Abstract
A crystal structure for a member of the AraC prokaryotic transcriptional activator family, MarA, in complex with its cognate DNA-binding site is described. MarA consists of two similar subdomains, each containing a helix–turn–helix DNA-binding motif. The two recognition helices of the motifs are inserted into adjacent major groove segments on the same face of the DNA but are separated by only 27 Å thereby bending the DNA by ≈35°. Extensive interactions between the recognition helices and the DNA major groove provide the sequence specificity.
The AraC family of prokaryotic transcriptional regulators includes >30 proteins from different microorganisms, 18 from Escherichia coli alone (1). Members of this family control expression of a variety of genes by binding to specific promoter sites as either monomers or dimers. For AraC, the first transcriptional activator discovered (2), there are functionally independent DNA-binding and dimerization domains (3). Here we report the crystal structure of the MarA–DNA complex, providing the first structural basis for DNA-binding by the AraC family activator.
MarA, a member of the AraC family, is a transcriptional activator of more than one dozen genes of the mar (multiple antibiotic resistance) regulon of E. coli (4–6). It consists of 129 amino acids, exists as a monomer in solution, does not contain a dimerization domain, and binds to an asymmetric, degenerate 20-bp DNA sequence (5, 7). This contrasts with other prokaryotic transcriptional regulators that generally act as dimers and bind tightly to unique direct or inverted repeat sequences (8–10).
METHODS
Selenomethionyl MarA with an N-terminal polyhistidine tag was expressed in an E. coli met− auxotropic strain, B834(DE3) (Novagen), and was purified as described for the native MarA (4). The purified MarA (≈30 mg/liter cell culture) was dissolved in a solution of 50% (vol/vol) glycerol, 50 mM Hepes (pH 8.0), and 0.5 M NaCl and was stored at −20°C for further use. The synthetic oligonucleotides (purchased from Keck Oligonucleotide Synthesis Facility of Yale University) were purified by reverse-phase HPLC (C4 column) by using a linear gradient of acetonitrile in 0.1 M triethylammonium acetate (pH 7.0). The MarA–DNA complex was prepared by first mixing equal molar amounts of the two complementary oligonucleotide strands at room temperature and then adding MarA to a solution of the duplex DNA at a 1:1.2 M ratio. Solubility of MarA was greatly enhanced by forming the complex with DNA. The MarA–DNA complex was dialyzed at 4°C against a buffer of 10 mM Hepes (pH 8.0) and 10 mM NaCl and was concentrated to 10 mg of MarA/ml of solution.
The crystals were grown at room temperature by the hanging drop method using the sparse matrix screen (11) from Hampton Research (Riverside, CA). The best diffracting crystals of a size 0.3 × 0.2 × 0.1 mm were obtained with a 22-bp double stranded DNA fragment (see Fig. 1B) by micro-seeding crystals in the presence of the mother liquor, 12% PEG 8000, 100 mM sodium cacodylate (pH 6.5), and 100 mM calcium acetate. The crystals were then transferred to the modified mother liquor with 15% PEG 8000 and 25% glycerol for cryoprotection and flash-frozen for storage.
Figure 1.
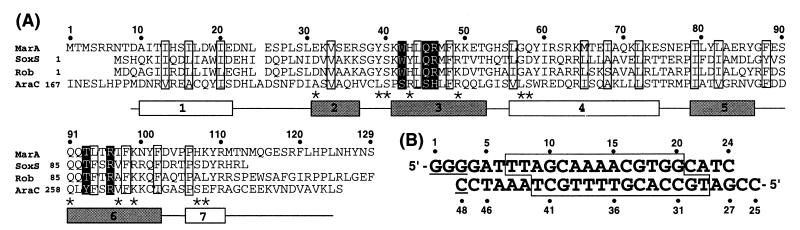
Sequence and secondary structure of MarA and the DNA oligomer used for crystallization. (A) Sequence of MarA aligned with SoxS, Rob, and AraC by using the pileup program (31). The secondary structural elements were assigned with the dssp program (32) and the HTH motifs represented by shaded boxes. Residues are enclosed in boxes according to their putative roles: white boxes for the hydrophobic core residues of the HTH motif and black-shaded boxes for the residues determining the sequence specificity. Asterisks mark MarA residues that interact with the phosphate backbone group of DNA. Residue numbers above the alignment correspond to the MarA sequence. (B) Double-stranded oligonucleotide used for crystallization. Sequences that are protected by DNase footprinting (5) are enclosed in a box. The underlined sequence is not a part of the mar promoter sequence but was required for crystal formation.
