is under circadian control. The connection of the circadian clock with ABA pathways was also proven in studies showing the overlap between the ABA-related signalling molecule cyclic adenosine diphosphate ribose (cADPR) and the circadian transcriptome (Dodd et al, 2007) and in studies in which a significant proportion of ABA-responsive genes was found to display diurnal oscillation (Mizuno and Yamashino, 2008). Together, these data imply that the ABA pathway, if considered as a clock output, should be altered when the circadian clock is not properly functioning. This is consistent with our studies showing a significant overlap among TOC1-regulated genes and ABA-related transcriptional networks.
Among the multiple roles of ABA along the plant life cycle, the hormone implications in plant responses to stressful environments are well established (Leung and Giraudat, 1998; Finkelstein et al, 2002; Zhu, 2002). Our studies show ABA-mediated drought phenotypes in TOC1-ox, TOC1 RNAi and toc1-2 mutant plants. The stomata phenotypes in toc1-2 and TOC1 RNAi plants were less severe than in TOC1-ox, suggesting a possible functional redundancy with other genes. This would be in consonance with a recent report showing that other members of the TOC1 family, the PRR9, 7 and 5 negatively regulate the biosynthetic pathways of ABA, chlorophyll, carotenoid and alpha-tocopherol (Fukushima et al, 2009). The physiological phenotypes of TOC1 mis-expressing plants establish a direct or indirect connection of TOC1 with plant responses to drought. In this sense, our results were in agreement with previous studies showing that proper rhythmic oscillation of stomatal opening is impaired in toc1-1 (Somers et al, 1998; Dodd et al, 2005). In these studies, it was proposed that the clock might allow stomata to anticipate dusk, contributing to increased water-use efficiency. The differential plant responses to drought, stomatal aperture and water-loss rates of TOC1 over-expressing and mutant plants were thereby consistent with previous studies and validated our microarray data.
Our studies show that ABAR is mis-regulated in TOC1-ox and TOC1 RNAi plants. A recent study has questioned the function of ABAR as a receptor, reporting that in barley, mutant plants do not display ABA phenotypes (Muller and Hansson, 2009). However, this issue was convincingly addressed in a recent study (Wu et al, 2009). Furthermore, our own studies with ABAR RNAi plants were consistent with the phenotypes reported in Arabidopsis, which are also in agreement with the presence of ABA-related motifs in the ABAR promoter and with the inclusion of ABAR as a gene regulated by ABA (Matsui et al, 2008). We found a significant hyposensitivity in ABAR RNAi plants to the ABA-mediated stomata closure, altered water-loss rates and decreased plant survival after dehydration. Furthermore, we observed an inverse correlation between ABAR mRNA abundance and water-loss rates, which highlights the important function of ABAR in the regulation of this plant response. ABAR function was also reinforced by our genetic studies in which the TOC1 RNAi phenotypes were completely reverted by the ABAR RNAi construct, which in addition to provide clues about TOC1 and ABAR genetic interaction, also assign a function for ABAR in the regulation of plant responses to drought.
