Abstract
Human DNA polymerase N (PolN) is an A-family nuclear DNA polymerase whose function is unknown. This study examines the possible role of PolN in DNA repair in human cells treated with PolN-targeted siRNA. HeLa cells with siRNA-mediated knockdown of PolN were more sensitive than control cells to DNA cross-linking agent mitomycin C (MMC), but were not hyper-sensitive to UV irradiation. The MMC hyper-sensitivity of PolN knockdown cells was rescued by the overexpression of DNA polymerase-proficient PolN but not by DNA polymerase-deficient PolN. Furthermore, in vitro experiments showed that purified PolN conducts low efficiency non-mutagenic bypass of a psoralen DNA interstrand cross-link (ICL), whose structure resembles an intermediate in the proposed pathway of ICL repair. These results suggest that PolN might play a role in translesion DNA synthesis during ICL repair in human cells.
Keywords: DNA interstrand cross-links, DNA polymerase N, TLS, Psoralen, Chemotherapeutics
E. coli PolI is a high fidelity DNA repair polymerase that conducts gap-filling DNA synthesis during nucleotide excision repair (NER), base excision repair (BER), and DNA interstrand cross-link (ICL) repair. E. coli PolI is the prototypical member of A-family DNA polymerases (1). Drosophila melanogaster Mus308 is an A-family nuclear DNA polymerase, the mutants of which are hyper-sensitive to DNA cross-linking agents but not to other DNA damaging agents (2–4). This suggests that Mus308 may play a role in ICL repair in Drosophila. Two nuclear A-family DNA polymerases, DNA polymerase N (PolN) and DNA polymerase Q (PolQ) were recently discovered (5, 6). It has been proposed that PolN and PolQ are mammalian orthologs of Mus308 and that they participate in ICL repair in mammalian cells. However, additional studies are needed to confirm the precise role(s) of PolN and PolQ in DNA repair in mammalian cells.
ICLs are generated endogenously as bi-functional products of lipid peroxidation and by exogenous exposure to DNA damaging agents, some of which are commonly used as cancer chemotherapeutic drugs (7, 8). Because ICLs covalently link the two complementary strands of duplex DNA, they prevent progression of the DNA replication fork and block RNA transcription. This property makes ICLs highly toxic to proliferating cells. The molecular mechanism of human ICL repair is poorly understood (7–10). The current model of mammalian ICL repair (7–10) suggests that when a DNA replication fork stalls at an ICL, it is recognized by an endonuclease that generates a double-strand break (DSB) in the vicinity of the ICL (11–14). Subsequently, XPF-ERCC1 unhooks the ICL, resulting in a gap across from the unhooked ICL (14–17), which cannot be sealed by replicative DNA polymerases δ or ε. Gaps generated during ICL repair are thought to be repaired by translesion DNA synthesis (TLS) or by homologous recombination using the homologous chromosome (the sister chromatid is not available). Once the gap is sealed, the DSB is repaired and the collapsed replication fork is coordinately restored by homology-directed DSB repair (13, 18). After DSB repair, the unhooked ICL is removed by NER (18). A recent biochemical study demonstrated that a triple-stranded structure with an unhooked ICL can occur during ICL repair in human cells, suggesting that a TLS polymerase is involved in ICL repair in human cells (18). However, the specific TLS DNA polymerase(s) that play roles in ICL repair in human and other mammalian cells have not yet been identified.
Human PolN is a recently identified, low fidelity A-family DNA polymerase that may facilitate TLS in human cells (19). Recent studies show that PolN bypasses thymine glycol DNA lesions in vitro (19). Nevertheless, the biological role of human PolN is not yet known. This study examined the biological function of PolN in human cells treated with PolN-targeted siRNA. The cellular sensitivity of the PolN-knockdown cells to various types of DNA damaging agents, including DNA cross-linking agents, was examined. Using an unhooked psoralen ICL, which is a structure that mimics an intermediate in the proposed pathway of ICL repair, we also characterized a potential TLS activity of PolN in vitro.
EXPERIMENTAL PROCEDURES
Cloning of human PolN and the construction of PolN-expression plasmids
PolN was amplified from human cDNA in two fragments. For the N-terminus, PCR was performed using the primers: 5′-GGCGCCATGGATCCGATGGAAAATTATGAGGCA and 5′-TGCTGACGTCTTCTCCATCTCCTC. For the C-terminus, PCR was done using the primers: 5′ GAGGAGATGGAGAAGACGTCAGCA and 5′- CAGCGTAAGCTTCTACAGACAAAATGAAGGCG. These primers introduced a BamHI site at the 5′ end and a HindIII site at the 3′ end of the full-length gene. The PCR products were cloned into the pCR4-TOPO vector (Invitrogen) and sequenced. The two fragments were ligated at the central AatII site in PolN to obtain the full-length gene. The PolN gene was inserted into pcDNA3.1 (Invitrogen) or pETDuet-1 (Novagen) to generate a N-terminal six His-tagged protein for expression in mammalian cells or E.coli, respectively.
A construct with a C-terminal truncation of the proline-rich region (PolNΔP) was also generated for better expression and solubility in E. coli, as described by Takata et al. (19). This deletion was reported not to affect PolN polymerase activity (5). The PolNΔP gene was ligated into pETDuet-1to add a His tag at the N-terminus and a FLAG tag at the C-terminus.
To generate a polymerase-defective mutant of PolN, the Asp (GAC) was changed to an Ala (GCC) at amino acid 624 by site-directed mutagenesis (5). Asp at amino acid 624 that is in a highly conserved polymerase motif 3 (also called motif A) was substituted to Ala to inactivate the DNA polymerase activity. The motif 3 forms a pocket for the incoming dNTP (20, 21). A substitution of the corresponding Asp in E. coli DNA polymerase I, which functions to coordinate the metal-mediated catalysis reaction, leading to the incorporation of the incoming nucleotide, abrogates the polymerase activity (20, 21). The D624A mutant form of PolN was shown to be inactive in polymerase activity (5). The PolN gene in pETDuet-1 was used for PCR with the primers: 5′-CACCTTTCTAGCAGCAGCCTTTTCACAGATTGAATTGCGGATTCT and 5′-AGAATCCGCAATTCAATCTGTGAAAAGGCTGCTGCTAGAAAGGTG (underlined nucleotides generate the mutation). A siRNA-resistant form of PolN was generated by site-directed mutagenesis using the PolN gene in pcDNA3.1. Seven silent mutations were introduced into the siRNA target region using the primers: 5′-CCATTACAGTTAAAGTCAATAGTACGTACGGGAACTCCTCAAGAAATATTGTG and 5′-CACAATATTTCTTGAGGAGTTCCCGTACGTACTATTGACTTTAACTGTAATGG (bold/italic letters are the siRNA target region and mutated nucleotides are underlined).
siRNA experiments
All the siRNA and transfection reagents used in this study were purchased from Dharmacon. Cells were seeded one day before transfection. The cells were transfected with siRNA (100 nM) in the presence of DharmaFECT1. One day after transfection, cells were plated or cell lysates were prepared. Protein was analyzed by Western blot and RNA was analyzed by RT-PCR. For co-transfection, the PolN-expression plasmid was added to the mixture of siRNA and DharmaFECT1. The efficiency of siRNA knockdown was monitored in 293 T-cells transiently expressing His-tagged PolN by Western blot with anti-His antibodies. Western blot data showed that two siRNAs suppressed the expression of His-PolN without suppressing the expression of the p70 subunit of RPA (data not shown). The siRNA knockdown of PolN in HeLa cells was analyzed by RT-PCR. RT-PCR of endogenous PolN and PolQ confirmed that PolN and PolQ are expressed in HeLa cells and that PolN-siRNA#1 and #2 suppressed PolN but not PolQ. The siRNA#2 (5’-GAACAGCACAUAUGGAAAU -3’) was selected for further experiments, because silent mutations could be introduced into its target sequence to generate a siRNA-resistant PolN for rescue experiments.
