Abstract
Gene expression profiles were examined using cDNA micro-array technology in human thyroid epithelial (Htori-3) cells exposed to a low, non-toxic dose (10 cGy) of radiation from HZE particles in the form of iron ions in the absence or presence of selenomethionine (SeM). A total of 215 genes were differentially regulated 2 h after exposure to a 10-cGy dose of iron-ion radiation. In the microarray analysis, SeM had profound effects on the radiation-induced expression of several specific genes, which includes PLAU, IGFBP3, FOLR1, B4GALT1 and COL1A1. Of particular interest to us was a gene cluster, “secreted proteins”, that was up-regulated after radiation exposure. Seven up-regulated genes of this gene cluster fall within the chemokine/cytokine gene cluster, namely, CXCL1, CXCL2, IL6, IL11, IL8, IL24 and TGFβ2. In microarray studies, the radiation-induced up-regulated expression of some these genes encoding cytokine/chemokine proteins was significantly decreased by SeM treatment. For IL8, TGFβ2, CXCL1 and CXCL2, these observations were validated by qPCR techniques. It is concluded that SeM can regulate ionizing radiation-induced gene expression and may serve as an effective countermeasure for some of the acute inflammatory/immune responses induced by low-dose HZE-particle radiation.
INTRODUCTION
It is assumed that ionizing radiation will be capable of causing adverse biological effects in astronauts during deep space travel. The risks may include carcinogenesis, cataracts and damage to the hematopoietic and central nervous systems. Evaluating and reducing these risks and identifying the underlying mechanism(s) involved in the causation of these effects at the cellular and molecular levels are the overall aims of our studies.
One type of space radiation includes high-atomic number (Z) and energy (HZE) particles, which are part of galactic cosmic radiation. The radiation from HZE particles is high-linear energy transfer (LET) radiation and therefore is likely to have greater potential for producing radiation-induced damage to biological tissues compared to low-LET space radiations. HZE particles transfer their energy to biological organisms through high-density ionizations and excitations along the particle track. The expected dose is non-uniform, and it is known that HZE-particle radiation can induce complex DNA damage, mutagenesis and carcinogenesis. Exposure to HZE-particle radiation does not occur on Earth and is therefore difficult to study; however, radiobiology investigations using HZE particles are possible at the NASA Space Research Laboratory (NSRL) at the Brookhaven National Laboratory, where beams of different HZE particles are produced and the biological effects can be evaluated in animal and tissue culture models.
Numerous studies in animal model systems have demonstrated cancer chemopreventive effects with selenium (1). Human studies have also suggested that selenium compounds may be able to prevent cancer (2–4). l-Selenomethionine (SeM) is a major component of dietary selenium that upon trans-sulferation is converted to selenocysteine, which is then incorporated into selenoproteins, including redox-active thioredoxin reductase and glutathione peroxidase (5). Previously, studies performed in our laboratory have demonstrated that supplementation with SeM protects against the following: (1) HZE-particle radiation-induced cell death in human breast epithelial cells (MCF-10) (6); (2) radiation-induced oxidative stress in vitro (6) and in vivo (7–9) (measured as changes in total antioxidant status in rat or mouse plasma or serum); and (3) radiation-induced transformation of human thyroid cells (6). These studies used cells of the HTori-3 cell line, which has been immortalized but is not tumorigenic (10). Because the gene profiles of astronauts exposed to galactic cosmic radiation are difficult to obtain, ground-based in vitro models simulating the space environment are studied using techniques such as cDNA microarray, with confirmation by qPCR.
Characterizing gene expression profiles after exposure to HZE-particle radiation provides a platform for detecting surrogate end-point biomarkers and potential countermeasures for adverse biological effects. To date, there are few reports available on HZE-particle radiation-induced changes in gene expression patterns. To investigate the distinct molecular biological effects of heavy ions at very low doses and to determine the effects of these low-fluence particles that astronauts encounter in the space radiation environment, we carried out cDNA microarray experiments using cells of a nontumorigenic human thyroid epithelial cell line. Additional experiments were conducted to determine gene expression profiles of irradiated cells in response to treatment with SeM, which was evaluated as a potential countermeasure for space radiation-induced biological effects. For these studies, a dose of 10 cGy was used; a previous study had shown, using cell survival analysis, that 10 cGy iron-ion radiation is non-toxic for HTori-3 cells (11).
MATERIALS AND METHODS
Cell Culture and Radiation Exposure
HTori-3 cells were maintained in T-25 tissue culture flasks for each condition for 3 days prior to irradiation in complete Dulbecco’s modified Eagle’s medium (DMEM). For each group, three biological replicates were prepared, representing three separate experiments. Twenty-four hours prior to radiation exposure, cells were exposed to medium supplemented or not with 5 µM SeM.
Cell exposure to HZE particles was performed at the NASA Space Radiation Laboratory (NSRL) Facility at the Brookhaven National Laboratory (BNL), Upton, NY. The measured doses were 0 or 10 cGy of radiation from 1.0 GeV/nucleon iron ions.
