Abstract
Background/Aims
Neuregulin 1 (NRG1) is a positional candidate gene in schizophrenia (SZ). Two major susceptibility loci in the NRG1 gene approximately one million nucleotides apart have been identified in genetic studies. Several candidate functional allelic variants have been described that might be involved in disease susceptibility. However, the findings are still preliminary. We recently mapped active promoters and other regulatory domains in several SZ and bipolar disorder (BD) candidate genes using ChIP-chip (chromatin immunoprecipitation hybridized to microarrays). One was the promoter for the NRG1 isoform, SMDF, which maps to the 3′ end of the gene complex. Analysis of the SNP database revealed several polymorphisms within the approximate borders of the region immunoprecipitated in our ChIP-chip experiments, one of which is rs7825588.
Methods
This SNP was analyzed in patients with SZ and BD and its effect on promoter function was assessed by electromobility gel shift assays and luciferase reporter constructs.
Results
A significant increase in homozygosity for the minor allele was found in patients with SZ (genotype distribution χ2 = 7.32, p = 0.03) but not in BD (genotype distribution χ2 = 0.52, p = 0.77). Molecular studies demonstrated modest, but statistically significant allele-specific differences in protein binding and promoter function.
Conclusion
The findings suggest that homozygosity for rs725588 could be a risk genotype for SZ.
Key Words: Bipolar disorder, Neuregulin, NRG1, rs725588, Schizophrenia, SMDF
Introduction
Neuregulin 1 (NRG1) was first identified as a candidate gene for schizophrenia (SZ) in Icelandic and Scottish families several years ago [1, 2]. Since then, the association has been replicated in other populations [3,4,5,6,7,8,9,10,11]. However, some studies fail to support a role for NRG1, consistent with the heterogeneity of SZ [12,13,14]. Most of the positive studies point to associations within a block of markers in the 5′ end of the gene referred to as the Icelandic haplotype.
In addition, investigators have provided evidence for associations to distal NRG1 markers in SZ, ∼1 Mb downstream, in Asians, African-Americans and some European populations [4,5,6, 8, 10,15,16,17,18,19,20,21]. This region contains the promoter for the heregulin isoforms, which map to ∼32,525,000, and the sensory and motor neuron-derived factor (SMDF) isoform at ∼32,624,000.
The various isoforms constituting the NRG1 family – at least 15 have been described – influence neurite outgrowth, adhesion, apoptosis, neuron migration, astrocyte differentiation and survival of oligodendrocytes by activating the receptor tyrosine kinase ErbB family [20,22,23,24,25,26]. NRG1 also has a post-development effect on neural function by mediating signaling pathways at NMDA receptor-rich postsynaptic densities [27]. There is also evidence that ErbB4 increases AMPA receptor-mediated synaptic currents, CA1 dendritic spine density and GABA release in response to depolarizing concentrations of potassium [28]. NRG1 proteins also increase α7 nicotinic acetylcholine receptors (nAchR), and modulate both hippocampal GABAergic interneurons and CA1 neurons [6, 29, 30]. A decrease in α7 nAchR expression correlated with an NRG1 promoter SNP (rs6994992) has been detected in the dorsolateral prefrontal cortex of SZ patients [31].
Mouse knockout studies also support a role for NRG1 in SZ pathogenesis. Mice heterozygous for either Nrg1 or ErbB4 have a deficit in prepulse inhibition, an SZ endophenotype [1].
Although NRG1 has been viewed as an SZ candidate gene since 2002, the functional allelic variants responsible for the positive association signals have not yet been unequivocally identified. However, several candidates have been targeted, including SNP8NRG243177, which maps to the Icelandic haplotype that appears to affect NRG1 type IV expression and is associated with cortical activation [11,32,33,34,35]. Recently, homozygosity for the minor allele of a nonsynonymous SNP at 32,572,900 (rs3924999) has been found to be associated with prepulse inhibition defects in patients with SZ, although the sample size was relatively small [9]. In addition, a mutation in the Ig-like domain, which cooperates with the EGF-like domain to bind ErbB receptors, was associated with impaired latent inhibition [36, 37].
Although these studies point to several functional SNPs as promising candidate alleles, it is likely, given the size of the NRG1 gene and the multitude of positive association and linkage findings in various populations to different portions of the NRG1 gene locus, as well as the small effect sizes so far found in suspected candidate variants and haplotypes, that many disease-causing functional mutations exist in the gene. If so, genetic variation in regulatory domains would be reasonable places to search for novel functional alleles.
We have recently begun to analyze NRG1 and other SZ and BD candidate genes using chromatin immunoprecipitates hybridized to microarrays – ChIP-chip – to identify potential regulatory elements. We screened tiled arrays containing the entire NRG1 gene locus with immunoprecipitates made from human fetal brain chromatin enriched for histone H3 acetylated at lysine 9 (H3K9Ac) and monomethylated at lysine 4 (H3K4me1), which are enriched in promoters and enhancers [38,39,40,41,42]. In so doing, we identified several potential regulatory domains in NRG1, DISC1, JARID2, DTNBP1, PDE4B, DAO, DAOA and the COMT/ARVCF locus on 22q11 [Pedrosa et al., in press]. One was the promoter of the NRG1 isoform, SMDF (also known as type III neuregulin). We analyzed an SNP in this region, rs7825588, in a cohort of patients with SZ and BD, and carried out molecular studies to assess its role in promoter function. Evidence is presented showing that homozygosity for the minor allele associates with SZ.
