Abstract
The fasting-activated longevity protein sirtuin 1 (SirT1, ref. 1) promotes gluconeogenesis in part, by increasing transcription of the key gluconeogenic genes pepck1 and g6pase2,3, through deacetylating PGC-1α and FOXO1 (ref. 4). In contrast, signal transducer and activator of transcription 3 (STAT3) inhibits glucose production by suppressing expression of these genes5,6. It is not known whether the inhibition of gluconeogenesis by STAT3 is controlled by metabolic regulation. Here we show that STAT3 phosphorylation and function in the liver were tightly regulated by the nutritional status of an animal, through SirT1-mediated deacetylation of key STAT3 lysine sites. The importance of the SirT1-STAT3 pathway in the regulation of gluconeogenesis was verified in STAT3-deficient mice in which the dynamic regulation of gluconeogenic genes by nutritional status was disrupted. Our results reveal a new nutrient sensing pathway through which SirT1 suppresses the inhibitory effect of STAT3, while activating the stimulatory effect of PGC-1α and FOXO1 on gluconeogenesis, thus ensuring maximal activation of gluconeogenic gene transcription. The connection between acetylation and phosphorylation of STAT3 implies that STAT3 may have an important role in other cellular processes that involve SirT1.
The transcription factor STAT3 participates in various critical cellular processes7. In the liver, STAT3 is known to suppress expression of the transcriptional co-activator of gluconeogenesis PGC-1α and to suppress gluconeogenic genes. Ectopic expression of STAT3 in leptin receptor mutant (lepr-/-) mice reduces PGC-1α transcription and reverses diabetes. This effect of STAT3 is abolished when Tyr 705 is mutated to a phenylalanine (Y705F; ref. 5). Other protein modifications of STAT3, such as acetylation, have recently been reported8-10. However, the functional significance of STAT3 acetylation remains ill-defined, and its relationship with STAT3 tyrosine phosphorylation remains unclear.
We hypothesized that STAT3 acetylation regulates physiological processes by mediating changes in the STAT3 phosphorylation status. We found that STAT3 acetylation was decreased after a 24-h fast and increased after feeding in the livers of C57BL/6J mice. STAT3 tyrosine phosphorylation directly correlated with the level of STAT3 acetylation, indicating that the reversible acetyl-modifications are functionally relevant (Fig. 1a). The observation that both acetylation and phosphorylation of STAT3 were evident in fed, but dramatically reduced in fasted, animals suggests that STAT3 acetylation and phosphorylation are actively downregulated on fasting. Overall, these observations signify that cellular metabolic status regulates STAT3 function in the liver, presumably owing to the sensitivity of the liver to the overall nutritional status of the organism11.
Figure 1.
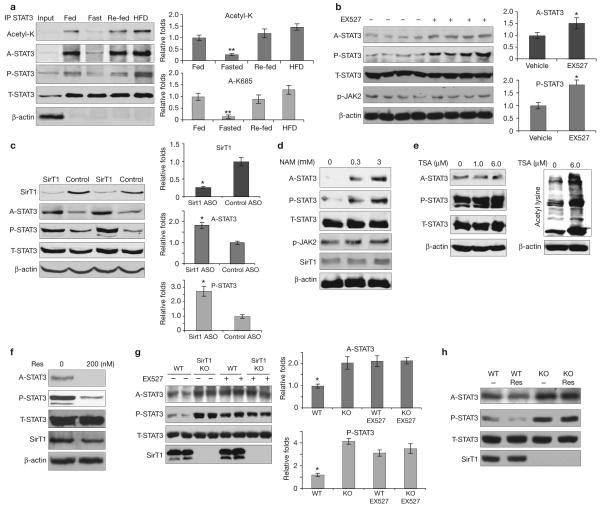
SirT1 is involved in regulating STAT3 acetylation. (a) STAT3 acetylation and phosphorylation changed in mouse livers under different nutritional conditions. Male C57Bl/6J mice were fed (normal laboratory chow), fasted (24 h), re-fed (24 h, normal laboratory chow after fasting) or fed with a high-fat diet (HFD, 48 h). The levels of STAT3 acetylation and phosphorylation in livers were determined by immunoprecipitation and/or western blot analysis. (b) EX527 induced acetylation and phosphorylation of STAT3. Fasted male C57Bl/6J mice were injected with EX527 (i.p. 10 mg per kg body weight) 6 h before being killed. (c) SirT1 ASO induced hepatic STAT3 acetylation and phosphorylation. C57Bl/6J mice were injected with ASO (i.p. 10 per kg body weight) five times in a 20-day period. (d-f) SV40-transformed mouse hepatic cells were treated with different doses of nicotinamide (NAM, d), trichostatin A (TSA, e) and resveratrol (Res, f). STAT3 acetylation levels were significantly increased by nicotinamide, but reduced by resveratrol. No change was observed with TSA. The significant increase of total protein acetylation shown by the pan-anti-acetylated lysine antibody indicated TSA was effective (f, left panel). (g, h) Decetylation of STAT3 is dependent on SirT1 in vitro. EX527 (10 μM for 6 h) increased STAT3 acetylation and phosphorylation in wild-type MEFs, but not in SirT1 KO MEFs (g). Data are mean ± s.e.m. of three repeated experiments, n = 2 cells. Resveratrol (100 nM for 6 h) decreased STAT3 acetylation and phosphorylation in wild-type MEFs, but not in SirT1 KO MEFs (h). Data are mean ± s.e.m., n = 5 mice * P < 0.05, ** P < 0.01 in a, b and c.