Our ChIP results suggest that regulation of ABAR expression by TOC1 occurs through direct binding of TOC1 to the ABAR promoter. A recent study has reported the physical association of TOC1 with chromatin, most likely through interaction with a TCP clock-associated factor-denominated CHE (Pruneda-Paz et al, 2009). It would be interesting to examine whether TOC1 forms protein complexes with CHE or other factors at the ABAR promoter. The mechanisms linking ABA with the circadian clock may also involve the ABA signalling factor ABI3, as it was shown earlier that ABI3 physically interacts with TOC1 (Kurup et al, 2000). In our studies, we found TOC1 binding in the absence of ABA, suggesting a role for TOC1 in the control of ABAR circadian expression. The binding itself is controlled by the clock, judging by the rhythmic oscillation found in TMG plants. Interestingly, treatment with ABA advanced the binding of TOC1 to the ABAR promoter and acutely repressed ABAR expression. Both advanced binding and ABAR repression occurred specifically during the subjective day, which is a highly relevant time in ABA-regulated processes. Indeed, the peak of ABA diurnal fluctuation, the maximal leaf transpiration rate and maximal effectiveness of ABA at closing stomata all occur at this specific time window (Robertson et al, 2009). Furthermore, TOC1 expression is acutely induced by ABA, and this induction is gated by the clock at this particular time. A plausible explanation for all these results is that plants use the circadian clock to coordinate TOC1 induction by ABA at a highly sensible phase. This induction rapidly shifts TOC1 binding to the ABAR promoter, which acutely advances ABAR repression. The effects of ABA treatment on the circadian expression of other clock-associated genes were examined earlier (Hanano et al, 2006). The study showed that ABA lengthens periodicity of bioluminescence in seedlings expressing the promoter of CHLOROPHYLL A/B-BINDING PROTEIN (CAB2/LHCB1*1), COLD- AND CIRCADIAN-REGULATED 2 (CCR2/AtGRP7) and CCA1 (Hanano et al, 2006) fused to the luciferase. The acute effects of ABA, which we observe on TOC1:LUC expression, might not affect the other genes examined in Hanano's study. It is also possible that due to the different experimental designs, the acute changes were missed. In agreement with this notion, we also found that after the acute induction of TOC1, the bioluminescence signals decreased in amplitude with a slight delay in the phase of TOC1:LUC expression. It cannot be ruled out, however, a possible saturation of the response because of the extended time with the high concentration of the hormone.
Interlocked feedback loops are common mechanisms for precise regulation of gene expression within the circadian oscillator (Rand et al, 2004). In the case of other metabolic or physiologic processes, feedback loops might also exert an important regulatory role. For instance, the ABA-insensitive 1(ABI1) and 2 (ABI2) phosphatases were found to function in a negative feedback regulatory loop within the ABA signalling pathway (Merlot et al, 2001). It was suggested that this loop might be responsible for resetting the ABA signalling cascade, thereby allowing the cell to continuously monitor the presence or absence of ABA (Merlot et al, 2001). A similar mechanistic explanation may underlie the reciprocal regulation between TOC1 and ABAR. It would be interesting to examine the mechanisms and identify other components implicated in this regulation. The existence of a feedback loop connecting the clock and ABA signalling pathways is also in line with previous studies showing that cADPR, which has an important function in molecular and physiological ABA responses (Juan-Pablo Sánchez, 2004), forms a feedback loop within the plant circadian clock (Dodd et al, 2007).
Our genetic studies are in agreement with the molecular data and functionally connect the reciprocal regulation of TOC1 and ABAR expression with plant tolerance to drought conditions. Although we could not obtain double ABAR-ox/TOC1-ox or ABAR-ox/TOC1 RNAi plants, the results showing that the phenotypes of TOC1 RNAi plants were reverted in the double ABAR/TOC1 RNAi plants indicating that TOC1 RNAi phenotypes are only evident in the presence of a functional ABAR. Conversely, the drought-related phenotypes of TOC1-ox plants were not significantly affected by decreasing ABAR abundance, reinforcing the negative role of TOC1 in the regulation of ABAR expression. Although our studies do not exclude the interaction of TOC1 with other ABA signalling components, the feedback loop between TOC1 and ABAR links a key oscillator component with drought-related responses. This mechanism might contribute to the clock-controlled gating of ABA activity at midday, a time when transpiration rate is higher and plants need a more precise control of hormone function if water supply is limited. Thus, the circadian gating of stomatal closure would ensure proper adjustments of guard cell closure in the heat of the afternoon, when this regulation is more needed. We propose that the reciprocal regulation between TOC1 and ABAR might function as a fine-tuned switch that helps to modulate the plant sensitivity to ABA, which in turn favours the temporal regulation of plant responses to dry environments (Figure 6E). The direct involvement of an essential clock component in these responses also provides a plausible explanation on how the circadian clock confers an adaptive advantage to plants (Dodd et al, 2005). Detailed characterization of the molecular components involved in clock interaction with other hormones such as auxin (Covington and Harmer, 2007) or cytokinins (Hanano et al, 2006; Salome et al, 2006; Zheng et al, 2006) might be useful to get further insights into the gating mechanisms synchronizing hormone signalling with plant growth and metabolism.
