Abstract
Sodium (23Na) MRI may provide unique information about the cellular and metabolic integrity of the brain. The quantification of tissue sodium concentration from 23Na images with nonzero echo time (TE) requires knowledge of tissue-specific parameters that influence the single-quantum sodium signal such as transverse (T2) relaxation times. We report the sodium (23Na) long component of the effective transverse relaxation time values obtained at 7 T in several brain regions from six healthy volunteers. A two-point protocol based on a gradient-echo sequence optimized for the least error per given imaging time was used (TE1 = 12 ms; TE2 = 37 ms; averaged N1 = 5; N2 = 15 times; pulse repetition time = 130 ms). The results reveal that long component of tissue sodium (mean ± standard deviation) varied between cerebrospinal fluid (54 ± 4 ms) and gray (28 ± 2 ms) and white (29 ± 2 ms) matter structures. The results also show that the long component increases as a function of the main static field B0, indicating that correlation time of sodium ion motion is smaller than the time-scale defined by the Larmor frequency. These results are a prerequisite for the quantification of tissue sodium concentration from 23Na MRI scans with nonzero echo time, will contribute to the design of future measurements (such as triple-quantum imaging), and themselves may be of clinical utility.
Keywords: brain, sodium, high magnetic field, MR imaging, transverse relaxation time
Brain tissue sodium concentration provides an indicator of cellular and metabolic integrity and ion homeostasis in pathologic conditions such as tumors and acute cerebral ischemia (1). However, the application of sodium MRI (23Na-MRI) is limited by the lower concentration and sensitivity of the 23Na nucleus compared to 1H. The increasing availability of magnets operating at 7 T for human imaging may help overcome these limitations by providing higher signal-to-noise ratio (SNR). The quantification of tissue sodium concentration has to take into account various instrumental- (2) acquisition- (3,4) and tissue-specific parameters that influence the single-quantum sodium signal such as longitudinal (T1) and transverse (T2*) relaxation times.
Long components have been measured in humans at lower fields (3,9,10); therefore, the aim of this study was to measure sodium long relaxation times in several human brain regions at 7 T using the method similar to that of Bartha and Menon (3). The short component maps of the sodium signal decay cannot be reliably measured due to low SNR (associated with fast signal decay) and technical limitations (3). Nonetheless, knowledge of the long sodium component will aid in the quantification of tissue sodium concentration and in the design of future quantitative measurements such as triple-quantum sodium mapping (11,12) and might become of diagnostic utility on its own.
MATERIALS AND METHODS
Six healthy volunteers (three males) with a mean age of 30 (range: 24–35) years were consecutively enrolled in this study. Approval for this study was obtained from the Institutional Board of Research Associates of New York University Medical Center, and informed consent was obtained from all subjects.
All experiments were performed on a whole-body 7-T Magnetom scanner (Siemens AG, Erlangen, Germany) with a custom-built, transmit-receive, dual-tuned, 1H/23Na head coil (XLR Imaging, London, ON, Canada). The coil comprises ∅28 × 21 cm 16-rung proton and ∅28 × 19 cm eight-rung sodium quadrature hybrid birdcage coils. Transmitter reference voltage (voltage needed to achieve 180° excitation pulse in 1 ms) for the sodium part of the coil varied subject to subject between 450 and 500 V. A vendor-provided shimming procedure based on a dual-echo steady state sequence was performed iteratively five to 10 times till it converged and yielded whole-head water line width of 34 ± 4 Hz full width at half maximum.