SirT1 can be induced to promote gluconeogenesis11,12 under conditions of fasting. Therefore, we asked whether SirT1 affects fed/fast-regulated STAT3 acetylation and phosphorylation. We injected the SirT1 inhibitor EX527, which has shown increased potency and specificity for SirT1 (ref. 13), into C57Bl/6J mice. EX527 increased acetylation and phosphorylation of hepatic STAT3 (Fig. 1c). These results are similar to those seen in animals treated with nicotinamide (Supplementary Information, Fig S1a), a less specific SirT1 inhibitor. EX527 also increased the acetylation of p53, a known SirT1 substrate, suggesting that a reduction of SirT1 function was achieved with EX527 treatment (Supplementary Information, Fig. S4b)1. In addition to EX527, we used an antisense oligionucleotide (ASO)14,15 to knockdown hepatic SirT1 on a chronic basis. SirT1 ASO induced significant STAT3 acetylation and tyrosine phosphorylation (Fig. 1c). Together, these results support the idea that SirT1 is critically involved in hepatic STAT3 regulation.
Next, we studied the effect of nicotinamide and resveratrol (a SirT1 activator) on STAT3 acetylation and phosphorylation in an SV40-transformed mouse hepatic cell line, previously used in gluconeogenic studies16,17. In cells treated with nicotinamide (0.3 mM and 3 mM), the levels of STAT3 acetylation and phosphorylation increased in a dose-dependent manner and were independent of the kinase JAK2 (Fig. 1d) and class I and II HDACs (other cells were treated with trichostatin A, TSA; Fig. 1e). Conversely, resveratrol18 (0.2 μM) reduced STAT3 acetylation and phosphorylation in the cultured hepatic cells (Fig. 1f). To determine whether deacetylation of STAT3 requires SirT1, we used SirT1 knockout (KO) and wild-type mouse embryonic fibroblasts19 (MEFs). First, we found that levels of STAT3 acetylation and phosophrylation were constitutively higher in the SirT1 KO MEFs than in the wild-type MEFs (Fig. 1g). Second, treatment with EX527 increased levels of STAT3 acetylation and phosphorylation in wild-type MEFs, but not in SirT1 KO MEFs (Fig. 1g). Moreover, resveratrol decreased STAT3 acetylation and phosphorylation in wild-type MEFs, but not SirT1 KO MEFs (Fig. 1h). Together, these results further indicate that deacetylation of STAT3 is dependent on SirT1.
To investigate STAT3 as a SirT1 substrate, we studied the role of ectopically expressed SirT1. We found that the level of STAT3 acetylation was greatly reduced by cotransfection of human (h) SirT1, in HEK293T cells (Fig. 2a). Moreover, the introduced hSirT1 was able to potently suppress p300/CBP-induced STAT3 acetylation, suggesting that both factors affected the same set of lysine residues of STAT3 (Fig. 2a). The deacetylation of STAT3 by hSirT1 was more effective than that by HDAC1, HDAC 3 (Fig. 2a) or HDAC2 (data not shown), which were previously implicated in STAT3 deacetylation10,20.
Figure 2.
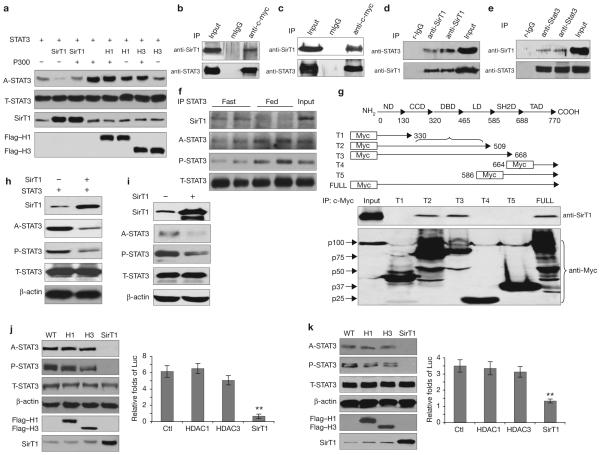
STAT3 phosphorylation and transactivation were downregulated by SirT1. (a) SirT1 deacetylates STAT3 in cultured cells. The effect of p300, SirT1 or HDACs on STAT3 acetylation was measured in transfected HEK293T cells (H1, HDCA1; H3, HDCA3). (b-e) SirT1 and STAT3 form complexes in vitro and in vivo. HEK293T cells were transfected with SirT1 and STAT3, or STAT3 alone (b, c). The physical interactions between exogenous STAT3 and exogenous (b) or endogenous (c) SirT1 were detected. In HEK293T cells, endogenous SirT1 and endogenous STAT3 were co-precipitated (d). STAT3 and SirT1 were co-precipitated by a STAT3 antibody in the livers of wild-type male mice (n = 4, e). (f) The physical interaction between STAT3 and SirT1 was enhanced in fasting livers. (g) Except for full-length STAT3, SirT1 was only precipitated by truncated STAT3-T2 and -T3, suggesting that both the DNA binding and the linker domains of STAT3 are involved in the interaction of STAT3 and SirT1. ND, N-terminal domain; CCD, coil-coil domain; DBD, DNA binding domain; LD, linker domain; SH2D, SH2 domain and TAD, transactivation domain. (h, i) SirT1-mediated deacetylation of STAT3 affects Y705-STAT3 phosphorylation. HEK293T cells were transfected with SirT1 and STAT3 (0.25 μg per well of each, h), or SirT1 alone (i), in 12-well-plates. (i) Each well was loaded with 100 μg of total protein to visually present the signals of endogenous A-STAT3 and P-STAT3. (j, k) The SirT1-mediated deacetylation of STAT3 affected STAT3 function (WT, wild-type). A2780 cells were treated with IL-6 (40 ng ml-1) for 12 h. A relatively low level of the SirT1, HDAC1 and HDAC3 plasmids (0.05 μg per well) were either transfected with STAT3 (0.1 μg per well) or untreated (control). The effects on either exogenous (20 μg per well, j) or endogenous (60 μg per well, k) STAT3 acetylation and phosphorylation were determined. STAT3 transactivation activities were detected by a STAT3 specific luciferase reporter (Luc). Data are mean ± s.e.m. of three repeated experiments, ** P < 0.01 in J and k.