Materials and methods
Plant material, growth conditions and bioluminescence analysis
The TOC1 RNAi, TMG-YFP (TMG), TOC1-YFP-ox (TOC1-ox) and the toc1-2 mutant plants (Strayer et al, 2000; Más et al, 2003a) were provided by Prof. Steve A Kay (University of California San Diego, La Jolla, CA, USA). The ABAR RNAi construct (Shen et al, 2006) was provided by Prof. Da-Peng Zhang (China Agricultural University, Beijing, China). The TOC1 RNAi/ABAR RNAi and TOC1-ox/ABAR RNAi plants were obtained after transforming the ABAR RNAi construct (Shen et al, 2006) by Agrobacterium tumefaciens-mediated DNA transfer (Clough and Bent, 1998). Seedlings were grown on Murashige and Skoog (MS) agar plates with 3% sucrose under controlled environmental conditions (50 μmol m−2 s−1 of cool white fluorescent light at 22°C). Luminescence analyses were performed as described earlier (Portolés and Más, 2007). In experiments of hormone treatments, (+) ABA (A4906; Sigma-Aldrich) was added to seedlings at the indicated concentration at different times during the diurnal and circadian cycle. For microarray experiments, WT (C24) and toc1-2 mutant seeds were stratified in the dark at 4°C for 4 days on MS plates with 3% sucrose. Plates were subsequently placed on a growing chamber under 12 h light:12 h dark cycles (LD) for 10 days followed by constant light conditions (LL). Samples were taken after 40 h in LL. WT (Col-0) and TOC1-ox samples were taken at Zeitgeber Time 16 (ZT-16) from plants synchronized under LD cycles for 10 days. Details of microarray hybridization and analysis are provided in Supplementary data. Microarray data were deposited at GEO (www.ncbi.nlm.nih.gov/geo; GEO accessions: GSE18001 and GSE18007).
ChIP assays
ChIP assays were performed as described earlier(Perales and Más, 2007). Specific details of the procedure are described in Supplementary data. For hormone treatment, seedlings were sprayed 4 h before sampling with 7.5 ml of 0.001% Triton X-100 aqueous solution containing 100 μM (±) ABA and samples were analysed at the indicated time points. Q-PCR was performed with the SYBR Premix Ex Taq (Takara) in a 96-well Lightcycler 480 system (Roche) and quantified with the Lightcycler 480 software (Version 1.5.0.39, Roche). A list of primers used for locus amplification is shown in Supplementary data.
Analysis of gene expression by northern blot and Q-PCR
Northern blot analysis of ABAR and TOC1 expression was performed with RNA from 12-day-old seedlings grown under the indicated conditions. For hormone treatment, seedlings were sprayed with 7.5 ml of 0.001% Triton X-100 aqueous solution containing 100 μM (±) ABA and samples analysed at the indicated time points. The northern blot of TOC1 expression shown in Figure 5D was obtained after spraying seedlings with the hormone at CT5 (so that can be compared with the waveform of TOC1∷LUC expression adding ABA at CT5 in Figure 5A). RNA was isolated using the Purelink Total RNA purification system (Invitrogen) and separated on 1.0% agarose/formaldehyde gels as described earlier (Perales and Más, 2007). ABAR probe was described earlier (Shen et al, 2006). TOC1 probe was described earlier (Más et al, 2003a). Quantification analysis was performed on a Bio-Rad phosphorimager using Quantity One software, version 6.1 (http://www.bio-rad.com/). For Q-PCR analysis, RNA was treated with RNAse-free DNase (Ambion) and the single-strand cDNA was synthesized using Superscript III (Invitrogen) and a mixture of oligo-dT18 and random hexamers (Invitrogen) following the manufacturer recommendation. The cDNA was diluted 10-fold with nuclease-free water and Q-PCR was performed with the SYBR Premix Ex Taq (Takara) in a 96-well Lightcycler 480 system (Roche). The IPP2 gene (isopentenyl pyrophosphate:dimethyl-allyl pyrophosphate isomerase) was used as a control (Hazen et al, 2005). Quantification was performed using Lightcycler 480 software (Version 1.5.0.39, Roche). Primers were designed using the PrimerExpress 2.0 software (Applied Biosystems) with lengths of 18–25 nucleotides, PCR amplicon lengths of 80–180 bp, 40–60% G:C content and 60–65°C. See Supplementary data for the list of primers used for Q-PCR assays.
