Abstract
In contrast to disorders of sexual differentiation caused by lack of androgen production or inhibited androgen action, defects affecting development of the bipotent genital anlagen have rarely been investigated in humans. We have previously documented that the transcription factor FOXF2 is highly expressed in human foreskin. Moreover, Foxf2 knockout mice present with cleft palate in combination with hypoplasia of the genital tubercle. We hypothesized that humans with disorders of sex development (DSD) in combination with cleft palate could have mutations in the FOXF2 gene. Eighteen children with DSD and cleft palate were identified in the Lübeck DSD database (about 1,500 entries). Genomic DNA sequence analysis of the FOXF2 gene was performed and compared with 10 normal female and 10 normal male controls, respectively. Two heterozygous DNA sequence variations were solely present in one single patient each but in none of the 20 normal controls: a duplication of GCC (c.97GCC[9]+[10]) resulting in an extra alanine within exon 1 and a 25∗G>A substitution in the 3′-untranslated region. Two patients carried a c.262G>A sequence variation predicting for an Ala88Thr exchange which was also detected in 2 normal controls. Two silent mutations, c.1272C>T (Ser424Ser) and c.1284T>C (Tyr428Tyr), respectively, occurred in the coding region of exon 2, again in both patients and normal controls. In conclusion, the majority of the detected sequence alterations were polymorphisms without obvious functional relevance. However, it cannot be excluded that the 2 unique DNA sequence alterations could have affected FOXF2 on the mRNA or protein level thus contributing to the observed disturbances in genital and palate development.
Key Words: Cleft palate, Disorders of sexual development, FOXF2, Genital development, Genital tubercle
Introduction
Human sexual development can be divided into sexual determination and sexual differentiation. Sex determination comprises development of the bipotent gonadal anlagen into either testes or ovaries (Hiort and Holterhus, 2000). Sexual differentiation comprises development of the internal and external genital organs under the control of sex hormones, i.e., the presence or absence of testosterone and Anti-Müllerian hormone (AMH) (Hiort and Holterhus, 2000). Disorders of sexual development (DSD) may occur at every single step due to interruption of the normal genetic and hormonal pathways leading to lack of androgens because of gonadal dysgenesis, defects in androgen biosynthesis, or inhibited androgen action in 46,XY males, eventually resulting in defective external masculinization.
A lot of children born with ambiguous genitalia lack a definite diagnosis because the mechanisms of normal and abnormal genital development in humans are still poorly deciphered. Interestingly, many DSD children present with associated malformations (Thyen et al., 2006). This suggests that so far unknown genetic events may interfere with specific early stages during embryogenesis potentially affecting development of both the genitalia and distinct somatic tissues. In a previous cDNA microarray based genome-wide study we identified that the transcription factor FOXF2 was significantly overexpressed in human foreskin fibroblasts from normal male individuals compared to labia majora fibroblasts from DSD patients with androgen insensitivity syndrome (Holterhus et al., 2003). Moreover, FOXF2 showed an almost identical expression pattern as HOXA13 (Holterhus et al., 2003) known to cause the hand-foot-genital syndrome, a combination of limb malformations and genital abnormalities (Mortlock and Innis, 1997). The FOXF2 gene belongs to the growing family of forkhead-box transcription factors characterized by a highly conserved ∼100-amino acid DNA binding domain. The human FOXF2 gene consists of 2 exons divided by an intron and is located at 6p25.3 (Blixt et al., 1998). Forkhead-box transcription factors play important roles in many aspects of embryonic development (Kaufmann and Knöchel, 1996). Foxf2 shows an early expression in the head mesenchym adjacent to the oropharynx and stomodeum during mouse embryogenesis (Ormestad et al., 2004), later on also in the respiratory system, urinary system, skeletal system, as well as the eye and inner ear (Aitola et al., 2000). Homozygous mutant Foxf2 knockout mice presented with cleft palate and an abnormal tongue and died shortly after birth (Wang et al., 2003). As described later, Foxf2 knockout mice also showed abnormalities of the external genitalia with a significant hypoplasia of the genital tubercle. We hypothesized that FOXF2 gene mutations could play a role in children with the combination of abnormalities of the external genitalia and cleft palate. In this study, we report the genomic DNA sequence analysis of the FOXF2 gene in DSD patients combined with cleft palate.
