Abstract
Our previous efforts have used in vitro selection to identify numerous Zn2+-dependent deoxyribozymes that ligate two RNA substrates with reaction at a 2′,3′-cyclic phosphate. Each deoxyribozyme creates one of several different RNA linkages, including native 3′–5′ and non-native 2′–5′ phosphodiester bonds as well as many unnatural linkages. In this report, we describe experiments to reveal design aspects of the selection strategy that favor site-selective and regioselective synthesis of native 3′–5′ RNA linkages. The results also reveal that an explicit selection pressure for RNA substrate sequence generality must be developed if the deoxyribozymes are to have practical generality.
Introduction
We previously established an in vitro selection strategy to identify deoxyribozymes (DNA enzymes)1,2 that ligate two RNA substrates.3 These substrates are one of two readily obtained combinations: 2′,3′-cyclic phosphate with 5′-hydroxyl (Scheme 1),3–5 or 2′,3′-diol with 5′-triphosphate.6–8 For both substrate combinations, multiple ligation junctions are possible, leading to several selectivity considerations. Through the experiments reported here, we sought to understand how to control both site-selectivity and regioselectivity in the deoxyribozyme-mediated RNA ligation reaction involving a 2′,3′-cyclic phosphate substrate. Our findings indicate that several subtle aspects of the selection design are important to providing native 3′–5′ RNA linkages in the ligation products.
Scheme 1.
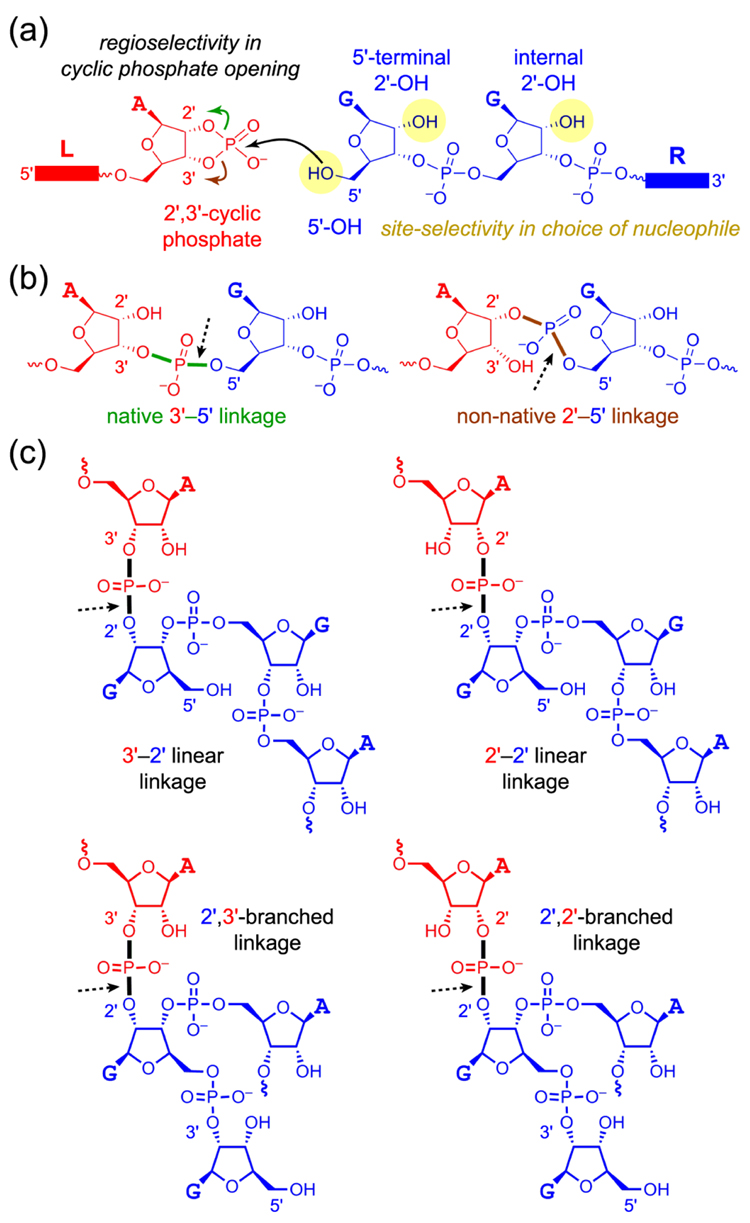
Potential DNA-catalyzed ligation reactions of a 2′,3′-cyclic phosphate RNA substrate. (a) The two RNA reaction partners. The strand providing the cyclic phosphate is designated as the ‘left-hand’ (L) substrate (red), and the strand providing the –OH nucleophile is designated as the ‘right-hand’ (R) substrate (blue). The 2′,3′-cyclic phosphate can be opened in either of two ways (green and brown arrows); controlling the regioselectivity of this opening is one main focus of this study. In addition, three potential –OH group nucleophiles are highlighted (yellow) on the R substrate; controlling the site-selectivity among these competing nucleophiles is another main focus of this study. (b) Nucleophilic attack by the 5′-OH of R with the 2′-O of L acting as the leaving group leads to the native 3′–5′ RNA linkage. When the 3′-O of L is instead the leaving group, the non-native 2′–5′ RNA linkage is formed. In each structure, the dashed arrow marks the bond formed between the 5′-OH and the P atom. (c) Various unnatural linkages are formed when either the 5′-terminal 2′-OH or the internal 2′-OH of R attacks the cyclic phosphate.