The MRI protocol included (a) a T1-weighted, magnetization-prepared, rapid-acquisition, gradient-echo sequence with echo time (TE)/pulse repetition time/inversion time: 2.6/2250/1100 ms; 192 1.0 mm slices, 256 × 256 matrix with 240 × 240 mm2 field of view; and (b) a three-dimensional gradient-recalled echo sequence with a nonselective excitation for single-quantum sodium images, with a field of view of 240 × 240 × 240 mm3, with 5 × 5 × 8 mm3 voxels. The maps were obtained using the Fleysher et al. method (13), assuming monoexponential relaxation optimized for a literature value of ~20 ms at 4 T (3). More specifically, two sodium images were acquired with TE values set to TE1 = 12 ms and TE1 = 37 ms. Images at each TE were averaged N1 = 5 and N2 = 15 times, respectively. Receiver bandwidth was set to 2.4 kHz. The pulse repetition time was set to 130 ms and the flip angle was set to 80° which was specific-absorption-rate-limit optimized (14) for a 1-ms nonselective excitation pulse and sodium T1 value of ~50 ms (5). At these settings, the scan time for the short TE was 15 min 36 sec and 46 min 48 sec for the long one, yielding total acquisition time just under 63 min. Note that even though the protocol was optimized for a guessed value, the error in the resultant measurements remains relatively constant over a wide range values between 15 and 50 ms (see for example Fig. 1 in Fleysher et al. (13)). The T1-weighted, magnetization-prepared, rapid-acquisition, gradient-echo images were resampled to 8-mm slice thickness to match the sodium images.
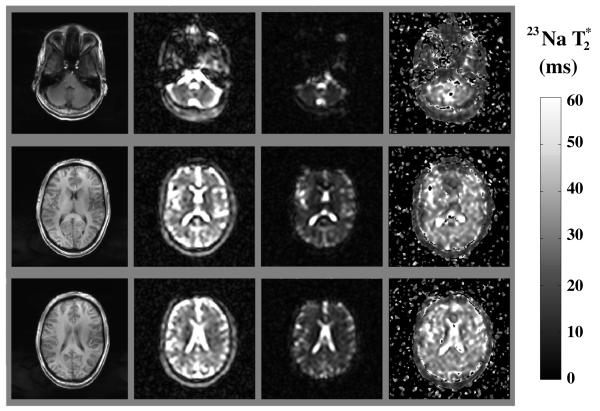
FIG. 1.
T1-weighted axial brain images (8 mm thick) from a healthy volunteer at 7 T (left column), single-quantum sodium images of the corresponding slices at TE = 12 ms and TE = 37 ms (center columns), and parametric map of the selected brain slices (right column). The short TE images were acquired once, while the long TE images were averaged three times, yielding total acquisition time of 63 min (see Materials and Methods section).
The sodium relaxation times were computed in vivo in each voxel using:
| [1] |
where S1 and S2 are the average image intensities at short (TE1) and long (TE2) TEs. were calculated in regions of interest in the following brain regions: cerebropsinal fluid (CSF) at the level of the lateral ventricles, thalamus, putamen, and occipital and frontal gray matter; splenium of the corpus callosum and periventricular, occipital, frontal, and cerebellar white matter. To minimize partial volume contamination, the regions of interest of variable size, as dictated by the anatomic region, were placed on the T1-weighted, magnetization-prepared, rapid-acquisition, gradient-echo images and overlaid automatically on the corresponding sodium parametric map using Image J (http://rsb.info.nih.gov/ij/). To exclude interobserver and to minimize intraobserver variability with respect to the region of interest placement, each subject data set was reviewed by two experienced observers at the same time. The average were recorded for each region bilaterally (except for CSF and corpus callosum) from the sodium parametric maps.
A special remark should be made regarding potential bias in the measurement. Since it is known that sodium signal from gray and white matter structures exhibits biexponential decay, ignorance of the short component in the measurement may lead to a biased estimate of the long component. Another contributor to the possible bias is the measurement noise. The bias due to both of these effects can be estimated using simulations. To do this, an ideal biexponential signal at TE = 12 ms was used as a parameter to Rice (15) distribution in 107 drawings. The second parameter of the Rice distribution was set to produce an SNR of 10 (as expected in the actual measurement). Then, the simulated data were processed as described above and compared with the true answer to estimate possible measurement bias. For expected physical parameters of and and the contribution to due to long component of 40% to the total signal, the bias in the measurement was less than 8%. For tissues where larger fractions of slow-decaying sodium are observed (5), this bias will be less than 3%. Similar evaluation can be done for expected bias in CSF where , which yielded bias of less than 2%. This expected accuracy in long measurement is similar to that provided by a multi-TE method (3).