Materials and Methods
The study was approved by the ethical committee of the University of Lübeck, Germany.
Patients and Normal Controls
Eighteen children were selected from 1,500 cases of the Lübeck DSD database because of the clinical combination of DSD and cleft palate. Table 1 summarizes the clinical, cytogenetic, molecular genetic, and hormonal findings. Ten normal female and 10 normal male individuals served as controls.
Table 1.
Summary of the patients
| Patient | Karyotype | External genitalia | Facial cleft | Associated malformations | Analysed genes without mutations | Hormone levelsa |
|---|---|---|---|---|---|---|
| 1 | 46,XY | micropenis, scrotum bipartitum, hypospadias | cleft palate | multiple anomalies including wide Aorta descendens, macrocephaly, hypertelorism, mental retardation | AR, SRY | testosterone basal: 0.285 nmol/l |
| 2 | 46,XY | micropenis, hypospadias | cleft palate | broad thumbs, prominent forehead, hypertelorism, dysplastic teeth and ears | AR | T baseline: 0.451 nmol/l, stimulated: 3.157 nmol/l; DHT: 0.7912 nmol/l; A: 2.094 nmol/l (age 5 years) [0.07–0.31 nmol/l] |
| 3 | 46,XY | scrotum bipartitum, perineal hypospadias | cleft palate | mental retardation, microcephaly, epilepsy, growth delay, iris coloboma | AR | T baseline: 0.662 nmol/l, stimulated: 5.812 nmol/l |
| 4 | 46,XY | hypospadias | cheilognathopalatoschisis, cleft larynx | growth delay, epicanthus, broad nasal root | AR, 5α-reductase II | T baseline: <0.7 nmol/l; DHT: 0.6536 nmol/l |
| 5 | 46,XX | uterus duplex, vaginal agenesis | cheilognatho-palatoschisis | aplastic kidney on the right, double-kidney on the left side, Meckel's diverticulum nmol/l] | AR, SRY | T baseline: 0.062 nmol/l (age 3 years) [0.07–0.35 |
| 6 | 46,XY | scrotum bipartitum, hypospadias | cleft palate with bifid uvula | AR, 5α-reductase II | T baseline: 0.937 nmol/l, stimulated: 7.388 nmol/l; DHT: 0.25 nmol/l | |
| 7 | 46,XY | micropenis, scrotal hypospadias | cleft palate | retrognathia, hypoplastic left kidney | AR, 5α-reductase II | |
| 8 | 46,XY | micropenis | cheilognatho-palatoschisis | deafness | no information | T baseline: <0.7 nmol/l |
| 9 | 46,XY | micropenis, hypospadias | cheilognatho- palatoschisis | Fallot's syndrome, iris coloboma | AR, 5α-reductase II | not documented |
| 10 | 46,XY | scrotal hypospadias | cleft palate | growth delay, facial abnormalities, vitium cordis | AR | not documented |
| 11 | 46,XY | hypospadias, scrotum bipartitum | cleft palate | hypoplastic rib, premature closure of the sagittal suture | AR, 5α-reductase II | not documented |
| 12 | not documented | micropenis, cryptorchidism | cheilognatho- palatoschisis | esophageal atresia Vogt IIIb, mental retardation, vitium cordis | AR: silent mutation, 5α-reductase II | not documented |
| 13 | 46,XY | hypospadias, cryptorchidism | cleft palate | mental delay, congenital hypothyroidism | AR, 5α-reductase II | not documented |
| 14 | 46,XY | perineal hypospadias, scrotum bipartitum, septal urorectale | cheilognatho- palatoschisis | hypertelorism, anal atresia, iris coloboma, dysplastic left kidney, accessoric left mammilla | no information | not documented |
| 15 | 46,XY | ambiguous genitalia | cheilognatho- palatoschisis | AR, 5α-reductase II | T baseline: 0.382 nmol/l, stimulated: 8.502 nmol/l; DHT: 0.206 nmol/l; A: 1.152 nmol/l | |
| 16 | 46,XY, partial trisomy 3q | scrotal hypospadias, ambigous genitalia with persistent vaginal anlagen | cleft palate | vitium cordis with ventricular septal defect, right sided aortic arch | AR | T baseline: 0.07 nmol/l, stimulated: 9.404 nmol/l (age 16 months) [0.07–0.35 nmol/l] |
| 17 | 45,X/46,XY | unclassified pseudo- hermaphroditism | cleft palate | no information | T baseline: 0.729 nmol/l, stimulated: 3.435 nmol/l; A: 2.792 nmol/l (age 7 years) [0.07–0.38 nmol/l] | |
| 18 | not documented | hypospadias | cheilognatho- palatoschisis | pyelectasis | no information | not documented |
(age at determination if documented) [normal baseline range of T related to age]; T: testosterone; DHT: dihydrotestosterone; A: androstenedione; normal stimulated testosterone in the hCG test >3.5 nmol/l (>100 ng/dl); normal stimulated T/DHT ratio in the hCG test <16; normal A/T ratio <1.