Our first report on RNA-ligating deoxyribozymes that use a 2′,3′-cyclic phosphate substrate revealed only formation of non-native 2′–5′ linkages rather than native 3′–5′ linkages.3 Subsequent experiments showed that the non-native connectivity is favored in several different contexts, for mechanistic reasons that remain unclear.4,5 All of these early findings used Mg2+ as the metal ion cofactor for the DNA enzymes. We later discovered that using Zn2+ in place of Mg2+ leads to deoxyribozymes that collectively create the various ligation products of Scheme 1, including formation of native 3′–5′ linkages (Scheme 1b).9 Such linkages are desirable for practical RNA ligation because no evidence remains in the RNA product that a ligation event had taken place.
In all of these cases, each deoxyribozyme creates only one particular RNA linkage and is therefore both site-selective and regioselective. The direction of site-selectivity for an individual deoxyribozyme is controlled by which functional group acts as the attacking nucleophile: the 5′-OH, the 2′-OH of the 5′-terminal nucleotide, or an internal 2′-OH of the second substrate. The direction of regioselectivity is determined by which O atom of the cyclic phosphate, 2′ or 3′, acts as the leaving group. During in vitro selection of our previously reported Zn2+-dependent deoxyribozymes, the directions of both site-selectivity and regioselectivity were controlled poorly, because examples were found of individual deoxyribozymes that collectively form at least five out of the 2 × 3 = 6 possible ligation products. This includes all four of the linkages formed by nucleophilic attack of either the 5′-OH or 2′-OH of the 5′-terminal nucleotide combined with both possible openings of the 2′,3′-cyclic phosphate.†
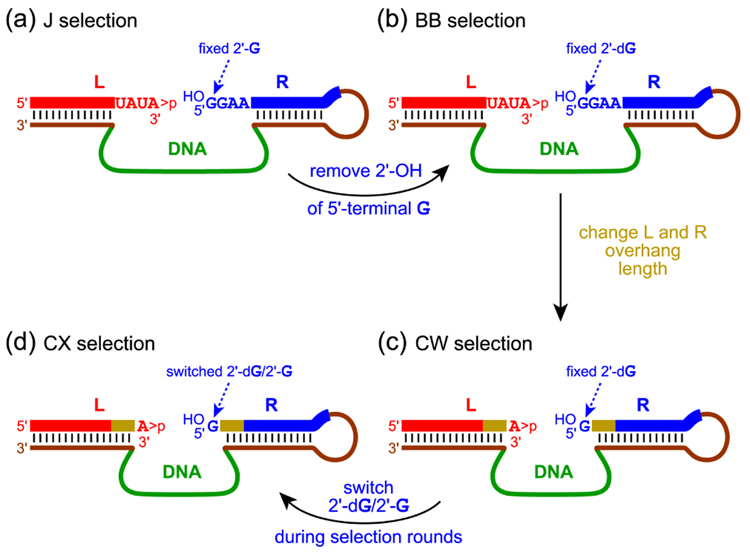
These Zn2+-dependent deoxyribozymes were identified using two selection strategies whose key steps are shown in Fig. 1a and 1b.9 These selections were designated as ‘J’ and ‘BB’ according to our laboratory’s ongoing alphabetical nomenclature. Two key aspects of the J selection design were as follows. (1) The presence of four-nucleotide RNA overhangs on both sides of the ligation junction, beyond which DNA:RNA base pairs were formed between the fixed deoxyribozyme binding arms and the RNA substrates. (2) An unmodified ‘right-hand’ (R) RNA substrate that reacts with the 2′,3′-cyclic phosphate of the ‘left-hand’ (L) RNA substrate. The J selection led to deoxyribozymes that use only the 5′-terminal guanosine 2′-OH group as the nucleophile, with two unnatural ligation products formed via both possible cyclic phosphate opening reactions (i.e., linear 3′–2′ and 2′–2′ linkages).† Thus, the J selection was site-selective (albeit for the undesired 5′-terminal 2′-OH nucleophile) but not regioselective.