RESULTS
In vivo T1-weighted anatomic images, together with corresponding sodium images and parametric maps from a single subject, are shown in Fig. 1. Even though only three slices are shown, 30 slices were acquired from each subject. The SNR in the brain tissue at TE1 was about 10, which theoretically provides 12% coefficient of variation for white matter structures and about 21% for the gray matter structures (13). The measured mean coefficients of variation for the tissue types with the corresponding standard errors of the mean were CVwhite matter = 14.3 ± 1.4% and CVgray matter = 20.3 ± 4.2%, respectively, which are in good agreement with the theoretically predicted values based on image SNRs. The from the studied regions are compiled in Table 1 and reveal similar values among the gray matter and white matter structures studied. The mean ± standard errors of the mean global values among the six subjects were 29 ± 2 ms for the white matter and 28 ± 2 ms for the gray and 54 ± 4 ms for CSF. Bias analysis was based on the assumptions that at least 40% of total sodium signal exhibits long decay and that leads to accuracy in gray/white matter values better than 9% and better than 2% in CSF.
Table 1.
Mean ± SEM Sodium Relaxation Times (Milliseconds) at 7 T in the Various Gray Matter (GM) and White Matter (WM) Brain Regionsa
|
(ms) |
||||
|---|---|---|---|---|
| Right | Left | Average | ||
| GM | Frontal cortex | 27 ± 3 | 28 ± 4 | 27 ± 2 |
| Occipital cortex | 29 ± 1 | 29 ± 1 | 29 ± 1 | |
| Putamen | 21± 2 | 24 ± 2 | 23 ± 2 | |
| Thalamus | 32 ± 3 | 32 ± 3 | 32 ± 2 | |
| Average | 28 ± 2 | |||
| WM | Frontal WM | 32 ± 1 | 31 ± 3 | 31 ± 1 |
| Occipital WM | 28 ± 1 | 29 ± 1 | 28 ± 1 | |
| Periventricular WM | 37 ± 2 | 32 ± 2 | 34 ± 1 | |
| Cerebellum | 25 ± 1 | 24 ± 1 | 24 ± 1 | |
| Splenium of CC | 28 ± 2 | |||
| Average | 29 ± 2 | |||
| CSF | 53 ± 5 | 56 ± 7 | 54 ± 4 | |
CC: corpus callosum.
DISCUSSION
It has been shown that reliable in vivo measurements of the short component of the sodium nuclear magnetic resonance signal are difficult due to low SNR and high line broadening (3). Therefore, the goal of this study was to quantify the long component of the decay rate in a healthy human brain at 7 T. For this purpose, we devised a protocol that provided the highest possible precision for measuring long components.
The measured values varied considerably between CSF and the gray and white matter. Since signal due to quadrupolar splitting from sodium ions in dilute water solutions is removed by “motion averaging”, sodium CSF transverse relaxation rate should be similar to that of saline. Indeed, the CSF value of 54 ± 4 (ms) found in this work is consistent with 54 (ms) saline T2 measurements at 8.3 T (9). The similarity of values for the gray (28 ± 2 ms) and white (29 ± 2 ms) matter suggests that the sodium ion environment in these two tissue types is similar despite the differences in the sodium concentrations. It is important to emphasize that even though statistically significant differences between values of the long component between gray and white matter or even among anatomic structures within a tissue type may exist, these differences alone are not likely to produce strong contrast on sodium images acquired with TE less than 20 ms.
We compared our measurements of sodium long values at 7T with those reported in the literature at 1.5 T and 4 T. Magnetic field dependence of the long and T2 component compiled from Bartha and Menon (3), Perman et al. (9), Winkler et al. (10), and this work is presented in Fig. 2. The plot shows that the long component increases in gray and white matter structures as a function of the main magnetic field, which indicates that neither macroscopic residual field gradients nor sodium diffusion has an appreciable contribution to the decay rate and that the correlation time of the random sodium ion motion is much smaller compared to the time scale defined by the Larmor frequency. Thus, according to the Bloembergen Purcell Pound theory (16) one could expect that sodium T2 value should be close to its T1. This is in agreement with previous sodium T1 and T2 measurements (5,9).