Genomic DNA Analyses
Genomic DNA was extracted from blood leukocytes according to standard procedures. The whole coding region of the FOXF2 gene including the exon-intron junctions was amplified by PCR. Nine primer pairs were selected using the software PrimerSelect 6.00 (DNAStar Inc. Madison, Wisconsin, USA) and they are listed in table 2. Exon 1 consists of 1,170 bp and was amplified in 8 overlapping fragments. Exon 2 was amplified as a whole. Amplification was performed on a programmable thermocycler (PTC-2000, My Research, Inc., Massachusetts, USA) using 100 ng of DNA in a 50 μl mixture including 20 pmol of each primer, 2 mM dNTPs (dATP, dGTP, sTTP, and dCTP), 1 U DNA-Polymerase (AmpliTaq, Roche), 20 mM Tris (pH 8–8.8), and 1.0–2.0 mM MgCl2. Exon 1 contains GC-rich areas. Therefore, to receive specific fragments, 5% DMSO was added for amplifying exon 1. Starting with an initial denaturing at 94°C for 5 min, the PCR program consisted of 34 cycles, denaturation at 98°C for 10 s, annealing at 49–66°C for 30 s, and elongation at 72°C for 2 min, and ended by a final elongation at 72°C for 5 min.
Table 2.
Primers used for amplification of FOXF2. S: sense, A: antisense.
| Fragment | Primer |
|---|---|
| Exon 1/1 | S: 5′-GGC GCT CGC AGG GCT TCT-3′ |
| A: 5′-TCA TCA GGG CGG CCT GGA-3′ | |
| Exon 1/2 | S: 5′-CCG GGT CCC AGA TGA CCA-3′ |
| A: 5′-GGG GCG CTG GCC GAA TTG-3′ | |
| Exon 1/3 | S: 5′-GTC GTC GTC CTC CGC CTC CT-3′ |
| A: 5′-TTG CTG GGC GAG CTC TGG AT-3′ | |
| Exon 1/4 | S: 5′-CTA CTC GTA CAT CGC GCT CAT-3′ |
| A: 5′-GAG GCC CTT AGG CAG CTT G-3′ | |
| Exon 1/5 | S: 5′-AGG GCT GGA AGA ACT CGG T-3′ |
| A: 5′-TAC ATG GGC TTG AGC GCC T-3′ | |
| Exon 1/6 | S: 5′-CGA CCC GGC CAG CGA GTT CAT-3′ |
| A: 5′-GGT ACC GGG CTG CTG CTG CTG TCC-3′ | |
| Exon 1/7 | S: 5′-CAG CCA CGC GCA CCC TCA-3′ |
| A: 5′-GCG AGT AGG AGG ACA TGC TGG A-3′ | |
| Exon 1/8 | S: 5′-GCG CCA TCG AAT GCC ACT-3′ |
| A: 5′-AGC CCT GGA GGC CTA GAA-3′ | |
| Exon 2 | S: 5′-CTG GAT GAC TTT GTT TCT GA-3′ |
| A: 5′-CGT GCA TGT GAC TTG AAT-3′ |