Patients and Methods
Subjects
Patients with BD from the Czech Republic were unrelated subjects recruited from in- and outpatient units at the Prague Psychiatric Center, Psychiatric Hospital Bohnice, Psychiatric Clinic (n = 167). Patients were diagnosed on the basis of either a Schedule for Affective Disorders and Schizophrenia-Lifetime Interview (n = 68) or by unstructured clinical interview modified from this Schedule using research diagnostic criteria for the diagnosis of either BD I or II (n = 99) [43, 44]. Control subjects from the Czech Republic were blood bank donors and patients hospitalized for medical reasons (n = 211). Seventy-one control subjects did not have underlying psychiatric illness based on a brief psychiatric clinical interview. In the remaining controls, all from blood bank donors, no formal testing procedure was used to screen for personal history of mental illness. However, the blood bank only accepted subjects who were not being treated for a psychiatric illness and had no family history of mental illness.
Patients with SZ (n = 176) were recruited from the Rockland State Hospital. Diagnosis was established by research diagnostic criteria using a Structured Clinical Interview for DSM or clinical interview. US controls (n = 175) were Caucasian blood bank donors. No formal testing procedure was used to screen these subjects to rule out individuals who had a personal or family history of mental illness, although the frequency of BD and SZ in a population of blood donors would be expected to be ≤1% for each. All patients signed an informed consent approved by the Ethical Committee on Clinical Investigation (Czech samples) and the Albert Einstein College of Medicine Institutional Review Board (US samples). In the SZ sample, 31% were female, with a mean age of 42 ± 10 years. In the control group for this sample, 45% were female, with a mean age of 48 ± 13 years. In the Czech bipolar cohort, 54% were female, with a mean age of 49 ± 17 years, while in the control group for this samples, 40% were female, with a mean age of 47 ± 16 years.
Polymorphism Detection and Genotyping
DNA was genotyped for rs7825588 using the TaqMan allelic discrimination technique according to the manufacturers instructions. Samples were amplified with PCR in 384-well plates using an Applied Biosystems Model 7900HT thermal cycler. Samples were genotyped twice.
Electromobility Gel Shift Assay (EMSA)
EMSA was performed according to published procedures [45]. Briefly, double-stranded oligonucleotide probes containing the polymorphic variants were constructed. The primers were annealed and end-labeled with 32P deoxynucleotides using Klenow polymerase to fill in 5′ overhangs, which generated double-stranded probes that were used for protein binding experiments. Nuclear protein extract was isolated from fetal (whole brain) and adult brains (parietal lobe). Protein (10 μm) was mixed with probe (1 ng, ∼106 counts) and incubated for 20 min at room temperature. Probes containing the different alleles were labeled simultaneously using the same amount of DNA and radioactive nucleotides. DNA-protein complexes were resolved by electrophoresis in a non-denaturing gel system containing 6% acrylamide and 1.6% glycerol. Specificity of the resulting binding activity was demonstrated by competition with non-radioactive probe added in 100-fold excess. Autoradiograms in the linear range of exposure were scanned and quantified by normalizing against unused probe. Differences between the two alleles for each sample were analyzed using a paired t test (two tailed).
EMSA primers:
SMDF-forward G agtgtgggtagagagcGgggagtgggggtt
SMDF-reverse G ccaacccccactcccCgctctctacccaca
SMDF-forward A agtgtgggtagagagcAgggagtgggggtt
SMDF-reverse A ccaacccccactcccTgctctctacccaca
Cloning of SMDF Promoter and Transfection in Neuroblastoma Cells
A 754-bp fragment from the SMDF promoter region containing the two different rs7825588 alleles (A or G) was cloned into the pGL2 basic plasmid (Promega), which contains the luciferase reporter gene. These were introduced into SH-SY5Y neuroblastoma cells using the liposomal transfection reagent, GeneCarrier-2 (3 μg/μg DNA; Epoch Biolabs; Sugar Land, Tex., USA). Following transfection, the cells were incubated for another 24 h, after which they were treated with 5 μM retinoic acid. Cells were harvested 24 h later and assayed for luciferase activity using a luciferase assay kit (Promega catalogue No. E4030); luminescence was measured in triplicate using a luminometer (Berthold Lumat LB9501). Experiments were carried out in three independent transfection assays. Prior to each transfection, plasmid integrity with respect to the relative amount of supercoiled material present in the preparation was separated by gel electrophoresis and quantified.
Statistical Analysis
A statistical program, StatXACT-5 (Cytel Software, Cambridge, Mass., USA), was used to compute the χ2 and Fisher statistics. The level of significance was set at p < 0.05. Hardy-Weinberg equilibrium (HWE) was computed using a goodness-of-fit χ2 determination.