FIG. 2.
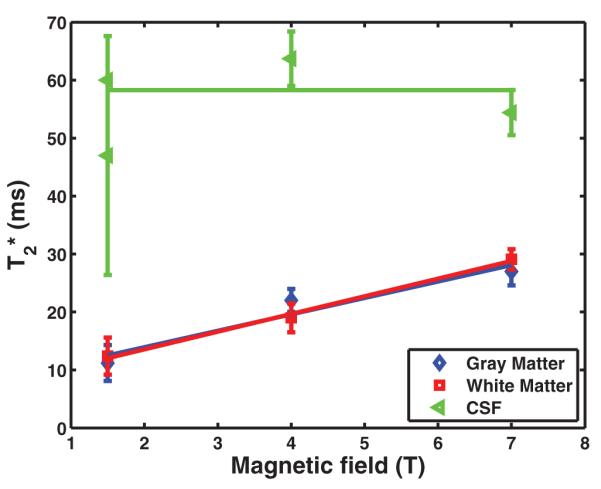
The plot shows the sodium long component dependency on magnetic field strength in gray matter, white matter, and CSF. Mean values and corresponding standard errors on the mean are reported for the three tissue types. The values at 1.5 T and 4 T field strengths are taken from the literature (3,9,10). values at 7 T are from Table 1.
Great care has been taken in designing the measurements so that the contributions due to random errors are minimized. A note should be made about the accuracy of the measurement and possible remaining biases which could have affected the measurement. For example, making TE1 small could lead to a considerable bias in measurement due to short component contribution. To “protect” against such an error, we selected a large TE1 value (we used TE1 = 12 ms). Setting TE1 too small would have led to an underestimation of values. On the other hand, TE1 cannot be made too long because images with TE2 (TE2 > TE1) could have very low SNR. Low SNR in TE2 images could shift values toward higher values. This is because noise distribution in the magnitude images is Rician and deviates significantly from the Gaussian for SNR less than 3 (15). Because the data were averaged several times, the SNR in these images was above 5, which indicates absence of the bias in the measurement. If this effect were appreciable, the true would be even higher than measured. Quantitatively, for the selected acquisition parameters possible bias in long component measurement was less than 9% for gray and white matter structures and less than 2% for CSF. These estimates of the bias were made assuming that the short component was less than 4 (ms) and that the long component contributed at least 40% to the total signal at zero TE. Thus, the accuracy in long measurement is similar to that provided by a multi-TE method (3). Based on these considerations, it is unlikely that the observed increase in values as a function of main magnetic field was caused by instrumental effects. Nonunique to this work, partial-volume effects could significantly degrade the quality of measurements (especially in the voxels with significant partial CSF volume).
CONCLUSIONS
Sodium values at 7 T were measured on a voxel-by-voxel basis using an optimal protocol that minimizes random errors and systematic biases for the fixed imaging time. The values of CSF were similar to previous reports at 1.5 and 4 T, while the values for gray and white matter structures increase as a function of the magnetic field strength. The obtained values will aid the quantification of sodium concentration in studies with nonzero TE values. In addition, future studies will investigate whether sodium long or T2 component maps have some diagnostic potential in pathologic conditions such as brain tumors, head trauma and multiple sclerosis.
ACKNOWLEDGMENTS
The authors thank Enzo Barberi and Robert Pinkerton for their help in hardware development.
Grant sponsor: National Institutes of Health; Grant numbers: R01NS051623, R01NS029029, and R01NS 39135.