To test whether SirT1 and STAT3 formed a complex in cells, HEK293T cells were co-transfected with the wild-type genes of hSirT1 and hSTAT3. Exogenous (Fig. 2b) and endogenous (Fig. 2c) hSirT1 proteins were detected in immunoprecipitation products, suggesting that STAT3 and hSirT1 formed complexes. Next, endogenous STAT3 was co-precipitated by endogenous hSirT1 (Fig. 2d). To determine whether this physical interaction between hSirT1 and STAT3 occurs in vivo, we examined their association in mouse liver. SirT1 was observed in the STAT3 immunoprecipitation products, suggesting an interaction between endogenous STAT3 and SirT1 in the liver of fasted animals (Fig. 2e), and less so in the liver of fed animals (Fig. 2f).
To determine the region(s) in STAT3 responsible for SirT1 binding, a set of five c-Myc tagged STAT3 deletion mutants (T1 to T5) were generated that were designed to test each of the domains in STAT3. The DNA binding and linker domain (from amino acid 330-590) of STAT3 (ref. 21) was found to be the key region involved in the STAT3-SirT1 interaction (Fig. 2g). The various truncated STAT3s that did not contain the DNA binding and linker domain failed to form complexes with SirT1, whereas the truncated STAT3s that contained this domain pulled down SirT1 (Fig. 2g). Moreover, the latter had greatly reduced acetylation and phosphorylation on cotransfection with SirT1 (data not shown), suggesting that a direct interaction between the STAT3 DNA binding and linker domain and SirT1 is required for the enzymatic function of SiT1 on STAT3.
We next analysed whether the acetylation state of STAT3 affects STAT3 phosphorylation and transactivation function. After hSirT1 overexpression, the phosphorylation of exogenous (Fig. 2h) and endogenous (Fig. 2i) STAT3 was again downregulated with high efficiency. Low levels of exogenous hSirT1 DNA (0.05 μg) resulted in a reduction of exogenous and endogenous STAT3 acetylation and phosphorylation (Fig. 2j, k), whereas transfection with HDAC1 and HDAC 3 had a limited effect (Fig. 2j, k). Consistently, an assay of STAT3-dependent luciferase reporter10 (p4x IRF-Luc) revealed that SirT1, but not HDAC1 or HDAC3, suppressed the transactivation function of STAT3 (Fig. 2j, k). However, at increased doses of plasmid DNA, HDAC3 appreciably reduced STAT3 activity (Supplementary Information, Fig. S2a); this may contribute to downregulation of STAT3 acetylation and phosphorylation under certain conditions and in selected cells8,20. From these results, we conclude that SirT1 specifically and effectively deacetylated STAT3 in cultured cells and in vivo, and that this modification is coupled with a downregulation of STAT3 phosphorylation and transactivation.
The direct interaction between SirT1 and STAT3, and its effect on STAT3 phosphorylation and function, suggest that the state of acetylation of STAT3 may directly regulate its phosphorylation. Acetylation in a limited number of lysine residues in STAT3 was reported8-10; however, the coupling of acetylation and phosphorylation through these sites was not established, suggesting more lysine acetylation sites are involved. To identify these, particularly in the carboxy-terminal region, which is crucial for STAT3 phosphorylation, we initially examined acetylation in the K685R-STAT3 mutant: Lys 685 was reported to be the only acetylation site at the C terminus of STAT3 (ref. 10). We used an anti-acetyl-STAT3 antibody and, although it detected a minor reduction in acetylation, significant signal was still detected (Fig. 3a). This signal was downregulated by SirT1, suggesting that other acetylation sites exist. Three new lysine-acetylation residues, K679, 707 and 709, were identified by tandem mass spectrometry analysis (Supplementary Information, Fig. S2). These sites are evolutionally conserved among mammalian STATs, (Fig. 3b) and located either in the end β-sheet structure of the SH2 domain21 (βH), or in the disordered signal stretch immediately after the SH2 domain. SH2 is known to specifically bind to phospho-tyrosine peptides22,23; therefore, it is critical to tyrosine signalling. Notably, the three new lysine residues are all in the vicinity of the Y705 of STAT3 (Fig. 3b), indicating that these sites may be pertinent to STAT3 phosphorylation7,24.
Figure 3.
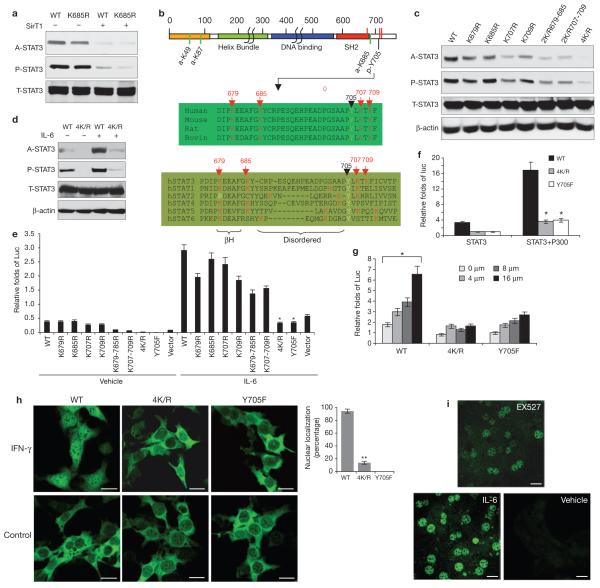
Critical novel acetylation sites regulate STAT3 phosphorylation and transactivation. (a) HEK293 cells were transfected with wild-type (WT) or K685R-STAT3 plasmids alone, or co-transfected with SirT1. STAT3 acetylation was determined. (b) A schematic representation of lysine acetylation sites identified in STAT3. Green lines represent the sites previously reported. Red lines represent a further three acetylation sites, identified by tandem mass spectrometry (Supplementary Information, Fig. S2b). (c, d) Mutations at four C-terminal lysine residues (4K/R) abolished STAT3 phosphorylation. A2780 cells were transfected with different K/R STAT3 mutants (c). Cells transfected with 4K/R or wild-type STAT3 were stimulated with 40 ng ml-1) for 6 h (d). (e-g) The 4K/R mutation abolished STAT3 transactivation function. A2780 cells were cotransfected with different K/R-STAT3 mutants (0.15 μg), hSirT1 (0.1 μg), STAT3-dependent luciferase reporter (0.1 μg) and an internal control reporter pRL-TK plasmid (0.01 μg, e). Cells were treated with IL-6 (40 ng ml-1) or left untreated. The transactivation function of STAT3 was assayed. The effect of p300 (0.35 μg per well, f) and various EX527 doses (g) on activating the transactivation function of wild-type or 4K/R- STAT3 were determined. (h, i) The 4K/R mutant disrupted STAT3 nuclear localization. STAT3 KO hepatoma cells were infected with retroviruses (pbabe-6Myc-STAT3 WT, Y705F and 4K/R) and stimulated with IFN-γ (50 ng ml-1) for 2 h. (i) Primary hepatocytes were prepared from fasted animals and cultured in low nutrient media for 12 h before treatment with EX527 (10 μM) for 6 h or IL-6 (40ng ml-1) for 1 h. The translocation of STAT3 was determined by immunofluorescence microscopy staining. Data are mean ± s.e.m. of three repeated experiments, * P < 0.005, ** P < 0.01 in e,f and g. Scale bars, 10 μm in h, 50 μm in i.