The crystal structure of the MarA–DNA complex was determined by the multiwavelength anomalous diffraction (MAD) method. MAD data were collected on a Raxis IV-imaging plate system at beam line X4A at the Brookhaven National Synchrotron Light Source. The diffraction data were collected at 95 K with the inverse beam method in 1.0° oscillation frames. Wavelength λ1, λ2, and λ3 near or at the K absorption edge of selenium were chosen so that the dispersive differences were maximized between λ1 and λ3 and that the anomalous differences were maximized at λ2. The collected data at three wavelengths were independently integrated and scaled by using the program hkl package (12). The crystal belongs to a space group P41212 with unit cell parameters a = b = 47.1, c = 298.2 Å and each asymmetric unit contains one molecule of MarA and a 22-bp DNA fragment. The data set collected at wavelength λ1 were considered as the native (Table 1). A program suite ccp4 package (13) was used for phasing. Se sites were located in difference Patterson maps and further verified by shelxs-97 (14). The initial positions of three Se sites were refined by using mlphare (13) with data of 20.0–2.7 Å, and the experimental phases were further extended by solvent flattening and histogram matching implemented in dm (13). The register of the DNA fragment in the pseudo-continuous DNA was further verified on difference Fourier maps by using iodo-substitution at the C5 methyl group of T7, T8, and T18 in the DNA fragment (Fig. 1B). The model for the MarA–DNA complex was built using o (15) and was refined to a resolution of 2.5 Å and later extended to 2.3 Å by using x-plor v3.851 (16) with the Engh and Huber parameters (17) for protein and the Parkinson et al. (18) parameters for DNA (Table 1). The data were anisotropic along the c axis, such that an anisotropic B-factor correction was applied to the data and then used for further refinements. Three rounds of simulated annealing at 3,000 K and manual rebuilding were followed by the assignment of water molecules. The stereochemistry of the final model was checked with procheck (19).
Table 1.
Data collection and refinement statics
| Data set | Se λ1 (native) | Se λ2 | Se λ3 |
|---|---|---|---|
| Wavelength, Å | 0.9793 | 0.9790 | 0.9679 |
| Resolution, Å | 20.0–2.3 | 20.0–2.5 | 20.0–2.5 |
| No. of observations | 121,112 | 107,252 | 108,117 |
| No. of unique reflections | 16,074 | 12,678 | 12,688 |
| Completeness,* % | 99.8 (99.8) | 99.8 (100.0) | 99.8 (100.0) |
| Rsym, % | 6.7 (56.2) | 6.4 (40.3) | 6.0 (41.5) |
| MAD analysis (20.0–2.7 Å) | |||
| Rcullis [Rcullis (ano)]† | (0.74) | 0.78 (0.68) | 0.67 (0.75) |
| Phasing power‡ | — | 0.97 | 1.51 |
| Mean FOM§ | 0.62 | ||
| Refinement statistics | |||
| Resolution, Å | 8.0–2.3 | ||
| Reflections, |F| > 3σ | 12,350 | ||
| Rfactor (Rfree)∥ | 0.225 (0.303) | ||
| No. of atoms | Protein, 975 | DNA, 978 | Water, 144 |
| Average B-factors, Å2 | Protein, 47.4 | DNA, 59.4 | Water, 59.2 |
| rms Deviations from ideal | |||
| Bond lengths 0.015 Å | |||
| Bond angles 1.44° |
Reflections with (I/σI ≥ −3.0) were included in data processing and values in parentheses are for the shell 2.30–2.55 Å for λ1, 2.50–2.62 Å for λ2, and λ3. Rsym = Σ|I − 〈I〉|/ΣI.
Rculles = Σ∥FPH ± FP| − FH|/Σ|FPH ± FP| where FP and FPH are the native and derivative observed structure amplitudes, respectively, and FH is the calculated heavy atom structure amplitude. Rculles (ano) = Σ|DPHobs − DPHcal |/ΣDPHobs where DPHobs and DPHcal are the observed and calculated anomalous differences for FPH.
Phasing power = rms (〈FH〉/E), where E is the residual lack of closure error.