RNA Isolation
Cells were harvested and pelleted 2 h postirradiation and immediately flash-frozen. Total RNA was isolated using RNeasy Mini Kit (Qiagen, Valencia, CA) following the vendor’s instructions.
cDNA Microarray
Gene chips and cDNA were prepared in-house by the Penn Microarray Facility at the University of Pennsylvania School of Medicine as described previously (12). Briefly, 1 µg of total RNA was converted to first-strand cDNA using Superscript II reverse transcriptase primed by a poly(T) oligomer that incorporated the T7 promoter. Second-strand cDNA synthesis was followed by in vitro transcription for linear amplification of each transcript and incorporation of biotinylated CTP and UTP. The cRNA products were fragmented to 200 nucleotides or less, heated for 5 min, and hybridized for 16 h to Affymetrix U133Av2 microarrays. The microarrays were then washed at low (6× SSPE) and high (100 mM MES, 0.1 M NaCl) stringency and stained with streptavidin-phycoerythrin. Fluorescence was amplified by adding biotinylated anti-streptavidin and an additional aliquot of streptavidin-phycoerythrin stain. A confocal scanner was used to collect the fluorescence signal at 3 µm resolution after excitation at 570 nm. The average signal from two sequential scans was calculated for each microarray feature.
Analysis of GeneChip Data
Affymetrix Microarray Suite 5.0 was used to quantify expression levels for targeted genes; default values provided by Affymetrix were applied to all analysis parameters. Border pixels were removed, and the average intensity of pixels within the 75th percentile was computed for each probe. The average of the lowest 2% of probe intensities occurring in each of 16 microarray sectors was set as background and subtracted from all features in that sector. Probe pairs were scored positive or negative for detection of the targeted sequence by comparing signals from the perfect match and mismatch probe features. The number of probe pairs meeting the default discrimination threshold (tau = 0.015) was used to assign a call of absent, present or marginal for each assayed gene, and a P value was calculated to reflect confidence in the detection call. A weighted mean of probe fluorescence (corrected for nonspecific signal by subtracting the mismatch probe value) was calculated using the one-step Tukey’s biweight estimate. This signal value, a relative measure of the expression level, was computed for each assayed gene. Global scaling was applied to allow comparison of gene signals across multiple microarrays. After exclusion of the highest and lowest 2%, the average total chip signal was calculated and used to determine what scaling factor was required to adjust the chip average to an arbitrary target of 150. All signal values from one microarray were then multiplied by the appropriate scaling factor.
Data analyses of statistical significance were performed using GeneSpring GX 7.3 software (Agilent Technologies, Palo Alto, CA). The median shift normalization to the 75th percentile and baseline transformation using the median of all samples was applied. ANOVA was used to compare the means of each condition (n = 3, representing three separate experiments). Cutoff ratios >1.50 and <−1.5 and P < 0.05 relative to the respective control group were selected for this study. The Benjamini and Hochberg false discovery rate (FDR) was 0.05 for multiple testing corrections.
Annotations of each gene in the GeneSpring software program were retrieved from GenBank, LocusLink and Unigene databases, providing gene symbol and gene ontology biological processes. Functional annotation of genes and clustering of gene sets were determined using the web-based DAVID Functional Annotation Clustering Tool (13, 14). The DAVID suite uses an enrichment analytic algorithm to provide the most relevant Gene Ontology terms associated with a given gene list, providing an EASE score or a modified Fisher’s exact P value.
Quantitative Real-Time RT-PCR
A two-step RT-PCR reaction was employed. cDNA was synthesized according to the protocol of Superscript II First-Strand cDNA Synthesis System (Invitrogen, Carlsbad, CA) using 2 µg total RNA. cDNA was diluted and frozen aliquots were stored at −20°C. The cDNA generated was amplified using Sybr® Advantage® Premix consisting of a full-length Taq™ polymerase with a hot-start Taq antibody and SYBR Green I (Clontech Laboratories, Inc.). All assays were performed on an ABI 7500 FastReal-Time PCR Detection System (Applied Biosystems). Primers were designed based on the relevant human nucleotide sequences as deposited on GenBank. Each primer set span across intron/exon boundaries and the sequence homology of selected oligomers were checked using a NCBI BLAST search to ensure sequence specificity to the target genes. The designed primers were prepared by Integrated DNA Technologies (Coralville, IA).
