Abstract
Schizophrenia and bipolar disorder are leading causes of morbidity across all populations, with heritability estimates of ∼80% indicating a substantial genetic component. Population genetics and genome-wide association studies suggest an overlap of genetic risk factors between these illnesses but it is unclear how this genetic component is divided between common gene polymorphisms, rare genomic copy number variants, and rare gene sequence mutations. We report evidence that the lipid transporter gene ABCA13 is a susceptibility factor for both schizophrenia and bipolar disorder. After the initial discovery of its disruption by a chromosome abnormality in a person with schizophrenia, we resequenced ABCA13 exons in 100 cases with schizophrenia and 100 controls. Multiple rare coding variants were identified including one nonsense and nine missense mutations and compound heterozygosity/homozygosity in six cases. Variants were genotyped in additional schizophrenia, bipolar, depression (n > 1600), and control (n > 950) cohorts and the frequency of all rare variants combined was greater than controls in schizophrenia (OR = 1.93, p = 0.0057) and bipolar disorder (OR = 2.71, p = 0.00007). The population attributable risk of these mutations was 2.2% for schizophrenia and 4.0% for bipolar disorder. In a study of 21 families of mutation carriers, we genotyped affected and unaffected relatives and found significant linkage (LOD = 4.3) of rare variants with a phenotype including schizophrenia, bipolar disorder, and major depression. These data identify a candidate gene, highlight the genetic overlap between schizophrenia, bipolar disorder, and depression, and suggest that rare coding variants may contribute significantly to risk of these disorders.
Introduction
The lifetime prevalence of both schizophrenia (MIM #181500) and bipolar disorder (MIM %125480) is about 1% and of major depressive disorder (MIM #608516) around 15%.1,2 With onset in young adulthood and lifelong duration, these illnesses impose a massive burden on individuals, their families, and healthcare providers.3 Although current symptom-based classifications define these as separate mental disorders, the boundaries between diagnoses are indistinct. Thus the incidence of depression and bipolar disorder is increased among relatives of probands with schizophrenia,2,4 and twin studies confirm genetic and environmental correlations between bipolar disorder and major depression.5 Estimated heritabilities of more than 80% for schizophrenia and bipolar disorder6,7 and 40% for recurrent major depression8 have driven the search for causative genes. Various approaches including linkage, gene association studies, investigation of cytogenetic abnormalities, and, recently, the detection of copy number variants (CNVs) have contributed to our understanding of genetic risk in these conditions but have, necessarily, only interrogated particular aspects of the continuum of human genetic variation. Recent genome-wide association studies (GWAS) that probe common variation at the whole genome scale in large case-control cohorts have found, and sometimes replicated, significant associations of a number of candidate genes with schizophrenia and bipolar disorder and have found substantial overlap of risk factors between these two illnesses.9–14 These studies have led to a reevaluation of models of the inheritance of schizophrenia and suggest that common polygenic variation may explain more than one third of the genetic contribution to schizophrenia.11 However, rare CNVs involving genes also make an important additional contribution to the total genetic variation associated with disease, and a parallel role for rare coding sequence variants seems likely.15–17 In particular, the role of rare mutations of large effect (genotype relative risk > 2) remains largely unexplored and is unlikely to be clarified by association strategies via common SNPs.
In the same way that CNVs have proved informative, the study of rare cytogenetic abnormalities can reveal major gene effects on disease risk for the individual and provide useful insights into etiological pathways relevant to illness in the wider population. The genes DISC1, PDE4B, GRIK4, and NPAS3 have each been identified at unique translocation breakpoints in single patients with schizophrenia and subsequently found to be associated with both schizophrenia and bipolar disorder in populations.18–25
In this paper we describe the discovery, through cytogenetics, of a candidate gene, ABCA13, for schizophrenia, bipolar disorder, and depression. ABCA13 is a member of the adenosine triphosphate (ATP)-binding cassette (ABC) super-family encoding transporters shuttling a variety of substrate/allocrites across cellular membranes.26 Gene mutations in members of the seven ABC subfamilies (A-G) contribute to human disorders with Mendelian or complex inheritance.27 In higher organisms, ABC proteins typically contain two transmembrane domains (TMDs) and two cytoplasmic nucleotide binding domains (NBDs), the latter containing characteristic ATP-binding Walker A and B motifs. We resequenced ABCA13 gene exons encoding these key functional domains in a “discovery” cohort of 100 cases with schizophrenia and 100 controls and identified 10 nonsynonymous rare coding variants. The frequency of these variants was then determined in a “test” sample of subjects with schizophrenia, bipolar disorder, major depression, and controls. Structural modeling and in silico prediction tools suggested pathological effects. We followed up affected and unaffected relatives of probands with mutations and demonstrated significant linkage of mutations with a phenotype that included schizophrenia, bipolar disorder, and depression. Finally, we determined that ABCA13 protein is expressed in mouse and human hippocampus and cortex, both regions relevant to schizophrenia and bipolar disorder.
Material and Methods
Clinical Description of the Individual Carrying the Complex Chromosomal Rearrangement inv(7)(p12.3;q21.11), t(7;8)(p12.3;p23)
The 48-year-old male carrying the complex chromosomal rearrangement inv(7)(p12.3;q21.11), t(7;8)(p12.3;p23) was identified by W.J.M. and was initially described in the hospital case notes as carrying a simple t(7;8) reciprocal translocation. The patient gave full consent to the study and was interviewed on separate occasions by a consultant psychiatrist (W.J.M.) and a psychiatric nurse.
The patient had a diagnosis of severe chronic schizophrenia, with continuous symptoms since his first admission to psychiatric hospital at the age of 16. From his records and discussions with ward staff, there was clear evidence of auditory hallucinations and bizarre delusions of a persecutory and grandiose nature in the past lasting more than 6 months. After an initial series of inpatient stays, he had been continuously in hospital for more than 25 years. On review he showed the classical negative signs of severe chronic schizophrenia with self-neglect to the extent that he needed complete support with self-care, and he showed formal thought disorder and exhibited motor stereotypies with restlessness and frequent pacing. All these symptoms had been constantly present for many years. He received clozapine and haloperidol as treatments for his schizophrenia and procycline to counteract side effects of these. He was on no other medications save a mild laxative. There had been no contact with any members of his family and details of family history were not available.
Clinical Description of Patient and Control Cohorts
The study was approved by the Central Office of Research Ethics Committees (COREC) in Scotland and all subjects gave informed written consent. Subjects were from the Northeast and Southeast of Scotland contacted through in-/out-patient services of hospitals in Scotland, and diagnoses were made according to DSM-IV criteria.28 All patients were interviewed by an experienced psychiatrist or psychiatric nurse via the “Schedule for Affective Disorders and Schizophrenia-Lifetime” version (SADS-L),29 supplemented by hospital case note review and information from carers and relatives. Diagnosis was reached by consensus between two psychiatrists.
A family study was designed to: (1) determine whether the rare variants were de novo or familial mutations; (2) describe the range of phenotypes in individuals with variants; and (3) test for linkage between variants and different phenotypes. Attempts were made to contact all first and second degree relatives of the probands who carried one of the ten variants selected for follow up. Some probands were members of large families previously recruited for linkage studies, and when possible these families were revisited so that diagnoses could be confirmed or updated. DNA samples were obtained from 106 relatives of 21 individuals carrying one of the rare mutations. DSM-IV diagnoses of family members were based on direct interview by a psychiatrist as described above.
Controls of similar North European ancestry were recruited through: (1) the South of Scotland Blood Transfusion Service and the Grampian Blood Transfusion service, (2) students taking part in an unrelated study, (3) hospital staff and social networks of the research team, and (4) the 1936 Lothian Birth Cohort (LBC1936).30 Controls were not screened for personal or family history of psychiatric illness.
These case-control individuals are part of a stable population that shows little evidence of stratification.11 Moreover, this cohort has been extensively genotyped in case-control studies of candidate genes producing both negative and positive results, suggesting a lack of cohort-dependent frequency bias.
Fluorescence In Situ Hybridization Analysis
BAC and fosmid clones mapping to 8p23.3 and 7p12.3 were selected with the UCSC genome browser database (Table S1 available online). Clone DNA prepared with the QIAGEN miniprep kit was labeled with Biotin dUTP or Digoxigenin dTTP by nick translation and hybridized to patient metaphase spreads with standard FISH methods.22 Slides counterstained with DAPI in Vectashield antifade solution were assessed on a Zeiss Axioskop2 fluorescence microscope with a chroma number 81000 multispectral filter and a Hamamatsu Photonics Orca-ER digital camera with SmartCapture2 software (Digital Scientific, UK).