For mutation screening, non-isotopic SSCP analysis (single strand conformation polymorphism analysis) was used. The fragment 1/6 of exon 1 was digested using the restriction enzyme PvuII prior to SSCP. PCR products were diluted 1.5:1 in 95% formamide, 89 mM Tris, 20 mM EDTA, 89 mM boric acid, 0.05% bromophenol blue, and 0.05% xylene cyanol. After a 5-min denaturation at 98°C, the PCR products were chilled on ice water and afterwards loaded onto a 6% polyacrylamide gel (0.8 mm thick) containing 5–10% glycerol. Electrophoresis was performed at 8–40 W for 4–17 h at room temperature. DNA bands were visualized using silver staining (Bassam et al., 1991). All fragments with aberrant mobility (band shifts) were sequenced using an automatic capillary sequencer (ABI PRISM 3100 Genetic Analyzer, Applied Biosystems, Foster City, CA, USA) according to standard procedures. In case of band shifts, the same PCR product was analysed in the 20 control samples as well. Since some exon 2 samples showed only minimal aberrations in migration patterns, this exon was directly sequenced in all 18 patients without prior SSCP.
Results
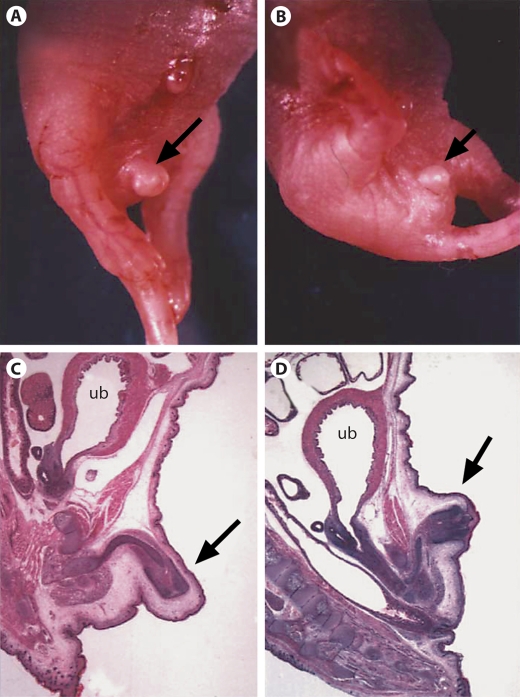
Previously we found that Foxf2–/– mice showed cleft palate (Wang et al., 2003). Recently, we also found that the genital tubercle was hypoplastic in these homozygous mice (fig. 1). Therefore, the Foxf2 knockout mice displayed both hypoplastic genitalia and cleft palate. The similarity of the DSD patients combined with cleft palate with the phenotypes of Foxf2 knockout mice prompted us to study the FOXF2 gene in DSD patients with cleft palate.
Fig. 1.
External genitalia of a normal control mouse and a Foxf2–/– knockout mouse at day P0 (postnatal day 0). A Macroscopic aspect of a normal control mouse; BFoxf2–/– mutant mouse depicting hypoplasia of the genital tubercle; C, D Underlying histological findings. The genital tubercle is marked by an arrow. ub = urinary bladder.
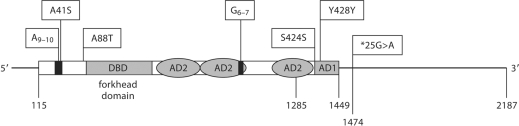
In patient 10 (table 3, fig. 2) we detected a heterozygous insertion of an additional GCC repeat (c.97GCC[9]+ [10]; accession no. ss105106973) predicting an insertion of an extra alanine in a polyalanine stretch within the N-terminus of FOXF2. This DNA sequence variation was neither detected in any of the 20 normal controls nor in any other patient and has not previously been described. In contrast, we found the heterozygous sequence variation c.121G>T affecting the same alanine repeat predicting for an A41S exchange on the protein level only in one normal female control but not in any of the patients. The affected alanine is the last residue of the 9 residues long polyalanine repeat.