Sequences were as follows: GAPDH forward primer 5′-GCCACATCGCTCAGACAC-3 and reverse primer 5′-GCCCAATACGACCAAATCC-3′; CXCL1 forward primer 5′-AGTGGCACTGCTGCTCCT-3′ and reverse primer 5′- TGGATGTTCTTGGGGTGAAT-3′; CXCL2 forward primer 5′-CTGCTCCTGCTCCTGGTG-3′ and reverse primer 5′-AGGGTCTGCAAGCACTGG-3′; IL6 forward primer 5′-GATGAGTACAAAAGTCCTGATCCa-3′ and reverse primer 5′- CTGCAGCCACTGGTTCTGT-3′; IL11 forward primer 5′-TGCACAGCTGAGGGACAA-3′ and reverse primer 5′-GCAGCCTTGTCAGCACAC-3′; and IL24 forward primer 5′-GCTGCAGCAGGAGGTTCT-3′ and reverse primer 5′-GCAGGGTGTGGACAAGGTAA-3′; IL8 forward primer 5′-CTGGCCGTGGCTCTCTTG-3′ and reverse primer 5′-CCTTGGCAAAACTGCACCTT-3′; TGFB2 forward primer 5′-CCAAAGGGTACAATGCCAAC-3′ and reverse primer 5′-CAGATGCTTCTGGATTTATGGTATT-3′. A cycle threshold (Ct) was assigned at the beginning of the logarithmic phase of PCR amplification. Data were analyzed by ABI software and gene expression was quantified using the 2−ΔΔCT, 2 (Delta Delta CT) method (15) normalized to the constitutively expressed housekeeping gene GAPDH. Relative changes were generated comparing the nontreated, sham-irradiated controls to the nontreated irradiated cells or the SeM-pretreated, sham-irradiated controls to the SeM pretreated, irradiated cells.
Three separate experiments were performed and results are given as means ± SD. qPCR figures and statistical analysis were constructed/determined using SigmaPlot version 10 software.
RESULTS
cDNA Microarray Analysis
The gene expression profiles of HTori-3 cells were analyzed 2 h after exposure to HZE-particle radiation. Using a cutoff of 1.5-fold or more, the expression levels of 215 genes were differentially regulated after radiation exposure at a dose of 10 cGy. A complete gene list of radiation responsive genes with the respective relative change in expression is included in the Supplementary Information. The gene expression values were determined by comparing the irradiated cells to the appropriate experimental nonirradiated controls.
Microarray data analysis using the DAVID Bioinformatics Database resulted in numerous clusters of genes categorized by the functional annotation of each gene. Examples of differentially expressed genes from the microarray data include “protein binding” with 121 genes in this cluster and an EASE score/P of 6.9 × 10−13, “cell differentiation”, with 46 genes and EASE score/P of 2.7 × 10−13, “transcription regulator activity”, with 33 genes and EASE score/P of 7.2 × 10−7, “protooncogene”, with 10 genes and EASE score/P of 4 × 10−4, “signal transduction, with 59 genes and an EASE score/P of 4.4 × 10−3, “secreted” cluster, with 20 genes and an EASE score/P of 9.4 × 10−3.
In response to physiological stress induced by ionizing radiation, it is expected that the expression of genes involved in pathways regulating the cell cycle or apoptosis may be differentially regulated. According to the Functional Annotation Analysis of DAVID, 2 h after HZE-particle exposure, 29 differentially expressed genes are clustered in the gene class “response to stress” with an EASE score/P = 2.9 ×10−5, 23 genes in the “cell cycle” cluster (EASE score/P = 4.6 ×10−4), and 25 genes in the “programmed cell death” cluster (EASE score/P = 9.0 × 10−6).
DAVID analysis also highlights several signaling pathway clusters. The signaling pathways that are affected by low-dose HZE-particle exposure include the transforming growth factor beta receptor signaling pathway, transmembrane receptor protein serine/threonine kinase signaling pathway, transmembrane receptor protein tyrosine kinase signaling pathway, JAK/STAT signaling pathway, MAPK signaling pathway, GnRH signaling pathway, T-cell receptor signaling pathway, and the TNFR1 signaling pathway.
Differential gene expression was analyzed in irradiated HTori-3 cells in the presence of SeM. Gene expression was compared between pretreated (with SeM), sham-irradiated cells and pretreated, irradiated cells. A total of 185 genes of the 215 genes that were differentially regulated after a 10-cGy dose of HZE-particle radiation responded to short-term SeM exposure. In other words, radiation responses in HTori-3 cells are different in the presence of SeM; specifically, the expression levels of 185 genes were affected when cells were pretreated with SeM and subsequently irradiated. The expression levels of these 185 genes changed from ≥1.5-fold to <1.5-fold or≤−1.5-fold to >−1.5-fold after pretreatment with SeM and radiation exposure. Those genes responding to SeM treatment with their corresponding relative change are included in the Supplementary Information.