DNA Resequencing
ABCA13 is a very large gene with 62 exons, so sequencing was restricted to selected exons encoding key functional domains: two NBDs, two TMDs, and a conserved hydrophobic membrane-dipping region (HDR). Oligonucleotide primers were designed with PRIMER3 software against exons 32–35, 37–40, 43, 50–53, and 54–59 of ABCA13 (NM_152701). PhredPhrap and Consed software were used for resequencing analysis. Confirmation of variants was obtained through independent resequencing reactions, restriction enzyme digests, cloning PCR products, and ABI TaqMan assays (all primers listed in Table S2).
Bioinformatics: Splicing, Sequence Alignment, Homology Modeling
Potential splice site variants were assessed with Berkeley Drosophila Genome Project splice site predication tool.31 Human ABCA13 orthologs were determined with a BLAST search against the translated nonredundant sequence database (tblastn), via the NCBI website with default parameters.32,33 Seven orthologous sequences were retrieved (GenBank identifiers): Homo sapiens (259013576), Pan troglodytes (114613334), Macaca mulatta (109066580), Mus musculus (30023642), Rattus norvegicus (158187532), Canis familiaris (73981662), Bos taurus (194666167), and Gallus gallus (50736039).
Sequences for all NBDs from ABCA transporters were retrieved directly from the SwissProt database.34 Because of their highly conservative sequences, the ortholog and ABCA-NBD multiple sequence alignments were generated automatically with ClustalX35 (Figure S1).
Numerous examples of experimentally determined ABC nucleotide-binding domains are deposited in the Protein Data Bank (PDB).36 For modeling human ABCA13-NBD1 (SwissProt Accession No: Q86UQ4; amino acid residues 3842–4074), three of its closest experimentally determined homologs, ascertained by a BLAST search against the PDB, were used as templates. These included X-ray structures from Thermatoga maritima (PDB ID: 1VPL; 2.1 Å) (Joint Center for Structural Genomics, unpublished), CysA from Alicyclobacillus acidocaldarius (PDB ID: 1Z47, chain A; 1.9 Å), and MalK from Escherichia coli (PDB IDs: 1Q12, chain A).37 The higher resolution 1Q12 (2.6 Å) structure of MalK E. coli was selected instead of 1Q1E (2.9 Å) and 1Q1B (2.8 Å). The target ABCA13-NBD1 shares 31%, 29%, and 30% sequence identity, respectively (∼52%–54% similarity), when compared with the templates from N-to-C terminus.
The alignment between the target and template sequences for modeling purposes was based on an initial multiple sequence alignment among them and a range of other-related protein sequences with the program MUMMALS38 (Figures S2 and S3).
Related sequences were first identified via the Network Protein Sequence Analysis server, with a BLASTsearch against the SwissProt database.34,39 After removing identical sequences from the returned output and considering only those sequences with at least 200 amino acids in length, 413 protein sequences were identified, and their corresponding homologous NBD-like protein regions were extracted (E-value cut-off ≤ 1e-10) and aligned with MUMMALS (Hidden Markov model option: HMM_1_3_1; Identity threshold: 0.6). The target-template alignment was further manually refined from the multiple sequence alignment in order to place gaps optimally guided by positioning of secondary structure elements, identified by STRIDE for the templates, and predicted by PsiPred version 2.5 for the target sequence.40–42
The program Modeler release 8 version 2 was used for model building based upon the aforementioned alignment.43 Twenty models were generated, and the one with the lowest objective function score was selected as the representative model. Nonidentical side chain residues (between target and templates) for the representative model were optimized with the side chain replacement program, SCWRL version 3.44,45 The model was then protonated under SYBYL version 6.9 (Tripos Associates, St. Louis, MO) and subject to brief energy minimization (30 steps steepest descent followed by 30 steps conjugate gradients) employing the Tripos forcefield to remove clashes and bad geometries.46 The model was finally checked for valid stereochemistry with PROCHECK version 3.5.4 (most favored regions, 89.8%; additional allowed regions, 9.8%; generously allowed regions, 0%; disallowed regions, 0.5%) and validated with the MetaMQAP II server47 (GDT_TS, 69; RMSD, 3.3 Å).47 All surface area calculations were performed with GETAREA version 1.1 via the following parameters: radius of water probe in Angstroms = 1.4; output level = Area per residue.48 The hydrogen-bonding network was analyzed with SYBYL version 6.9 (Tripos Associates) and the Protein Interactions Calculator.49
Statistical Analysis
Variants for further study were selected from resequencing “discovery” set data. However, all statistics relating to individual and global variant influence were based solely on TaqMan genotyping data derived from the secondary “test stage.” This approach ensured unbiased estimates of effect sizes. For each variant in the test stage, allele frequencies were compared with Fisher's exact test. A combined association test across all nonsynonymous mutations was performed by comparing the number of variants in cases to the number in controls. To account for variable sample size across the total, sample size was adjusted to N = n / (Σ(1/Ni)), where Ni is the samples size at the ith variant and n is the number of variants. The number of observed variants was adjusted as Σ(pi)∗N, where pi is the frequency of the ith variant. The population attributable risk (PAR) was calculated as follows: PAR(%) = 100 ∗ P(any ns variant) ∗ (OR − 1) / [1 + P(any ns variant) ∗ (OR − 1)] with P(any ns variant) = the probability of having any of the ns variants, calculated from the controls. This probability (= 0.024) is the same for all groups because it is calculated from the controls.
Nonparametric linkage analysis was conducted with Merlin50 to calculate allele sharing between all sets of affected relatives in the 14 families informative for linkage with two or more affected relatives, under both a “narrow” phenotype definition (schizophrenia and bipolar disorder) and a “broad” phenotype (schizophrenia, bipolar disorder, and major depression). For each SNP, the allele frequency on which identity-by-descent is estimated was specified to be 0.0025 (average mutation rate in controls). The LOD score and p value were calculated via the exponential model described by Kong and Cox.51
Immunofluorescence
Two separate Abca13 antibodies, NC1 and 807, were raised in rabbits via conjugated peptide immunization. Frozen adult mouse brain sections (10 μm thickness) were fixed in ice-cold acetone and blocked, incubated with primary antibody, and washed in phosphate-buffered saline + 0.1% Tween. After Alexa Fluor fluorescent donkey anti-rabbit secondary antibodies incubation and washing, slides were mounted in DAPI/antifade solution. Images were captured as described for FISH.
Results
Cytogenetics
We characterized a chromosomal rearrangement, inv(7)(p12.3;q21.11), t(7;8)(p12.3;p23), comprising a pericentric inversion of chromosome 7 coupled with a translocation between chromosomes 7 and 8 in a patient with chronic schizophrenia. FISH confirmed only one breakpoint, at 7p12.3, directly disrupted a gene, ABCA13 (Figures 1A and 1B; Table S1). We hypothesize that ABCA13 disruption causes illness in this individual through haploinsufficiency that commonly occurs in similarly truncated transcripts through nonsense-mediated decay.
Figure 1.
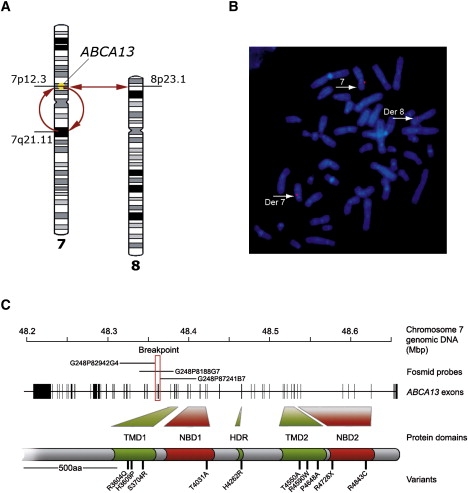
Cytogenetic Disruption of the ABCA13 Gene and Location of Functional Domain-Encoding Exons Selected for Resequencing
(A) A pericentric inversion of chromosome 7 and reciprocal translocation with chromosome 8 identified in a patient with schizophrenia. The location of the disrupted ABCA13 gene is indicated on the short arm of chromosome 7.
(B) Fluorescence in situ hybridization image showing patient metaphase chromosomes hybridized with breakpoint fosmid G248P8188G7. Three hybridization signals are located on normal chromosome 7 and derived chromosomes 7 and 8.
(C) Schematic of the ABCA13 locus. The positions of gene exons and the fosmid probes positioning the breakpoint (red box) are indicated. The relationship between resequenced exons and ABCA13 protein functional domains is highlighted together with the approximate positions of the ten rare variants studied in greater detail.