Fig. 1.
Selection designs for deoxyribozyme-catalyzed RNA ligation using a 2′,3′-cyclic phosphate RNA substrate. In each case, the two RNA substrates (L and R) and the deoxyribozyme are shown poised for the key selection step (see Fig. 2). (a) and (b) The previously reported J and BB selections, in which four overhanging RNA nucleotides were present on both the L and R substrates.9 The 5′-terminal nucleotide of R was fixed as either G with a 2′-OH (2′-G; J selection) or G with a 2′-H (2′-dG; BB selection). (c) The new CW selection, which is the same as BB except only one overhanging RNA nucleotide was present on both L and R. (d) The new CX selection, which differs from BB via both the overhang length and in switching of 2′-dG/2′-G in various selection rounds.
Because both of the products formed by deoxyribozymes from the J selection were unnatural RNA linkages, we changed the selection strategy in an attempt to favor formation of native 3′–5′ linkages. The ensuing BB selection was identical to the J selection except that the R substrate had a 2′-deoxy-G nucleotide at its 5′-terminus rather than a standard G ribonucleotide (i.e., 2′-dG rather than 2′-G). This change was made to prevent emergence of deoxyribozymes that require nucleophilic reactivity of the 5′-terminal 2′-OH group of R (as was observed in the J selection), because this functional group was consistently absent during the BB selection process. Indeed, the BB selection led to deoxyribozymes that use either the 5′-terminal 5′-OH or the internal 2′-OH of the second R nucleotide (but not the 5′-terminal 2′-OH) as the nucleophile, again with both cyclic phosphate openings.† Although one particular product among this collection was the desired native 3′–5′ linkage, overall the redesigned selection experiment did not control either the site-selectivity or the regioselectivity of the RNA ligation process.
In the present work, we explored how the selection design can be further modified to control the direction of site-selectivity and regioselectivity during DNA-catalyzed RNA ligation. We focused on strategically changing two particular aspects of the J/BB selection design. First, in the new CW 55 selection (Fig. 1c) we altered the RNA overhang length from four nucleotides to only one nucleotide. The reduced overhang length should help to suppress nucleophilic reactivity by the internal 2′-OH of R, because this nucleotide is now physically constrained within a DNA:RNA Watson-Crick base pair; this change should therefore contribute to controlling the site-selectivity of ligation. Another benefit of reducing the RNA overhang length on both substrates is an anticipated increase in RNA substrate sequence generality of the resulting deoxyribozymes, because we have observed that long single-stranded regions of the RNA substrates often lead to deoxyribozymes that have undesired requirements for specific RNA nucleotides in these non-duplex regions.3,6,9–11 RNA substrate generality is an important practical feature of useful RNA-ligating deoxyribozymes.