Mean figure of merit (FOM) = Σ|Fbest|/F.
∥ R = Σ|Fc − Fo|/ΣFo Rfree is the same as R, but for 10% of the data that was not used for the refinement.
RESULTS
Overall Structure of the MarA–DNA Complex.
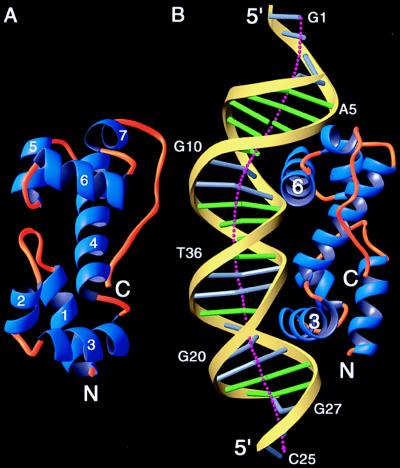
MarA was cocrystallized with a double-stranded 22-bp DNA fragment with 5′-overhanging bases (Fig. 1B) containing the DNA-binding site of MarA found in the mar promoter (5). Most of the residues in MarA and all of the nucleotides in the DNA are readily traceable in the initial electron density map, with the exception of some terminal residues in MarA (residues 1–8 and 125–129) (Fig. 1A). The crystal structure of the complex reveals that MarA has two helix–turn–helix (HTH) motifs and binds as a monomer to adjacent segments of the major groove, and that the DNA is in canonical B-form but bent by ≈35° (Fig. 2).
Figure 2.
The overall structure of MarA and its complex with DNA. (A) Ribbon diagram of MarA as seen from the DNA. Helix-1, -2, -3 and a part of helix-4 form the N subdomain and the remainder of the helices form the C subdomain. The two recognition helices, helix-3 and -6, which protrude above the plane, are not parallel but are tilted 30° relative to each other. The two subdomains are superimposable, with rms deviations of 1.6 Å for corresponding Cα atoms (33). (B) A ribbon diagram of the MarA–DNA complex showing the insertion of the recognition helices into the adjacent DNA major grooves and the overall bending of DNA. This orientation is obtained by a rotation of ≈90° along the DNA helical axis in the view of A. The N subdomain is bound to the downstream sequences of the binding site in the mar promoter and positioned closer to the transcription start site, whereas the C subdomain is bound to the upstream sequence. Different color codes are used for the DNA: yellow ribbon for the sugar-phosphate backbone, green cylinder for the A–T base pairs, and silver cylinder for the G–C base pairs. The dotted line shows the overall DNA helical axis calculated by the curves program (22). Clearly, both ends are curved more than the central part of DNA (bases 10–20). These figures were prepared by using the ribbons program (34).
MarA: Bipartite HTH Motif.
MarA is composed of seven α-helices and folds into two structurally similar subdomains with a long C-terminal loop (Figs. 1A and 2A). The N and C subdomains (residues 10–61 and 62–110) contain an HTH DNA-binding motif (residues 31–52 and 79–102), and are connected by helix-4. The two subdomains interact with each other noncovalently by hydrogen bonds and van der Waals contacts between residues in the loop joining helix-1 and -2 (mainly Glu-25 and Ser-26) and in the helix-5 region (mainly Arg-85 and Tyr-86). The spatial arrangement of the two subdomains has unique consequences. Because the recognition helices (helix-3 and -6) in the HTH motif protrude from the same face of the protein, MarA binds to one face of the DNA. This binding distorts the DNA because the two recognition helices are separated by 27 Å whereas the pitch of B-form DNA is 34 Å.
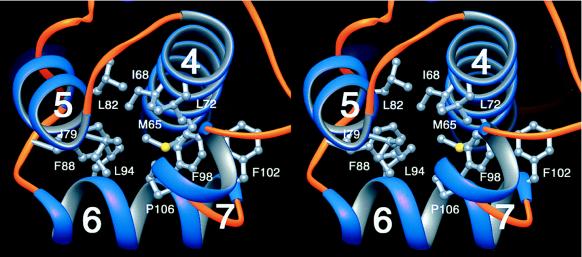
Structural comparisons using the program dali (20) indicated that MarA has a unique overall fold although its subdomains share structural similarity with other HTH DNA-binding domains (21). Helices 1 and 2 (helices 4 and 5) are antiparallel to one another and almost perpendicular to helix-3 (helix-4). Inside of these triangularly oriented three helices, there are patches of hydrophobic residues protruding from one side of each helix that form a hydrophobic core (Fig. 3). The fourth helix of each subdomain, the N-terminal region of helix-4 in the N subdomain and helix-7 in the C subdomain, closes off the hydrophobic core. The presence of the fourth helix as well as its orientation are quite different from those of other DNA-binding proteins.