Results
Genotyping Analysis of rs7825588
In a previous ChIP-chip analysis of several candidate genes for SZ and BD using antibodies to histone H3 acetylated at lysines 9 and 14 (H3K9/14Ac) and monomethylated at lysine 4 (H3K4me1), a number of active promoters and putative enhancers were identified in fetal brain (in press). One was on chromosome 8 between ∼32,623,000–32,626,000, which contains the promoter for the NRG1 isoform SMDF (transcription start site, 32,624,071) [46]. The ChIP-chip peak encompassed the TATA-less and GC-rich promoter, the 5′ untranslated region, and exon 1, and includes 10 conserved transcription factor binding sites (TFBS track, UCSC); TFBS are often found in conserved regulatory elements. Among the TFBS in this promoter are several that are developmentally regulated, including the homeodomain-containing transcription factor Nkx and the forkhead transcription factor Fox04, as well as Cdc5, which has been implicated in dopaminergic and glutamatergic signaling [47]. Six known SNPs are found within the approximate boundaries of the peaks, including rs7825588 at 32,623,942, which is 128 bp upstream of the SMDF transcription start site and 837 bp from the translation start, rs4389886, a rare variant in the 5′ UTR, and three relatively rare nonsynonymous SNPs found primarily in African-Americans [rs34918173 (A34E); rs34822181 (P127A); rs35641374 (L133V)], and one, rs3735774 (A46G), found in Asian populations. In this study, we genotyped a cohort of European Caucasian patients from the New York Metropolitan area and controls, as well as a cohort of patients with BD from the Czech Republic and controls for rs7825588. We chose this SNP primarily because it is the most informative in our population. In addition, a potential binding site for the MZF1 transcription factor is found in the G-allele for rs7825588 (TFSEARCH) [48].
In the BD and control cohorts from the Czech Republic, allele and genotype frequencies were not significantly different (table 1), and the frequencies were similar to those reported for the HapMap-CEU population. In addition, the genotype distribution did not deviate from that expected in the sample assuming HWE. By contrast, there was a significant difference in the genotype distribution between patients with SZ and controls in the US sample (χ2 = 7.32, p = 0.03), although there was no significant difference in allele frequencies. When a Bonferroni correction is applied to the data to account for multiple testing (analysis of SZ and BD data sets), the association falls just short of significance at the p = 0.025 level (0.05 divided by 2 tests). There was a significant deviation from the HWE in the patient sample due to the excessive number of homozygotes for the minor allele (HWE, p = 0.04). Since deviation from HWE can be caused by genotyping error, we reanalyzed the samples; no genotyping errors were detected. Copy variation is another cause for HWE deviation. However, none has been reported in this region by array analysis (Database for Genomic Variants: http://projects.tcag.ca/variation/) or by SNP arrays in >1,000 controls [Dr. Tamim Shaikh, submitted and pers. commun.].
Table 1.
Analysis of SMDF promoter SNP rs7825588
| Genotypes |
Alleles |
||||
|---|---|---|---|---|---|
| GG | GA | AA | G | A | |
| Controls | 131 (0.76) | 42 (0.24) | 0 | 304 (0.88) | 42 (0.12) |
| SZ | 129 (0.77) | 32 (0.19) | 6 (0.04) | 290 (0.86) | 44 (0.13) |
| Controls | 172 (0.82) | 35 (0.17) | 2 (0.01) | 379 (0.91) | 39 (0.09) |
| BD | 127 (0.79) | 31 (0.19) | 2 (0.01) | 285 (0.89) | 35 (0.11) |
BD vs. controls: allele frequency, χ2 = 0.52, p = 0.47; genotype distribution, χ2 = 0.52, p = 0.77; HWE controls, p = 0.69; BD, p = 1.0. SZ vs. controls: allele frequency, χ2 = 0.16, p = 0.68; genotype distribution, χ2 = 7.32, p = 0.03; HWE controls, p = 0.08; SZ, p = 0.04.
EMSA
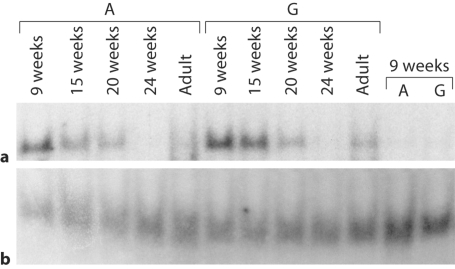
As a first step towards determining whether rs7825588 has functional significance, EMSA experiments were carried out. Crude nuclear protein extracts from fetal brain samples were annealed to allele-specific, double-stranded oligonucleotide probes and separated by non-denaturing gel electrophoresis. As seen in figure 1, a DNA-protein complex was detected in the 9-week-old fetus for both the A- and G-containing probes [fig. 1a, bands in the first lanes of each set (A, G); bottom bands in fig. 1bshow an unused probe]. The binding was specific since the signal was markedly attenuated when unlabeled competitor was added during the annealing phase (last two lanes labeled 9 weeks). As seen in figure 1, there is a decrease in signal intensity during fetal development (15–24 weeks) in adult brain tissue (adult lanes). We scanned bands in the linear range of exposure and found a 30.3% decrease in signal intensity for the oligonucleotide probe containing the minor allele (A) compared with the G-allele (p = 0.0271, t test).
Fig. 1.
Assessment of rs7825588 function by EMSA. Crude nuclear extracts from fetal brains and an adult were combined with labeled oligonucleotides containing rs7825588 (A is minor allele; G is major allele). a DNA-protein complexes (exposure time: 8 h). b Unused probe (exposure time: 30 s). Last two lanes show competition experiment with 100-fold excess of unlabeled probe added to protein extracted from 9-week brain tissue. There is a 30.3% decrease in signal intensity for the oligonucleotide probe containing the minor allele (A) compared with the G allele (p = 0.0271, t test).