RT-PCR
RNA was isolated with Trizol reagent. The cDNA was prepared using SuperScript II RT (Invitrogen). Random hexamers (100 ng) were annealed to 2.5 µg total RNA and incubated at 65°C for 5 min and cooled on ice for 1 min. The primed RNA was added to 1X RT buffer, 5 nM MgCl2, 10 mM DTT, 1 µl RNaseOUT Inhibitor, and 50 units of SuperScript II RT. The reaction was incubated at 25°C for 10 min and 42°C for 50 min. The reaction was terminated at 70°C for 15 min and 1 µl of RNase H was added. For amplification of PolN, the primers were: 5′-CCAAGCACCCAATTCAGATT and 5′-GCGTACACCACCTTCTTGGT. For amplification of PolQ the primers were: 5′-CGAACTATCTGGGTGACTGG and 5′-GCCCTTTTCACTAACCACAC (22). GAPDH primers were: 5′-ACCACAGTCCATGCCATCAC and 5′-TCCACCACCCTGTTGCTGTA. The PCR reaction (20 µl) contained 1X PCR buffer, 0.25 µM of each primer, 0.2 mM of each dNTP, and 0.4 µl Taq. The program for PCR was: 94°C for 2 min and 25 cycles of: 1) 95°C for 5 sec, 2) 59°C for 10 sec, and 3) 72°C for 1.5 min. PCR products were analyzed by agarose gel.
Colony-forming assay
One day after siRNA transfection, cells were seeded at 750 cells/100 mm dish and treated with DNA damaging agents on the following day. HeLa cells were treated with 0.01, 0.03, or 0.05 µg/ml and U2OS cells were treated with 0.05, 0.1, and 0.5 µg/ml MMC. Cells were incubated at 37°C for 2 h, washed twice with PBS and fresh media was added. For cisplatin treatment, the cells were exposed to 0.01, 0.1, and 1 µM of cisplatin for 2 hr, washed twice with PBS and fresh media was added. For UV irradiation, the media was removed, cells were exposed to 4 or 8 J/m2, and fresh media was added. For IR treatment, cells were exposed to 1 or 3 Gy. Treated cells were incubated at 37°C for 7–10 days. Colonies were fixed with ethanol, stained with Giemsa solution and counted. The surviving fraction was calculated by dividing the number of colonies on treated plates by the number on untreated plates.
Purification of PolNΔP from E. coli
Full-length PolN was poorly expressed and had poor solubility, and thus could not be purified in large amounts from E. coli for biochemical experiments (data not shown). However, PolNΔP, which lacks the C-terminal proline region, had much higher solubility and higher expression. Therefore, PolNΔP was purified from E. coli for biochemical characterization in vitro. Importantly, PolNΔP rescued the DNA repair defect in PolN-siRNA-treated U2OS cells as efficiently as PolN did, indicating that the C-terminal proline-rich region is dispensable for the DNA repair activity of PolN in human cells (Fig. s3).
PolN ΔP in the pETDuet-1 vector was transformed into E. coli Rosetta 2 (DE3) pLysS. A single colony was grown at 37°C in 20 ml LB media with 34 µg/ml chloramphenicol and 100 µg/ml ampicillin. The culture was transferred to 2 L LB with 34 µg/ml chloramphenicol and 100 µg/ml ampicillin and grown at 37°C until the OD600 reached 0.5. The culture was cooled on ice for 30 min and then induced with 1 mM IPTG at 16 °C for 16 h. Cells were harvested by centrifugation at 5000 × g for 15 min, washed with PBS, and the pellet was frozen. The pellet was thawed on ice and resuspended in 10 volumes of lysis buffer (50 mM Tris-HCl pH 7.5, 10% glycerol, 5 mM EDTA, 0.1% Triton X, 0.1 mg/ml BSA, and EDTA-free protease inhibitors from Roche Applied Science). The resuspended pellet was sonicated on ice (20 cycles of 10 sec with a 20 sec pause) and incubated with 0.25% polyethyleneimine for 15 min at 4°C with rocking. The lysate was clarified by centrifugation at 13,000 rpm for 30 min. The supernatant was incubated with 2 mM ATP and 10 mM MgSO4 for 10 min at 37°C. NaCl was added to a final concentration of 100 mM and the lysate was applied to a 6 ml DEAE column. The flow through, containing PolN, was collected and was added to a 50 ml Phosphocellulose 11 column. The column was washed with 10 column volumes of a low salt buffer (50 mM Tris-HCl pH 7.5, 0.1 M NaCl, 10% glycerol) and PolN was eluted with a high salt buffer (50 mM Tris-HCl pH 7.5, 1 M NaCl, 10% glycerol, protease inhibitors). Peak fractions, as determined by silver stain, were combined (45 ml) and incubated with 1 ml TALON resin for 3 h at 4°C with rocking. The resin was washed with 200 column volumes of the high salt buffer containing 10 mM imidazole. Proteins were eluted with an imidazole gradient (10 mM– 100 mM) and run on a 10% SDS-PAGE gel to determine purity. Immunoblots were performed with anti-His polyclonal antibody (H-15, Santa Cruz) and anti-FLAG M2 monoclonal antibody (Sigma).
Substrate preparation (23)
Primer oligonucleotides (100 pmol) were labeled using T4 polynucleotide kinase and γ-32P ATP (1 mCi) and purified from a 10% sequencing gel. A 12-mer oligonucleotide (5'-GAAGCTACGAGC-3') with a psoralen furan-side mono adduct at the T, was generous gift from Dr. J. E. Hearst. This 12-mer (100 pmol) was phosphorylated with cold ATP by T4 polynucleotide kinase, and annealed to 100 pmol of a 74-mer oligonucleotide (5’-CCTGCTGCAGCCCAAGCTTGGCGCTCGCTCGTAGCTTCTCAGGGTGGCCAGCTGGCGCAGATCTGGCTCGAGGA-3') (underlined portion complementary to 12-mer). The partially duplexed DNA was exposed to UVA (366 nm) for 10 min to convert the mono-adduct to an ICL. The cross-linked substrate was purified from a 10% sequencing gel and annealed to a 5’-32P-labeled 36-mer primer (5'-TCCTCGAGCCAGATCTGCGCCAGCTGG CCACCCTGA-3') to generate a nicked substrate. A 5’-32P-labeled 22-mer primer (5'-TCCTCGAGCCAGATCTGCGCCA-3') was used as a primer for the gapped substrate. The substrate was then purified from a 6% native gel. Under these conditions, the cross-linked substrate is well separated from the non-cross-linked substrate. Typically, less than 0.3 % of the substrate was non-cross-linked (23). For the undamaged substrate, a 32P-labeled 22-mer primer was annealed to the 74-mer template, or a 32P- labeled 18-mer primer was annealed to a 31-mer template.