Resequencing Functional Domain Exons
The ABCA13 locus had not previously been associated with psychiatric disorders. To investigate whether rare functional variants in this gene contribute to psychiatric disorders, we resequenced 19 out of the total of 62 exons in the discovery cohort of 100 unrelated cases with schizophrenia (the same diagnosis as the patient with the chromosome abnormality) and 100 controls. These exons were selected to encode key functional domains: two NBDs, two TMDs, and a conserved hydrophobic membrane-dipping region (HDR) (Figure 1C).
We identified 32 variants not previously recorded in dbSNP: 20 in exons and 12 within flanking intronic sequences (Table 1; Figure S4). Of the 20 exonic variants, 14 were nonsynonymous, of which 8 were identified initially in schizophrenia cases, 3 in schizophrenia and controls, and 3 in controls only.
Table 1.
All ABCA13 Variants with Provisional dbSNP Identifiers Identified during the Resequencing Discovery Stage of This Study
| Domain | Chr7 Location bp NCBI Build 36.1 | Base Change | dbSNP Reference | AA Change | Substitution Type | MAF Cases | MAF controls | Discovery Populationa |
|---|---|---|---|---|---|---|---|---|
| TMD1 | 48,382,318 | G→A | ss136294996 | R3604Q | missense | A: 0.0003 | A: 0.0005 | case |
| TMD1 | 48,382,333 | A→C | ss136295002 | H3609P | missense | C: 0.0085 | C: 0.0043 | both |
| TMD1 | 48,382,337 | A→C | ss136295006 | P3610P | synonymous | C: 0.0096 | C: 0.0000 | case |
| TMD1 | 48,382,337 | A→T | ss136295010 | P3610P | synonymous | T: 0.0048 | T: 0.0037 | both |
| TMD1 | 48,382,619 | T→G | ss136295014 | S3704R | missense/splice variant | G: 0.0003 | G: 0.0000 | case |
| TMD1 | 48,384,364 | C→T | ss136295018 | intronic | T: 0.0095 | T: 0.0160 | both | |
| TMD1 | 48,384,474 | A→G | ss136295022 | intronic | G: 0.0045 | G: 0.0000 | case | |
| TMD1 | 48,384,623 | T→C | ss136295026 | intronic | C: 0.0048 | C: 0.0053 | both | |
| TMD1 | 48,398,156 | G→A | ss136295030 | intronic | A: 0.0523 | nd | case | |
| TMD1 | 48,398,232 | G→C | ss136295034 | intronic | C: 0.0048 | nd | case | |
| NBD1 | 48,399,292 | C→G | ss136295038 | L3861L | synonymous | G: 0.0000 | G: 0.0052 | control |
| NBD1 | 48,399,322 | T→C | ss136295042 | T3871T | synonymous | C: 0.0000 | C: 0.0052 | control |
| NBD1 | 48,413,738 | C→T | ss136295046 | intronic | T: 0.0092 | nd | case | |
| NBD1 | 48,420,643 | C→T | ss136295050 | intronic | T: 0.1651 | T: 0.2067 | both | |
| NBD1 | 48,420,683 | A→G | ss136295054 | T4031A | missense | G: 0.0009 | G: 0.0000 | case |
| NBD1 | 48,420,731 | C→G | ss136295058 | L4047V | missense (>1% frequency) | G: 0.0169 | G: 0.0104 | both |
| NBD1 | 48,420,834 | C→G | ss136295062 | intronic | G: 0.1651 | G: 0.2067 | both | |
| HDR | 48,465,394 | T→C | ss136295066 | P4260P | synonymous | C: 0.0000 | C: 0.0052 | control |
| HDR | 48,465,399 | A→G | ss136295070 | H4262R | missense | G: 0.0003 | G: 0.0000 | case |
| TMD2 | 48,518,027 | C→T | ss136295074 | R4454C | missense (>1% frequency) | T: 0.0142 | T: 0.0156 | both |
| TMD2 | 48,518,057 | C→A | ss136295078 | L4464M | missense (>1% frequency) | A: 0.0095 | A: 0.0208 | both |
| TMD2 | 48,526,874 | A→G | ss136295082 | T4550A | missense | A: 0.0056 | A: 0.0014 | case |
| TMD2 | 48,526,994 | C→T | ss136295086 | R4590W | missense | T: 0.0044 | T: 0.0019 | control |
| TMD2 | 48,527,112 | C→G | ss136295090 | intronic | G: 0.0286 | G: 0.0161 | both | |
| TMD2 | 48,530,327 | C→G | ss136295094 | P4648A | missense | G: 0.0000 | G: 0.0004 | control |
| NBD2 | 48,534,459 | A→G | ss136295098 | R4707R | synonymous | G: 0.0500 | G: 0.0526 | both |
| NBD2 | 48,534,520 | C→T | ss136295102 | R4728X | nonsense | T: 0.0025 | T: 0.001 | case |
| NBD2 | 48,538,544 | C→T | ss136295106 | intronic | T: 0.0000 | T: 0.0153 | control | |
| NBD2 | 48,538,545 | C→T | ss136295110 | intronic | T: 0.0000 | T: 0.0052 | control | |
| NBD2 | 48,597,311 | A→G | ss136295114 | I4841V | missense | G: 0.0040 | G: 0.0000 | case |
| NBD2 | 48,597,317 | C→T | ss136295118 | R4843C | missense | T: 0.0050 | T: 0.0031 | control |
| NBD2 | 48,604,841 | C→A | ss136295122 | intronic | A: 0.0519 | A: 0.0789 | both |
The genomic and functional domain location of the variants is displayed together with mutation class and frequency in the discovery set. The ten variants selected for further genotyping are shown in bold.
Indicates the resequencing cohort in which the variant was originally discovered.
The Frequency of Variants in Cases and Controls
Ten variants were selected for further analysis in a test cohort of subjects with schizophrenia (1019), bipolar disorder (680), major depression (365), and 2270 controls, based on the criteria: (1) present in no more than one of 100 controls, (2) causing a nonconservative amino acid change, and (3) showing cross-species conservation of the wild-type residue (Table 2). Genotyping assays designed against these 10 mutations were used to screen cases and controls. Despite relatively small numbers, three variants (H3609P, T4031A, and T4550A) were significantly more frequent in bipolars than controls and R4843L was more common in schizophrenia (Table 2), although none remained significant after correction for multiple testing of 10 variants in three phenotypes. A global comparison of the frequencies of all ten variants between cases and controls in this test stage showed a significant increase in the frequency of mutations in schizophrenia (p = 0.0057 OR = 1.93 95% CI 1.24) and bipolar disorder (p = 0.00007 OR 2.71 95% CI 1.73) but only a trend in major depression (p = 0.096 OR 1.67 95% CI 0.88). Similar significance was obtained when all case diagnoses were grouped (p = 0.0004 OR = 2.1 95% CI 1.45). The population attributable risks of the variants were 2.21%, 4.00%, and 2.61% for schizophrenia, bipolar disorder, and across all diagnoses, respectively.
Table 2.