Some of the genes encoding “secreted” proteins resulted in gene expression changes in response to treatment with SeM and radiation exposure (Fig. 1). Specifically, within the “secreted” cluster is the cluster named “cytokine/C-X-C” genes. The expression of seven genes in this cluster was up-regulated after exposure to HZE-particle radiation. These genes are transforming growth factor beta 2 (TGFβ2), interleukin 8 (IL8), interleukin 6 (IL6), interleukin 24 (IL24), interleukin 11 (IL11), chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha; CXCL1), and chemokine (C-X-C motif) ligand 2 (CXCL2) (Fig. 1). In response to radiation exposure when the cells were pretreated with SeM, the expression levels of these genes do not reflect the radiation-induced up-regulation observed when SeM was not present (Table I). These data suggest that SeM pretreatment may abrogate radiation-induced up-regulation of cytokine/chemokine gene expression.
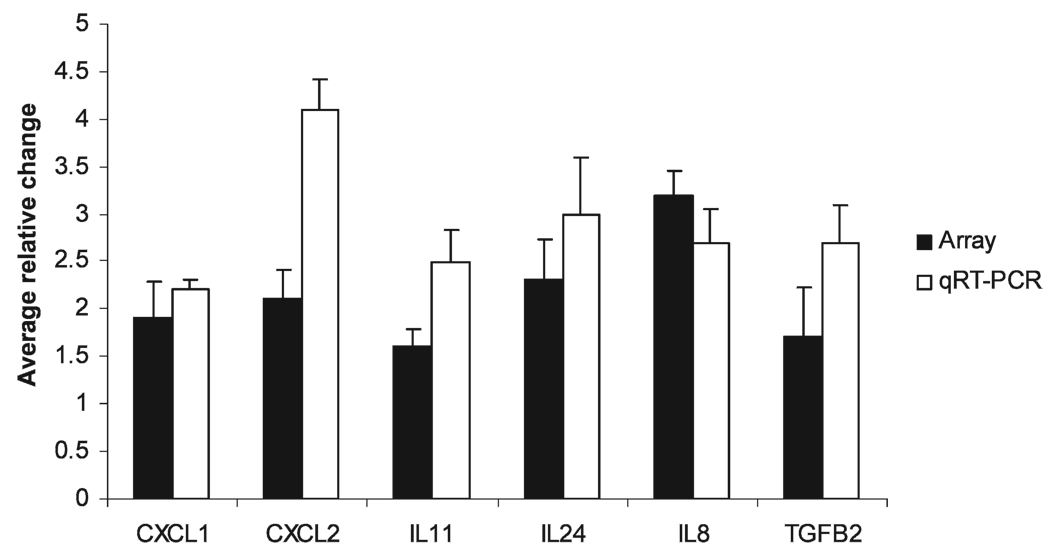
FIG. 1.
Effects of HZE-particle radiation and SeM supplementation in HTori-3 cells. Data generated from the microarray were analyzed with GeneSpring software (Agilent Technologies). Differentially expressed genes exhibiting statistically significant >1.5-fold changes or <−1.5-fold changes with P < 0.05 (determined by ANOVA, with the Benjamini and Hochberg’s FDR correction procedure set at the P-value cutoff of 0.05) relative to the respective control group were classified into functional networks. Expression of 19 genes encoding “secreted” proteins is shown in the absence and presence of SeM and is shown as mean relative change ± SD.
TABLE 1.
Effects of SeM Supplementation on Radiation-Induced Cytokine/Chemokine Gene Expression, Determined by Microarray Analysis
| Gene name | Abbreviated gene name | Accession number | Average relative change in response to radiation |
Average relative change in response to SeM + radiation |
|---|---|---|---|---|
| Chemokine (C-X-C motif) ligand 1 | CXCL1 | NM_001511 | 1.9 | 1.0 |
| Chemokine (C-X-C motif) ligand 2 | CXCL2 | NM_002089 | 2.1 | 1.1 |
| Interleukin 11 | IL11 | NM_000641 | 1.6 | 1.2 |
| Interleukin 24 | IL24 | NM_006850 | 2.3 | 1.4 |
| Interleukin 6 | IL6 | NM_000600 | 1.5 | 1.1 |
| Interleukin 8 | IL8 | NM_000584 | 3.2 | 1.3 |
| Transforming growth factor beta 2 | TGFB2 | NM_001135599 | 1.7 | 1.2 |
Other genes in Fig. 1 show remarkable responses to radiation in the presence of SeM. For example, plasminogen activator, urokinase (PLAU), insulin-like growth factor binding protein 3 (IGFBP3), folate receptor 1 (FOLR1), glycoprotein-4-beta-galactosyltransferase 2 (B4GALT1), and collagen, type I, alpha 1 (COL1A1) show complete reversal of radiation-induced gene expression changes only in the presence of SeM.