The second altered aspect of the selection design was to repeatedly switch the 5′-terminal nucleotide of the R substrate between 2′-dG and 2′-G during the iterated rounds of the new CX selection (Fig. 1d). By switching this nucleotide, we expected to continue to suppress nucleophilic reactivity of the corresponding 5′-terminal 2′-OH of R. Importantly, we also expected this switching to provide a direct pressure for the resulting deoxyribozymes to retain substantial ligation activity when the 2′-OH is present on the RNA substrate. The previous BB deoxyribozymes, which were identified by selection with the fixed 2′-dG substrate, catalyzed RNA ligation relatively poorly with the 2′-G substrate when compared with the 2′-dG substrate.9
By assaying a collection of the Zn2+-dependent deoxyribozymes that emerged from both of the new CW and CX selection experiments, we determined how both altered aspects of the selection design influenced the site-selectivity and regioselectivity of DNA-catalyzed RNA ligation. We were pleased to find that changing both design aspects simultaneously (in the CX selection) led to control of both site-selectivity and regioselectivity, because nearly all of the resulting deoxyribozymes create solely native 3′–5′ RNA linkages. We also examined the RNA substrate sequence generality of deoxyribozymes that form native 3′–5′ linkages from both new selections. The observation of nontrivial sequence requirements near the ligation junction suggests that further experiments to obtain highly general deoxyribozymes will require the introduction of a selection pressure specifically designed to induce such generality.
Results
In vitro selection of new deoxyribozymes
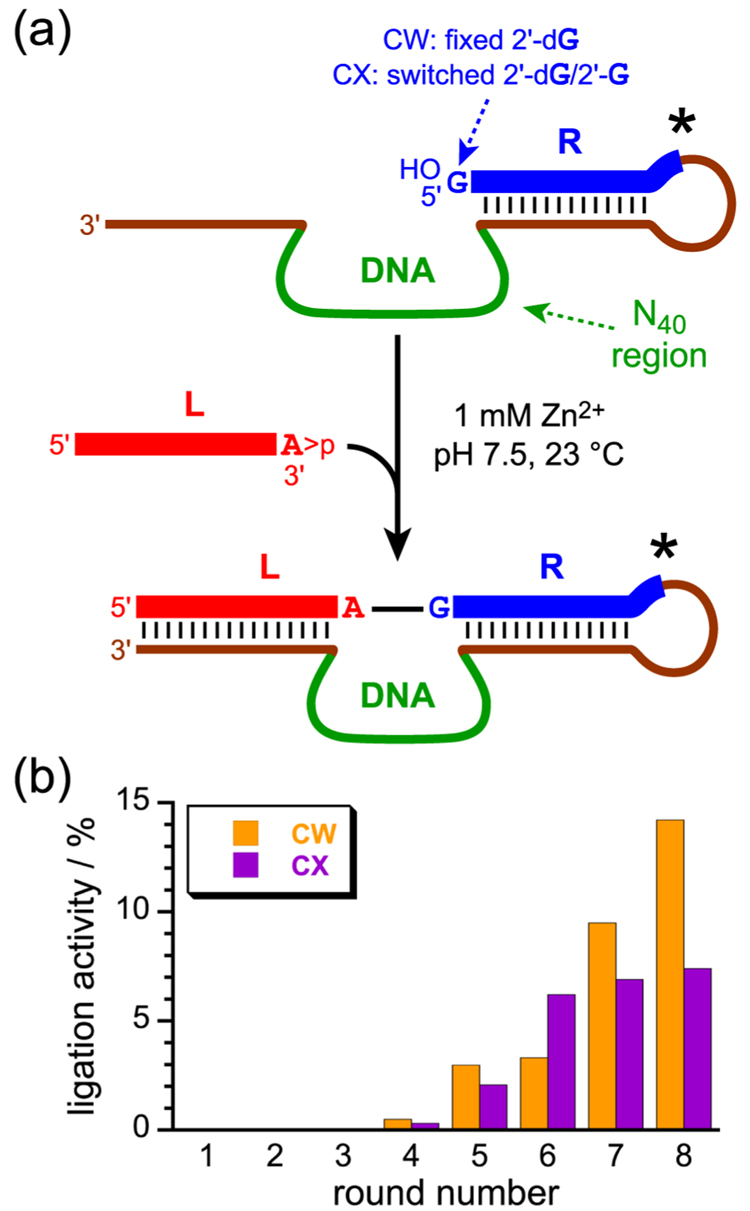
For both of the new selection experiments designated CW and and CX (Fig. 1), the procedure was analogous to that employed previously for the J and BB selections.9 The R substrate was attached covalently to the deoxyribozyme strand using T4 RNA ligase. Then, in the key selection step (Fig. 2a), the L and R substrates were provided the opportunity to become ligated in 70 mM Tris, pH 7.5, 150 mM NaCl, 2 mM KCl, and 1 mM ZnCl2 by incubation for 3 h at 23 °C. Those DNA sequences that successfully joined L and R were separable by 8% polyacrylamide gel electrophoresis (PAGE) due to the pre-existing covalent connection between R and the deoxyribozyme (i.e., L+R+DNA was readily separated from R+DNA). Finally, the DNA pool (now enriched in catalytically active sequences) was regenerated by PCR, and the two single strands were separated by denaturing PAGE; the undesired strand was longer than the desired strand due to the presence of a nonamplifiable spacer within the DNA primer that initiates synthesis of the undesired strand. This process was iterated for eight rounds, with the activity progression shown in Fig. 2b.