The Properties of Ten Rare ABCA13 Variants Genotyped in Large Test Cohorts of Cases and Controls
|
Representation in Discovery Set |
Representation in Test Set |
SCZ |
BP |
MDD |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variant | Domain | Exon | Genomic Position | Base Change | WT Residue Conservation | Residue Change | Type | SCZ (n = 100) | Controls (n = 100) | Cases (total) | Cases by Diagnosis (Total) | Controls (Total) | p Value | Odds Ratio | p Value | Odds Ratio | p Value | Odds Ratio |
| 1 | TMD1 | 33 | 48,382,318 | G→A | RH, D, R, M, | R3604Q | missense | 1 | 0 | 0 (1666) | 0/841 SCZ, 0/494 BP, 0/331 MDD | 1 (951) | ||||||
| 2 | TMD1 | 33 | 48,382,333 | A→C | RH, D, R, M, Ch | H3609P | missense | 3 | 1 | 25 (1607) | 10/857 SCZ, 10/488 BP, 5/262 MDD | 8 (939) | 0.334 | 1.37 | 0.050 | 2.43 | 0.130 | 2.62 |
| 3 | TMD1 | 33 | 48,382,619 | T→G | RH, D, R, M, Ch | S3704R | miss./splice | 1 | 0 | 0 (1606) | 0/851 SCZ, 0/496 BP, 0/259 MDD | 0 (939) | ||||||
| 4 | NBD1 | 40 | 48,420,683 | A→G | RH, D, R, M, Ch | T4031A | missense | 1 | 0 | 3 (2072) | 0/1019 SCZ, 3/678 BP, 0/365 MDD | 0 (2262) | 0.012 | infinity | ||||
| 5 | HDR | 43 | 48,465,399 | A→G | RH, R, M, Ch | H4262R | missense | 1 | 0 | 0 (1717) | 0/837 SCZ, 0/562 BP, 0/318 MDD | 0 (980) | ||||||
| 6 | TMD2 | 52 | 48,526,874 | A→G | RH, D, R, Ch | T4550A | missense | 1 | 0 | 18 (1608) | 9/861 SCZ, 8/482 BP, 1/265 MDD | 3 (938) | 0.054 | 3.29 | 0.0097 | 5.25 | 0.63 | 1.18 |
| 7 | TMD2 | 52 | 48,526,994 | C→T | RH, D, R, M, Ch | R4590W | missense | 0 | 1 | 17 (1846) | 6/874 SCZ, 7/622 BP, 4/350 MDD | 4 (971) | 0.38 | 1.5 | 0.087 | 2.75 | 0.14 | 2.78 |
| 8 | TMD2 | 53 | 48,530,327 | C→G | RH, D, R, M, Ch | P4648A | missense | 0 | 1 | 0 (1488) | 0/869 SCZ, 0/619 BP | 0 (959) | ||||||
| 9 | NBD2 | 54 | 48,534,520 | C→T | RH, D, R, M, Ch | R4728X | nonsense | 1 | 0 | 10 (2058) | 4/1004 SCZ, 5/680 BP, 1/374 MDD | 5 (2270) | 0.370 | 1.51 | 0.057 | 3.35 | 0.600 | 1.21 |
| 10 | NBD2 | 57 | 48,597,317 | C→T | RH, D, R, M, Ch | R4843C | missense | 0 | 1 | 19 (1770) | 12/815 SCZ, 5/619 BP, 2/336 MDD | 6 (1025) | 0.047 | 2.54 | 0.400 | 1.38 | 0.630 | 1.02 |
| Global significance of all variants by diagnosis (95% CI lower limit shown for odds ratios): | 5.70E-03 | 1.93 (1.24) | 7.42E-05 | 2.71 (1.73) | 9.66E-02 | 1.67 (0.88) | ||||||||||||
| Population attributable risk by diagnosis: | 2.21% | 4.00% | 0.02% | |||||||||||||||
| p Value | Odds Ratio | |||||||||||||||||
| Global significance of all variants across all diagnoses (95% CI lower limit shown for odds ratio): | 2.60E-04 | 2.1 (1.45) | ||||||||||||||||
| Population attributable risk across all diagnoses: | 2.61% | |||||||||||||||||
The genomic location, nature of the nucleotide mutation, resulting amino acid change, and cross-species conservation of the wild-type residue are listed. The statistical significance (significant p values highlighted in bold type), odds ratio, and population attributable risk for individual and grouped (global assessment) variants are detailed in relation to their frequency in the individual and combined case and control groupings. In conjunction with pathological prediction and family data, only P4648A failed to show evidence for a causal role in psychiatric illness. Genomic position based on NCBI build 36.1. p values calculated by Fisher's exact test (one-tailed) excluding variant counts from the discovery schizophrenia and control sets. CI, confidence interval. Abbreviations: SCZ, schizophrenia; BP, bipolar disorder; MDD, major depressive disorder; NBD, nucleotide binding domain; TMD, transmembrane domain clusters; HDR, hydrophobic dipping region. Conservation abbreviations: RH, rhesus monkey; D, dog; R, rat; M, mouse; Ch, chicken.
Study in Families of Individuals with Mutations
Relatives of probands carrying eight of the ten rare mutations gave consent to take part in the family study designed to investigate the phenotypes associated with mutations and to determine whether these were de novo or familial mutations. 106 relatives in 21 families were genotyped (Figure 2). Families ranged in size from 2 to 16 persons and the diagnoses in family members (including probands) carrying one of the variants were schizophrenia (14), bipolar disorder (10), major depression (19), minor depression, anxiety, or alcoholism (3), and no psychiatric diagnosis (19). No variants were confirmed as de novo mutations.
Figure 2.
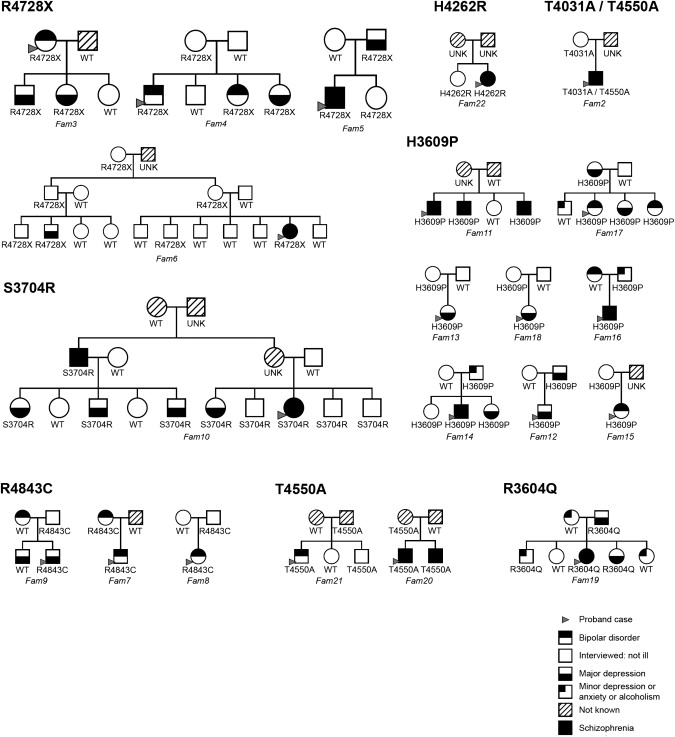
Cosegregation of ABCA13 Variants with Illness in Families
These families were all included in the linkage analysis. Images created with CraneFoot application.83
Nonparametric single point linkage analysis with MERLIN50 to estimate identity by descent (IBD) between all pairs of affected relatives in the 14 multiply affected families informative for linkage gave a combined Z score of 4.18 and a Kong and Cox51 LOD score of 4.38 (p = 3.5 × 10−6), establishing significant linkage of ABCA13 mutations with a “broad” phenotype that included cases with schizophrenia, bipolar disorder, and major depression. A LOD score of 2.1 (p = 0.0009) was achieved when the analysis was performed under the more restrictive phenotype definition including only cases with schizophrenia and bipolar disorder.
Genetic Properties and Clinical Phenotypes Associated with Individual Rare Variants
Data on each variant are presented in Table 2. Figure 3 highlights the sequence conservation and structural consequences of the potential null mutations R4728X and S3704R and missense mutation T4031A. Figure S5 shows the location of the nine predicted pathological variants on a topological representation of ABCA13 protein.
Figure 3.
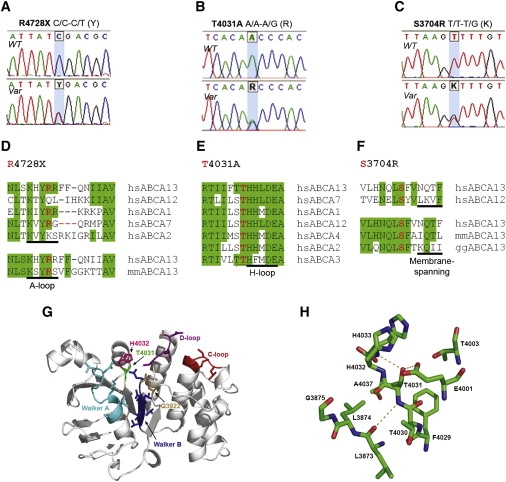
Characterization of the R4728X, T4031A, and S3704R Rare Variants
(A–C) Sequence of wild-type (top) and heterozygous (bottom) alleles for variants R4728X (A), T4031A (B), and S3704R (C).
(D–F) The wild-type residues are highly conserved within the ABCA family of proteins and between species orthologs for T4031A and S3704R (E, F) but less so for R4728X (D), which most likely reflects its different mode of pathological action (hs, Homo sapiens; mm, Mus musculus; gg, Gallus gallus).
(G) Location of T4031 and ATP-binding site on the 3D model of ABCA13-NBD1. The side chains of ATP-binding motifs are shown (Walker A, cyan; Walker B, blue; Q-loop, light brown; H-loop, pink; D-loop, magenta; C-loop, red) and mutated residue T4031A (green). Secondary structure was assigned by STRIDE and schematic representation of the 3D model produced with PyMol.