Validation of Microarray Results by Quantitative RT-PCR
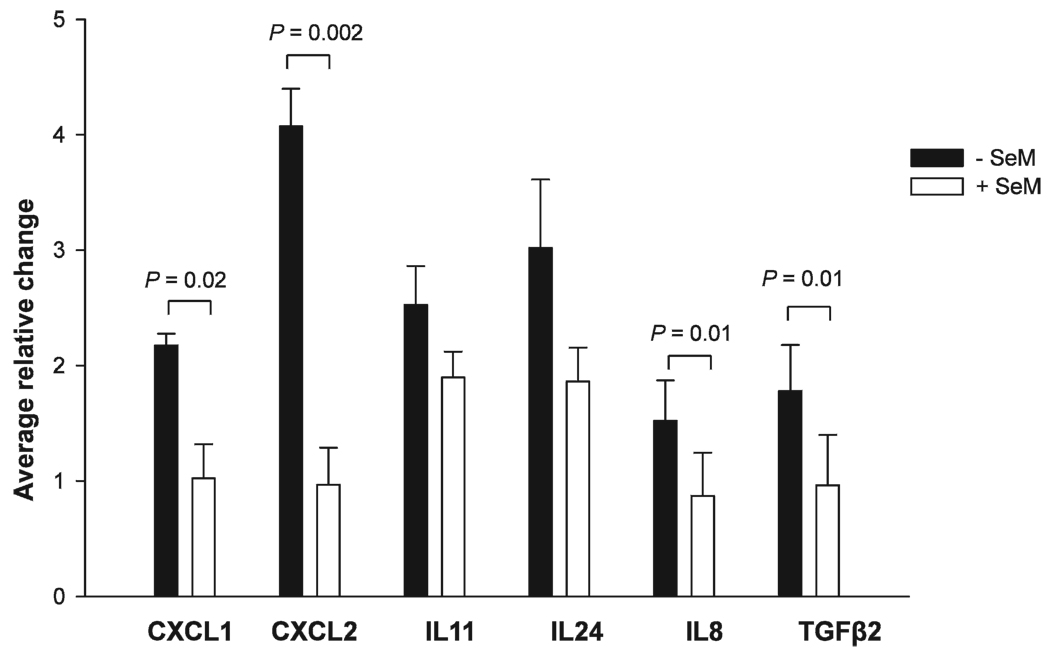
To confirm regulation of gene expression induced by iron-ion radiation, qPCR validation was performed. Altered mRNA expression of the inflammatory cytokine/chemokine genes was chosen for qPCR validation because this group of genes displayed up-regulated expression in the presence of radiation and down-regulated expression with SeM supplementation (compared to radiation alone) in a consistent manner in the microarray analyses. The average qPCR relative induction resulting from three independent experiments was determined by the Delta Delta CT method (15), normalized to GAPDH. Radiation exposure resulted in an up-regulation of CXCL1 (2.2-fold), CXCL2 (4.1-fold), IL8 (2.7-fold), IL11 (2.5-fold), IL24 (3.0-fold), and TGFβ2 (2.7-fold) compared to sham-irradiated controls, as determined by qPCR. The results of the qRT-PCR experiments are in reasonable agreement with the microarray experiments for six of the seven genes identified from the microarray results (Fig. 2). The observed HZE-particle radiation-induced up-regulation of IL6 (from the microarray results) was not reproducible by qPCR. However, consistent with the microarray data, supplementation with SeM decreased radiation-induced expression of CXCL1, CXCL2, IL24, IL11, IL8 and TGFβ2 (Fig. 3); statistically significant reductions were observed for CXCL1, CXCL2, IL8 and TGFβ2. Thus supplementation with SeM reduces the radiation-induced increases in expression levels of most of the genes encoding chemokine/cytokine proteins. Supplementation with SeM in nonirradiated cells resulted in a slight reduction of IL8 mRNA levels that was not statistically significant (data not shown).
FIG. 2.
Comparison of the HZE-particle radiation-induced up-regulated gene expression patterns in HTori-3 cells by microarray analysis and qRT-PCR. Data shown represent statistically significant gene expression results from the microarray experiments (black bars) and the qRT-PCR experiments (gray bars).
FIG. 3.
Effects of SeM supplementation on gene expression profiles in HZE-particle-irradiated HTori-3 cells. Gene expression was quantified by qRT-PCR and data are normalized to the reference gene GAPDH. Changes in gene expression are normalized to the sham-irradiated controls. Analyses to determine whether there were statistically significant differences between the untreated (− SeM) and pretreated (+ SeM) irradiated groups were performed, and P values were determined by the Student’s t test. P values are indicated for the comparisons exhibiting statistically significant differences between treatment groups.