Fig. 2.
Key selection step and progression of the new selection experiments. (a) The key selection step, in which the L and R substrates are joined by the action of the deoxyribozyme that is attached to R (asterisk). Note that this covalent attachment is required only during the selection process itself; L and R can be provided to the deoxyribozyme as separate substrates for analysis of ligation activity after the selection process is completed. (b) Ligation activities of the CW and CX selections by round number.
For the CW selection, the only difference from the prior BB selection was that only one overhanging RNA nucleotide (rather than four) was present on both L and R. As in the BB selection, the 2′-dG R substrate was used in all CW rounds. For the CX selection, only one overhanging nucleotide was present on both L and R; in addition, the 5′-terminal nucleotide of the R substrate was switched between 2′-dG and 2′-G in various rounds. The CX selection began in round 1 with the 2′-dG substrate and used this substrate in rounds 2 and 4; the alternative 2′-G substrate was used in rounds 3 and 5–8.
Deoxyribozymes from the CX selection
We first analyzed individual deoxyribozymes from the CX selection, in which both aspects of the selection design (overhang length and switching of 2′-dG/2′-G) were altered relative to the previous BB selection. The ligation activity of the uncloned CX pool was ca. 7% (Fig. 2b), which is relatively low. Nonetheless, we have obtained highly active deoxyribozymes from such pools in many past cases (note that individual DNA enzymes can have much higher ligation yield than the pool average). We cloned deoxyribozymes from round 7CX using standard procedures that were adapted in straightforward fashion from our past selection experiments.3,12 Briefly, these procedures involved (1) insertion of the double-stranded PCR product (purified on agarose gel) into a TA cloning vector; (2) screening of ‘miniprep’ DNA from individual E. coli colonies by restriction digestion to verify the presence of the expected insert; (3) PCR using the miniprep DNA as a template to prepare individual deoxyribozyme clones; and (4) trimolecular RNA ligation assays for each clone using the L and R substrates, with the L substrate bearing a 5′-32P radiolabel.
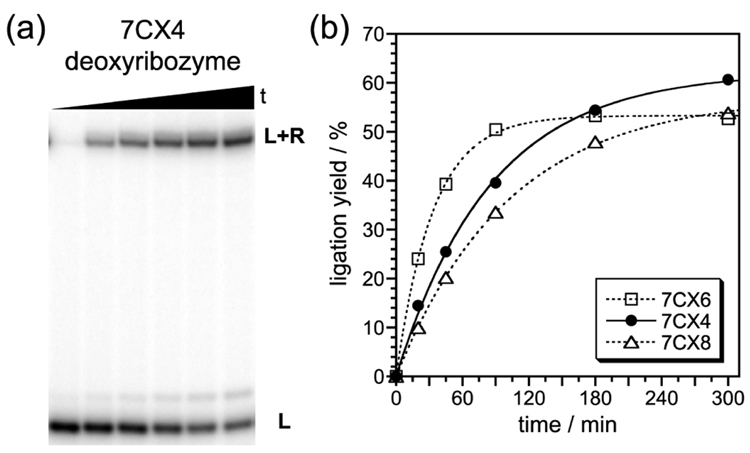
Seven 7CX clones with promising ligation activities were sequenced,† and the deoxyribozymes were prepared independently by solid-phase DNA synthesis. Quantitative RNA ligation assays (Fig. 3) revealed that six out of seven 7CX deoxyribozymes have ligation yield in the 45–60% range, reaching maximal yield with t1/2 of 20–60 min (kobs 0.01–0.03 min−1). Because the ligation reaction of a 2′,3′-cyclic phosphate is inherently reversible, the reverse cleavage reaction can occur, and this likely reduces the net ligation yield below the maximal 100%.