(H) Magnified view of T4031 hydrogen bonds indicated by dashed orange lines. The buried side chain hydroxyl group of T4031 forms a potential “loop-stabilizing” H-bond with the backbone nitrogen of H4033 and additionally with the side chain of the catalytic Walker B amino acid E4001, both absent in the Ala variant. The backbone-backbone H-bond between T4031 and L3873 is also shown. Only amino acid residues within a 5 Å sphere radius to T4031 are shown (color code: carbon atoms, green; oxygen atoms, red; nitrogen atoms, blue; hydrogen atoms not shown for clarity). The proximity of functional residues H4032 (H-loop), G3875 (Walker A), and E4001 (Walker B) is evident.
1. R3604Q was detected in a single discovery case with schizophrenia and one of 951 controls. A pathological effect was predicted by pmut and Panther online utilities (Table S3).52–55 In the family of the schizophrenia proband (Fam19), the mutation was present in a parent and two siblings and all three relatives had affective disorder.
2. H3609P was detected in three subjects with schizophrenia and one control in the discovery cohort and was more frequent in bipolars than controls (p = 0.05 OR 2.43) in the test cohort. A pathological effect was predicted by pmut. Eight families were followed up and the mutation was detected in four cases of bipolar illness or depression in Fam17, three siblings with schizophrenia in Fam11, two cases of schizophrenia, bipolar disorder, or depressions in each of families 12, 14, and 16, and in an unaffected parent in families 13, 15, and 18. Three bipolar cases were compound heterozygotes carrying both the T4550A and H3609P mutations and one person with severe chronic schizophrenia was homozygous for H3609P whereas all controls carrying variants were monoallelic, reinforcing the possibility of interactive effects of rare variants on these phenotypes.
3. S3704R, located in an extracellular loop of the first TMD (Figures 3C and 3F) was found in a subject with schizophrenia in the discovery cohort and cosegregated with schizophrenia (one case) and depression (four cases) in the proband's family (Fam10). The mutation was not detected in a further 1606 cases or in 939 controls. A pathological effect was predicted by pmut. In addition to its amino acid substitution, this mutation introduces a novel cryptic splice donor site, potentially resulting in a transcript with a downstream frameshift and premature stop codon. Hence, S3704R displays the features of a rare Mendelian, highly penetrant, deleterious mutation.
4. T4031A, a missense variant located in NBD1 (Figures 3B and 3E), was identified in three individuals with bipolar disorder in a sample of 2072 cases and was absent in 2262 controls (p = 0.012). A pathological effect was predicted by Panther. As with R4728X (see below), the missense variant occurred on a distinct haplotype (Table S4), implying that present-day carriers of the mutation are likely to be descended from a common European ancestor.
T4031A is strictly conserved across species and among all human ABCA NBD1s (Figure 3E; Figure S1), and we evaluated the consequence of the mutation on protein structure and function by means of a comparative three-dimensional model (Material and Methods; Figures 3G and 3H). T4031 lies within the protein core immediately adjacent to the functional H-loop histidine residue (H4032) and in proximity to other functional motif residues from Walker A (G3875) and Walker B (E4001). Substitution to Ala likely alters H-loop conformation and, in turn, catalytic activity. The adjacent H-loop histidine residue (H4032) is essential for ATP hydrolysis in NBDs56,57 and the equivalent mutation, H2128R, in NBD2 of human ABCA4 has been implicated in Stargardt disease (MIM #248200), a lipid deposition disorder.58
5. H4262R, located distal to the HDR, was detected in a single person with schizophrenia and an unaffected sibling (Fam22) but was absent from control, bipolar disorder, and depression cohorts. A pathological effect was predicted by pmut and PolyPhen.
6. T4550A is a missense variant found more frequently in bipolars than controls (p = 0.0097 OR 5.52) with a similar trend in schizophrenia (p = 0.054 OR 3.29) but not depression (p = 0.63 OR 1.18). Two families were followed up and the mutation was detected in a second sibling with schizophrenia in one family (Fam20) and an unaffected sibling in the other (Fam21).
7. R4590W was discovered in a control and in the test stage was relatively frequent and found in 17 cases and 4 controls. A pathological effect was predicted by pmut and PolyPhen. No families were available for follow-up.
8. P4648A was discovered in a single control and was not detected in the test samples. Controls were not screened for current or past episodes of mental illness and family details are not available. We consider this variant nonpathological.
9. R4728X, a nonsense mutation located at the start of NBD2 (Figures 3A and 3D), was found in four cases with schizophrenia, five with bipolar disorder, one with depression, and five controls (not screened for personal or family history of psychiatric illness). Four of the mutation carriers had relatives who agreed to take part in the study. In two of these families (Fam3 and Fam4), the mutation segregated with bipolar disorder and depression and in two (Fam5 and Fam6) with schizophrenia and depression (Figure 2). The mutation showed incomplete penetrance and was found on a distinct haplotype background (as T4031A, see above) (Table S4). The functional significance of the mutation is not known but transcripts containing nonsense mutations are commonly degraded, resulting in reduced gene dosage. Moreover, an equivalent mutation, R1275X, within the conserved A-loop motif in the human ABCC6 gene is pathological, causing pseudoxanthoma elasticum (MIM #264800).59
10. R4843C was initially discovered in a control, but in the test stage was more frequent in cases. A pathological effect was predicted by pmut. The comparison of schizophrenia (12/815) with controls (6/1025) was significant (p = 0.047 OR 2.54). Follow-up was possible with three families where the mutation was present in a bipolar parent in Fam7, an unaffected parent in Fam8, and a sibling pair with depression Fam9. A bipolar parent of the proband with unipolar depression in Fam9 did not have the mutation but we have no diagnosis for the other parent who has the variant.
Compound Heterozygosity
Strikingly, five cases were found to be compound heterozygotes and one homozygous, whereas all controls carrying variants were monoallelic, suggesting possible additive effects on risk. Two of the four cases with T4031A, one with schizophrenia and one bipolar, were compound heterozygotes also carrying the variant T4550A. Only one relative of the two compound heterozygotes was available for study and this unaffected parent of the proband with schizophrenia carried only T4031A. Two bipolar cases were compound heterozygotes carrying both the H3609P and R4590W mutations, one bipolar case carried H3609P and T4550A, and one person with severe chronic schizophrenia was homozygous for H3609P.
Abca13 Expression in Human and Mouse Brain
RT-PCR confirmed earlier reports26,60 of ABCA13 mRNA transcript expression in brain and indicated specific expression in human hippocampus and cortex (data not shown). Immunological detection of Abca13 protein by two independently raised antibodies showed low-to-moderate expression in most adult mouse brain regions and higher expression in dentate gyrus granule cells and ventricular areas including choroid plexus and ependymal cells (Figure 4).
Figure 4.
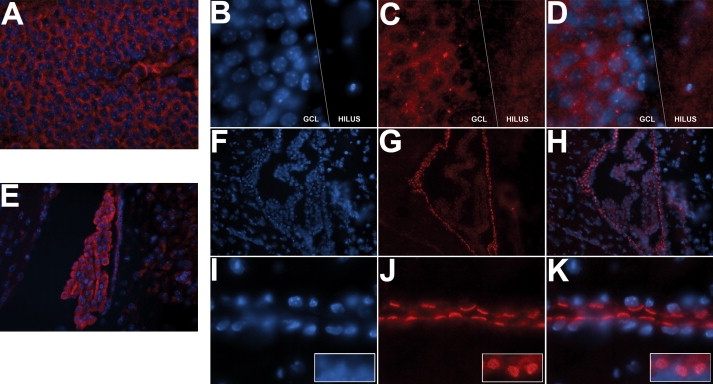
Abca13 Protein Expression in the Adult Mouse Brain
Polyclonal rabbit Abca13 antibodies NC1 (A, E) and 807 (B–D, F–K) were used to detect protein expression in frozen coronal brain sections. The pattern of cytoplasmic expression in the dentate gyrus granule cell layer (GCL) and choroid plexus was suggestive of both golgi body/endoplasmic reticulum localization (confirmed by colocalization with calnexin endoplasmic reticulum marker) (data not shown) and centrosomal localization (A–D, where B, C, and D are DAPI nuclear staining, Abca13 expression, and merged image, respectively). Strong expression was observed in ventricular regions, particularly the choroid plexus (E–H) and the ciliated ependymal cells lining the ventricular system (I–K). (F) and (I), (G) and (J), and (H) and (K) are DAPI nuclear staining, Abca13 expression, and merged image, respectively. Images (I), (J), and (K) also contain insets showing the Abca13-positive ependymal cell cilia face on.
Discussion
We have identified a cytogenetic disruption of the ABCA13 gene and 10 rare nonsynonymous mutations of which 9 demonstrated evidence consistent with a role in the etiology of schizophrenia and bipolar disorder. Association of ABCA13 coding variants with schizophrenia and bipolar disorder was significant for all variants taken together and for specific individual variants. The population attributable risk of the 10 rare variants detected was 2.2% for schizophrenia and 4.0% for bipolar, suggesting that this gene may have an important role in a subgroup of patients and an influence that crosses traditional diagnostic categories. To our knowledge, this is the first instance where multiple and combined coding variants in a single gene have been associated with risk of schizophrenia and bipolar disorder.