DISCUSSION
In these studies we observed that low-dose, HZE-particle radiation exposure affects global gene expression patterns in non-tumorigenic human epithelial cells. We used cDNA microarray analyses to investigate gene expression patterns after exposure to HZE-particle radiation in HTori-3 cells to determine whether SeM affects the biological responses to radiation. HTori-3 cells cultured in the absence or presence of SeM were exposed to a low dose (10 cGy) of iron-ion radiation and then harvested 2 h after exposure. We designated changes in gene expression as statistically significant alterations at levels of 1.5-fold or more; numerous genes were found to be differentially regulated after irradiation. The microarray results indicated that SeM had profound effects on the radiation-induced expression of several specific genes, including PLAU, IGFBP3, FOLR1, B4GALT1 and COL1A1. Of particular interest to us was the effect of SeM on the genes of the “secreted proteins” gene cluster. The microarray results indicated that iron-ion exposure results in the up-regulation of seven genes representing cytokines/chemokines and the qPCR analysis confirmed these results for six out of seven of these genes (CXCL1, CXCL2, IL8, IL11, IL24 and TGFβ2). The microarray analysis indicated that SeM pretreatment of the irradiated cells reduced the up-regulated gene expression levels of all seven genes in this cluster; the qPCR analysis confirmed the microarray results for four of these genes (CXCL1, CXCL2, IL8 and TGFβ2). Thus our results suggest that exposure to iron-ion radiation results in increased levels of genes in the chemokine/cytokine gene cluster and that SeM pretreatment of irradiated cells can potentially mitigate HZE-particle radiation-induced increases in the expression patterns of some of the genes.
Ionizing radiation produces a cascade of chemokines and cytokines that activate and modulate a number of signaling pathways and cellular functions (16–19). These secreted proteins additionally play a vital role in the cancer microenvironment and in tumor pathogenesis and progression. We found that CXCL1, CXCL2 and IL6 were up-regulated in human epithelial cells exposed to 10 cGy HZE-particle radiation 2 h postirradiation; these transcripts were also up-regulated in normal human fibroblasts in response to a low dose of X rays (1 cGy) 1 h after exposure (20).
We also observed an up-regulation of IL11, IL24, IL8 and TGFβ2 in human epithelial cells after exposure to HZE-particle radiation that is mitigated by SeM pretreatment. IL11 is a known inducer of proliferation in a number of cancers, including colorectal, gastric, prostate and ovarian cancers (21–24); thus a suppression of expression of IL11 by SeM can be viewed as a potential anti-carcinogenic effect. IL24 is distinct from other interleukin cytokines in that it has proven to be an effective tumor suppressor (25, 26). This is the first report of increased IL24 transcript levels after exposure to ionizing radiation, and therefore the role of radiation-induced IL24 transcription needs further investigation. It is noteworthy that IL6, IL11 and IL24 are all activators of the Janus kinase/signal transducers and activators of a transcription (JAK/STAT) pathway, which regulates principal cell fate decisions involving processes such as cell proliferation, differentiation and apoptosis. From the microarray analysis, some additional genes that are involved in the JAK/STAT pathway showed differential gene expression patterns as a result of exposure to low-dose HZE-particle radiation, including IFNGR1 (interferon gamma receptor 1) and STAT2 (signal transducer and activator of transcription 2). These data suggest that low-dose exposure to iron-ion radiation affect the regulation of transcription, and particularly that involved in the JAK/STAT pathway.
We verified gene expression changes for two central cytokines, IL8 and TGFβ2. We report here that SeM pretreatment in cells abrogates HZE-particle-induced gene transcription of IL8 and TGFβ2, as indicated by microarray analysis and verified by qPCR. High-LET radiation, in the form of α particles, was shown by Narayanan et al. (27) to increase IL8 mRNA levels; in these studies, secreted IL8 levels were increased in parallel with elevated production of reactive oxygen species (ROS). Those authors also showed inhibition of IL8 levels by treatment with antioxidants. TGFβ is a widely studied cytokine that is expressed in a variety of tissues. Studies by Iyer et al. (28) suggest that α-particle radiation-induced TGFβ production is linked with increased intracellular ROS in human fibroblasts. If indeed elevated ROS levels contribute to high-LET radiation-induced increases in the transcript levels of IL8 and TGFβ, this would suggest a potential mechanism by which SeM treatment could reduce the radiation-induced transcript levels of IL8 and TGFβ in the studies reported here. As noted in the Introduction, SeM is expected to protect against free radical-induced biological effects, because it is converted to selenocysteine and then incorporated into the well-established antioxidant enzymes thioredoxin reductase and glutathione peroxidase (5). In the experiments described by Iyer et al. (28), when neutralizing TGFβ antibodies were added to the supernatants of previously irradiated cells and transferred to nonirradiated cells, the ROS by-stander response was completely inhibited (28). It has been hypothesized that TGFβ plays an important role in radiation-induced bystander effects (28, 29). Other cytokines thought to play a role in radiation-induced bystander effects include IL8, IL2 and tumor necrosis factor alpha (TNFα) (27, 30). In our studies, we did not observe radiation-induced regulation of the IL2 gene or TNFα. Other genes that are members of the TNF superfamily, however, did show differential expression 2 h after iron-ion exposure: MAP3K1 (mitogen-activated protein kinase kinase kinase 1; down-regulated 1.5-fold), JUN (v-jun sarcoma virus 17 oncogene homolog; up-regulated 1.7-fold), CASP2 (caspase 2, apoptosis-related cysteine peptidase; down-regulated 2.1-fold), BAG4 (silencer of death domains; down-regulated 1.7-fold), TNFAIP3 (tumor necrosis factor inducible protein 3; up-regulated 1.5-fold) and TNFSF9 (tumor necrosis factor soluble factor 9; down-regulated 1.5-fold).