Reporter Gene Transfection Assay to Assess rs7825588 Function
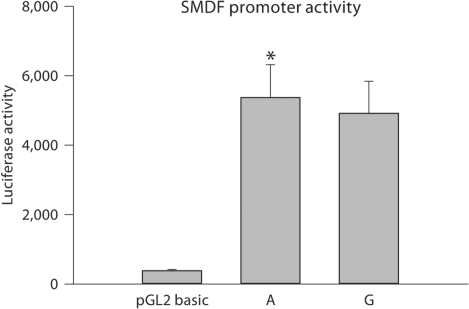
In order to determine whether the different rs7825588 alleles affect SMDF promoter activity, a 754-bp segment of the promoter containing one or the other allele was cloned into a luciferase reporter plasmid and transfected into SH-SY5Y neuroblastoma cells. As seen in figure 2, promoter activity increased ∼17- to 18-fold compared with that found in pGL2-basic, which has low intrinsic promoter activity. A 9.8% increase in promoter activity was detected for the minor allele. Although the difference between the two alleles was small, it was statistically significant (two-tailed paired t test, p = 0.02 for fold increase in A-allele compared with G-allele).
Fig. 2.
Assessment of rs7825588 using luciferase promoter assay. Fragments (754 bp) from the SMDF promoter region containing the two different rs7825588 alleles (A or G) were cloned into pGL2. Low basal activity for pGL2 is seen on the left. Luciferase activity for promoters containing the ‘A’ and ‘G’ alleles are shown. Experiments were carried out 3 times on separate cell passages in triplicate. Means ± SE. * p = 0.02 vs. G-containing promoters (paired t test; two tailed).
Discussion
The different isoforms produced by transcription initiation at various NRG1 promoters have distinct roles in neuronal and glial function. What accounts, therefore, for the positive association findings spanning the entire ∼1.2-Mb NRG1 gene locus, which suggest that several isoforms may influence disease pathogenesis? The initial genetic finding, subsequently confirmed by several groups, showed an association to the Icelandic haplotype, which encompasses the GGF2 (glial growth factor-2 or type II neuregulin) promoter, the most 5′ NRG1 coding element. However, more distal association signals have been identified in other studies ∼one million bases 3′ to the Icelandic haplotype, which incorporates the promoters and 5′ exons for several isoforms, including SMDF. One possibility is that despite distinct roles for NRG1 isoforms in synaptogenesis, neuronal development, migration, Schwann cell and oligodendrocyte differentiation and myelination, there is convergence on a common functional or structural pathway. One possibility is glutamatergic signaling mediated by the NMDA complex, which may be disrupted in SZ [49,50,51,52,53]. Thus, genetic variation affecting any one of several different isoforms could conceivably have a common effect on glutamate transmission and behavioral phenotype, albeit through different molecular pathways.
Thus, our finding of a modest association between a functional SMDF promoter variant in SZ is compatible with the positive association signals that implicate more 5′ regions of the NRG1 gene complex. In addition, our findings could explain other positive association signals that point to this NRG1 region. In 2005, Petryshen et al. [6], for example, demonstrated an association to several markers in the 3′ end of NRG1, including one, rs2466058, which is only ∼2 kb from rs7825588. Also, in 2003, Yang et al. [4] identified several individual SNPs and 3′ haplotypes in a Han Chinese SZ population, including rs2954041, which is only ∼18 kb from the SMDF promoter. Finally, in 2007, Thomson et al. [10] found haplotypes associated with both SZ and BD in a Scottish cohort that incorporates the SMDF promoter region. These findings could be due to linkage disequilibrium with rs7825588. Our results are also compatible with the finding that differences in leukocyte SMDF expression exist in SZ-discordant siblings [6].
SMDF regulates Schwann cell membrane growth and differentiation, and triggers myelination in sensory and motor neurons [23, 54]. Recent work suggests that SMDF influences myelination in the CNS too [55]. Altered SMDF expression and myelination in the CNS in SZ would be compatible with imaging studies, which show abnormalities in subcortical white matter density in patients compared with controls [34, 56]. In addition, oligodendrocyte and myelin abnormalities have been suggested to occur in SZ on the basis of array expression, in situ hybridization and morphological studies [7,57,58,59,60]. A role for SMDF is also consistent with recent findings showing that mice heterozygous for a disruption in type III Nrg1 have enlarged lateral ventricles, decreased dendritic spine density on subicular pyramidal neurons, hypofunction in the medial prefrontal cortex, impaired performance on delayed alternation memory tasks and deficits in prepulse inhibition [61]. Finally, SMDF was found to affect α7 nAChR-mediated synaptic responses and targeting of the receptor on presynaptic membranes; α7 nAChR affects cognitive function and has been implicated in SZ [62,63,64]. Stimulation of NRG1 type III increased the surface levels of axonal α7 nAChR from a preexisting intracellular pool via a phosphatidylinositol 3-kinase signaling pathway [63]. Interestingly, treatment with a partial α7 nAChR agonist was recently shown to improve negative symptoms and working memory in a phase 2 clinical trial [65].
These studies and our findings suggest that SMDF dysregulation may be an underlying pathogenic process in a subset of patients with SZ.