Next, a systematic site-mutagenesis of these lysine sites of interest was performed. Mutation of all four lysine to arginine abolished the acetylation signals of STAT3 (Fig. 3c). Single or double lysine-to-arginine mutations had a limited effect on STAT3 phosphorylation, whereas changes of all four lysines to arganine (4K/R) largely abolished STAT3 phosphorylation. This effect is specific to the C-terminal acetylation sites, as mutations of the amino-terminal acetylation sites (K49/87R) did not affect the phosphorylation of STAT3 (Supplementary Information, Fig. S3a, b). These results imply that multiple lysine-acetylation sites adjacent to Y705 are vital for STAT3 phosphorylation. In contrast, the dominant-negative Y705F mutation did not affect the acetylation of STAT3 (Supplementary Information, Fig. S3c). More importantly, the 4K/R-STAT3 mutant was no longer sensitive to stimulation by IL-6 (interleukin) and IFN-γ (interferon-γ) which otherwise acetylated and phosphorylated STAT3. This suggests that the newly identified lysine acetylation sites are critical for cytokine stimulation of STAT3 (Fig. 3d).
To test whether the mutations of these acetylation sites affect STAT3 transcriptional function, STAT3-dependent luciferase reporter assays were performed in HEK293T cells and human ovarian cancer A2780 cells25. Consistent with the phosphorylation results, the 4K/R mutation largely abolished IL-6-induced STAT3 activation and had a dominant-negative-like effect, comparable to that of the Y705F mutation (Fig. 3e); whereas individual mutations had limited effects on STAT3 transactivation. Moreover, 4K/R-STAT3 was not capable of activating a known endogenous STAT3 target (hAGT; ref. 20) in HepG2 cells (Supplementary Information, Fig. S3d). Finally, ectopic expression of p300 (ref. 10) and treatment with EX527, which increased STAT3 acetylation and phosphorylation in vivo13, failed to activate 4K/R-STAT3, whereas the same treatments increased the transactivation of wild-type STAT3 (Fig. 3f, g). These results suggest that the acetylation of the cluster of C-terminal lysine sites is up or downregulated by p300 or SirT1, respectively, and is crucial for STAT3 phosphorylation and transactivation. Next, we asked if the 4K/R mutations affect STAT3 localization, as phosphorylation is crucial for STAT3 nuclear translocation7. We introduced wild-type-, 4K/R- or Y705F-STAT3 into liver-STAT3 knockout (STAT3 LKO) hepatoma cells using a retrovirus (pbabe-6xcMyc-STAT3 containing wild-type-, Y705F- and 4K/R- STAT3) to test STAT3 nuclear translocation by immunofluorescence microscopy staining. Similarly to Y705F-STAT3, the mutant 4K/R-STAT3 significantly disrupted the nuclear translocation of STAT3 (Fig. 3h). This acetylation-related STAT3 localization was further supported by experiments in primary hepatocytes treated with EX527 (Fig. 3i). In addition, we found that the efficiency of 4K/R-STAT3 dimerization was greatly reduced (Supplementary Information, Fig. S3e).
If SirT1 promotes gluconeogenesis, in part through the suppression of STAT3, the knockout of STAT3 in liver should mimic the effect of SirT1 and increase gluconeogenesis independent of SirT1 activity. In normal chow fed STAT3 LKO mice, we observed a significant upregulation of pepck1 and g6pase, but not of the control genes (cytochrome-c and gk; Fig. 4a-c). However, the further upregulation of pepck1 and g6pase genes on fasting26 was limited (Fig. 4a). The lack of repression of gluconeogenic gene expression after feeding, was due to the absence of STAT3 bound to the promoter region of pepck1, as shown by chromatin immunopreciptiation (ChIP) assay (Supplementary Information, Fig. S3f).
Figure 4.