In vitro DNA polymerase assay
DNA polymerase assays for PolNΔP or DNA polymerase eta (PolH, purchased from Enzymax) were performed as follows. Reaction mixtures (17 mM Tris-HCl pH 7.5, 3.3% glycerol, 17 mM NaCl, 10 mM MgCl2, 5 fmol of the substrate, and 77 µM of each dNTP in 30 µl) were incubated for 1 min at 37 °C before the addition of polymerase. After adding the indicated amount of PolNΔP or PolH, the mixtures were incubated at 37°C for the indicated time. The amount of PolN or PolH used was determined to give similar primer-extension efficiency on an undamaged template (Fig. s4). The reactions were terminated by phenol/chloroform extraction. The reaction products were isolated by ethanol precipitation and analyzed on an 8% sequencing gel. The dried gel was exposed to a PhosphorImage screen, an image was obtained by scanning the screen with the Typhoon 9410 (GE Healthcare), and the products were quantified by ImageQuant. The amount of 74 nt full-length product was used to calculate the percent bypass activity. The sum of products 37 – 42 nt in length was used to calculate the percent strand displacement activity. The 42 nt product is caused by inhibition of polymerase activity one nucleotide before the ICL.
RESULTS
siRNA-mediated PolN knockdown causes sensitivity to MMC
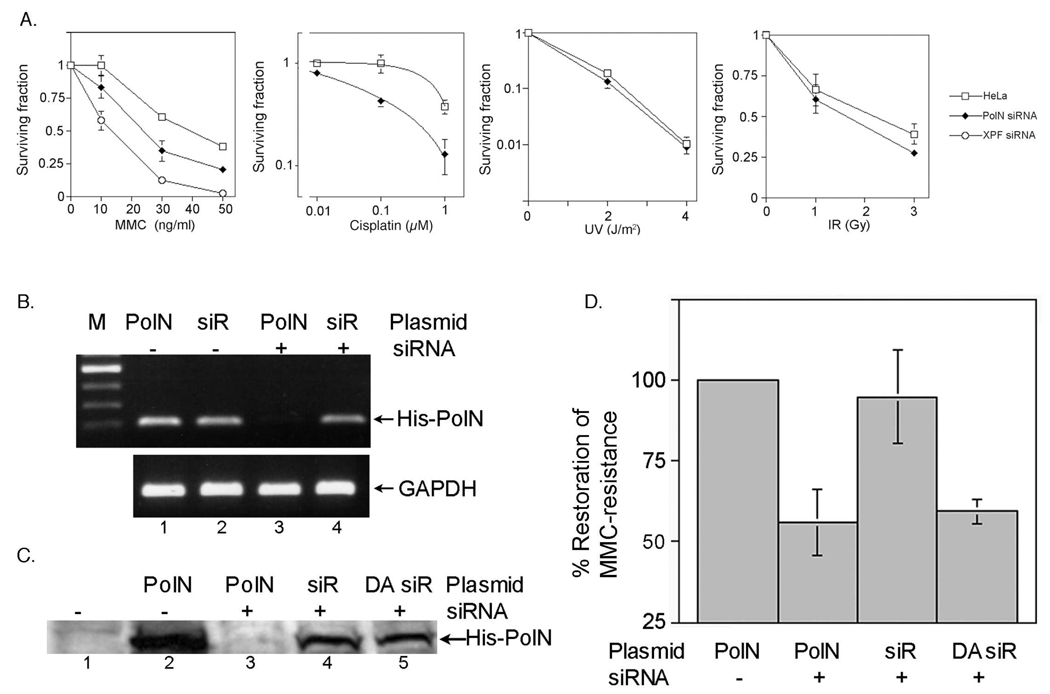
The biological role of PolN was investigated by knocking down PolN expression in HeLa cells with siRNA. For this purpose, four siRNA were purchased from Dharmacon and screened for selective and efficient knockdown of PolN expression (Fig. s1). One of these siRNAs, PolN-siRNA#2, was selected for further study, and its effects on PolN expression were confirmed by RT-PCR (Fig. 1B) and Western blot (Fig. 1C). As a positive control, HeLa cells were treated with XPF-siRNA, which efficiently knocks down the expression of the NER gene XPF (Fig. s2). HeLa cells treated with or without PolN-siRNA or XPF-siRNA were exposed to MMC, cisplatin, UV- or γ-irradiation. Survival was measured using a colony-forming assay (Fig. 1A). After exposure to MMC, the survival of cells treated with PolN-siRNA was lower than wild-type HeLa cells but higher than cells treated with XPF-siRNA. Cells treated with PolN-siRNA were also sensitive to another DNA cross-linking agent, cisplatin, while they were not sensitive to UV irradiation and were moderately sensitive to γ-irradiation at a high dose. Similar results were observed in U2OS cells treated with PolN-siRNA (Fig. 2A). Suppression of endogenous PolN in the U2OS cells led to cellular MMC-sensitivity. These results demonstrate that PolN is required for resistance to DNA cross-linking agents, MMC and cisplatin, in HeLa cells, the first cellular phenotype associated with the knockdown of PolN.
Fig. 1.
DNA damage sensitivity of HeLa cells treated with PolN-siRNA. (A) Suppression of endogenous PolN in HeLa cells results into cellular sensitivity specifically to DNA cross-linking agents. HeLa cells were incubated with PolN-siRNA or XPF-siRNA for 24 h, and then treated with 0.05 µg/ml of MMC for 2 h or irradiated with the indicated dose of UV (254 nm) or IR. Cells were incubated for 10 days and cell number was measured by colony-forming assay. The bards in the graph represent standard deviation from six independent experiments. (B–D) Rescue experiments using siRNA-resistant PolN. (B) An RNAi-resistant PolN was generated by introducing seven silent point mutations at the target sequences of PolN-siRNA in pcDNA-His-PolN (siRPolN). PolN-siRNA was co-transfected into HeLa cells with either wild type pcDNA-His-PolN (PolN, lanes 1 and 3) or siRPolN (siR, lanes 2 and 4). After 24 h, the expression level of His-PolN was determined by RT-PCR (lanes 1–4). GAPDH was used as control. (C) The siRPolN or RNAi-resistant polymerase defective PolN mutant (siRPolN-DA) was co-transfected with PolN-siRNA#2. After 24 h, the expression level of His-PolN was determined by Western blot with anti-His antibodies. (D) DNA polymerase activity of PolN is required for MMC-resistance. Cellular sensitivity to MMC (50 ng/ml) was measured as described in Figure 1A. The graph shows MMC-resistance normalized to the control cells (defined as “100% MMC-resistance”). The expression of a siRNA-resistant PolN (siR) restored the MMC-resistance to the level of the control (third column), while the expression of a siRNA-resistant, polymerase deficient PolN (DA siR) failed to fully restore MMC-resistance (fourth column). Data shown are the average ± standard deviation (error bars) based on data from the three independent experiments.