A significant limitation of this study is the relatively small sample size available to us for the case-control association analysis. Thus, type I error is a distinct possibility and the findings need to be replicated in a larger data set. In addition, more extensive analysis of rs7825588 is indicated. The EMSA findings strongly suggest that binding to an unknown protein is affected by the polymorphism. However, the promoter assay was equivocal showing statistically significant, albeit marginal allele-specific differences. It should be noted that while promoter activity was higher with the disease-associated ‘A’ allele, there was a decrease in signal in the DNA-protein complex seen on EMSA. One possibility is that the unknown protein binding to the SNP may be a transcriptional repressor (i.e. less binding, higher transcription). Another is that the minor allele leads to an increase in transcription in neuroblastoma cells, but a decrease in the developing brain. The latter is more consistent with the findings by Chen et al. [61], who showed that a decrease in type III expression in a knockout model had behavioral and structural changes that mimicked clinical SZ.
Despite these limitations, the findings presented here, as well as other studies implicating a defect in myelination in SZ, suggest that rs7825588 and other genetic variants affecting SMDF expression should be viewed as plausible candidates in SZ susceptibility.
Acknowledgements
We would like to thank Hana Fridrichova for her help in recruiting patients and her administrative support for the Czech BD cohort, the Departments of Human Genetics and Molecular Genetics of the Albert Einstein College of Medicine for their assistance, and Dr. Joseph Locker of the Department of Pathology for discussions related to the luciferase assay. We are also grateful to Dr. Tamim Shaikh for providing unpublished data. P.S. was supported by a research grant from IGA MZ CR NR8564, and H.M.L. by the National Institute of Mental Health (R01MH073164) and by the Juvenile Bipolar Research Foundation.
References
- 1.Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, et al. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71:877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stefansson H, Sarginson J, Kong A, Yates P, Steinthorsdottir V, Gudfinnsson E, et al. Association of neuregulin 1 with schizophrenia confirmed in a Scottish population. Am J Hum Genet. 2003;72:83–87. doi: 10.1086/345442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams NM, Preece A, Spurlock G, Norton N, Williams HJ, Zammit S, et al. Support for genetic variation in neuregulin 1 and susceptibility to schizophrenia. Mol Psychiatry. 2003;8:485–487. doi: 10.1038/sj.mp.4001348. [DOI] [PubMed] [Google Scholar]
- 4.Yang JZ, Si TM, Ruan Y, Ling YS, Han YH, Wang XL, et al. Association study of neuregulin 1 gene with schizophrenia. Mol Psychiatry. 2003;8:706–709. doi: 10.1038/sj.mp.4001377. [DOI] [PubMed] [Google Scholar]
- 5.Li T, Stefansson H, Gudfinnsson E, Cai G, Liu X, Murray RM, et al. Identification of a novel neuregulin 1 at-risk haplotype in Han schizophrenia Chinese patients, but no association with the Icelandic/Scottish risk haplotype. Mol Psychiatry. 2004;9:698–704. doi: 10.1038/sj.mp.4001485. [DOI] [PubMed] [Google Scholar]
- 6.Petryshen TL, Middleton FA, Kirby A, Aldinger KA, Purcell S, Tahl AR, Morley CP, McGann L, Gentile KL, Rockwell GN, Medeiros HM, Carvalho C, Macedo A, Dourado A, Valente J, Ferreira CP, Patterson NJ, Azevedo MH, Daly MJ, Pato CN, Pato MT, Sklar P. Support for involvement of neuregulin 1 in schizophrenia pathophysiology. Mol Psychiatry. 2005;10:366–374. doi: 10.1038/sj.mp.4001608. [DOI] [PubMed] [Google Scholar]
- 7.Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. [DOI] [PubMed] [Google Scholar]
- 8.Li D, Collier DA, He L. Meta-analysis shows strong positive association of the neuregulin 1 (NRG1) gene with schizophrenia. Hum Mol Genet. 2006;15:1995–2002. doi: 10.1093/hmg/ddl122. [DOI] [PubMed] [Google Scholar]
- 9.Hong LE, Wonodi I, Stine OC, Mitchell BD, Thaker GK. Evidence of missense mutations on the neuregulin 1 gene affecting function of prepulse inhibition. Biol Psychiatry. 2008;63:17–23. doi: 10.1016/j.biopsych.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomson PA, Christoforou A, Morris SW, Adie E, Pickard BS, Porteous DJ. Association of Neuregulin 1 with schizophrenia and bipolar disorder in a second cohort from the Scottish population. Mol Psychiatry. 2007;12:94–104. doi: 10.1038/sj.mp.4001889. [DOI] [PubMed] [Google Scholar]
- 11.Georgieva L, Dimitrova A, Ivanov D, Nikolov I, Williams NM, Grozeva D, Zaharieva I, Toncheva D, Owen MJ, Kirov G, O'Donovan MC. Support for neuregulin 1 as a susceptibility gene for bipolar disorder and schizophrenia. Biol Psychiatry. 2008;64:419–427. doi: 10.1016/j.biopsych.2008.03.025. [DOI] [PubMed] [Google Scholar]