Fig. 3.
Catalytic activities of the 7CX deoxyribozymes. (a) PAGE image using 7CX4 as a representative example. (b) Kinetic plot for 7CX6, 7CX4, and 7CX8 (kobs = 0.030, 0.012, and 0.010 min−1).
Our key interest in the new deoxyribozymes is the junction within their RNA ligation products. Because of the reversible nature of 2′,3′-cyclic phosphate ring opening, the principle of microscopic reversibility enables the rapid determination of the ligation junction formed by each new DNA enzyme via assaying for cleavage of suitably linked RNA ‘product’ strands. For example, any 7CX deoxyribozyme that joins the L and R substrates with formation of a native 3′–5′ RNA linkage should also be observed to cleave the 3′–5′ linkage of a separately synthesized product strand. Similarly, if the same deoxyribozyme does NOT create the non-native 2′–5′ linkage (i.e., is highly selective for 3′–5′ linkage formation), then it should not be observed to cleave the analogous product strand that has a 2′–5′-linkage at the cleavage site. The opposite experimental outcome, cleavage of the 2′–5′ but not 3′–5′ product strand, would indicate a deoxyribozyme that creates 2′–5′ linkages. Finally, cleavage of neither the 3′–5′ nor 2′–5′ product strand would suggest a deoxyribozyme that creates an unnatural RNA linkage formed by attack of a nucleophile other than the 5′-OH of the R substrate (i.e., the terminal or internal 2′-OH; see Scheme 1a). Because this third outcome would constitute negative data (unlike for successful cleavage of either the 3′–5′-linked or 2′–5′-linked product strands), further experimental confirmation of the unnatural RNA linkage assignment would be necessary.
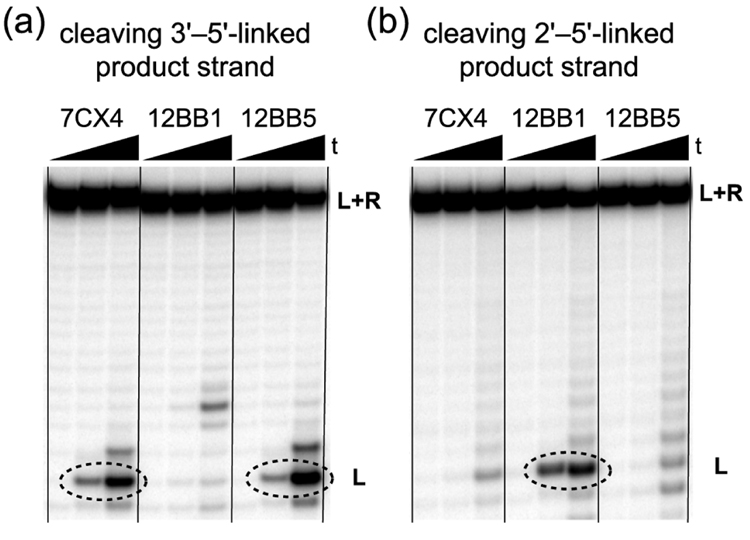
We tested each of the seven 7CX deoxyribozymes for their ability to cleave product strands that have either a 3′–5′ or a 2′–5′ linkage directly at the ligation site (Fig. 4). In all seven cases, the 7CX deoxyribozyme cleaved only the 3′–5′-linked strand, as illustrated for 7CX4 along with assays of the previously reported9 12BB1 (here a positive control for cleaving the 2′–5′-linked product) and 12BB5 (a positive control for cleaving the 3′–5′-linked product). On the basis of these data, we concluded that all seven of the tested 7CX deoxyribozymes create native 3′–5′ RNA linkages. This finding is in sharp contrast to the results from the previous BB selection, in which only two out of six tested 12BB deoxyribozymes formed 3′–5′ linkages.9 Therefore, changing both aspects of the selection design (overhang length and switching of 2′-dG/2′-G; Fig. 1) lead to a striking extent of control over both the site-selectivity and regioselectivity in the DNA-catalyzed RNA ligation process, in favor of synthesizing the native 3′–5′ linkage.