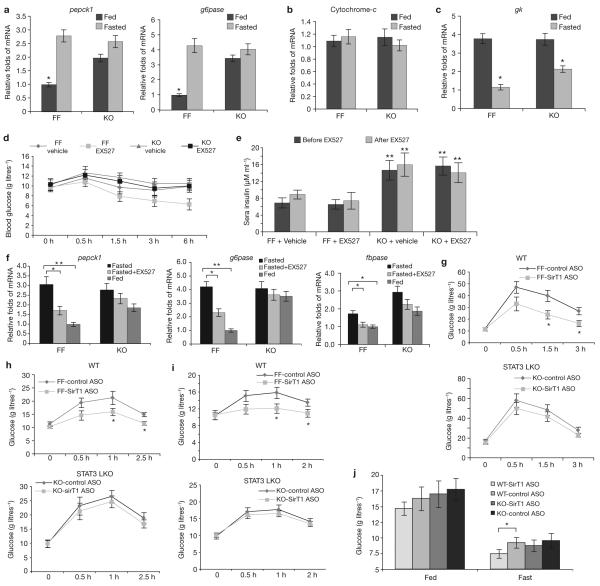
Liver-STAT3 deficiency disrupted fasting/SirT1 controlled gluconeogenesis. (a) Liver-STAT3 deficiency mimicked the effect of SirT1 in promoting gluconeogenic gene expression, independently of nutritional status and SirT1 activity. The mRNA levels of pck1 and g6pase in livers were determined in fed and fasted STAT3f/ f (FF) and STAT3 LKO (KO) mice using qrtPCR analysis (mean ± s.e.m., n = 5 livers). (b, c) Expression of the mitochondrial gene cytochrome-c as a control (b), and the glycolytic gene gk (c), were not altered by the absence of STAT3. Mean ± s.e.m., *P < 0.05, n = 5 livers in b and c. (d) STAT3 deficiency disrupted fasting/SirT1-induced hypoglycemia. The STAT3f/f and STAT3 LKO mice were fasted overnight, and then injected with EX527 (i.p. 10 mg per kg body weight). Plasma glucose levels were determined at 0, 30, 90, 180 and 360 min. (e) The same treatment did not alter the plasma insulin levels, which were determined before and after treatment with EX527. In d and e, data are mean ± s.e.m., n = 6 mice, ** P < 0.01. (f) STAT3 deficiency in the livers disrupted fasting/SirT1 induced gluconeogenic gene expression. The transcripts of the key gluconeogenic genes pepck1, g6pase and fbpase in the livers were detected by qrt-PCR (mean ± s.e.m., n = 5 * P < 0.05, ** P < 0.01). (g-i) The liver STAT3 deficiency impaired SirT1 controlled liver glucose production as assessed by a glucose tolerance test (GTT, g), pyruvate tolerance test (PTT, h) and glucagon-stimulation test (GST, Glucagon, i). Data are mean ± s.e.m., n = 6 mice in g-i. (j) STAT3 LKO-impaired SirT1 knockdown induced a reduction in glucose in animals on a high-fat diet. STAT3f/f and STAT3 LKO mice were fed a high-fat diet for two- and-a-half weeks. SirT1 ASO or control ASO was administered five times during a two week period at a dose of i.p. 10 mg per kg body weight. The levels of plasma glucose levels were measured under conditions of feeding and overnight fasting (mean ± s.e.m., n = 7 mice, * P = 0.022).
Next, we asked whether STAT3 is involved in the SirT1-mediated induction of hepatic gluconeogenesis. EX527 was used to inhibit SirT1 activity in STAT3 LKO mice and littermate controls after a 24-h fast. As reported previously6, STAT3 LKO mice maintained normal glucose levels under such conditions, despite increasing gluconeogenic gene expression and plasma insulin levels (Fig. 4a, e). EX527 (i.p. 10 mg per kg body weight) reduced plasma glucose levels in fasting wild-type mice, whereas the plasma glucose levels of STAT3 LKO mice were unchanged (Fig. 4d), suggesting that STAT3 deficiency disrupted the ability of EX527 to lower fasting glucose levels independent of insulin concentration (Fig. 4e). Consistent with the reduction of plasma glucose levels in wild-type animals after EX527 treatment, the expression of the gluconeogenic genes was reduced in the livers of wild-type mice but was blunted in STAT3 LKO mice (Fig 4f). Next, we studied the effect of SirT1 knockdown on glucose homeostasis in the STAT3 LKO model. The levels of hepatic STAT3 acetylation and phosphorylation were significantly increased with SirT1 ASO in wild-type mice (Fig. 1d; Supplementary Information, Fig. S4a). Moreover, SirT1 ASO decreased hepatic gluconeogenic gene transcription (pepck1, g6pase and fabpase) in wild-type, but not STAT3 LKO, mice (data not shown).
Finally, a glucose tolerance test (Fig. 4g), a pyruvate tolerance test (Fig. 4h) and a glucagon-stimulation test (Fig. 4i) were conducted to evaluate various aspects of glucose homeostasis in wild-type and STAT3 LKO mice, with or without SirT1 knockdown. An overall reduction in glucose production was indicated in the SirT1 ASO-treated control animals. SirT1 ASO treatment of STAT3 LKO mice had little effect on the parameters measured by all three tests (Fig. 4g-i). These data led us to conclude that SirT1 promotes gluconeogenesis, in part, by suppressing the inhibitory effect of STAT3 on the expression of gluconeogenic genes. To test the effect of hepatic insulin resistance on the interaction of SirT1 with STAT3, all groups of mice were fed with a high-fat diet (Supplementary Information, Fig. S5a). Blood glucose levels on fasting were decreased in SirT1 ASO-treated wild-type mice, but unchanged in SirT1-ASO treated STAT3 LKO mice (Fig. 4j; Fig. S5b). This observation indicates that the loss of hepatic STAT3 is critical for SirT1 function. In addition, our results are consistent with the SirT1 LKO model12, but differ from the SirT1 transgenic mouse model.3,27. However, gluconeogenic gene expression was increased in isolated primary SirT1 transgenic hepatocytes treated with cAMP and deprived of insulin treatment, suggesting that insulin-mediated inhibition of gluconeogenesis may be independent of the SirT1 pathway3.