- 12.Ikeda M, Takahashi N, Saito S, Aleksic B, Watanabe Y, Nunokawa A, Yamanouchi Y, Kitajima T, Kinoshita Y, Kishi T, Kawashima K, Hashimoto R, Ujike H, Inada T, Someya T, Takeda M, Ozaki N, Iwata N. Failure to replicate the association between NRG1 and schizophrenia using Japanese large sample. Schizophr Res. 2008;101:1–8. doi: 10.1016/j.schres.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 13.Munafò MR, Attwood AS, Flint J. Neuregulin 1 genotype and schizophrenia. Schizophr Bull. 2008;34:9–12. doi: 10.1093/schbul/sbm129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanders AR, Duan J, Levinson DF, Shi J, He D, Hou C, et al. No significant association of 14 candidate genes with schizophrenia in a large European ancestry sample: implications for psychiatric genetics. Am J Psychiatry. 2008;165:497–506. doi: 10.1176/appi.ajp.2007.07101573. [DOI] [PubMed] [Google Scholar]
- 15.Ling YS, Han YH, Wang XL, Zhou M, Zhang HY, Kong QM, Liu C, Zhang DR, Yu YQ, Liu SZ, Ju GZ, Shu L, Ma DL, Zhang D, Yang JZ, Si TM, Ruan Y. Association study of neuregulin 1 gene with schizophrenia. Mol Psychiatry. 2003;8:706–709. doi: 10.1038/sj.mp.4001377. [DOI] [PubMed] [Google Scholar]
- 16.Fukui N, Muratake T, Kaneko N, Amagane H, Someya T. Supportive evidence for neuregulin 1 as a susceptibility gene for schizophrenia in a Japanese population. Neurosci Lett. 2006;396:117–120. doi: 10.1016/j.neulet.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 17.Lachman HM, Pedrosa E, Nolan KA, Glass M, Ye K, Saito T. Analysis of polymorphisms in AT-rich domains of neuregulin 1 gene in schizophrenia. Am J Med Genet Neuropsychiatr Genet. 2006;141B:102–109. doi: 10.1002/ajmg.b.30242. [DOI] [PubMed] [Google Scholar]
- 18.Walss-Bass C, Liu W, Lew DF, Villegas R, Montero P, Dassori A, et al. A novel missense mutation in the transmembrane domain of neuregulin 1 is associated with schizophrenia. Biol Psychiatry. 2006;60:548–553. doi: 10.1016/j.biopsych.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 19.Addington AM, Gornick MC, Shaw P, Seal J, Gogtay N, Greenstein D, Clasen L, Coffey M, Gochman P, Long R, Rapoport JL. Neuregulin 1 (8p12) and childhood-onset schizophrenia: susceptibility haplotypes for diagnosis and brain developmental trajectories. Mol Psychiatry. 2007;12:195–205. doi: 10.1038/sj.mp.4001906. [DOI] [PubMed] [Google Scholar]
- 20.Benzel I, Bansal A, Browning BL, Galwey NW, Maycox PR, McGinnis R, Smart D, St Clair D, Yates P, Purvis I. Interactions among genes in the ErbB-Neuregulin signalling network are associated with increased susceptibility to schizophrenia. Behav Brain Funct. 2007;3:31. doi: 10.1186/1744-9081-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turunen JA, Peltonen JO, Pietiläinen OP, Hennah W, Loukola A, Paunio T, Silander K, Ekelund J, Varilo T, Partonen T, Lönnqvist J, Peltonen L. The role of DTNBP1, NRG1, and AKT1 in the genetics of schizophrenia in Finland. Schizophr Res. 2007;91:27–36. doi: 10.1016/j.schres.2006.11.028. [DOI] [PubMed] [Google Scholar]
- 22.Buonanno A, Fischbach GD. Neuregulin and ErbB receptor signaling pathways in the nervous system. Curr Opin Neurobiol. 2001;11:28–96. doi: 10.1016/s0959-4388(00)00210-5. [DOI] [PubMed] [Google Scholar]
- 23.Michailov GV, Sereda MW, Brinkmann BG, Fischer TM, Haug B, Birchmeier C, Role L, Lai C, Schwab MH, Nave KA. Axonal neuregulin-1 regulates myelin sheath thickness. Science. 2004;304:700–703. doi: 10.1126/science.1095862. [DOI] [PubMed] [Google Scholar]
- 24.Britsch S. The neuregulin-I/ErbB signaling system in development and disease. Adv Anat Embryol Cell Biol. 2007;190:1–65. [PubMed] [Google Scholar]
- 25.Mei L, Xiong WC. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat Rev Neurosci. 2008;9:437–452. doi: 10.1038/nrn2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roy K, Murtie JC, El-Khodor BF, Edgar N, Sardi SP, Hooks BM, Benoit-Marand M, Chen C, Moore H, O'Donnell P, Brunner D, Corfas G. Loss of erbB signaling in oligodendrocytes alters myelin and dopaminergic function, a potential mechanism for neuropsychiatric disorders. Proc Natl Acad Sci USA. 2007;104:8131–8136. doi: 10.1073/pnas.0702157104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li B, Woo RS, Mei L, Malinow R. The neuregulin-1 receptor erbB4 controls glutamatergic synapse maturation and plasticity. Neuron. 2007;54:583–597. doi: 10.1016/j.neuron.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woo RS, Li XM, Tao Y, Carpenter-Hyland E, Huang YZ, Weber J, Neiswender H, Dong XP, Wu J, Gassmann M, Lai C, Xiong WC, Gao TM, Mei L. Neuregulin-1 enhances depolarization-induced GABA release. Neuron. 