ABCA13 had not previously been implicated as a risk factor in major psychiatric disorders although a suggestive role for common ABCA13 gene variants in schizophrenia recently emerged from the International Schizophrenia Consortium GWAS data set where a significance level of p < 0.001 was reached at a single SNP within ABCA13 and a cluster of SNPs within a few kb of the gene.10
The family study included affected and unaffected relatives from 21 families, and in all cases the variants were familial with no instances of de novo mutation. Association with major depressive disorder was not significant in the case-control study but this was a small and conservative group because controls were not screened for personal or family history of psychiatric illness and the control groups would be expected to include some individuals with depression that has a lifetime prevalence of ∼15%. However, a striking finding was that the same mutation was associated with schizophrenia, bipolar disorder, as well as major depression in separate families. The nonsense mutation R4728X was detected in cases with bipolar disorder and depression in Fam3 and Fam4 and with schizophrenia and major depression in Fam5 and Fam6. A similar picture was found for H3609P, which was present in three siblings with schizophrenia in Fam11 and two cases with bipolar disorder and two with major depression in Fam17. The family data also demonstrated incomplete penetrance for seven of the eight variants because these were detected in unaffected relatives. This variable penetrance and range of phenotypes in families with rare ABCA13 mutations is similar to the pattern of illnesses in relatives carrying a translocation disrupting the gene DISC1 who were diagnosed with schizophrenia, depression, and bipolar disorder.61
Rare variants in other members of the ABCA subfamily also give rise to variable disease phenotypes, for example ABCA162 (two lipid-related conditions: Tangier disease [MIM #205400] and familial hypoalphalipoproteinaemia [MIM #604091]) and ABCA463 (four forms of degenerative retinal dystrophy: Stargardt disease, retinitis pigmentosa [MIM #601718], cone rod dystrophy [MIM #604116], and age-related macular degeneration [MIM #153800]), and it has been suggested that precise clinical presentation may be determined by the site of the mutation.62,64 However, in the present study both missense and null mutations contribute to a full range of phenotypes with no clear effect of the site or class of mutation on clinical outcome. The discovery of five compound heterozygotes and one homozygous mutation carrier suggests that additive and interactive combinations of mutations may contribute to these complex phenotypes.
All ABCA proteins functionally characterized to date shuttle lipid molecules across cell membranes.65 More than 1000 different lipid species exist in eukaryotic cells and in addition to being structural components of cellular membranes, different species have key roles in vesicular trafficking, signal transduction, and transcriptional regulation.66,67 Although the precise allocrite for the ABCA13 transporter has not been identified, the presence of a conserved C-terminal motif known to be shared among the lipid transporting group suggests that it is likely to be lipid related.68 Abnormalities in lipid metabolism have been strongly implicated in psychiatric illness with the most consistent finding being depletion of essential fatty acids in tissues of schizophrenic and bipolar patients.69–74 The oligodendrocyte lineage transcription factor 2 (OLIG2) showed association with schizophrenia,75,76 and a growing body of evidence based on imaging studies, microarray analysis, and metabolic and transcriptome investigation has implicated oligodendroglial pathophysiology and myelin biosynthesis in psychiatric disorders.77–79 Consistent with these discoveries are the findings that membrane phospholipids and the expression of genes encoding enzymes important in sphingolipid metabolism are significantly altered in postmortem brain tissue from schizophrenic patients and upregulated in response to antidepressant drugs.80,81 Disruption of lipid membrane structures and lipid biosynthetic pathways has the potential to affect many important biological processes including intracellular signaling pathways and neurotransmitter function.
Rare sequence variants are known to have an important role in the inheritance of common diseases, and different categories of “rare variant” have been described. Classical rare deleterious familial mutations typically show strong familial aggregation, have high penetrance, and low frequencies. A second type of variant (mostly missense mutations), have higher frequencies of up to 2%–3% in the case population, ORs of 2 or more, and tend to show less familial aggregation.82 In the present study S3704R conforms to the first class of null private mutation with high penetrance and low frequency whereas the missense mutations H3609P, T4550A, and R4590W show characteristics more typical of the second class by virtue of also being detected in controls, having moderate ORs, and higher observed frequencies. These data lend support to a genetic architecture of schizophrenia, bipolar disorder, and depression comparable to other common genetically complex and heterogeneous diseases and provide a novel biological pathway for investigation.
Supplemental Data
Supplemental Data include five figures and four tables and can be found with this article online at http://www.cell.com/AJHG.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Cranefoot pedigree drawing, http://www.finndiane.fi/software/cranefoot/
Network Protein Sequence Analysis server, http://npsa-pbil.ibcp.fr/
NCBI website, http://www.ncbi.nlm.nih.gov/BLAST/
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
PhredPhrap and Consed software, http://bozema.mbt.washington.edu/index.html
PRIMER3, http://workbench.sdsc.edu
Protein Data Bank (PDB), http://www.rcsb.org/pdb
PyMol, http://www.pymol.org/
UCSC genome browser database, http://genome.ucsc.edu
Acknowledgments
We would like to dedicate this paper to Walter Muir whose sudden death happened soon after this paper was written. His astute clinical observation and cytogenetic skills were the starting point for studying ABCA13 and his deep understanding of patients with psychosis combined with a formidable knowledge of psychiatric genetics made him an inspiring colleague, mentor, and friend.
We are indebted to the patients and their families who have contributed to these studies. H.M.K. was supported by a University of Edinburgh PhD Scholarship; B.S.P. held a Sim Fellowship from the Royal College of Physicians in Edinburgh. D.C.S. acknowledges funding from the Scottish Research Development Grant. The work was supported by grants from The Wellcome Trust, London and the Chief Scientist Office of the Scottish Government, and Research Into Ageing and Help the Aged. D.J.P., I.J.D., J.M.S., and P.M.V. are members of the University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology, which is supported by BBSRC, EPSRC, ESRC, and MRC as part of the Lifelong Health and Wellbeing Initiative. A.F.McR. and P.M.V. are supported by the Australian National Health and Medical Research Council. DNA extraction and genotyping was performed by the Wellcome Trust Clinical Research Facility Genetics Core, Western General Hospital, Edinburgh.
References
- 1.Kessler R.C., Berglund P., Demler O., Jin R., Koretz D., Merikangas K.R., Rush A.J., Walters E.E., Wang P.S. The epidemiology of major depressive disorder: results from the National Comorbidity Survey Replication (NCS-R) JAMA. 2003;289:3095–3105. doi: 10.1001/jama.289.23.3095. [DOI] [PubMed] [Google Scholar]
- 2.Lichtenstein P., Yip B.H., Bjork C., Pawitan Y., Cannon T.D., Sullivan P.F., Hultman C.M. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373:234–239. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO. (2001). World Health Report http://www.who.int/whr/2001/en.