We found that low-dose HZE-particle radiation induces differential gene expression in human thyroid epithelial cells and that pretreatment with SeM potentially prevents or attenuates some of the observed changes. The results of the microarray analyses reported here provide a basis of knowledge about the changes in gene expression that might be expected from cellular exposure to low doses of high-LET radiation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ms. Ying-Hui Ko for technical assistance in the qPCR experiments, Dr. Jeffrey Ware for assistance with cell irradiation, and the staff at BNL for facilitating the radiation work. This research was supported by NASA Grant NAG9-1517 and NIH Training Grant 2T32CA009677.
Footnotes
SUPPLEMENTARY INFORMATION
Complete list of genes that were differentially expressed 2 h after exposure to low-dose HZE-particle radiation (10 cGy) in the absence and presence of l-selenomethionine (SeM). http://dx.doi.org/10.1667/RR1363.1.S1
REFERENCES
- 1.Greeder GA, Milner JA. Factors influencing the inhibitory effect of selenium on mice inoculated with Ehrlich ascites tumor cells. Science. 1980;209:825–827. doi: 10.1126/science.7406957. [DOI] [PubMed] [Google Scholar]
- 2.Medina D, Thompson H, Ganther H, Ip C. Semethylselenocysteine: a new compound for chemoprevention of breast cancer. Nutr. Cancer. 2001;40:12–17. doi: 10.1207/S15327914NC401_5. [DOI] [PubMed] [Google Scholar]
- 3.Krishnan K, Campbell S, Abdel-Rahman F, Whaley S, Stone WL. Cancer chemoprevention drug targets. Curr. Drug Targets. 2003;4:45–54. doi: 10.2174/1389450033347028. [DOI] [PubMed] [Google Scholar]
- 4.Klein EA, Lippman SM, Thompson IM, Goodman PJ, Albanes D, Taylor PR, Coltman C. The selenium and vitamin E cancer prevention trial. World J. Urol. 2003;21:21–27. doi: 10.1007/s00345-002-0314-z. [DOI] [PubMed] [Google Scholar]
- 5.Allan CB, Lacourciere GM, Stadtman TC. Responsiveness of selenoproteins to dietary selenium. Annu. Rev. Nutr. 1999;19:1–16. doi: 10.1146/annurev.nutr.19.1.1. [DOI] [PubMed] [Google Scholar]
- 6.Kennedy AR, Ware JH, Guan J, Donahue JJ, Biaglow JE, Zhou Z, Stewart J, Vazquez M, Wan XS. Selenomethionine protects against adverse biological effects induced by space radiation. Free Radic. Biol. Med. 2004;36:259–266. doi: 10.1016/j.freeradbiomed.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 7.Guan J, Wan XS, Zhou Z, Ware J, Donahue JJ, Biaglow JE, Kennedy AR. Effects of dietary supplements on space radiation-induced oxidative stress in Sprague-Dawley rats. Radiat. Res. 2004;162:572–579. doi: 10.1667/rr3249. [DOI] [PubMed] [Google Scholar]
- 8.Guan J, Stewart J, Ware JH, Zhou Z, Donahue JJ, Kennedy AR. Effects of dietary supplements on the space radiation-induced reduction in total antioxidant status in CBA mice. Radiat. Res. 2006;165:373–378. doi: 10.1667/rr3523.1. [DOI] [PubMed] [Google Scholar]
- 9.Kennedy AR, Guan J, Ware JH. Countermeasures against space radiation induced oxidative stress in mice. Radiat. Environ. Biophys. 2007;46:201–203. doi: 10.1007/s00411-007-0105-4. [DOI] [PubMed] [Google Scholar]
- 10.Lemoine NR, Mayall ES, Jones T, Sheer D, McDermid S, Kendall-Taylor P, Wynford-Thomas D. Characterisation of human thyroid epithelial cells immortalised in vitro by simian virus 40 DNA transfection. Br. J. Cancer. 1989;60:897–903. doi: 10.1038/bjc.1989.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kennedy AR, Zhou Z, Donahue JJ, Ware JH. Protection against adverse biological effects induced by space radiation by the Bowman-Birk inhibitor and antioxidants. Radiat. Res. 2006;166:327–332. doi: 10.1667/RR3599.1. [DOI] [PubMed] [Google Scholar]
- 12.Stewart J, Ware J, Fortina P, Breaux J, Gulati S, Kennedy A. l-Selenomethionine modulates high LET radiation-induced alterations of gene expression in cultured human thyroid cells. Oncol. Rep. 2006;16:569–574. [PubMed] [Google Scholar]