2007;54:599–610. doi: 10.1016/j.neuron.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 29.Yang X, Kuo Y, Devay P, Yu C, Role L. A cysteine-rich isoform of neuregulin controls the level of expression of neuronal nicotinic receptor channels during synaptogenesis. Neuron. 1998;20:255–270. doi: 10.1016/s0896-6273(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Ford B, Mann MA, Fischbach GD. Neuregulins increase α7 nicotinic acetylcholine receptors and enhance excitatory synaptic transmission in GABAergic interneurons of the hippocampus. J Neurosci. 2007;21:5660–5669. doi: 10.1523/JNEUROSCI.21-15-05660.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathew SV, Law AJ, Lipska BK, Dávila-García MI, Zamora ED, Mitkus SN, Vakkalanka R, Straub RE, Weinberger DR, Kleinman JE, Hyde TM. α7 nicotinic acetylcholine receptor mRNA expression and binding in postmortem human brain are associated with genetic variation in neuregulin 1. Hum Mol Genet. 2007;16:2921–2932. doi: 10.1093/hmg/ddm253. [DOI] [PubMed] [Google Scholar]
- 32.Hall J, Whalley HC, Job DE, Baig BJ, McIntosh AM, Evans KL, et al. A neuregulin 1 variant associated with abnormal cortical function and psychotic symptoms. Nat Neurosci. 2006;9:1477–1478. doi: 10.1038/nn1795. [DOI] [PubMed] [Google Scholar]
- 33.Law AJ, Lipska BK, Weickert CS, Hyde TM, Straub RE, Hashimoto R, et al. Neuregulin 1 transcripts are differentially expressed in schizophrenia and regulated by 5′ SNPs associated with the disease. Proc Natl Acad Sci USA. 2006;103:6747–6752. doi: 10.1073/pnas.0602002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McIntosh AM, Moorhead TW, Job D, Lymer GK, Muñoz Maniega S, McKirdy J, Sussmann JE, Baig BJ, Bastin ME, Porteous D, Evans KL, Johnstone EC, Lawrie SM, Hall J. The effects of a neuregulin 1 variant on white matter density and integrity. Mol Psychiatry. 2008;13:1054–1059. doi: 10.1038/sj.mp.4002103. [DOI] [PubMed] [Google Scholar]
- 35.Tan W, Wang Y, Gold B, Chen J, Dean M, Harrison PJ. Molecular cloning of a brain-specific, developmentally regulated neuregulin 1 (NRG1) isoform and identification of a functional promoter variant associated with schizophrenia. J Biol Chem. 2007;282:24343–24351. doi: 10.1074/jbc.M702953200. [DOI] [PubMed] [Google Scholar]
- 36.Rimer M, Barrett DW, Maldonado MA, Vock VM, Gonzalez-Lima F. Neuregulin-1 immunoglobulin-like domain mutant mice: clozapine sensitivity and impaired latent inhibition. Neuroreport. 2005;16:271–275. doi: 10.1097/00001756-200502280-00014. [DOI] [PubMed] [Google Scholar]
- 37.Eto K, Eda K, Kanemoto S, Abe S. The immunoglobulin-like domain is involved in interaction of Neuregulin1 with ErbB. Biochem Biophys Res Commun. 2006;350:263–271. doi: 10.1016/j.bbrc.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 38.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 39.Roh TY, Cuddapah S, Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 2005;19:542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roh TY, Wei G, Farrell CM, Zhao K. Genome-wide prediction of conserved and nonconserved enhancers by histone acetylation patterns. Genome Res. 2007;17:74–81. doi: 10.1101/gr.5767907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 42.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;9:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 43.Endicott J, Spitzer RL. A diagnostic interview: the schedule for affective disorders and schizophrenia. Arch Gen Psychiatry. 1978;35:837–844. doi: 10.1001/archpsyc.1978.01770310043002. [DOI] [PubMed] [Google Scholar]
- 44.Spitzer RL, Endicott J, Robins E. Research diagnostic criteria: rationale and reliability. Arch Gen Psychiatry. 1978;35:773–782. doi: 10.1001/archpsyc.1978.01770300115013. [DOI] [PubMed] [Google Scholar]
- 45.Hope BT, Kelz MB, Duman RS, Nestler EJ. Chronic electroconvulsive seizure (ECS) treatment results in expression of a long-lasting AP-1 complex in brain with altered composition and characteristics. J Neurosci. 1994;14:4318–4328. doi: 10.1523/JNEUROSCI.14-07-04318.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steinthorsdottir V, Stefansson H, Ghosh S, Birgisdottir B, Bjornsdottir S, Fasquel AC. Multiple novel transcription initiation sites for NRG1. Gene. 2004;342:97–105. doi: 10.1016/j.gene.2004.07.029. [DOI] [PubMed] [Google Scholar]
- 47.Chirgui K, Svenningsson P, Greengard P. Cyclin-dependent kinase 5 regulates dopaminergic and glutamatergic transmission in the striatum. Proc Natl Acad Sci USA. 2004;101:2191–2196. doi: 10.1073/pnas.0308652100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lenny N, Westendorf JJ, Hiebert SW. Transcriptional regulation during myelopoiesis. Mol Biol Rep. 1997;24:157–168. doi: 10.1023/a:1006859700409. [DOI] [PubMed] [Google Scholar]