- 4.Maier W., Lichtermann D., Minges J., Hallmayer J., Heun R., Benkert O., Levinson D.F. Continuity and discontinuity of affective disorders and schizophrenia. Results of a controlled family study. Arch. Gen. Psychiatry. 1993;50:871–883. doi: 10.1001/archpsyc.1993.01820230041004. [DOI] [PubMed] [Google Scholar]
- 5.McGuffin P., Rijsdijk F., Andrew M., Sham P., Katz R., Cardno A. The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch. Gen. Psychiatry. 2003;60:497–502. doi: 10.1001/archpsyc.60.5.497. [DOI] [PubMed] [Google Scholar]
- 6.Kieseppa T., Partonen T., Haukka J., Kaprio J., Lonnqvist J. High concordance of bipolar I disorder in a nationwide sample of twins. Am. J. Psychiatry. 2004;161:1814–1821. doi: 10.1176/ajp.161.10.1814. [DOI] [PubMed] [Google Scholar]
- 7.Sullivan P.F., Kendler K.S., Neale M.C. Schizophrenia as a complex trait: Evidence from a meta-analysis of twin studies. Arch. Gen. Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 8.Sullivan P.F., Neale M.C., Kendler K.S. Genetic epidemiology of major depression: Review and meta-analysis. Am. J. Psychiatry. 2000;157:1552–1562. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- 9.Ferreira M.A., O'Donovan M.C., Meng Y.A., Jones I.R., Ruderfer D.M., Jones L., Fan J., Kirov G., Perlis R.H., Green E.K. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat. Genet. 2008;40:1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Donovan M.C., Craddock N., Norton N., Williams H., Peirce T., Moskvina V., Nikolov I., Hamshere M., Carroll L., Georgieva L. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat. Genet. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 11.Purcell S.M., Wray N.R., Stone J.L., Visscher P.M., O'Donovan M.C., Sullivan P.F., Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stefansson H., Ophoff R.A., Steinberg S., Andreassen O.A., Cichon S., Rujescu D., Werge T., Pietilainen O.P., Mors O., Mortensen P.B. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sullivan P.F., de Geus E.J., Willemsen G., James M.R., Smit J.H., Zandbelt T., Arolt V., Baune B.T., Blackwood D., Cichon S. Genome-wide association for major depressive disorder: A possible role for the presynaptic protein piccolo. Mol. Psychiatry. 2009;14:359–375. doi: 10.1038/mp.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.WTCCC Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.ISC Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirov G., Gumus D., Chen W., Norton N., Georgieva L., Sari M., O'Donovan M.C., Erdogan F., Owen M.J., Ropers H.H. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum. Mol. Genet. 2008;17:458–465. doi: 10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- 17.Stefansson H., Rujescu D., Cichon S., Pietilainen O.P., Ingason A., Steinberg S., Fossdal R., Sigurdsson E., Sigmundsson T., Buizer-Voskamp J.E. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamnasaran D., Muir W.J., Ferguson-Smith M.A., Cox D.W. Disruption of the neuronal PAS3 gene in a family affected with schizophrenia. J. Med. Genet. 2003;40:325–332. doi: 10.1136/jmg.40.5.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Millar J.K., Pickard B.S., Mackie S., James R., Christie S., Buchanan S.R., Malloy M.P., Chubb J.E., Huston E., Baillie G.S. Dros. Inf. Serv.C1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science. 2005;310:1187–1191. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- 20.Millar J.K., Wilson-Annan J.C., Anderson S., Christie S., Taylor M.S., Semple C.A., Devon R.S., Clair D.M., Muir W.J., Blackwood D.H. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum. Mol. Genet. 2000;9:1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- 21.Pickard B.S., Malloy M.P., Christoforou A., Thomson P.A., Evans K.L., Morris S.W., Hampson M., Porteous D.J., Blackwood D.H., Muir W.J. Cytogenetic and genetic evidence supports a role for the kainate-type glutamate receptor gene, GRIK4, in schizophrenia and bipolar disorder. Mol. Psychiatry. 2006;11:847–857. doi: 10.1038/sj.mp.4001867. [DOI] [PubMed] [Google Scholar]
- 22.Pickard B.S., Malloy M.P., Porteous D.J., Blackwood D.H., Muir W.J. Disruption of a brain transcription factor, NPAS3, is associated with schizophrenia and learning disability. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2005;136:26–32. doi: 10.1002/ajmg.b.30204. [DOI] [PubMed] [Google Scholar]
- 23.Pickard B.S., Christoforou A., Thomson P.A., Fawkes A., Evans K.L., Morris S.W., Porteous D.J., Blackwood D.H., Muir W.J. Interacting haplotypes at the NPAS3 locus alter risk of schizophrenia and bipolar disorder. Mol. Psychiatry. 2008;14:874–884. doi: 10.1038/mp.2008.24. [DOI] [PubMed] [Google Scholar]
- 24.Pickard B.S., Knight H.M., Hamilton R.S., Soares D.C., Walker R., Boyd J.K., Machell J., Maclean A., McGhee K.A., Condie A. A common variant in the 3′UTR of the GRIK4 glutamate receptor gene affects transcript abundance and protects against bipolar disorder. Proc. Natl. Acad. Sci. USA. 2008;105:14940–14945. doi: 10.1073/pnas.0800643105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pickard B.S., Thomson P.A., Christoforou A., Evans K.L., Morris S.W., Porteous D.J., Blackwood D.H., Muir W.J. The PDE4B gene confers sex-specific protection against schizophrenia. Psychiatr. Genet. 2007;17:129–133. doi: 10.1097/YPG.0b013e328014492b. [DOI] [PubMed] [Google Scholar]
- 26.Prades C., Arnould I., Annilo T., Shulenin S., Chen Z.Q., Orosco L., Triunfol M., Devaud C., Maintoux-Larois C., Lafargue C. The human ATP binding cassette gene ABCA13, located on chromosome 7p12.3, encodes a 5058 amino acid protein with an extracellular domain encoded in part by a 4.8-kb conserved exon. Cytogenet. Genome Res. 2002;98:160–168. doi: 10.1159/000069852. [DOI] [PubMed] [Google Scholar]
- 27.Uitto J. The gene family of ABC transporters—novel mutations, new phenotypes. Trends Mol. Med. 2005;11:341–343. doi: 10.1016/j.molmed.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 28.American Psychiatric Association . American Psychiatric Association; Washington, DC: 2004. Diagnostic and Statistical Manual of Mental Disorders. [Google Scholar]
- 29.Endicott J., Spitzer R.L. A diagnostic interview: The schedule for affective disorders and schizophrenia. Arch. Gen. Psychiatry. 1978;35:837–844. doi: 10.1001/archpsyc.1978.01770310043002. [DOI] [PubMed] [Google Scholar]
- 30.Deary I.J., Gow A.J., Taylor M.D., Corley J., Brett C., Wilson V., Campbell H., Whalley L.J., Visscher P.M., Porteous D.J. The Lothian Birth Cohort 1936: A study to examine influences on cognitive ageing from age 11 to age 70 and beyond. BMC Geriatr. 2007;7:28. doi: 10.1186/1471-2318-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reese M.G., Eeckman F.H., Kulp D., Haussler D. Improved splice site detection in Genie. J. Comput. Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 32.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 33.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boeckmann B., Bairoch A., Apweiler R., Blatter M.C., Estreicher A., Gasteiger E., Martin M.J., Michoud K., O'Donovan C., Phan I. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheffel F., Demmer U., Warkentin E., Hulsmann A., Schneider E., Ermler U. Structure of the ATPase subunit CysA of the putative sulfate ATP-binding cassette (ABC) transporter from Alicyclobacillus acidocaldarius. FEBS Lett. 2005;579:2953–2958. doi: 10.1016/j.febslet.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 38.Pei J., Grishin N.V. MUMMALS: Multiple sequence alignment improved by using hidden Markov models with local structural information. Nucleic Acids Res. 2006;34:4364–4374. doi: 10.1093/nar/gkl514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Combet C., Blanchet C., Geourjon C., Deleage G. NPS@: Network protein sequence analysis. Trends Biochem. Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 40.Frishman D., Argos P. Knowledge-based protein secondary structure assignment. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 41.Jones D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 42.McGuffin L.J., Bryson K., Jones D.T. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 43.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 44.Bower M.J., Cohen F.E., Dunbrack R.L. Prediction of protein side-chain rotamers from a backbone-dependent rotamer library: A new homology modeling tool. J. Mol. Biol. 1997;267:1268–1282. doi: 10.1006/jmbi.1997.0926. [DOI] [PubMed] [Google Scholar]
- 45.Canutescu A.A., Shelenkov A.A., Dunbrack R.L. A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 2003;12:2001–2014. doi: 10.1110/ps.03154503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clark M., Cramer R.D., Vanopdenbosch N. Validation of the general-purpose Tripos 5.2 force-field. J. Comput. Chem. 1989;10:982–1012. [Google Scholar]