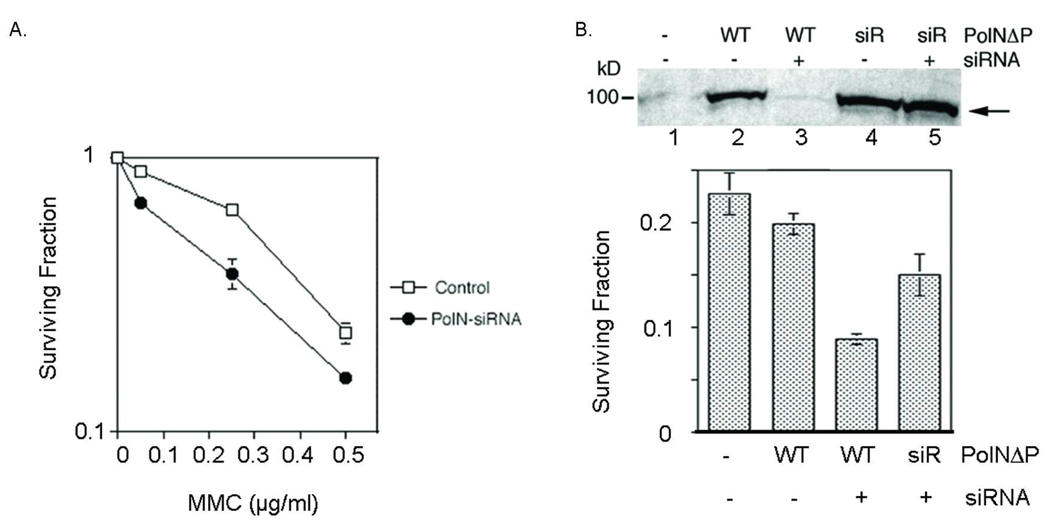
Fig. 2.
PolN-depleted U2OS cells are sensitive to MMC. (A) U2OS cells were transfected with PolN-siRNA. After 24 hrs, cells were plated for a colony-forming assay as described in Fig. 1A. PolN-depleted cells had increased sensitivity to MMC compared to mock-treated U2OS cells. Error bars represent the standard deviation of three independent experiments. (B) U2OS cells stably expressing siR PolNΔP are resistant to PolN siRNA and restore resistance to MMC. U2OS cells stably expressing WT PolNΔP or siR PolNΔP were generated by selection with G418. U2OS cells stably expressing PolNΔP or siRPolNΔP were transfected with the PolN-siRNA and treated with MMC. A colony-forming assay was used to determine survival. (Top) Western blot analysis with the anti-His antibody. PolNΔP and siRPolNΔP were stably expressed in U2OS cells (lanes 2 and 4). With the addition of siRNA PolN, PolNΔP expression was suppressed by more than 95%, while siRPolNΔP was resistant to the siRNA treatment (lanes 3 and 5). (Bottom) Sensitivity of PolN-depleted cells to MMC. Mock-treated U2OS cells are sensitive to MMC (0.5 µg/ml) (first column). It was noted that overexpression of PolNΔP did not increase resistance to MMC (second column). By the treatment with siRNA, PolNΔP expressing U2OS cells became sensitive to MMC (third column), while siR PolNΔP cells restored the MMC-resistance similar to that of mock-treated cells (fourth column). Error bars represent the standard deviation of three independent experiments.
The DNA polymerase activity of PolN is required for MMC-resistance in human cells
To confirm the specificity of PolN-siRNA, PolN expression plasmids were constructed to express recombinant wild-type PolN or siRNA-resistant PolN (siRPolN) (see Materials and Methods). Furthermore, to examine whether the DNA polymerase activity of PolN is required for resistance to MMC, a plasmid expressing polymerase-deficient PolN (siRPolN-DA) was also constructed. This plasmid expresses PolN with a D624A mutation that abolishes PolN polymerase activity (Fig. s3) (5), and expression of PolN-DA from this plasmid is resistant to siRNA knockdown (Fig. 1C, lane 5). Fig. 1B–D shows that the expression of siRPolN rescued sensitivity of HeLa cells to MMC (Fig. 1D, third column), but the expression of siRPolN-DA failed to rescue MMC-sensitivity in PolN knockdown cells (Fig. 1D, fourth column). Western blot confirmed that siRPolN and siRPolN-DA are expressed at similar levels in these cells (Fig. 1C, lanes 4 and 5). In addition, overexpression of siR PolNΔP, which is a variant form of PolN used for in vitro experiments in this study, restored wild type resistance to MMC in the U2OS cells treated with PolN-siRNA (Fig. 2B). These results suggest that the DNA polymerase activity of PolN is required for resistance to MMC in human cells.
In vitro bypass of an unhooked psoralen ICL by PolNΔP
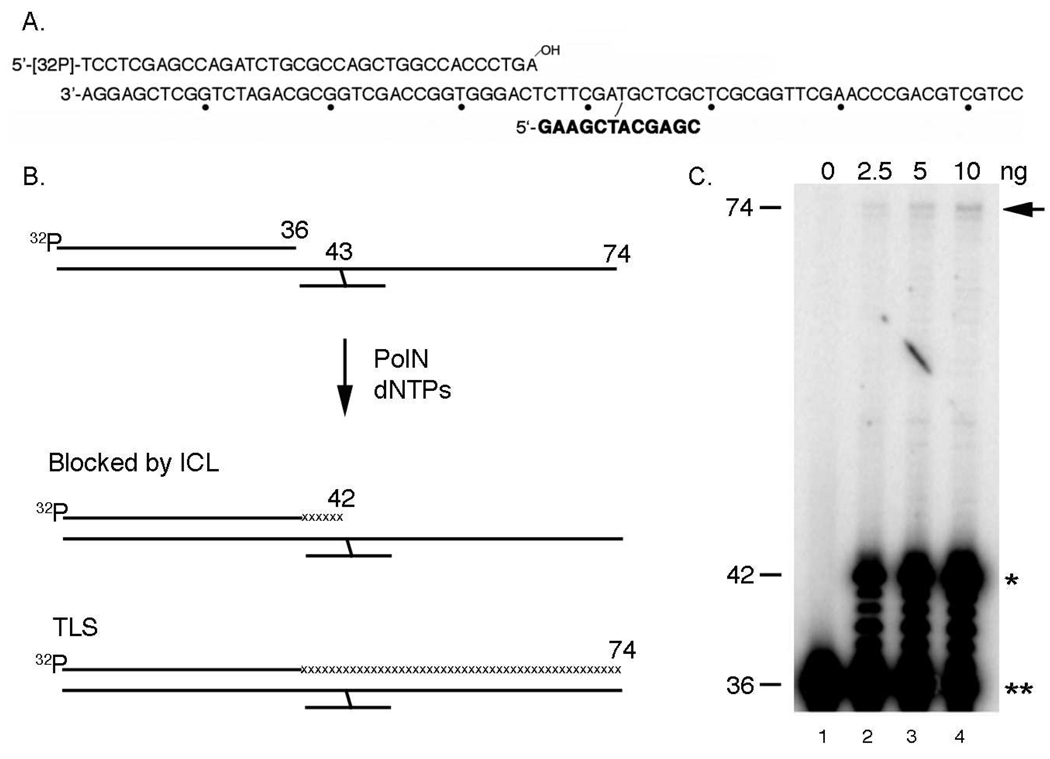
The results presented above suggest that PolN-catalyzed DNA synthesis may contribute to repair of ICLs induced by MMC in HeLa cells (Fig. 1). Previous studies suggest that ICLs are repaired during the S phase in mammalian cells (11, 13, 14), and that this reaction involves TLS (18). In vitro studies also showed that purified PolNΔP, which lacks the C-terminal proline-rich PolN region, acts as a TLS DNA polymerase on a DNA substrate containing thymine glycol (19). Importantly, the expression of PolNΔP rescued the MMC sensitivity in PolN knockdown cells as efficiently as full-length PolN, indicating that the C-terminal proline-rich region is dispensable for the DNA repair activity of PolN in human cells (Fig. 2). The ability of PolNΔP to function as a TLS polymerase was examined using a psoralen ICL. The substrate contains a 12-mer oligonucleotide attached to a 74-mer oligonucleotide via a psoralen ICL (see Materials and Methods), which resembles the "unhooked" intermediate in the proposed pathway of ICL repair (Fig. 3Aand 3B) (23). The 74-mer/12-mer was hybridized to a complementary upstream 36-mer or 22-mer, to generate either a nicked or gapped DNA substrate (23).