Figure 3.
A stereo diagram of the relative orientations of the four helices and the hydrophobic core in the C subdomain. The view, from the top of Fig. 2A, shows the antiparallel orientations of helix-4 and -5 and the perpendicular location of helix-6 relative to the preceding helices. The putative hydrophobic residues from one face of each helix are indicated with their side chains. This figure was prepared by using the ribbons program (34).
Structure of DNA and its Interactions with MarA.
MarA binds to DNA by inserting separate recognition helices into the two adjacent segments of the major groove with the helical axes of the recognition helices almost parallel to the DNA base pairs (Fig. 2B). In the complex, the overall structure of the DNA is close to the canonical B-form with an average helical twist of 34.4° and an average rise of 3.3 Å per base pair (22), but there are significant changes in global and local DNA conformation. Two kinks in the DNA are observed near A9–Gl0 and G20–C21 and result in an overall bend of ≈35° in the DNA toward MarA. The observed global bending of DNA is consistent with changes in DNA local parameters: (i) the central base pairs (nucleotide number 11–16) have a narrow minor groove (width of ≈3Å relative to the average of 5.4Å), and (ii) abrupt changes in the angles between successive base pairs are observed near the kinks.
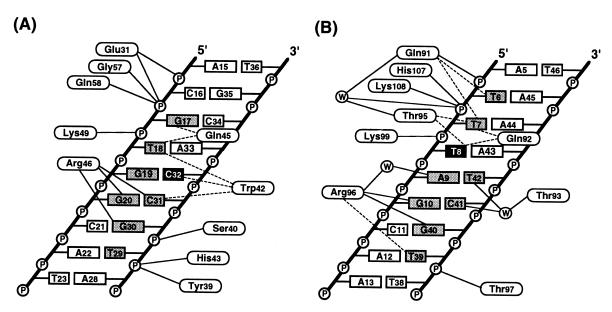
In general, hydrogen bonds and charge and shape complementarity are the primary sources of specificity between DNA-binding proteins and the bases in the major groove (9–10). Fig. 4 shows schematically these two classes of interactions: the direct and water-mediated indirect hydrogen bonds (solid lines) and van der Waals contacts (dashed lines) between the residues in MarA and the DNA. The binding of MarA to DNA buried a total MarA surface area of ≈930 Å2, similar to the typical buried surface area observed in the binding of antibodies to antigens (23). The interacting DNA bases are consistent with the DNase protection data (see Fig. 1B), and there are no interactions with the bases in the DNA minor groove. Although the overall binding scheme is similar in the two subdomains, there are two distinct differences in the interactions with DNA between the N and C subdomains. First, there are minor differences in the docking orientations of the recognition helices with DNA. Second, there are several water molecules bound at the interface between the C subdomain and DNA. Some of these water molecules mediate hydrogen bonds between the residues in the recognition helix and the major groove bases as well as the backbone phosphate groups.
Figure 4.
A schematic representation of MarA interactions with DNA. The interactions are for the N subdomain (A) and the C subdomain (B) of MarA. Dashed and solid lines correspond respectively to van der Waals interactions (interatomic distance <4.0 Å) between the nonpolar atoms and hydrogen bonds (interatomic distance <3.5 Å) between the polar atoms. Water molecules are observed only in the C subdomain and are enclosed in circles. The bases buried by the binding of MarA are shaded in gray and two bases, C32 and T8, that may have roles in the sequence specificity are in black (see text for more details). Except for Gly-57, Gln-58, His-107, and Lys-108, all of the other indicated residues use their side chains to make hydrogen bonds to the phosphate backbone groups and the bases. The average interatomic distance for hydrogen bonds is ≈2.9 Å. Although the side chain of Lys-99 is 3.7 Å distant from the phosphate group, Lys-99 was considered as a hydrogen bond donor due to its relatively blurred electron density at the ɛ-amino group. Other possible hydrogen bonds between Gln-92 and the O4 of T7 and between Trp-42 and the N4 of C32 are not shown here due to relatively long interatomic distances of 3.5 Å.