Measurements of guard cell dimensions, stomatal conductance and plant survival to drought conditions
Detached rosette leaves from 3-week-old plants grown under ShD conditions (8 h light:16 h dark) were incubated for 2 h in a buffer containing 50 mM KCl and 10 mM MES, pH 6.15 with cool white fluorescent light (50 μmol m−2 s−1). ABA was subsequently added in the same buffer solution at different concentrations (0, 1 and 5 μM) and leaves were incubated for 2 more hours with the hormone. Abaxial epidermis were carefully taken with tweezers and mounted with the same buffer solution. Epidermis strips were observed with a Zeiss AxioPhot microscope with a × 63 objective. Approximately 100–200 stomata per sample were photographed using an Olympus DP70 camera attached to the microscope. Stomatal aperture was scored as width:length pore ratio using CellD software (Olympus). For stomatal conductance measurements, plants were grown under ShD for 5 weeks and 60–65% relative humidity. A Licor 6400 photosynthesis system with a 6400-15 Arabidopsis leaf chamber attachment (Licor, NE) was used for gas exchange measurements. Temperature was maintained at 22°C, humidity to 62–67% and CO2 to 400 μmol mol−1. Stomatal conductance was monitored every 20 s until stabilization. For water-loss assays, rosette leaves of comparable size from 4-week-old plants grown under ShD were detached, placed abaxial side up on a Petri dish and weighted at different times after detachment. Assays were performed at 22°C and 60–65% relative humidity. For dehydration experiments and plant survival assays, 2-week-old plants grown on agar plates under ShD were transferred to filter paper on Petri dishes and left un-watered for 10–12 h under LL. Survival percentages were scored 2 days after re-watering the plants. Greenish, healthy-looking plants were counted as surviving. Experiments were repeated at least twice with about 20–40 plants per treatment and genotype. Statistical significance was evaluated using one- and two-way ANOVA.
Supplementary Material
Supplementary Data
Review Process File
Acknowledgments
We thank Dr M Pagès, Dr T Stratmann, Dr M Yanovsky and members of P Más lab for helpful comments on the experiments and the manuscript. We are grateful to Dr DP Zhang (China Agricultural University, Beijing, China) for the ABAR RNAi construct and Dr T Altabella for help with the stomatal conductance measurements. This work was supported by grants to PM from the Spanish Ministry of Science and Education (MEC) (BIO2007-66068), from the EMBO YIP program and from EUROHORCS (European Heads Of Research Councils) and European Science Foundation (ESF) through the EURYI Award. TL is a recipient of a JAE-CSIC pre-doctoral fellowship; JCC is supported by a post-doctoral contract trough the CRAG.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ (2005) Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Gen 6: 544–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busk PK, Pagès M (1998) Regulation of abscisic acid-induced transcription. Plant Mol Biol 37: 425–435 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Correia MJ, Pereira JS, Chaves MM, Rodrigues ML, Pacheco CA (1995) ABA xylem concentrations determine maximum daily leaf conductance of field-grown Vitis vinifera L. plants. Plant Cell Environ 18: 511–521 [Google Scholar]
- Covington M, Maloof J, Straume M, Kay SA, Harmer S (2008) Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol 9: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington MF, Harmer SL (2007) The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol 5: e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd AN, Gardner MJ, Hotta CT, Hubbard KE, Dalchau N, Love J, Assie J-M, Robertson FC, Jakobsen MK, Goncalves J, Sanders D, Webb AAR (2007) The Arabidopsis circadian clock incorporates a cADPR-based feedback loop. Science 318: 1789–1792 [DOI] [PubMed] [Google Scholar]
- Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, Hibberd JM, Millar AJ, Webb AA (2005) Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633 [DOI] [PubMed] [Google Scholar]