Fig. 3.
In vitro ICL-bypass activity of PolNΔP. (A) Substrate DNA used in this study. A 12-mer with a single psoralen furan-side monoadduct (shown in bold) was annealed to a 74-mer. The partially duplexed DNA was exposed to UVA light to generate an ICL. The 5′-32P-labeled primers were annealed to the cross-linked template. Dots indicate every tenth nucleotide. (B) Scheme of in vitro TLS assay. A 32P-labeled 36-mer was annealed adjacent to the 12-mer cross-linked fragment, leaving a nick. The primer can only be extended by strand displacement activity. When in vitro chain elongation is inhibited at the site of a psoralen ICL, a 43 nt fragment is produced. When the elongation is inhibited one nucleotide before an ICL, a 42 nt fragment is generated. A full TLS reaction gives a 74 nt fragment. (C) ICL-bypass by PolNΔP. Purified PolNΔP (2.5, 5, or 10 ng) was incubated with a nicked substrate containing an unhooked ICL for 1 h. The extension of the primer of this template requires strand-displacement activity. In lanes 2–4, percent bypass was 0.03%, 0.05%, or 0.09%, respectively, in lanes 2 to 4. The arrow indicates the fully bypassed product (74 nt in length), the single asterisk represents the termination product at one nucleotide before the ICL (43 nt in length) and the double asterisk shows the primer (36 nt in length).
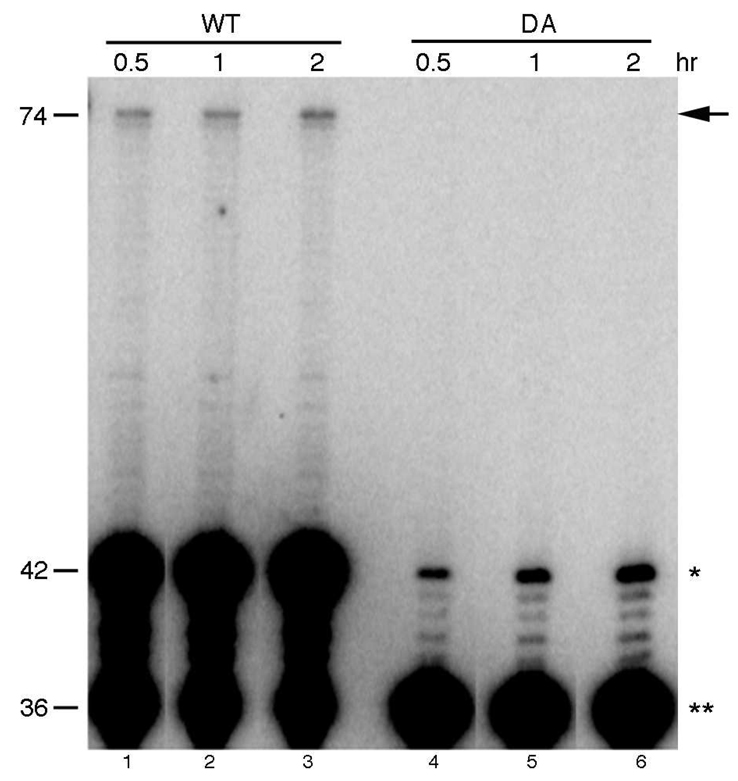
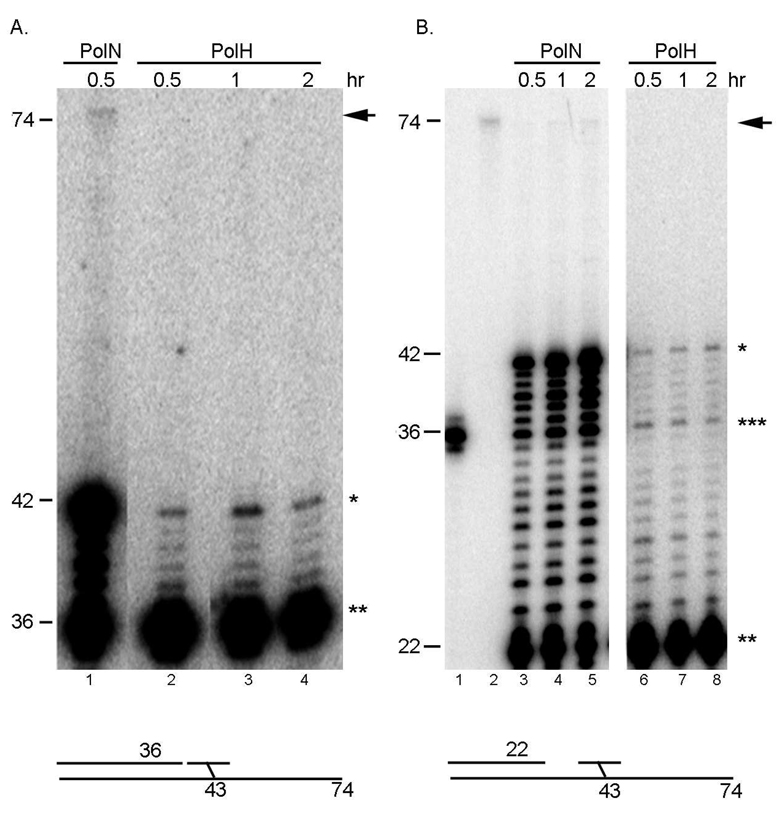
The nicked DNA substrate was incubated in vitro with recombinant purified PolNΔP or polymerase-deficient PolNΔP-DA (Fig. 3 and Fig. 4). Purified PolNΔP showed strong strand displacement activity 5' to the ICL (Fig. 3C, lanes 2–4; Fig. 4, lanes 1–3); however, DNA synthesis by PolNΔP was strongly inhibited by the unhooked ICL, and proceeded beyond the DNA lesion at 42 nt with very low efficiency (Fig. 3C, lanes 2–4; Fig. 4, lanes 1–3). It is estimated that PolNΔP bypassed the DNA lesion ~ 0.1% of the input DNA substrate (Fig. 4, lane 3). However, the low-level bypass activity detected in this assay is intrinsic to PolNΔP, because it is not detected in reactions with polymerase-deficient PolNΔP - DA (Fig. 4, lanes 4–6). For comparison, a Y-family TLS polymerase, DNA polymerase eta (PolH) was also incubated with the unhooked ICL in vitro. PolH is a well-characterized TLS polymerase implicated in the error-prone repair of ICLs in mammalian cells (24, 25). This error-prone ICL repair pathway is mediated by nucleotide excision repair (NER). NER is proposed to initiate the unhooking of an ICL, generating a similar structure to the substrate used in this study. PolH is believed to bypass the unhooked ICL with the expense of induced mutations (24, 25). Purified PolH showed very weak strand displacement activity and was unable to bypass the unhooked ICL in the nicked (Fig. 5A, lanes 2–4) or gapped DNA substrate (Fig. 5B, lanes 6–8). In contrast, PolH DNA polymerase activity was comparable to PolNΔP on an undamaged DNA substrate (Fig. s4). These results indicate that PolN is capable of low efficiency bypass of an unhooked ICL in vitro, which is consistent with a biochemical role for PolN polymerase activity in the repair of ICLs in human cells.