Hydrogen Bonds and van der Waals Interactions of MarA with the DNA Major Grooves.
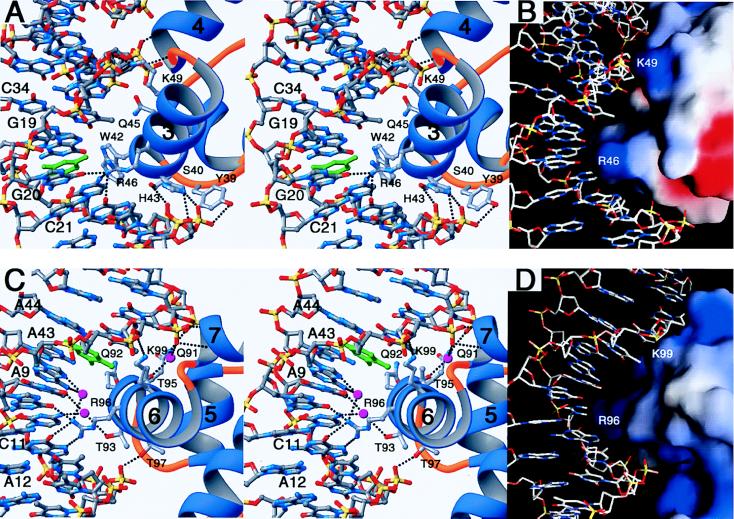
Details of the interactions of the N and C subdomain residues with the DNA of the major groove are shown in Figs. 5 A and C, respectively. In both subdomains, the backbone phosphate groups in the two DNA strands make extensive hydrogen bonds with the residues in the HTH motif (see Fig. 4) and also with the main chain NH groups of the N-terminal residues in the fourth helices (Gly-57 and Gln-58 in helix-4 and His-107 and Lys-108 in helix-7). The positive helix dipole at the N terminus of the fourth helices may contribute to neutralizing the negatively charged phosphate groups of the DNA backbone. Only a few sequence-specific direct hydrogen bonds are present between arginine in the central region of a recognition helix and the bases in the major groove. In the N subdomain, the guanidinium group of Arg-46 penetrates into the central major groove and makes hydrogen bonds with the O6 of G30, the N4 of C31, and the O6 of G20. In the C subdomain, Arg-96 makes hydrogen bonds to both the O6 and N7 atoms of G40. In addition, it interacts directly with the O6 of G10 and interacts via water with the N6 and N7 of A9. Thr-93 makes water-mediated sequence-specific hydrogen bonds with the N4 of C41 and the O4 of T42.
Figure 5.
Stereo close-up view of the interface between DNA and MarA. The interactions are for the N subdomain (A) and the C subdomain (C) of MarA. This orientation is similar to that in Fig. 2B. Hydrogen bonds in Fig. 4 are represented with black dashed lines between the interacting atoms (red for oxygen, blue for nitrogen, yellow for phosphorous, and silver for carbon) and the C32 and T8 bases are colored green. Water molecules at the interface of the C subdomain correspond to balls in magenta. Molecular surfaces of the N subdomain (B) and the C subdomain (D) of MarA are shown docked into the major groove of DNA. Arg-46 and Arg-96 protrude from the recognition helices and penetrate into the major groove. Figures A and C were prepared by using ribbons (34), and B and D were prepared by using grasp (35).
The molecular surfaces of the N and C subdomains of MarA are complementary in shape to the DNA (Fig. 5 B and D). Clearly, the protruding side chains of residues in helix-3 and -6 are docked into the concave DNA major groove and result in tight packing against several bases in the major groove. In the N subdomain, the side chain of Trp-42 makes van der Waals contacts mainly with C32 (green-colored base in Fig. 5A; see Fig. 4), within 3.6 Å distance from the C5 atom. Replacements of cytosine with thymine at this position, thereby introducing a methyl group on C5, would result in unfavorable interactions with MarA. This structural situation would be reversed at T18, where the 5-methyl group makes van der Waals interactions with the side chain of Gln-45 and provides additional binding energy for MarA. Therefore, MarA may have a preference for C over T at position 32 but T over C at position 18. In the C subdomain, Gln-91 and Thr-95 are within van der Waals interaction distances of the C5 atoms of T7 and T8 (green-colored base in Fig. 5C), and therefore a thymine base would be preferred over a cytosine at this position as was predicted for T18. However, other bases buried by the interaction with MarA such as T6, C41, and T39 (see Fig. 4) are either not a part of the interface or far beyond van der Waals interaction distances with the interface residues—>6.0 Å—and thus would not contribute to the sequence specificity.