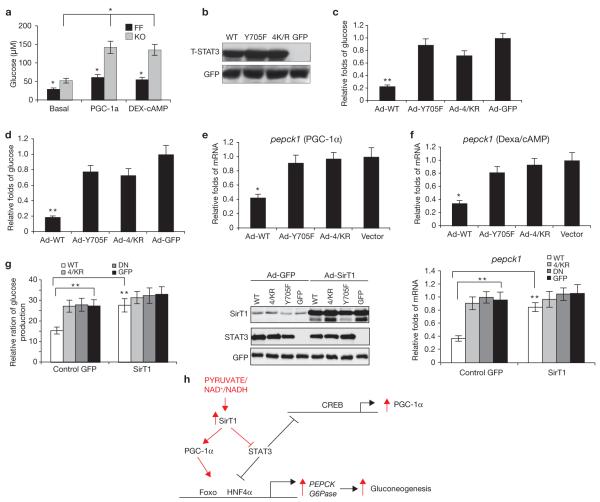
If the C-terminal cluster of acetylation lysine sites are crucial for STAT3 transactivation, the C-terminal 4K/R mutant should disrupt the suppressive effect of STAT3 on hepatic glucose production. First, we found that the glucose production in STAT3 LKO primary hepatocytes was increased, compared with that in wild-type cells (Fig. 5a). To further test the inhibitory effect of STAT3 on glucose production in hepatocytes, we promoted cellular glucose production in these cells, either by introducing PGC-1α or by treating cells with dexamethasone (50 nM) and 8-bromo-cAMP (2 μM; refs 2, 26). Equivalent amounts of STAT3 total proteins were detected in STAT3-/- cells in which wild-type STAT3, 4K/R- or Y705F-STAT3 had been re-introduced (Fig. 5b). Wild-type STAT3 suppressed hepatic glucose production (Fig. 5c, d), whereas, 4K/R- or Y705F-STAT3 had little effect. Consistent with the data on glucose production, we observed a strong reduction in expression of the gluconeogenic pepck1 transcripts by wild-type-STAT3, but not by 4K/R-STAT3, in STAT3 LKO hepatocytes ectopically expressing PGC-1α or treated with dexamethasone/cAMP (Fig. 5e, f).
Figure 5.
The 4K/R mutant STAT3 is defective in suppressing hepatic gluconeogenesis. (a) Basal-, PGC-1α- or dexa/cAMP-stimulated glucose production in primary hepatocytes was significantly increased in the absence of STAT3 (* P < 0.05; FF, STAT3f/f; KO, STAT3 LKO). (b) Wild-type-(WT), 4K/R- and Y705F-STAT3 were introduced into primary hepatocytes by adenoviruses (Ad), and an equal amount of each STAT3 protein was detected. (c, d) Wild-type STAT3, but not 4K/R- or Y705F- STAT3, effectively inhibited the promotion of glucose production by either PGC-1α (c) or dexa/cAMP (d), in primary hepatocytes (** P < 0.01). (e, f) Similarly, the wild-type-STAT3, but not 4K/R- or Y705F-STAT3, inhibited the expression of pepck1, when promoted by PGC-1α (e) or dexa/cAMP (f, * P < 0.05). (g) Ectopic SirT1 disrupted the effect of wild-type-STAT3 on suppressing glucose production. STAT3 KO primary hepatocytes were co-infected by adenoviruses, with adeno-SirT1, adeno-wild-type-STAT3, mutant-STAT3 or GFP control (** P < 0.01). The middle panel shows the level of protein expression of SirT1 and STAT3s; the right panel shows the level pepck1 mRNA. (h) A schematic representation of the nutrient sensing pathway through which SirT1 regulates hepatic gluconeogenesis by both suppressing STAT3 and activating PGC-1α /Foxo1. Data are mean ± s.e.m. of three repeated experiments in a-g.
To determine whether the change in hepatic glucose production by re-introduced wild-type-STAT3 (but not 4K/R-STAT3) is mediated by SirT1, we co-infected STAT3 LKO primary hepatocytes with both adeno-SirT1 and adeno-STAT3s (wild type, 4K/R, Y705F and GFP). Ectopic SirT1 blunted the suppression of glucose production by wild-type-STAT3; however, this effect was limited in 4K/R-STAT3- or Y705F-STAT3-expressing cells (Fig. 5g). These results further support the idea that hepatic glucose production is dependent on SirT1-mediated downregulation of STAT3.
Our findings reveal a new molecular mechanism whereby SirT1 suppresses the inhibitory effect of STAT3 on gluconeogenesis, while activating PGC-1α and Foxo1 (ref. 4) to stimulate gluconeogenesis in the liver, in response to nutrient signals. This dual function of SirT1 has a central role in preventing concurrent activation of the two counter-mechanisms of gluconeogenesis regulation (Fig. 5h). These findings have implications for defining the basic pathways of energy homeostasis, diabetes and lifespan.
METHODS
Animals
All the mice used in this study were on a C57BL/6J background. Twelve-week-old male wild-type mice (Jackson Laboratory) were fed ad libitum, fasted for 24 h, fasted and re-fed for 24 h or fed on a high-fat diet (45 Kcal% fat) for 48 h. STAT3 LKO mice were generated by crossing STAT3 f/f28 and Alb-Cre transgenic mice, B6.Cg-Tg (Alb-cre) 21Mgn/J (Jackson Laboratory). Various treatments were applied depending on the experiment, including: fasting, feeding, re-feeding, feeding with a high-fat diet and administration of various chemical compounds. EX527 (6-Chloro-2, 3, 4, 9-tetrahydro-1H-carbazole-1-carboxamide; Tocris; i.p.10 mg per kg body weight) and nicotinamide (Sigma; i.p. 50 mg per kg body weight or 150 mg per kg body weight) were introduced for 6 h. Animals were killed and the liver, white adipose, muscles, kidney, heart, brain and spleen tissues were subjected to western blotting or quantitative real-time PCR (qrt-PCR) analysis. The sera were collected through tail-vein puncture at different times to test glucose, insulin and liver function (Alanine Aminotransferase, ALT; assays performed in the Yale Mouse Metabolic Phenotyping Center). All procedures were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and under the approval of the Yale Medical School Animal Care and Use Committee.