- 13.Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 14.Hosack DA, Dennis G, Jr., Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Okunieff P, Xu J, Hu D, Liu W, Zhang L, Morrow G, Pentland A, Ryan JL, Ding I. Curcumin protects against radiation-induced acute and chronic cutaneous toxicity in mice and decreases mRNA expression of inflammatory and fibrogenic cytokines. Int. J. Radiat. Oncol. Biol. Phys. 2006;65:890–898. doi: 10.1016/j.ijrobp.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 17.Derradji H, Bekaert S, De Meyer T, Jacquet P, Abou-El-Ardat K, Ghardi M, Arlette M, Baatout S. Ionizing radiation-induced gene modulations, cytokine content changes and telomere shortening in mouse fetuses exhibiting forelimb defects. Dev. Biol. 2008;322:302–313. doi: 10.1016/j.ydbio.2008.07.032. [DOI] [PubMed] [Google Scholar]
- 18.Muller K, Meineke V. Radiation-induced alterations in cytokine production by skin cells. Exp. Hematol. 2007;35:96–104. doi: 10.1016/j.exphem.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 19.Neta R. Modulation with cytokines of radiation injury: suggested mechanisms of action. Environ. Health Perspect. 1997;105 Suppl. 6:1463–1465. doi: 10.1289/ehp.97105s61463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujimori A, Okayasu R, Ishihara H, Yoshida S, Eguchi-Kasai K, Nojima K, Ebisawa S, Takahashi S. Extremely low dose ionizing radiation up-regulates CXC chemokines in normal human fibroblasts. Cancer Res. 2005;65:10159–10163. doi: 10.1158/0008-5472.CAN-05-2015. [DOI] [PubMed] [Google Scholar]
- 21.Yamazumi K, Nakayama T, Kusaba T, Wen CY, Yoshizaki A, Yakata Y, Nagayasu T, Sekine I. Expression of interleukin-11 and interleukin-11 receptor alpha in human colorectal adenocarcinoma; immunohistochemical analyses and correlation with clinicopathological factors. World J. Gastroenterol. 2006;12:317–321. doi: 10.3748/wjg.v12.i2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson CB, Judd LM, Menheniott TR, Kronborg I, Dow C, Yeomans ND, Boussioutas A, Robb L, Giraud AS. Augmented gp130-mediated cytokine signalling accompanies human gastric cancer progression. J. Pathol. 2007;213:140–151. doi: 10.1002/path.2218. [DOI] [PubMed] [Google Scholar]
- 23.Campbell CL, Jiang Z, Savarese DM, Savarese TM. Increased expression of the interleukin-11 receptor and evidence of STAT3 activation in prostate carcinoma. Am. J. Pathol. 2001;158:25–32. doi: 10.1016/S0002-9440(10)63940-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campbell CL, Guardiani R, Ollari C, Nelson BE, Quesenberry PJ, Savarese TM. Interleukin-11 receptor expression in primary ovarian carcinomas. Gynecol. Oncol. 2001;80:121–127. doi: 10.1006/gyno.2000.6064. [DOI] [PubMed] [Google Scholar]
- 25.Sieger KA, Mhashilkar AM, Stewart A, Sutton RB, Strube RW, Chen SY, Pataer A, Swisher SG, Grimm EA, Chada S. The tumor suppressor activity of MDA-7/IL-24 is mediated by intracellular protein expression in NSCLC cells. Mol. Ther. 2004;9:355–367. doi: 10.1016/j.ymthe.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 26.Mhashilkar AM, Schrock RD, Hindi M, Liao J, Sieger K, Kourouma F, Zou-Yang XH, Onishi E, Takh O, Chada S. Melanoma differentiation associated gene-7 (mda-7): a novel anti-tumor gene for cancer gene therapy. Mol. Med. 2001;7:271–282. [PMC free article] [PubMed] [Google Scholar]
- 27.Narayanan PK, LaRue KE, Goodwin EH, Lehnert BE. Alpha particles induce the production of interleukin-8 by human cells. Radiat. Res. 1999;152:57–63. [PubMed] [Google Scholar]
- 28.Iyer R, Lehnert BE, Svensson R. Factors underlying the cell growth-related bystander responses to alpha particles. Cancer Res. 2000;60:1290–1298. [PubMed] [Google Scholar]
- 29.Barcellos-Hoff MH, Brooks AL. Extracellular signaling through the microenvironment: a hypothesis relating carcinogenesis, bystander effects and genomic instability. Radiat. Res. 2001;156:618–627. doi: 10.1667/0033-7587(2001)156[0618:esttma]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 30.Facoetti A, Ballarini F, Cherubini R, Gerardi S, Nano R, Ottolenghi A, Prise KM, Trott KR, Zilio C. Gamma ray-induced bystander effect in tumour glioblastoma cells: a specific study on cell survival, cytokine release and cytokine receptors. Radiat. Prot. Dosimetry. 2006;122:271–274. doi: 10.1093/rpd/ncl431. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.