Fig. 4.
A DNA polymerase activity deficient PolN (DA) cannot support TLS of the unhooked ICL in vitro. Wild type PolNΔP (5 ng) and a DNA polymerase activity deficient variant DA (10 ng) were incubated at 37°C with the nicked ICL substrate for the indicated times and the products were analyzed on an 8% sequencing gel. Products longer than 36 nt are due to strand displacement by PolNΔP. Bypass products are longer than 42 nt and the full-length product is at 74 nt. The arrow indicates the fully bypassed product, the single and double asterisks represent the termination product at one nucleotide before the ICL (43 nt in length) and the primer (36 nt in length), respectively. Percent bypass was 0.05% (lane 1), 0.07% (lane 2), and 0.11% (lane 3). In lanes 1–3, percent strand displacement was 55%, 64%, and 82%, respectively.
Fig. 5.
PolH is unable to bypass the unhooked ICL in vitro. We used 40 ng of PolH and 2.5 ng of PolN, which give a similar level of polymerase activity on undamaged DNA under the conditions used (Fig. s4). (A) ICL bypass activity with the nicked substrate. PolNΔP was incubated with the substrate for 30 min (lane 1). PolH was incubated with the substrate for the indicated time at 37°C (lanes 2–5). Percent bypass was 0.06% and 62% of the primer was extended in lane 1. No bypass activity was detected in lanes 2–5. Percentage of the extended primer by PolH was 8% (lane 2), 12% (lane 3) and 12% (lane 4). The arrow indicates the fully bypassed products (74 nt in length), the single and double asterisks represent the termination product at one nucleotide before the ICL (43 nt in length) and the primer (36 nt in length), respectively. (B) ICL bypass activity with the gapped substrate. Percent bypass was 0.03% in lane 5 and 53%, 66%, and 83% of the primer was extended in lanes 3, 4, and 5, respectively. No bypass activity was detected in lanes 6–8. Percentage of the extended primer by PolH was 17% (lane 6), 22% (lane 7) and 26% (lane 8). Lane 1; 36 mer marker, and lane 2; 74 mer marker. Products longer than 36 nt (indicated by the triple asterisks) are due to strand displacement by PolNΔP. The arrow indicates the fully bypassed products, and the single and double asterisks represent the termination product at one nucleotide before the ICL (43 nt in length) and the primer (22 nt in length), respectively.
To examine the accuracy of bypass by PolNΔP, the DNA sequence of the 74 nt bypass products was determined by PCR-based DNA sequencing (23). This method was employed previously to sequence bypass products of abasic lesions by Dpo4 DNA polymerase from S. solfataricus (26, 27). First, labeled 74 nt fragments were purified from an 8% denaturing DNA sequencing gel and amplified for 10 cycles using primers that hybridize to the 5' and 3' ends of the DNA substrate. PCR products were cloned directly into the pCR-vector using the TA-cloning system, and 31 clones were selected for DNA sequencing. The results revealed that there were no point or frameshift mutations in the 31 clones (> 97% accuracy). Regardless of the mechanism, PolNΔP conducts low efficiency non-mutagenic bypass of an unhooked ICL in vitro.
DISCUSSION
This study provides genetic evidence for the involvement of PolN in resistance to MMC and repair of ICLs in human cells and biochemical evidence that PolN may act as a TLS polymerase during ICL repair. These conclusions are supported by the fact that siRNA-mediated knockdown of PolN reduces resistance to MMC in HeLa cells, and that wild type resistance to MMC is restored by DNA polymerase-proficient PolN but not by DNA polymerase-deficient PolN. In vitro studies also demonstrate that purified PolNΔP has a weak, but measurable ability to bypass an unhooked psoralen ICL and that ICL bypass is not mutagenic under the conditions tested.
HeLa cells treated with PolN-siRNA were moderately sensitive to MMC, but less sensitive to MMC than HeLa cells treated with XPF-siRNA (Fig. 1). This may reflect the presence of residual PolN activity that escaped suppression by PolN-siRNA. Alternatively, other DNA polymerase(s) expressed in HeLa cells could promote ICL repair in the absence of PolN. In fact, other DNA polymerases have been identified, whose inactivation renders cells sensitive to DNA cross-linking agents. These include DNA polymerase theta (PolQ) and DNA polymerase zeta (PolZ). Like PolN, PolQ is a nuclear DNA polymerase with an A-family DNA polymerase domain (6, 28, 29). PolQ knockout mice have been generated and PolQ(−/−) ES cells are moderately sensitive to MMC and IR (30); thus, PolQ knockout mouse cells have a phenotype similar to PolN-knockdown human cells, described here for the first time. These results suggest that PolQ may have overlapping function(s) with PolN in ICL repair. PolZ is a TLS polymerase that has homology to the B-family DNA polymerase domain (31). PolZ knockout MEF cells are sensitive to a variety of DNA damaging agents including UV irradiation and DNA cross-linking agents (32). Significantly, PolZ is required for the mutagenic effects of these DNA damaging agents (32). Systematic genetic studies are needed to investigate potential redundancy in the function of PolN, PolQ and PolZ during ICL repair in human cells.
Interestingly, PolN orthologs have only been identified in vertebrate species thus far. Recently, a PolN-knockout was generated in chicken DT40 cells (33). These PolN knockout cells, the only PolN-knockout cell line reported to date, lack a detectable phenotype. In contrast, orthologs of PolQ exist in lower eukaryotes including D. melanogaster (3), Caenorhabditis elegans (34) and Arabidopsis thaliana (35). Mutation in Drosophila Mus308, a PolQ ortholog, results in severe cellular sensitivity to DNA cross-linking agents (4). However, PolQ knockout DT40 cells are not sensitive to MMC and display sensitivity to H2O2, a similar phenotype to DNA polymerase beta knockout DT40 cells (33). In addition, genetic and biochemical studies demonstrated that chicken PolQ participates in BER of oxidative DNA damage, suggesting that the biological roles of PolQ and DNA polymerase beta may overlap (33). These results clearly suggest complex and overlapping role(s) for nuclear A-family DNA polymerases in DNA repair. Nonetheless, the results presented here strongly suggest that human PolN acts as a repair DNA polymerase during ICL repair in HeLa cells.