- Edwards KD, Anderson PE, Hall A, Salathia NS, Locke JC, Lynn JR, Straume M, Smith JQ, Millar AJ (2006) FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell 18: 639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SSL, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14: S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17: 3470–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima A, Kusano M, Nakamichi N, Kobayashi M, Hayashi N, Sakakibara H, Mizuno T, Saito K (2009) Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Proc Natl Acad Sci USA 106: 7251–7256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RM, Tingay S, Wang ZY, Tobin EM (2002) Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol 129: 576–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanano S, Domagalska MA, Nagy F, Davis SJ (2006) Multiple phytohormones influence distinct parameters of the plant circadian clock. Genes Cells 11: 1381–1392 [DOI] [PubMed] [Google Scholar]
- Harmer SL (2009) The circadian system in higher plants. Annu Rev Plant Biol 60: 357–377 [DOI] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- Hazen SP, Naef F, Quisel T, Gendron J, Chen H, Ecker J, Borevitz J, Kay S (2009) Exploring the transcriptional landscape of plant circadian rhythms using genome tiling arrays. Genome Biol 10: R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA (2005) LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci USA 102: 10387–10392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K (2007) Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci 12: 343–351 [DOI] [PubMed] [Google Scholar]
- Hotta CT, Gardner MJ, Hubbard KE, Baek SJ, Dalchau N, Suhita D, Dodd AN, Webb AAR (2007) Modulation of environmental responses of plants by circadian clocks. Plant Cell Environ 30: 333–349 [DOI] [PubMed] [Google Scholar]
- Ingram J, Bartels D (1996) The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 377–403 [DOI] [PubMed] [Google Scholar]
- Juan-Pablo Sánchez PDN-HC (2004) ABA activates ADPR cyclase and cADPR induces a subset of ABA-responsive genes in Arabidopsis. Plant J 38: 381–395 [DOI] [PubMed] [Google Scholar]
- Kurup S, Jones HD, Holdsworth MJ (2000) Interactions of the developmental regulator ABI3 with proteins identified from developing Arabidopsis seeds. Plant J 21: 143–155 [DOI] [PubMed] [Google Scholar]
- Leung J, Giraudat J (1998) Abscisic acid signal transduction. Annu Rev Plant Physiol Plant Mol Biol 49: 199–222 [DOI] [PubMed] [Google Scholar]
- Locke JC, Kozma-Bognar L, Gould PD, Feher B, Kevei E, Nagy F, Turner MS, Hall A, Millar AJ (2006) Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol Syst Biol 2: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Makino S, Matsushika A, Kojima M, Yamashino T, Mizuno T (2002) The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: Characterization with APRR1-overexpressing plants. Plant Cell Physiol 43: 58–69 [DOI] [PubMed] [Google Scholar]
- Más P (2005) Circadian clock signaling in Arabidopsis thaliana: from gene expression to physiology and development. Int J Dev Biol 49: 491–500 [DOI] [PubMed] [Google Scholar]
- Más P (2008) Circadian clock function in Arabidopsis thaliana: time beyond transcription. Trends Cell Biol 18: 273–281 [DOI] [PubMed] [Google Scholar]
- Más P, Alabadí D, Yanovsky MJ, Oyama T, Kay SA (2003a) Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15: 223–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P, Kim WJ, Somers DE, Kay SA (2003b) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis. Nature 426: 567–570 [DOI] [PubMed] [Google Scholar]
- Matsui A, Ishida J, Morosawa T, Mochizuki Y, Kaminuma E, Endo TA, Okamoto M, Nambara E, Nakajima M, Kawashima M, Satou M, Kim J-M, Kobayashi N, Toyoda T, Shinozaki K, Seki M (2008) Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol 49: 1135–1149 [DOI] [PubMed] [Google Scholar]
- McClung C (2008) Comes a time. Curr Opin Plant Biol 11: 514–520 [DOI] [PubMed] [Google Scholar]
- Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J (2001) The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J 25: 295–303 [DOI] [PubMed] [Google Scholar]
- Michael TP, Breton G, Hazen SP, Priest H, Mockler TC, Kay SA, Chory J (2008a) A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol 6: e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, McClung CR (2003) Enhancer trapping reveals widespread circadian clock transcriptional control in Arabidopsis. Plant Physiol 132: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, Hazen SP, Shen R, Priest HD, Sullivan CM, Givan SA, Yanovsky M, Hong F, Kay SA, Chory J (2008b) Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet 4: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Salome PA, Yu HJ, Spencer TR, Sharp EL, McPeek MA, Alonso JM, Ecker JR, McClung CR (2003) Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302: 1049–1053 [DOI] [PubMed] [Google Scholar]