Fig. 4.
Cleavage assays to rapidly establish the RNA ligation junction connectivities created by the new deoxyribozymes. The diagnostic bands are circled. Microscopic reversibility requires that deoxyribozymes which are able to cleave a given linkage (e.g., 3′–5′ or 2′–5′) must form that linkage. Assays used either (a) the 3′–5′-linked ‘product’ strand, or (b) the 2′–5′-linked ‘product’ strand, with incubation in 70 mM Tris, pH 7.5, 150 mM NaCl, 2 mM KCl, 1 mM ZnCl2 at 23 °C (t = 0, 1, 20 h). In both cases illustrated here, the 7CX4 deoxyribozyme was tested along with the 12BB1 and 12BB5 DNA enzymes from our previous report.9 These latter deoxyribozymes form the non-native 2′–5′-linked and native 3′–5′-linked product, respectively, and therefore serve as positive and negative controls for these cleavage assays. The data unambiguously indicate that 7CX4 creates a native 3′–5′ RNA linkage. Other 7CX, 8CW, and 12BB clones were tested in similar fashion.
With an eye towards efficiently assaying additional DNA enzyme clones, we showed that the linkages in the RNA products from individual deoxyribozymes can be determined without prior sequencing and solid-phase synthesis of each clone. For this purpose, we chose eleven additional 7CX clones for which single-stranded DNA was available by PCR using miniprep DNA isolated during the cloning process. Without knowing the sequences of these eleven additional 7CX clones, the catalytically active DNA strands were obtained by PCR, purified by PAGE, and used in the ‘product’ strand cleavage assays of Fig. 4. Ten of these additional eleven 7CX clones create native 3′–5′ linkages, and one clone creates non-native 2′–5′ linkages. Sequencing then showed that of the ten additional deoxyribozymes that create 3′–5′ linkages, six unique sequences are represented; the lone deoxyribozyme that forms 2′–5′ linkages is also unique. Therefore, in the final analysis we identified a total of 14 unique 7CX deoxyribozymes, 13 of which form native 3′–5′ linkages. Sequences of all clones are listed in the supplementary information.†
To verify the 3′–5′ and 2′–5′ linkage assignments made for 7CX deoxyribozymes on the basis of the Fig. 4 assays, in several cases we used the deoxyribozymes to synthesize their RNA ligation products. We then directly determined the linkages using our previously established approaches, in which 3′–5′ linkages are cleaved selectively by the 8–17 deoxyribozyme whereas 2′–5′ linkages are cleaved in the presence of the complementary DNA template at 100 mM Mg2+ and pH 9.0.3 In all tested cases, the results were clearly in accord with the Fig. 4 assay (data not shown), confirming the linkage assignments.
Deoxyribozymes from the CW selection
We also examined DNA enzymes from the CW selection, for which only the overhang length was changed relative to the BB selection (Fig. 1). The ligation activity of the uncloned CW pool was ca. 14% at round 8 (Fig. 2b), and the 8CW clones were examined. The ligation junctions formed by individual 8CW clones were assayed by the same approach as used for the final eleven 7CX clones; i.e., as PCR products from miniprep DNA without prior sequencing or solid-phase synthesis. For the 8CW deoxyribozymes, 20 active clones were analyzed, leading to ten clones that form native 3′–5′ linkages and ten that form non-native 2′–5′ linkages.§ Sequencing revealed that these 20 clones can be grouped into 16 unique classes, of which eight create native 3′–5′ linkages and eight form non-native 2′–5′ linkages.† The lack of unnatural linkages in the products indicates that the CW selection design led to improved control of site-selectivity in favor of 5′-OH nucleophilicity when compared with the BB selection. In contrast, the CW selection design (unlike the CX selection design) led to very poor control over regioselectivity in 2′,3′-cyclic phosphate opening during DNA-catalyzed RNA ligation.