Our in vitro experiments indicate that PolNΔP bypasses an unhooked psoralen ICL with very low efficiency (Fig. 2). One explanation for this result is that a PolN accessory factor, which stimulates PolNΔP’s bypass activity, is lacking from the in vitro reaction performed here. In fact, our preliminary data suggest the existence of such accessory factors in nuclear extract from HeLa cells (Fig. s5). The addition of the nuclear extract stimulated the PolNΔP -mediated TLS of the unhooked ICL, while there was no significant effect on the PolH-mediated TLS. Identification of these factors is currently under way. As a candidate approach, PCNA and RPA, which stimulate several other DNA polymerases (36, 37), were tested for ability to stimulate human PolNΔP. However, recombinant PCNA and RPA (purified from E. coli) did not stimulate ICL bypass by PolNΔP under the conditions used here (data not shown). Post-translational modifications of these factors (38, 39) and/or other accessory factors might be required for efficient bypass of an unhooked ICL by PolNΔP. Alternatively, coordination with other DNA polymerase(s) could enhance the bypass activity of PolNΔP. In fact, it has been reported that the efficient bypass of some DNA lesions in vitro requires two DNA polymerases (37, 40). Many Y-family DNA polymerases, including PolH and Rev1, insert a nucleotide opposite a DNA lesion with relatively high efficiency, but extend the primer from the lesion with low efficiency (inserter DNA polymerases). In contrast, PolZ extends the primer from the DNA lesion with relatively higher efficiency (an extender DNA polymerase). Thus, a combination of a Y-family DNA polymerase and PolZ accomplishes bypass of DNA damage efficiently in vitro (37, 40). This “inserter-extender” mechanism may also be relevant for efficient bypass of an unhooked ICL.
Displacement of short oligonucleotides 5' or 3' to an ICL might also be required for bypass of an unhooked ICL. Our results indicate that PolNΔP is capable of displacing a DNA oligonucleotide on the 5' side of an ICL with high efficiency. Interestingly, PolQ has ssDNA-dependent ATPase activity (6) and it was also reported recently that PolQ is an "extender" (41). Thus, it is possible that more than two TLS polymerases might be required for bypass of an unhooked ICL, including PolN, PolQ, PolZ and Rev1.
Interestingly, PolNΔP appears to carry out non-mutagenic bypass of the unhooked psoralen ICL in vitro. Although we cannot formally eliminate the possibility that the fully bypassed products were the results of polymerization of undamaged DNA existed in the substrate preparation, this suggests that PolNΔP may only extend primers with dAMP incorporated opposite the unhooked ICL. It was reported that PolNΔP conducts efficient error-free bypass of thymine glycol in vitro, preferentially incorporating dAMP opposite the DNA lesion (19) and extending the primer. E. coli DNA polymerase I Klenow fragment, the prototype A-family DNA polymerase, also incorporates dAMP opposite a psoralen ICL (23) or thymine glycol (42, 43). Thus, preferential incorporation of dAMP opposite these DNA lesions and efficient extension of the primer may be characteristic of A-family DNA polymerases. In future studies, it will be important to determine whether PolNΔP carries out non-mutagenic bypass of cisplatin ICLs, which are guanine-derived DNA lesions.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Dr. John E. Hearst for a psoralen monoaddcuted 12-mer.
We thank to the Eppley Molecular Biology Core Facility (supported by the Eppley Cancer Center Support Grant, P30CA036727) for the synthesis of oligonucleotides used in this study and the UNMC DNA Sequencing Core Facility (supported by P20 RR016469 from the INBRE Program of the National Center for Research Resources) for sequencing analysis. This work was supported by RO1 CA95291 (to T.B.), RO1 GM080458 (to T.B.), and 5T32 CA09476 from the National Institutes of Health (to L.Z.).
Footnotes
Supporting Information Available
Supporting information: Supplemental Figures 1–5
REFERENCES
- 1.Pavlov YI, Shcherbakova PV, Rogozin IB. Roles of DNA polymerases in replication, repair, and recombination in eukaryotes. Int Rev Cytol. 2006;255:41–132. doi: 10.1016/S0074-7696(06)55002-8. [DOI] [PubMed] [Google Scholar]
- 2.Sekelsky JJ, Burtis KC, Hawley RS. Damage control: the pleiotropy of DNA repair genes in Drosophila melanogaster. Genetics. 1998;148:1587–1598. doi: 10.1093/genetics/148.4.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris PV, Mazina OM, Leonhardt EA, Case RB, Boyd JB, Burtis KC. Molecular cloning of Drosophila mus308, a gene involved in DNA cross-link repair with homology to prokaryotic DNA polymerase I genes. Mol Cell Biol. 1996;16:5764–5771. doi: 10.1128/mcb.16.10.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd JB, Sakaguchi K, Harris PV. mus308 mutants of Drosophila exhibit hypersensitivity to DNA cross-linking agents and are defective in a deoxyribonuclease. Genetics. 1990;125:813–819. doi: 10.1093/genetics/125.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marini F, Kim N, Schuffert A, Wood RD. POLN, a nuclear PolA family DNA polymerase homologous to the DNA cross-link sensitivity protein Mus308. J Biol Chem. 2003;278:32014–32019. doi: 10.1074/jbc.M305646200. [DOI] [PubMed] [Google Scholar]
- 6.Seki M, Marini F, Wood RD. POLQ (Pol theta), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 2003;31:6117–6126. doi: 10.1093/nar/gkg814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dronkert ML, Kanaar R. Repair of DNA interstrand cross-links. Mutat Res. 2001;486:217–247. doi: 10.1016/s0921-8777(01)00092-1. [DOI] [PubMed] [Google Scholar]
- 8.Noll DM, Mason TM, Miller PS. Formation and repair of interstrand cross-links in DNA. Chem Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niedernhofer LJ, Lalai AS, Hoeijmakers JH. Fanconi anemia (cross)linked to DNA repair. Cell. 2005;123:1191–1198. doi: 10.1016/j.cell.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 10.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 11.Akkari YM, Bateman RL, Reifsteck CA, Olson SB, Grompe M. DNA replication is required To elicit cellular responses to psoralen-induced DNA interstrand cross-links. Mol Cell Biol. 2000;20:8283–8289. doi: 10.1128/mcb.20.21.8283-8289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bessho T. Induction of DNA replication-mediated double strand breaks by psoralen DNA interstrand cross-links. J Biol Chem. 2003;278:5250–5254. doi: 10.1074/jbc.M212323200. [DOI] [PubMed] [Google Scholar]