Table 3.
Sequence variants found in the study
| dbSNP accession | nt sequence variation | AA sequence variation | Patient | No. of controls |
|---|---|---|---|---|
| ss105106971 | c.121G>T (heterozygous) | p.A41S | – | 1 |
| ss105106973 | c.97GCC[9]+[10] (heterozygous) | p.A33[9]+[10] | 10 | – |
| ss107795095 | c.262G>A (heterozygous) | p.A88T | 3, 14 | 2 |
| ss105106972 | c.1272C>T (heterozygous) | p.S424S (silent) | 15 | 1 |
| rs2293783 | c.1284T>C (heterozygous) | p.Y428Y (silent) | 2, 4, 6, 7, 10, 11, 17 | 9 |
| rs2293783 | c.1284T>C (homozygous) | p.Y428Y (silent) | 5 | – |
| rs58230522 | c.904GGC[5] (homozygous) | p.G301[6] | 9, 13, 15 | 6 |
| rs58230522 | c.904GGC[5]+[6] (heterozygous) | p.G301[6]+[7] | 1, 2, 3, 5, 6, 7, 12, 14, 16, 18 | 12 |
| rs58230522 | c.904GGC[6] (homozygous) | p.G301[7] | 4, 8, 10, 11, 17 | 2 |
| rs45600838 | ∗25G>A (heterozygous) | 3′ UTR | 12 | – |
Fig. 2.
Summary of the position of detected sequence variations in the FOXF2 mRNA (NM_001452). nt.1–nt.114: 5′-untranslated region; nt.115–nt.117: Startcodon; nt.1285: end of exon 1; nt.1447–nt.1449: Stopcodon; nt.1450–nt.2187: 3′-untranslated region; nt.1474: G/A: SNP rs45600838. DBD: DNA binding domain, forkhead domain; AD1: activation domain 1; AD2: 3 regions of the non contiguous activation function 2. Upper boxes indicate amino acid changes by SNPs within the FOXF2 reading frame. A41S (SNP ss105106971); A9–10: homopolymer stretch of 9–10 alanine residues (SNP ss105106973); A88T (SNP ss107795095); G6–7: homopolymer stretch of 6–7 glycine residues (SNP rs58230522); S424S (SNP ss105106972); Y428Y (SNP rs2293783).
Two additional novel SNPs were detected in the coding region of FOXF2 during our analyses. Two patients (patients 3 and 14) as well as 2 male controls showed a heterozygous nucleotide substitution c.262G>A (accession no. ss107795095) predicting a conversion of alanine to threonine at codon position 88 (A88T) (table 3, fig. 2). The second polymorphism was a silent mutation in the coding region of exon 2. A heterozygous exchange of cytosine by thymine at position 1272 (c.1272C>T; S424S; accession no. ss105106972) was found in patient 15 and in 1 male individual of the control group. A frequent heterozygous exchange of thymine by cytosine was found at position 1284 (c.1284T>C; Tyr428Tyr), a previously known FOXF2 sequence variation (rs2293783). It was detected in patients 2, 4, 6, 7, 10, 11, 17 as well as in 9 normal controls (4 males and 5 females). Patient 5 carried the same sequence variation in a homozygous state which was not found in any of the normal controls. The PCR fragment 6 of exon 1 contained 5 or 6 GGC repeats coding for glycine from position 904 onwards (c.904GGC(5_6); rs58230522) in our set of individuals. The GGC repeat is preceded by a GGG codon also coding for a glycine leading to a repeat of 6 or 7 glycine residues (fig. 2). In contrast to the alanine repeat in the N-terminus, the glycine repeat is more variable. Of our 18 patients, 3 were homozygous for 5 GGC repeats, 10 were heterozygous for 5 and 6 repeats, and 5 were homozygous for 6 GGC repeats. GGC repeats were distributed similarly in the group of control individuals. Patient 12 showed a heterozygous replacement of guanine to adenine at the 3′-untranslated region (25∗G>A; rs45600838) of the FOXF2 gene. This nucleotide exchange was neither found in any of the 20 control individuals nor in any of the other 17 patients.