SirT1 knockdown (ASO)
Control and STAT3 LKO mice (2-4 months old) were divided into control ASO and SirT1 ASO groups. The mice received food ab libitum and were housed on a 12-h dark/light cycle. SirT1 ASO 5′-ATACCATTCTTTGGTCTAGA-3′ (ASO # 384856) or control ASO (ASO# 141923; ISIS Pharmaceuticals) was administered five times during a 2-3 week period (i.p. 10 mg per kg body weight). ASO solutions were sterilized through a 0.44-μm filter before injection. The ASO targets the 3′UTR of SirT1 mRNA and has no significant crossreactivity to other sirtuin family members. The control ASO had the same chemistry as SirT1 ASO, and had a scrambled oligonucleotide sequence. Both ASOs were prepared in normal saline.
Measurement of glucose metabolism
The glucose tolerance test (glucose 2 g per kg body weight i.p.), pyruvate tolerance test (Pyruvate 2 g per kg body weight i.p.) and glucagon-stimulation test (Glucagon 200 μg per kg body weight i.p.) were conducted to evaluate various aspects of glucose production and metabolism.
Constructs
The pcDNA3-6×Myc-mSTAT3 and its K685R mutant expression vectors were from the laboratory of Y. E. Chin10. pcDNA3-6×cMyc-mSTAT3 K49R, K87R, K49-87R, K679R, K685R, K707R, K709R, K685-679R, K707-709R, K679-685-707-709R and Y705F were derived from the wild-type pcDNA3-Myc-mSTAT3 vector using a site-mutagenesis kit (Stratagene). The deletion mutants of STAT3 were constructed through PCR, and the schematic map of deletions is shown in Fig. 2g. The sequences of the oligonucleotide primers are: STAT3-F, 5′-cgaattcc ATG GCT CAG TGG AAC CAG-3′; S134-R, 5′-cctcgag tca AGC TGT TGG GTG GTT GG-3′; S323-R, 5′-cctcgagtcaCAC CAC GAA GGC ACT CTT-3′; S590-R, 5′-cctcgagtcaGCT GAT GAA ACC CAT GAT G-3′; S668-R, 5′-cctcgagtcaTGG AGA CAC CAG GAT GTT G-3′; S770-R, 5′-cctcgag TCA CAT GGG GGA GGT AGC-3′; S586-F, 5′-cgaattcc ATG GGT TTC ATC AGC AAG G-3′; S664-F, 5′-cgaattccATC CTG GTG TCT CCA CTT G-3′.
Retroviruses pBaBe-6×cMyc-STAT3 (wild-type, Y705F, K49-87R and K679-685-707-709R) were subcloned from pcDNA3-xcMyc-STAT3s, by switching restriction endonuclease (SalI). All constructs were verified by nucleotide sequencing in Yale KECK facilities. pTOPO-hSirT1 and its inactivated form H363Y were from Wei Gu (Columbia University, New York). HA-p300 constructs were provided Tsi-Pang Yao (Duke University, Carolina). Flag-HDAC1, Flag-HDAC2 and Flag-HDAC3 constructs were originally provided by Edward Seto (H. Lee Moffitt Cancer Center and Research Institute, Florida). P4xIRF-1-Luc is a STAT3 specific Luciferase reporter. APRE-luciferase reporter29 was a gift from David Levy (New York University School of Medicine). A PRE-TK plasmid was used as an internal control for transfection efficiency.
Cell culture, plasmid transfection, drug treatment and primary hypatocytes
A2780 cells (Sigma) were cultured in RPMI 1640 medium with 10% fetal bovine serum (FBS) supplemented with glutamine (2 mM). The SV40-immobilized mouse hepatocytes (from Domenico Accili, Columbia University, New York) were maintained in modified Eagle’s medium containing 10% FBS. SirT1 KO MEFs and wild-type MEFs were gifts from Leonard Guarente (Harvard University, Massachusetts). MEFs and HEK293T cells were maintained in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum. Cells were transiently transfected using LipofectAMINE 2000 (Invitrogen), according to the manufacturer’s protocol. Cells were collected and washed with cold PBS for experiments and cells received one of the following treatments: (i) 200 nM Resveratrol (Sigma) for 4 h; (ii) 0.3-3 mM Nicotinamide (Sigma) for 4 h; (iii) 1-6 μM TSA (Sigma) for 6-12 h; (iv) 40 ng ml-1 IL-6 (Sigma) for 6-12 h; (v) 50 ng ml-1 IFN-γ (Sigma) for 2-6 h; (vi) 10 mM EX527 (Tocris) or (vii) 50 nM Dexamathasone (Sigma) + 2 μM 8-bromo-cAMP (Sigma).
Mouse primary hepatocytes were prepared in the Hepatocyte Isolation Core of the Yale Liver Center (Yale University School of Medicine, New Haven). Details are shown in supplementary methods.
Immunoprecipitation
Protein-protein interactions in cells were also analysed by co-immunoprecipitation. The details of these experiments have been described previously30. Ectopic Myc-STAT3 or SirT1 were expressed in HEK293T cells by transient transfection with PcDNA3cMyc-STAT3 or PcDNA3.1/V5-his-SirT1. The interaction of ectopic STAT3 with ectopic SirT1, or ectopic STAT3 with endogenous SirT1, was tested using an anti-c-Myc antibody. Co-IP of endogenous STAT3 and SirT1 from HEK293T cells and liver tissues was performed using an anti-STAT3 antibody (Santa Cruz). Cell pellets and mouse liver tissues were sonicated in modified IP buffer, and pre-cleaned with normal rabbit or mouse IgG-conjugated nProtein A Sepharose 4 Fast Flow beads (Amersham) for 2 h at 4 °C. The pre-cleaned lysates were then mixed with c-Myc, the anti-STAT3 antibody and normal rabbit or mouse IgG (negative control)-conjugated nProtein A Sepharose 4 Fast Flow beads. Immunoprecipitation products were separated by SDS-PAGE, and blotted using a SirT1 antibody (Millipore), and β-actin (Sigma), which was used as an internal control.