REFERENCES
- 1.Thulborn KR, Davis D, Snyder J, Yonas H, Kassam A. Sodium MR imaging of acute and subacute stroke for assessment of tissue viability. Neuroimaging Clin N Am. 2005;15:639–653. xi–xii. doi: 10.1016/j.nic.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Oh CH, Hilal SK, Cho ZH, Mun IK. Radio frequency field intensity mapping using a composite spin-echo sequence. Magn Reson Imaging. 1990;8:21–25. doi: 10.1016/0730-725x(90)90207-i. [DOI] [PubMed] [Google Scholar]
- 3.Bartha R, Menon RS. Long component time constant of 23Na relaxation in healthy human brain. Magn Reson Med. 2004;52:407–410. doi: 10.1002/mrm.20144. [DOI] [PubMed] [Google Scholar]
- 4.Christensen JD, Barrere BJ, Boada FE, Vevea JM, Thulborn KR. Quantitative tissue sodium concentration mapping of normal rat brain. Magn Reson Med. 1996;36:83–89. doi: 10.1002/mrm.1910360115. [DOI] [PubMed] [Google Scholar]
- 5.Winter PM, Bansal N. TmDOTP(5-) as a (23)Na shift reagent for the subcutaneously implanted 9L gliosarcoma in rats. Magn Reson Med. 2001;45:436–442. doi: 10.1002/1522-2594(200103)45:3<436::aid-mrm1057>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 6.Seshan V, Sherry AD, Bansal N. Evaluation of triple quantum-filtered 23Na NMR spectroscopy in the in situ rat liver. Magn Reson Med. 1997;38:821–827. doi: 10.1002/mrm.1910380519. [DOI] [PubMed] [Google Scholar]
- 7.Stobbe R, Beaulieu C. In vivo sodium magnetic resonance imaging of the human brain using soft inversion recovery fluid attenuation. Magn Reson Med. 2005;54:1305–1310. doi: 10.1002/mrm.20696. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein M, King K, Zhou X. Handbook of MRI pulse sequences. Elsevier Academic Press; Burlington, MA: 2004. p. 1040. [Google Scholar]
- 9.Perman WH, Turski PA, Houston LW, Glover GH, Hayes CE. Methodology of in vivo human sodium MR imaging at 1.5 T. Radiology. 1986;160:811–820. doi: 10.1148/radiology.160.3.3737922. [DOI] [PubMed] [Google Scholar]
- 10.Winkler SS, Thomasson DM, Sherwood K, Perman WH. Regional T2 and sodium concentration estimates in the normal human brain by sodium-23 MR imaging at 1.5 T. J Comput Assist Tomogr. 1989;13:561–566. doi: 10.1097/00004728-198907000-00002. [DOI] [PubMed] [Google Scholar]
- 11.Hancu I, Boada FE, Shen GX. Three-dimensional triple-quantum-filtered (23)Na imaging of in vivo human brain. Magn Reson Med. 1999;42:1146–1154. doi: 10.1002/(sici)1522-2594(199912)42:6<1146::aid-mrm20>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 12.LaVerde G, Nemoto E, Jungreis CA, Tanase C, Boada FE. Serial triple quantum sodium MRI during non-human primate focal brain ischemia. Magn Reson Med. 2007;57:201–205. doi: 10.1002/mrm.21087. [DOI] [PubMed] [Google Scholar]
- 13.Fleysher R, Fleysher L, Gonen O. The optimal MR acquisition strategy for exponential decay constants estimation. Magn Reson Imaging. 2008;26:433–435. doi: 10.1016/j.mri.2007.08.014. Epub 2008 Feb 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stobbe R, Beaulieu C. Sodium imaging optimization under specific absorption rate constraint. Magn Reson Med. 2008;59:345–355. doi: 10.1002/mrm.21468. [DOI] [PubMed] [Google Scholar]
- 15.Rice S. Mathematical analysis of random noise. Bell Syst Tech J. 1945;24:146–156. [Google Scholar]
- 16.Bloembergen N, Purcell E, Pound R. Relaxation effects in nuclear magnetic resonance absorption. Phys Rev. 1948;73:679–715. [Google Scholar]
- 17.Hubbard P. Nonexponential nuclear magnetic relaxation by quadrupole interactions. J Chem Phys. 1970;53:985–987. [Google Scholar]