Discussion
Here we report for the first time on a genomic DNA sequence analysis of the FOXF2 gene in 18 patients with DSD combined with cleft palate without a currently established endocrine or genetic diagnosis as based on the files available in our database. We identified 4 different DNA sequence alterations of the FOXF2 gene which also occurred in the group of normal controls at a comparable frequency thus excluding their relevance for the observed phenotypes. However, 2 additional heterozygous FOXF2 sequence variations, an expanded GCC repeat in exon 1, leading to an extra alanine (A9–10) in the N-terminus of FOXF2 and a heterozygous G>A exchange in the 3′-untranslated region, were present in only 1 patient each (patient 10 and 12, respectively) but neither in any of the 20 controls nor in any of the other 17 patients.
There are published biological examples in other endocrine diseases suggesting that the type of the two identified unique FOXF2 sequence alterations in our DSD patients could play a functional role in the phenotype. In the first, a recently reported insertion of an extra CTG in the polyleucine repeat in exon 1 of the GNAS1 gene causing Albright's hereditary osteodystrophy (AHO) (Lim et al., 2002) has considerable similarities with the exon 1 mutation of FOXF2 observed in our patient 10 extending the polyalanine repeat (fig. 2). Secondly, the replacement of guanine with adenine at the 3′-untranslated region (25∗G>A; rs45600838) of the FOXF2 gene in patient 12 has similarities with a previously described silent mutation within the very end of exon 8 of the androgen receptor gene causing aberrant splicing of the androgen receptor mRNA inducing partial androgen insensitivity syndrome (Hellwinkel et al., 2001). Therefore, it cannot be excluded that the 25∗G>A could modify FOXF2 mRNA transcription and thus contribute to the observed phenotype. The importance of an adequate gene dose for sexual differentiation processes has been well documented for many genes, namely SF-1(NR5A1) (Achermann et al., 2002), WNT4 (Jordan et al., 2001), and DAX1(NR0B1) (Parma et al., 1997; Iyer and McCabe, 2004). Taken together, it may be hypothesized that the two unique FOXF2 gene alterations as detected in patients 10 and 12 could have functional consequences on the mRNA or the protein level resulting in reduced gene function or gene dosage at critical times during genital development, thus being involved in expression of the underlying phenotypes. Unfortunately, DNA of the parents and of potential siblings was not available. Also primary tissues like cultured genital fibroblasts were not available for functional studies. Therefore, the exact role of these 2 FOXF2 sequence alterations remains uncertain at this point.
Due to the combination of cleft palate and impaired genital tubercle outgrowth in the Foxf2 knockout mice (fig. 1) and due to the identical expression pattern of FOXF2 and HOXA13 in our genome wide study (Holterhus et al., 2003), we speculated that FOXF2 would potentially act on the level of development of the bipotent genital anlagen but not on the level of androgen production or androgen action. Therefore, one would expect normal baseline and stimulated testosterone levels in affected patients. Unfortunately, hormonal data were incomplete and not available in each of the 18 cases of our DNA database (table 1). In particular, there was no hormonal data available in patients 10 and 12, the ones with the unique sequence alterations. In some patients, hCG-induced rise of testosterone was only marginal, e.g., patient 2: 91 ng/dl; patient 17: 99 ng/dl. Since the latter patient carried a chromosomal mosaicism 45,X/46,XY, it is likely that this patient has gonadal dysgenesis as the underlying cause of genital undervirilization with cleft palate occurring for other currently unknown reasons.
The combination of hypospadias and cleft palate is not rare and has been described as part of different syndromes. In none of the 18 patients, a definitive diagnosis had so far been established. Patient 2 had clinical characteristics compatible with the Robinow syndrome which is characterized by dwarfism, genital hypoplasia like micropenis and reduced clitoral size, spinal abnormalities, and renal tract abnormalities. Facial features include hypertelorism with midfacial hypoplasia and a broad and prominent forehead combined with a short nose. Oral features contain a tented upper lip. A cleft palate is not a commonly described criterion (Patton and Afzal, 2002). In patient 2 only very few features are fulfilled (table 1).