- 13.De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol Cell Biol. 2000;20:7980–7990. doi: 10.1128/mcb.20.21.7980-7990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rothfuss A, Grompe M. Repair kinetics of genomic interstrand DNA cross-links: evidence for DNA double-strand break-dependent activation of the Fanconi anemia/BRCA pathway. Mol Cell Biol. 2004;24:123–134. doi: 10.1128/MCB.24.1.123-134.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher LA, Bessho M, Bessho T. Processing of a Psoralen DNA Interstrand Cross-link by XPF-ERCC1 Complex in Vitro. J Biol Chem. 2008;283:1275–1281. doi: 10.1074/jbc.M708072200. [DOI] [PubMed] [Google Scholar]
- 16.Kuraoka I, Kobertz WR, Ariza RR, Biggerstaff M, Essigmann JM, Wood RD. Repair of an interstrand DNA cross-link initiated by ERCC1-XPF repair/recombination nuclease. J Biol Chem. 2000;275:26632–26636. doi: 10.1074/jbc.C000337200. [DOI] [PubMed] [Google Scholar]
- 17.Zhang N, Lu X, Zhang X, Peterson CA, Legerski RJ. hMutSbeta is required for the recognition and uncoupling of psoralen interstrand cross-links in vitro. Mol Cell Biol. 2002;22:2388–2397. doi: 10.1128/MCB.22.7.2388-2397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cipak L, Watanabe N, Bessho T. The role of BRCA2 in replication-coupled DNA interstrand cross-link repair in vitro. Nat Struct Mol Biol. 2006;13:729–733. doi: 10.1038/nsmb1120. [DOI] [PubMed] [Google Scholar]
- 19.Takata K, Shimizu T, Iwai S, Wood RD. Human DNA polymerase N (POLN) is a low fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J Biol Chem. 2006;281:23445–23455. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- 20.Patel PH, Loeb LA. DNA polymerase active site is highly mutable: evolutionary consequences. Proc Natl Acad Sci U S A. 2000;97:5095–5100. doi: 10.1073/pnas.97.10.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel PH, Loeb LA. Getting a grip on how DNA polymerases function. Nat Struct Biol. 2001;8:656–659. doi: 10.1038/90344. [DOI] [PubMed] [Google Scholar]
- 22.Kawamura K, Bahar R, Seimiya M, Chiyo M, Wada A, Okada S, Hatano M, Tokuhisa T, Kimura H, Watanabe S, Honda I, Sakiyama S, Tagawa M, J OW. DNA polymerase theta is preferentially expressed in lymphoid tissues and upregulated in human cancers. Int J Cancer. 2004;109:9–16. doi: 10.1002/ijc.11666. [DOI] [PubMed] [Google Scholar]
- 23.Zietlow L, Bessho T. DNA polymerase I-mediated translesion synthesis in RecA-independent DNA interstrand cross-link repair in E. coli. Biochemistry. 2008;47:5460–5464. doi: 10.1021/bi702343y. [DOI] [PubMed] [Google Scholar]
- 24.Mogi S, Butcher CE, Oh DH. DNA polymerase eta reduces the gamma-H2AX response to psoralen interstrand crosslinks in human cells. Exp Cell Res. 2008;314:887–895. doi: 10.1016/j.yexcr.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng H, Wang X, Warren AJ, Legerski RJ, Nairn RS, Hamilton JW, Li L. Nucleotide excision repair- and polymerase eta-mediated error-prone removal of mitomycin C interstrand cross-links. Mol Cell Biol. 2003;23:754–761. doi: 10.1128/MCB.23.2.754-761.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiala KA, Hypes CD, Suo Z. Mechanism of abasic lesion bypass catalyzed by a Y-family DNA polymerase. J Biol Chem. 2007;282:8188–8198. doi: 10.1074/jbc.M610718200. [DOI] [PubMed] [Google Scholar]
- 27.Fiala KA, Suo Z. Sloppy bypass of an abasic lesion catalyzed by a Y-family DNA polymerase. J Biol Chem. 2007;282:8199–8206. doi: 10.1074/jbc.M610719200. [DOI] [PubMed] [Google Scholar]
- 28.Sharief FS, Vojta PJ, Ropp PA, Copeland WC. Cloning and chromosomal mapping of the human DNA polymerase theta (POLQ), the eighth human DNA polymerase. Genomics. 1999;59:90–96. doi: 10.1006/geno.1999.5843. [DOI] [PubMed] [Google Scholar]
- 29.Seki M, Masutani C, Yang LW, Schuffert A, Iwai S, Bahar I, Wood RD. High-efficiency bypass of DNA damage by human DNA polymerase Q. Embo J. 2004;23:4484–4494. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shima N, Munroe RJ, Schimenti JC. The mouse genomic instability mutation chaos1 is an allele of Polq that exhibits genetic interaction with Atm. Mol Cell Biol. 2004;24:10381–10389. doi: 10.1128/MCB.24.23.10381-10389.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gan GN, Wittschieben JP, Wittschieben BO, Wood RD. DNA polymerase zeta (pol zeta) in higher eukaryotes. Cell Res. 2008;18:174–183. doi: 10.1038/cr.2007.117. [DOI] [PubMed] [Google Scholar]
- 32.Wittschieben JP, Reshmi SC, Gollin SM, Wood RD. Loss of DNA polymerase zeta causes chromosomal instability in mammalian cells. Cancer Res. 2006;66:134–142. doi: 10.1158/0008-5472.CAN-05-2982. [DOI] [PubMed] [Google Scholar]
- 33.Yoshimura M, Kohzaki M, Nakamura J, Asagoshi K, Sonoda E, Hou E, Prasad R, Wilson SH, Tano K, Yasui A, Lan L, Seki M, Wood RD, Arakawa H, Buerstedde JM, Hochegger H, Okada T, Hiraoka M, Takeda S. Vertebrate POLQ and POLbeta cooperate in base excision repair of oxidative DNA damage. Mol Cell. 2006;24:115–125. doi: 10.1016/j.molcel.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muzzini DM, Plevani P, Boulton SJ, Cassata G, Marini F. Caenorhabditis elegans POLQ-1 and HEL-308 function in two distinct DNA interstrand cross-link repair pathways. DNA Repair (Amst) 2008;7:941–950. doi: 10.1016/j.dnarep.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 35.Inagaki S, Suzuki T, Ohto MA, Urawa H, Horiuchi T, Nakamura K, Morikami A. Arabidopsis TEBICHI, with helicase and DNA polymerase domains, is required for regulated cell division and differentiation in meristems. Plant Cell. 2006;18:879–892. doi: 10.1105/tpc.105.036798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maga G, Villani G, Crespan E, Wimmer U, Ferrari E, Bertocci B, Hubscher U. 8-oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature. 2007;447:606–608. doi: 10.1038/nature05843. [DOI] [PubMed] [Google Scholar]
- 37.Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 38.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 39.Andersen PL, Xu F, Xiao W. Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res. 2008;18:162–173. doi: 10.1038/cr.2007.114. [DOI] [PubMed] [Google Scholar]
- 40.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 41.Seki M, Wood RD. DNA polymerase theta (POLQ) can extend from mismatches and from bases opposite a (6-4) photoproduct. DNA Repair (Amst) 2008;7:119–127. doi: 10.1016/j.dnarep.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ide H, Kow YW, Wallace SS. Thymine glycols and urea residues in M13 DNA constitute replicative blocks in vitro. Nucleic Acids Res. 1985;13:8035–8052. doi: 10.1093/nar/13.22.8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayes RC, LeClerc JE. Sequence dependence for bypass of thymine glycols in DNA by DNA polymerase I. Nucleic Acids Res. 1986;14:1045–1061. doi: 10.1093/nar/14.2.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.