- Mizuno T, Yamashino T (2008) Comparative transcriptome of diurnally oscillating genes and hormone-responsive genes in Arabidopsis thaliana: insight into circadian clock-controlled daily responses to common ambient stresses in plants. Plant Cell Physiol 49: 481–487 [DOI] [PubMed] [Google Scholar]
- Muller AH, Hansson M (2009) The barley magnesium chelatase 150-kD subunit is not an abscisic acid receptor. Plant Physiol 150: 157–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH (1998) Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA 95: 8660–8664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-Y, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow T-fF, Alfred SE, Bonetta D, Finkelstein R, Provart NJ, Desveaux D, Rodriguez PL, McCourt P, Zhu J-K, Schroeder JI, Volkman BF et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales M, Más P (2007) A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell 19: 2111–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portolés S, Más P (2007) Altered oscillator function affects clock resonance and is responsible for the reduced day-length sensitivity of CKB4 overexpressing plants. Plant J 51: 966–977 [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Breton G, Para A, Kay SA (2009) A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DA, Shulgin BV, Salazar D, Millar AJ (2004) Design principles underlying circadian clocks. J R Soc Interface 1: 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson F, Skeffington A, Gardner M, Webb A (2009) Interactions between circadian and hormonal signalling in plants. Plant Mol Biol 69: 419–427 [DOI] [PubMed] [Google Scholar]
- Salome PA, To JPC, Kieber JJ, McClung CR (2006) Arabidopsis response regulators ARR3 and ARR4 play cytokinin-independent roles in the control of circadian period. Plant Cell 18: 55–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E (2001) Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell 13: 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229 [DOI] [PubMed] [Google Scholar]
- Shen Y-Y, Wang X-F, Wu F-Q, Du S-Y, Cao Z, Shang Y, Wang X-L, Peng C-C, Yu X-C, Zhu S-Y, Fan R-C, Xu Y-H, Zhang D-P (2006) The Mg-chelatase H subunit is an abscisic acid receptor. Nature 443: 823–826 [DOI] [PubMed] [Google Scholar]
- Somers DE, Webb AAR, Pearson M, Kay SA (1998) The short-period mutant toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 125: 485–494 [DOI] [PubMed] [Google Scholar]
- Strayer CA, Oyama T, Schultz TF, Raman R, Somers DE, Más P, Panda S, Kreps JA, Kay SA (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50: 571–599 [DOI] [PubMed] [Google Scholar]
- Verslues PE, Zhu J-K (2007) New developments in abscisic acid perception and metabolism. Curr Opin Plant Biol 10: 447–452 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Wijnen H, Young MW (2006) Interplay of circadian clocks and metabolic rhythms. Annu Rev Genet 40: 409–448 [DOI] [PubMed] [Google Scholar]
- Wu F-Q, Xin Q, Cao Z, Liu Z-Q, Du S-Y, Mei C, Zhao C-X, Wang X-F, Shang Y, Jiang T, Zhang X-F, Yan L, Zhao R, Cui Z-N, Liu R, Sun H-L, Yang X-L, Su Z, Zhang D-P (2009) The Mg-chelatase H subunit binds abscisic acid and functions in abscisic acid signaling: new evidence in Arabidopsis. Plant Physiol 150: 1940–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeilinger MN, Farre EM, Taylor SR, Kay SA, Doyle FJ (2006) A novel computational model of the circadian clock in Arabidopsis that incorporates PRR7 and PRR9. Mol Syst Biol 2: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D-P, Wu Z-Y, Li X-Y, Zhao Z-X (2002) Purification and identification of a 42-kilodalton abscisic acid-specific-binding protein from epidermis of broad bean leaves. Plant Physiol 128: 714–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Deng Y, Mu J, Ji Z, Xiang T, Niu Q-W, Chua N-H, Zuo J (2006) Cytokinin affects circadian-clock oscillation in a phytochrome B- and Arabidopsis response regulator 4-dependent manner. Physiol Plant 127: 277–292 [Google Scholar]
- Zhu J-K (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data
Review Process File