Patient 12 had some clinical features related to the autosomal dominant CHARGE syndrome. Three major clinical criteria reported by Verloes (2005) are ocular coloboma, choanal atresia, and hypoplasia of semicircular canals; minor criteria are rhombencephalic dysfunction, hypothalamo-hypophyseal dysfunction, malformation of the internal or external ear, malformation of the mediastinal organs, and mental retardation. In a typical CHARGE syndrome, 3 major or 2 major and 2 minor criteria must be fulfilled (Verloes, 2005). In patient 12, none of the three majors is reported, and therefore, a typical CHARGE syndrome is excluded.
Until today, investigation of genes potentially involved in the development of the early bipotent genital anlagen outside the gonads and outside androgen-dependent sexual differentiation has been performed predominantly in mouse models (Haraguchi et al., 2000). In a more recent study, Beleza-Meireles et al. (2007) did not find mutations of clear pathogenetic relevance in the promising candidate genes FGFR2, FGF8, FGF10, and BMP7 in patients with hypospadias. The latter are part of a complex developmental network of gene regulation and epithelial-mesenchymal interactions involving the urethral plate epithelium and the genital tubercle (Morgan et al., 2003). Nevertheless, it is important for the first step towards a better understanding of normal and aberrant human genitalia development to investigate genomic mutations and variations of promising new candidate genes of early genital anlagen development in human since many DSD patients lack a well-defined molecular diagnosis yet.
Acknowlegement
This study was part of a German Clinical Research Group project funded by the DFG (German Research Council) (Grant No. KFO111 to O.H. and to P.M.H.). R.W., A.R.U., O.H., and P.M.H. are members of the German national BMBF (Ministry of Education and Science)-funded DSD-network. The authors thank Claudia Havel for her excellent technical assistance.
References
- 1.Achermann JC, Ozisik G, Ito M, Orun UA, Harmanci K, et al. Gonadal determination and adrenal development are regulated by the orphan nuclear receptor steroidogenic factor-1, in a dose-dependent manner. J Clin Endocrinol Metab. 2002;87:1829–1833. doi: 10.1210/jcem.87.4.8376. [DOI] [PubMed] [Google Scholar]
- 2.Aitola M, Carlsson P, Mahlapuu M, Enerbäck S, Pelto-Huikko M. Forkhead transcription factor FoxF2 is expressed in mesodermal tissues involved in epithelio-mesenchymal interactions. Dev Dyn. 2000;218:136–149. doi: 10.1002/(SICI)1097-0177(200005)218:1<136::AID-DVDY12>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 3.Bassam BJ, Caetano-Anolles G, Gresshoff PM. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem. 1991;196:80–83. doi: 10.1016/0003-2697(91)90120-i. [DOI] [PubMed] [Google Scholar]
- 4.Beleza-Meireles A, Lundberg F, Lagerstedt K, Zhou X, Omrani D, et al. FGFR2, FGF8, FGF10 and BMP7 as candidate genes for hypospadias. Eur J Hum Genet. 2007;15:405–410. doi: 10.1038/sj.ejhg.5201777. [DOI] [PubMed] [Google Scholar]
- 5.Blixt A, Mahlapuu M, Bjursell C, Darnfors C, Johannesson T, et al. The two-exon gene of the human forkhead transcription factor FREAC-2 (FKHL6) is located at 6p25.3. Genomics. 1998;53:387–390. doi: 10.1006/geno.1998.5451. [DOI] [PubMed] [Google Scholar]
- 6.Haraguchi R, Suzuki K, Murakami R, Sakai M, Kamikawa M, et al. Molecular analysis of external genitalia formation: the role of fibroblast growth factor (Fgf) genes during genital tubercle formation. Development. 2000;127:2471–2479. doi: 10.1242/dev.127.11.2471. [DOI] [PubMed] [Google Scholar]