DISCUSSION
Sequence Recognition by MarA.
Detailed genetic and biochemical studies of the MarA binding sites in ten mar regulon promoters provide an extended consensus sequence for MarA binding (7, 24). This consensus sequence is numbered as in Fig. 1B: y (C or T), r (A or G), and n (any nucleotide) with invariant nucleotides underlined.
4 r r y T T r r y n r y n y r T G C y r T 23
47 y y r A A y y r n y r n r y A C G r y A 28
Comparison of this sequence with the oligonucleotide used in the current study suggests that specificity is achieved largely by shape complementarity of the binding sites. The bases that interact directly via hydrogen bonds with Arg-46 and Arg-96 are not strictly conserved (see Fig. 4), whereas the highly conserved or invariant bases (T7, T8, T18, and C32) are involved in van der Waals interactions with the corresponding amino acids. The contribution to the specificity by the two arginine residues in the recognition helices is still unclear and awaits further biochemical analyses.
AraC Family of Transcriptional Regulatory Proteins.
Alignment of MarA, Rob (24), SoxS (25), and the DNA-binding domain of the AraC protein (Fig. 1A) strongly suggests that these AraC family proteins have folds similar to that of MarA. In each case, there are highly conserved or invariant residues for the potential hydrophobic core of the HTH motif and for interaction with the backbone phosphate groups of DNA and the major groove bases. Because Rob and SoxS also bind as monomers to MarA-binding sites in the mar regulon, it is not surprising that MarA shares with these two activators the invariant residues for the sequence specific hydrogen bonds (Arg-46, Thr-93, and Arg-96) and van der Waals interactions (Trp-42 and Gln-45) but not with the DNA-binding domain of AraC, which has a distinct DNA-binding site (Fig. 1A). These alignments support the general proposal that the regulatory proteins of the AraC family bind the target DNA site in a manner similar to MarA and that sequence specificity is derived mainly from interactions of the side chains of the recognition helices with bases in the major groove.
Comparisons of MarA with Other Transcriptional Regulatory Proteins with Bipartite HTH Motifs.
Whereas some eukaryotic transcriptional regulatory factors contain two HTH domains per subunit (26–30), MarA is the first prokaryotic transcriptional activator with a bipartite HTH motif. MarA differs from these eukaryotic proteins in its structure and the manner of binding to DNA. First, the two subdomains in MarA are structurally homologous and are linked with a helix. Second, the linker helix serves as a portion of the HTH units and is involved in interactions with the DNA backbone phosphate groups. Third, the linker helix imposes the orientation and distance restraints on the two subdomains for proper binding, so that the two HTH motifs bind in tandem to the same face of the target DNA and thereby dictate the extent of bending of the DNA. The eukaryotic transcription factors with two HTH domains have highly flexible loops as linkers, so that the two motifs can bind to DNA in various orientations relative to one another (either on opposite sides of the DNA, on perpendicular sides of the DNA, or along the major groove of the DNA) with a parallel or antiparallel arrangement of the two recognition helices.
Acknowledgments
We thank Fred Dyda for help with data collection and discussion and Craig Ogata for advice in data collection at the Howard High Beam line X4A.
ABBREVIATIONS
- mar
multiple antibiotic resistance
- HTH
helix–turn–helix
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (PDB ID code 1BL0).