- 49.Moghaddam B. Bringing order to the glutamate chaos in schizophrenia. Neuron. 2003;40:881–884. doi: 10.1016/s0896-6273(03)00757-8. [DOI] [PubMed] [Google Scholar]
- 50.Hahn CG, Wang HY, Cho DS, Talbot K, Gur RE, Berrettini WH, Bakshi K, Kamins J, Borgmann-Winter KE, Siegel SJ, Gallop RJ, Arnold SE. Altered neuregulin 1/ErbB4 signaling contributes to NMDA receptor hypofunction in schizophrenia. Nat Med. 2006;12:824–828. doi: 10.1038/nm1418. [DOI] [PubMed] [Google Scholar]
- 51.Coyle JT. Glutamate and schizophrenia: beyond the dopamine hypothesis. Cell Mol Neurobiol. 2006;26:365–384. doi: 10.1007/s10571-006-9062-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Javitt DC. Glutamate and schizophrenia: phencyclidine, N-methyl-d-aspartate receptors, and dopamine-glutamate interactions. Int Rev Neurobiol. 2007;78:69–108. doi: 10.1016/S0074-7742(06)78003-5. [DOI] [PubMed] [Google Scholar]
- 53.Carter CJ. Schizophrenia susceptibility genes converge on interlinked pathways related to glutamatergic transmission and long-term potentiation, oxidative stress and oligodendrocyte viability. Schizophr Res. 2006;86:1–14. doi: 10.1016/j.schres.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 54.Nave KA, Salzer JL. Axonal regulation of myelination by neuregulin 1. Curr Opin Neurobiol. 2006;16:492–500. doi: 10.1016/j.conb.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 55.Taveggia C, Thaker P, Petrylak A, Caporaso GL, Toews A, Falls DL, Einheber S, Salzer JL. Type III neuregulin-1 promotes oligodendrocyte myelination. Glia. 2008;56:284–293. doi: 10.1002/glia.20612. [DOI] [PubMed] [Google Scholar]
- 56.Winterer G, Konrad A, Vucurevic G, Musso F, Stoeter P, Dahmen N. Association of 5′ end neuregulin-1 (NRG1) gene variation with subcortical medial frontal microstructure in humans. Neuroimage. 2008;40:712–718. doi: 10.1016/j.neuroimage.2007.12.041. [DOI] [PubMed] [Google Scholar]
- 57.Walterfang M, Wood SJ, Velakoulis D, Pantelis C. Neuropathological, neurogenetic and neuroimaging evidence for white matter pathology in schizophrenia. Neurosci Biobehav Rev. 2006;30:918–948. doi: 10.1016/j.neubiorev.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 58.McCullumsmith RE, Gupta D, Beneyto M, Kreger E, Haroutunian V, Davis KL, Meador-Woodruff JH. Expression of transcripts for myelination-related genes in the anterior cingulate cortex in schizophrenia. Schizophr Res. 2007;90:15–27. doi: 10.1016/j.schres.2006.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moorhead TW, Job D, Lymer GK, Muñoz Maniega S, McKirdy J, Sussmann JE, Baig BJ, Bastin ME, Porteous D, Evans KL, Johnstone EC, Lawrie SM, Hall J. The effects of a neuregulin 1 variant on white matter density and integrity. Mol Psychiatry. 2008;13:1054–1059. doi: 10.1038/sj.mp.4002103. [DOI] [PubMed] [Google Scholar]
- 60.Konrad A, Winterer G. Disturbed structural connectivity in schizophrenia primary factor in pathology or epiphenomenon? Schizophr Bull. 2008;34:72–92. doi: 10.1093/schbul/sbm034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen YJ, Johnson MA, Lieberman MD, Goodchild RE, Schobel S, Lewandowski N, Rosoklija G, Liu RC, Gingrich JA, Small S, Moore H, Dwork AJ, Talmage DA, Role LW. Type III neuregulin-1 is required for normal sensorimotor gating, memory-related behaviors, and corticostriatal circuit components. J Neurosci. 2008;28:6872–6883. doi: 10.1523/JNEUROSCI.1815-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Young JW, Crawford N, Kelly JS, Kerr LE, Marston HM, Spratt C, Finlayson K, Sharkey J. Impaired attention is central to the cognitive deficits observed in alpha 7 deficient mice. Eur Neuropsychopharmacol. 2007;17:145–155. doi: 10.1016/j.euroneuro.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 63.Hancock ML, Canetta SE, Role LW, Talmage DA. Presynaptic type III neuregulin1-ErbB signaling targets alpha7 nicotinic acetylcholine receptors to axons. J Cell Biol. 2008;181:511–521. doi: 10.1083/jcb.200710037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhong C, Du C, Hancock M, Mertz M, Talmage DA, Role LW. Presynaptic type III neuregulin 1 is required for sustained enhancement of hippocampal transmission by nicotine and for axonal targeting of α7 nicotinic acetylcholine receptors. J Neurosci. 2007;28:9111–9116. doi: 10.1523/JNEUROSCI.0381-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Freedman R, Olincy A, Buchanan RW, Harris JG, Gold JM, Johnson L, Allensworth D, Guzman-Bonilla A, Clement B, Ball MP, Kutnick J, Pender V, Martin LF, Stevens KE, Wagner BD, Zerbe GO, Soti F, Kem WR. Initial phase 2 trial of a nicotinic agonist in schizophrenia. Am J Psychiatry. 2008;165:1040–1047. doi: 10.1176/appi.ajp.2008.07071135. [DOI] [PMC free article] [PubMed] [Google Scholar]