- 7.Hellwinkel OJ, Holterhus PM, Struve D, Marschke C, Homburg N, Hiort O. A Unique exonic splicing mutation in the human androgen receptor gene indicates a physiologic relevance of regular androgen receptor transcript variants. J Clin Endocrinol Metab. 2001;86:2569–2575. doi: 10.1210/jcem.86.6.7543. [DOI] [PubMed] [Google Scholar]
- 8.Hiort O, Holterhus PM. The molecular basis of male sexual differentiation. Eur J Endocrinol. 2000;142:101–110. doi: 10.1530/eje.0.1420101. [DOI] [PubMed] [Google Scholar]
- 9.Holterhus PM, Hiort O, Demeter J, Brown PO, Brooks JD. Differential gene-expression patterns in genital fibroblasts of normal males and 46,XY females with androgen insensitivity syndrome: evidence for programming involving the androgen receptor. Genome Biol. 2003;4:R37. doi: 10.1186/gb-2003-4-6-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iyer A, McCabe E. Molecular mechanisms of DAX1 action. Mol Genet Metab. 2004;83:60–73. doi: 10.1016/j.ymgme.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 11.Jordan B, Mohammed M, Ching S, Delot E, Chen X, et al. Up-regulation of WNT-4 signaling and dosage-sensitive sex reversal in humans. Am J Hum Genet. 2001;68:1102–1109. doi: 10.1086/320125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaufmann E, Knöchel W. Five years on the wings of fork head. Mech Dev. 1996;57:3–20. doi: 10.1016/0925-4773(96)00539-4. [DOI] [PubMed] [Google Scholar]
- 13.Lim SH, Poh LK, Cowell CT, Tey BH, Loke KY. Mutational analysis of the GNAS1 exons encoding the stimulatory G protein in five patients with pseudohypoparathyroidism type 1a. J Pediatr Endocrinol Metab. 2002;15:259–268. doi: 10.1515/jpem.2002.15.3.259. [DOI] [PubMed] [Google Scholar]
- 14.Morgan EA, Nguyen SB, Scott V, Stadler HS. Loss of Bmp7 and Fgf8 signaling in Hoxa13-mutant mice causes hypospadia. Development. 2003;130:3095–3109. doi: 10.1242/dev.00530. [DOI] [PubMed] [Google Scholar]
- 15.Mortlock D, Innis J. Mutation of HOXA13 in hand-foot-gential syndrome. Nat Genet. 1997;15:179–180. doi: 10.1038/ng0297-179. [DOI] [PubMed] [Google Scholar]
- 16.Ormestad M, Astorga J, Carlsson P. Differences in the embryonic expression patterns of mouse Foxf1 and -2 match their distinct mutant phenotypes. Dev Dyn. 2004;229:328–333. doi: 10.1002/dvdy.10426. [DOI] [PubMed] [Google Scholar]
- 17.Parma P, Pailhoux E, Puissant C, Cotinot C. Porcine Dax-1 gene: isolation and expression during gonadal development. Mol Cell Endocrinol. 1997;135:49–58. doi: 10.1016/s0303-7207(97)00189-5. [DOI] [PubMed] [Google Scholar]
- 18.Patton MA, Afzal AR. Robinow syndrome. J Med Genet. 2002;39:305–310. doi: 10.1136/jmg.39.5.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thyen U, Lanz K, Holterhus PM, Hiort O. Epidemiology and initial management of ambiguous genitalia at birth in Germany. Horm Res. 2006;66:195–203. doi: 10.1159/000094782. [DOI] [PubMed] [Google Scholar]
- 20.Verloes A. Updated diagnostic criteria for CHARGE syndrome: a proposal. Am J Med Genet. 2005;133A:306–308. doi: 10.1002/ajmg.a.30559. [DOI] [PubMed] [Google Scholar]
- 21.Wang T, Tamakoshi T, Uezato T, Shu F, Kanzaki-Kato N, et al. Forkhead transcription factor Foxf2 (LUN)-deficient mice exhibit abnormal development of secondary palate. Dev Biol. 2003;259:83–94. doi: 10.1016/s0012-1606(03)00176-3. [DOI] [PubMed] [Google Scholar]