References
- 1.Gallegos M-T, Michán C, Ramos J L. Nucleic Acids Res. 1993;21:807–810. doi: 10.1093/nar/21.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Englesberg E, Wilcox G. Annu Rev Genet. 1974;8:219–242. doi: 10.1146/annurev.ge.08.120174.001251. [DOI] [PubMed] [Google Scholar]
- 3.Schleif R. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, editor. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1300–1309. [Google Scholar]
- 4.Jair K-W, Martin R G, Rosner J L, Fujita N, Ishihama A, Wolf R E. J Bacteriol. 1995;177:7100–7104. doi: 10.1128/jb.177.24.7100-7104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin R G, Jair K-W, Wolf R E, Rosner J L. J Bacteriol. 1996;178:2216–2223. doi: 10.1128/jb.178.8.2216-2223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen S P, Hächler H, Levy S B. J Bacteriol. 1993;175:1484–1492. doi: 10.1128/jb.175.5.1484-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fawcett W P, Wolf R E. J Bacteriol. 1995;177:1742–1750. doi: 10.1128/jb.177.7.1742-1750.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrison S C, Aggarwal A K. Annu Rev Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- 9.Pabo C O, Sauer R T. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 10.Steitz T A. Q Rev Biophys. 1990;23:205–280. doi: 10.1017/s0033583500005552. [DOI] [PubMed] [Google Scholar]
- 11.Jancarik J, Kim S H. J Appl Crystallogr. 1991;24:409–411. [Google Scholar]
- 12.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 13.Dodson E J, Winn M, Ralph A. Methods Enzymol. 1997;277:620–633. doi: 10.1016/s0076-6879(97)77034-4. [DOI] [PubMed] [Google Scholar]
- 14.Sheldrick G M. Acta Crystallogr A. 1990;46:467–473. [Google Scholar]
- 15.Jones T A, Zou J Y, Cowan S W, Kjelgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 16.Brünger A T. x-plor. Yale University, New Haven, CT: Molecular Biophysics and Biochemistry; 1996. , Version 3.8. [Google Scholar]
- 17.Engh R A, Huber R. Acta Crystallogr A. 1991;47:392–400. [Google Scholar]
- 18.Parkinson G, Vojtechovsky J, Clowney L, Brünger A T, Berman H M. Acta Crystallogr D. 1996;52:57–64. doi: 10.1107/S0907444995011115. [DOI] [PubMed] [Google Scholar]
- 19.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 20.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 21.Wintjens R, Rooman M. J Mol Biol. 1996;262:294–313. doi: 10.1006/jmbi.1996.0514. [DOI] [PubMed] [Google Scholar]
- 22.Lavery R, Sklenar H. J Biomol Struct Dyn. 1989;6:655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- 23.Davies D R, Cohen G H. Proc Natl Acad Sci USA. 1996;93:7–12. doi: 10.1073/pnas.93.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ariza R R, Li Z, Ringstad N, Demple B. J Bacteriol. 1995;177:1655–1661. doi: 10.1128/jb.177.7.1655-1661.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z, Demple B. J Biol Chem. 1994;269:18371–18377. [PubMed] [Google Scholar]
- 26.Ogata K, Morikawa S, Nakamura H, Sekikawa A, Inoue T, Kanai H, Sarai A, Ishii S, Nishimura Y. Cell. 1994;79:639–648. doi: 10.1016/0092-8674(94)90549-5. [DOI] [PubMed] [Google Scholar]
- 27.Klemm J D, Rould M A, Aurora R, Herr W, Pabo C O. Cell. 1994;77:21–32. doi: 10.1016/0092-8674(94)90231-3. [DOI] [PubMed] [Google Scholar]
- 28.Xu W, Rould M A, Jun S, Desplan C, Pabo C O. Cell. 1995;80:639–650. doi: 10.1016/0092-8674(95)90518-9. [DOI] [PubMed] [Google Scholar]
- 29.König P, Giraldo R, Chapman L, Rhodes D. Cell. 1996;85:125–136. doi: 10.1016/s0092-8674(00)81088-0. [DOI] [PubMed] [Google Scholar]
- 30.Jacobson E M, Li P, Keon-del-Rio A, Rosenfeld M G, Aggarwal A K. Genes Dev. 1997;11:198–212. doi: 10.1101/gad.11.2.198. [DOI] [PubMed] [Google Scholar]
- 31.Genetics Computer Group. Wisconsin Package. Madison, WI: Genetics Computer Group; 1996. , Version 9.1. [Google Scholar]
- 32.Kabsch W, Sander C. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 33.Cohen G H. J Appl Crystallogr. 1997;30:1160–1161. [Google Scholar]
- 34.Carson M. J Appl Crystallogr. 1991;24:958–961. [Google Scholar]
- 35.Nicholls A. grasp Graphical Representation and Analysis of Surface Properties. New York: Columbia University; 1993. [Google Scholar]