Identification of STAT3 acetylation sites by mass spectrometry
The STAT3 complexes were immunoprecipitated, separated by SDS-PAGE and stained with SYPRO Ruby (Bio-Rad). The visible bands were excised. Gel pieces were subjected to a modified in-gel trypsin digestion procedure, and the peptides were subjected to liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) analysis (Taplin Biological Mass Spectrometry Facility). The most abundant ions were obtained, and the MS/MS spectra was directly searched against the non-redundant protein database of the National Center for Biotechnology Information with the SEQUEST database search algorithm.
ChIP assays
The ChIP Assay Kit and protocol (Upstate Biotechnology) were used. P200 (region -226 to -24 of the mouse C/EBPδ promoter containing STAT3 binding sites), was used for STAT3 positive control primers31, see Supplementary Information, Methods for details.
Statistical analysis
Results are expressed as the mean ± s.e.m., and statistical analysis was performed by one-way or two-way ANOVA analysis of variance and Student’s t-test. A P < 0.05 was considered significant.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. Shanabrough for her technical support and careful revision of this manuscript. Some constructs were obtained from Y. E. Chin, W. Gu, P. Yao, E. Seto and D. Levy. SirT1, PGC-1α adenovirus was a gift from P. P. Puigserver. SirT1 KO MEFs and wild-type MEFs were a gift from L. P. Guarente. Part of this work was supported by an ADA grant to Q. G. (1-08-RA-54) and NIH grants to T. L. H. (DK-08000 and DK-060711), G. I. S. (DK-40936 and DK-076169) and J.L.B. (DK-P30-34989). The preparation of primary hepatocytes was performed in the Liver Center of Yale University School of Medicine. SirT1 ASO and Control ASO were provided by ISIS Pharmaceuticals, Inc.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Luo J, et al. Negative control of p53 by Sir2α promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 2.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 3.Banks AS, et al. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008;8:333–341. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frescas D, Valenti L, Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J. Biol. Chem. 2005;280:20589–20595. doi: 10.1074/jbc.M412357200. [DOI] [PubMed] [Google Scholar]
- 5.Inoue H, et al. Role of STAT-3 in regulation of hepatic gluconeogenic genes and carbohydrate metabolism in vivo. Nature Med. 2004;10:168–174. doi: 10.1038/nm980. [DOI] [PubMed] [Google Scholar]
- 6.Inoue H, et al. Role of hepatic STAT3 in brain-insulin action on hepatic glucose production. Cell Metab. 2006;3:267–275. doi: 10.1016/j.cmet.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 7.Zhong Z, Wen Z, Darnell JE., Jr Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 8.Ray S, Boldogh I, Brasier AR. STAT3 NH2-terminal acetylation is activated by the hepatic acute-phase response and required for IL-6 induction of angiotensinogen. Gastroenterology. 2005;129:1616–1632. doi: 10.1053/j.gastro.2005.07.055. [DOI] [PubMed] [Google Scholar]
- 9.Wang R, Cherukuri P, Luo J. Activation of Stat3 sequence-specific DNA binding and transcription by p300/CREB-binding protein-mediated acetylation. J. Biol. Chem. 2005;280:11528–11534. doi: 10.1074/jbc.M413930200. [DOI] [PubMed] [Google Scholar]
- 10.Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 11.Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. Metabolic adaptations through the PGC-1α and SIRT1 pathways. FEBS Lett. 2008;582:46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen D, et al. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22:1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Solomon JM, et al. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol. Cell. Biol. 2006;26:28–38. doi: 10.1128/MCB.26.1.28-38.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan JH, Lim S, Wong WS. Antisense oligonucleotides: from design to therapeutic application. Clin. Exp. Pharmacol. Physiol. 2006;33:533–540. doi: 10.1111/j.1440-1681.2006.04403.x. [DOI] [PubMed] [Google Scholar]
- 15.Savage DB, et al. Reversal of diet-induced hepatic steatosis and hepatic insulin resistance by antisense oligonucleotide inhibitors of acetyl-CoA carboxylases 1 and 2. J. Clin. Invest. 2006;116:817–824. doi: 10.1172/JCI27300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puigserver P, et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1α interaction. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 17.Nakae J, Park BC, Accili D. Insulin stimulates phosphorylation of the forkhead transcription factor FKHR on serine 253 through a Wortmannin-sensitive pathway. J. Biol. Chem. 1999;274:15982–15985. doi: 10.1074/jbc.274.23.15982. [DOI] [PubMed] [Google Scholar]
- 18.Howitz KT, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 19.Li X, et al. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol. Cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 20.Ray S, Lee C, Hou T, Boldogh I, Brasier AR. Requirement of histone deacetylase1 (HDAC1) in signal transducer and activator of transcription 3 (STAT3) nucleocytoplasmic distribution. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becker S, Groner B, Muller CW. Three-dimensional structure of the Stat3β homodimer bound to DNA. Nature. 1998;394:145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 22.Koch CA, Anderson D, Moran MF, Ellis C, Pawson T. SH2 and SH3 domains: elements that control interactions of cytoplasmic signaling proteins. Science. 1991;252:668–674. doi: 10.1126/science.1708916. [DOI] [PubMed] [Google Scholar]
- 23.Pawson T, Gish GD. SH2 and SH3 domains: from structure to function. Cell. 1992;71:359–362. doi: 10.1016/0092-8674(92)90504-6. [DOI] [PubMed] [Google Scholar]
- 24.Minami M, et al. STAT3 activation is a critical step in gp130-mediated terminal differentiation and growth arrest of a myeloid cell line. Proc. Natl Acad. Sci. USA. 1996;93:3963–3966. doi: 10.1073/pnas.93.9.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maloney A, et al. Gene and protein expression profiling of human ovarian cancer cells treated with the heat shock protein 90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 2007;67:3239–3253. doi: 10.1158/0008-5472.CAN-06-2968. [DOI] [PubMed] [Google Scholar]
- 26.Yoon JC, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.