- 47.Laskowski R.A., Macarthur M.W., Moss D.S., Thornton J.M. Procheck—A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 48.Fraczkiewicz R., Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J. Comput. Chem. 1998;19:319–333. [Google Scholar]
- 49.Tina K.G., Bhadra R., Srinivasan N. PIC: Protein interactions calculator. Nucleic Acids Res. 2007;35:W473–W476. doi: 10.1093/nar/gkm423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abecasis G.R., Cherny S.S., Cookson W.O., Cardon L.R. Merlin—Rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 51.Kong A., Cox N.J. Allele-sharing models: LOD scores and accurate linkage tests. Am. J. Hum. Genet. 1997;61:1179–1188. doi: 10.1086/301592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferrer-Costa C., Gelpi J.L., Zamakola L., Parraga I., de la Cruz X., Orozco M. PMUT: A web-based tool for the annotation of pathological mutations on proteins. Bioinformatics. 2005;21:3176–3178. doi: 10.1093/bioinformatics/bti486. [DOI] [PubMed] [Google Scholar]
- 53.Ramensky V., Bork P., Sunyaev S. Human non-synonymous SNPs: Server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sunyaev S., Ramensky V., Bork P. Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends Genet. 2000;16:198–200. doi: 10.1016/s0168-9525(00)01988-0. [DOI] [PubMed] [Google Scholar]
- 55.Thomas P.D., Campbell M.J., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nikaido K., Ames G.F. One intact ATP-binding subunit is sufficient to support ATP hydrolysis and translocation in an ABC transporter, the histidine permease. J. Biol. Chem. 1999;274:26727–26735. doi: 10.1074/jbc.274.38.26727. [DOI] [PubMed] [Google Scholar]
- 57.Zaitseva J., Jenewein S., Jumpertz T., Holland I.B., Schmitt L. H662 is the linchpin of ATP hydrolysis in the nucleotide-binding domain of the ABC transporter HlyB. EMBO J. 2005;24:1901–1910. doi: 10.1038/sj.emboj.7600657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fishman G.A., Stone E.M., Grover S., Derlacki D.J., Haines H.L., Hockey R.R. Variation of clinical expression in patients with Stargardt dystrophy and sequence variations in the ABCR gene. Arch. Ophthalmol. 1999;117:504–510. doi: 10.1001/archopht.117.4.504. [DOI] [PubMed] [Google Scholar]
- 59.Bergen A.A., Plomp A.S., Hu X., de Jong P.T., Gorgels T.G. ABCC6 and pseudoxanthoma elasticum. Pflugers Arch. 2007;453:685–691. doi: 10.1007/s00424-005-0039-0. [DOI] [PubMed] [Google Scholar]
- 60.Barros S.A., Tennant R.W., Cannon R.E. Molecular structure and characterization of a novel murine ABC transporter, Abca13. Gene. 2003;307:191–200. doi: 10.1016/s0378-1119(03)00465-7. [DOI] [PubMed] [Google Scholar]
- 61.Blackwood D.H., Fordyce A., Walker M.T., St Clair D.M., Porteous D.J., Muir W.J. Schizophrenia and affective disorders—Cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: Clinical and P300 findings in a family. Am. J. Hum. Genet. 2001;69:428–433. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaminski W.E., Piehler A., Wenzel J.J. ABC A-subfamily transporters: Structure, function and disease. Biochim. Biophys. Acta. 2006;1762:510–524. doi: 10.1016/j.bbadis.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 63.Kitiratschky V.B., Grau T., Bernd A., Zrenner E., Jagle H., Renner A.B., Kellner U., Rudolph G., Jacobson S.G., Cideciyan A.V. ABCA4 gene analysis in patients with autosomal recessive cone and cone rod dystrophies. Eur. J. Hum. Genet. 2008;16:812–819. doi: 10.1038/ejhg.2008.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Slatter T.L., Jones G.T., Williams M.J., van Rij A.M., McCormick S.P., Frikke-Schmidt R., Tybjaerg-Hansen A., Morison I.M. Novel rare mutations and promoter haplotypes in ABCA1 contribute to low-HDL-C levels. Clin. Genet. 2008;73:179–184. doi: 10.1111/j.1399-0004.2007.00940.x. [DOI] [PubMed] [Google Scholar]
- 65.Albrecht C., Viturro E. The ABCA subfamily—Gene and protein structures, functions and associated hereditary diseases. Pflugers Arch. 2007;453:581–589. doi: 10.1007/s00424-006-0047-8. [DOI] [PubMed] [Google Scholar]
- 66.D'Angelo G., Vicinanza M., De Matteis M.A. Lipid-transfer proteins in biosynthetic pathways. Curr. Opin. Cell Biol. 2008;20:360–370. doi: 10.1016/j.ceb.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 67.van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fitzgerald M.L., Okuhira K., Short G.F., Manning J.J., Bell S.A., Freeman M.W. ATP-binding cassette transporter A1 contains a novel C-terminal VFVNFA motif that is required for its cholesterol efflux and ApoA-I binding activities. J. Biol. Chem. 2004;279:48477–48485. doi: 10.1074/jbc.M409848200. [DOI] [PubMed] [Google Scholar]
- 69.Doris A.B., Wahle K., MacDonald A., Morris S., Coffey I., Muir W., Blackwood D. Red cell membrane fatty acids, cytosolic phospholipase-A2 and schizophrenia. Schizophr. Res. 1998;31:185–196. doi: 10.1016/s0920-9964(98)00016-4. [DOI] [PubMed] [Google Scholar]
- 70.Glen A.I., Glen E.M., Horrobin D.F., Vaddadi K.S., Spellman M., Morse-Fisher N., Ellis K., Skinner F.S. A red cell membrane abnormality in a subgroup of schizophrenic patients: Evidence for two diseases. Schizophr. Res. 1994;12:53–61. doi: 10.1016/0920-9964(94)90084-1. [DOI] [PubMed] [Google Scholar]
- 71.McNamara R.K., Jandacek R., Rider T., Tso P., Hahn C.G., Richtand N.M., Stanford K.E. Abnormalities in the fatty acid composition of the postmortem orbitofrontal cortex of schizophrenic patients: Gender differences and partial normalization with antipsychotic medications. Schizophr. Res. 2007;91:37–50. doi: 10.1016/j.schres.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McNamara R.K., Jandacek R., Rider T., Tso P., Stanford K.E., Hahn C.G., Richtand N.M. Deficits in docosahexaenoic acid and associated elevations in the metabolism of arachidonic acid and saturated fatty acids in the postmortem orbitofrontal cortex of patients with bipolar disorder. Psychiatry Res. 2008;160:285–299. doi: 10.1016/j.psychres.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peet M., Laugharne J., Rangarajan N., Horrobin D., Reynolds G. Depleted red cell membrane essential fatty acids in drug-treated schizophrenic patients. J. Psychiatr. Res. 1995;29:227–232. doi: 10.1016/0022-3956(95)00001-l. [DOI] [PubMed] [Google Scholar]
- 74.Yao J.K., van Kammen D.P., Gurklis J. Red blood cell membrane dynamics in schizophrenia. III. Correlation of fatty acid abnormalities with clinical measures. Schizophr. Res. 1994;13:227–232. doi: 10.1016/0920-9964(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 75.Georgieva L., Moskvina V., Peirce T., Norton N., Bray N.J., Jones L., Holmans P., Macgregor S., Zammit S., Wilkinson J. Convergent evidence that oligodendrocyte lineage transcription factor 2 (OLIG2) and interacting genes influence susceptibility to schizophrenia. Proc. Natl. Acad. Sci. USA. 2006;103:12469–12474. doi: 10.1073/pnas.0603029103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sims R., Hollingworth P., Moskvina V., Dowzell K., O'Donovan M.C., Powell J., Lovestone S., Brayne C., Rubinsztein D., Owen M.J. Evidence that variation in the oligodendrocyte lineage transcription factor 2 (OLIG2) gene is associated with psychosis in Alzheimer's disease. Neurosci. Lett. 2009;461:54–59. doi: 10.1016/j.neulet.2009.05.051. [DOI] [PubMed] [Google Scholar]
- 77.Segal D., Koschnick J.R., Slegers L.H., Hof P.R. Oligodendrocyte pathophysiology: A new view of schizophrenia. Int. J. Neuropsychopharmacol. 2007;10:503–511. doi: 10.1017/S146114570600722X. [DOI] [PubMed] [Google Scholar]
- 78.Sokolov B.P. Oligodendroglial abnormalities in schizophrenia, mood disorders and substance abuse. Comorbidity, shared traits, or molecular phenocopies? Int. J. Neuropsychopharmacol. 2007;10:547–555. doi: 10.1017/S1461145706007322. [DOI] [PubMed] [Google Scholar]
- 79.Tkachev D., Mimmack M.L., Huffaker S.J., Ryan M., Bahn S. Further evidence for altered myelin biosynthesis and glutamatergic dysfunction in schizophrenia. Int. J. Neuropsychopharmacol. 2007;10:557–563. doi: 10.1017/S1461145706007334. [DOI] [PubMed] [Google Scholar]
- 80.Narayan S., Head S.R., Gilmartin T.J., Dean B., Thomas E.A. Evidence for disruption of sphingolipid metabolism in schizophrenia. J. Neurosci. Res. 2008;87:278–288. doi: 10.1002/jnr.21822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schmitt A., Wilczek K., Blennow K., Maras A., Jatzko A., Petroianu G., Braus D.F., Gattaz W.F. Altered thalamic membrane phospholipids in schizophrenia: A postmortem study. Biol. Psychiatry. 2004;56:41–45. doi: 10.1016/j.biopsych.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 82.Bodmer W., Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat. Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Makinen V.P., Parkkonen M., Wessman M., Groop P.H., Kanninen T., Kaski K. High-throughput pedigree drawing. Eur. J. Hum. Genet. 2005;13:987–989. doi: 10.1038/sj.ejhg.5201430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