Analysis of additional 12BB deoxyribozymes
Only six 12BB clones were examined in our previous report, leading to two deoxyribozymes that create native 3′–5′ linkages, two that form non-native 2′–5′ linkages, and two that form unnatural linkages by nucleophilic attack of the internal 2′-OH of the R substrate.9 Here we analyzed 21 additional 12BB clones to improve the statistical coverage of the distribution of ligation products. Of these additional clones, seven, nine, and five form 3′–5′, 2′–5′, or unnatural linkages, respectively.§ Sequencing then indicated that these 21 clones can be grouped into 13 unique classes, of which five each create 3′–5′ and 2′–5′ linkages, and three form unnatural linkages.† Combining these clones with the six unique 12BB deoxyribozymes identified previously,9 the final ratio of the three possible types of products (native, non-native, unnatural) is 7:7:5. Therefore, the BB selection was confirmed to have poor control over the direction of both site-selectivity and regioselectivity.
Evaluating identities of unnatural linkages
The identities of the unnatural linkages created by three of the newly identified 12BB deoxyribozymes (12BB23, 30, and 32) were probed by additional experiments. In all three cases, partial alkaline hydrolysis of the ligation product verified that the linkage is unnatural (neither native 3′–5′ nor non-native 2′–5′), because unusually rapid cleavage at the ligation site was observed upon brief incubation under partial alkaline hydrolysis conditions of pH 9.2 and 90 °C (data not shown; similar to the findings for unnatural linkages in our previous report9). In contrast, several of the products from clones that form either 3′–5′ or 2′–5′ linkages led to conventional cleavage ladders upon partial alkaline hydrolysis. Also in all three cases, the ligation reaction was completely suppressed by using an R substrate that has a 2′-deoxy modification at the second nucleotide position, consistent with reaction of this 2′-OH as a nucleophile.9 Finally, cleavage of each new unnatural product by 12BB8 proceeded with both kobs and yield that are approximately the same as for 12BB8-catalyzed cleavage of the 12BB8 product itself.9 Considered together, these data strongly suggest that all three of the new deoxyribozymes synthesize the same unnatural linkage, which (as is the case for 12BB8) is either the 2′,3′-branched or 2′,2′-branched linkage.†
RNA substrate generality of the 7CX and 8CW deoxyribozymes
One important motivation for pursuing RNA-ligating deoxyribozymes is their potential utility for practical RNA ligation. For the 2′,3′-diol and 5′-triphosphate RNA substrate combination, we have reported several deoxyribozymes that join two substrates via native 3′–5′ linkages and that simultaneously have useful rates, yields, and substrate sequence generalities.8 However, for the 2′,3′-cyclic phosphate and 5′-OH substrate combination, no such deoxyribozymes have yet been found. Here we performed initial assays on several of the new 7CX and 8CW deoxyribozymes to explore their generality for the two RNA substrate sequences.
On both RNA substrates, the selection design leaves either four overhanging unpaired RNA nucleotides (J and BB selections) or one overhanging unpaired RNA nucleotide (CW and CX selections; Fig. 1). Because RNA-ligating deoxyribozymes that require numerous unpaired RNA substrate nucleotides typically have substantial sequence requirements for those nucleotides,3,6,9–11 we did not consider any of the 12BB deoxyribozymes that create native 3′–5′ linkages as likely candidates for high RNA substrate generality. In contrast, because a smaller number of overhanging RNA nucleotides has been more compatible with substrate sequence generality, the new 7CX and 8CW deoxyribozymes (which have only one-nucleotide overhangs) are more reasonable candidates for generality. We therefore examined systematic changes in both the L and R RNA substrate nucleotides, while always making corresponding changes to the deoxyribozyme binding arms to maintain Watson-Crick complementarity.
We focused our attention on the set of 7CX and 8CW deoxyribozymes that had potentially useful (45–60%) ligation yields with the ‘parent’ RNA substrate sequences and also form native 3′–5′ linkages. This set constituted all 13 of the relevant 7CX clones and six out of eight of the relevant 8CW clones. The generality assays were performed in two phases, in all cases using deoxyribozymes prepared by solid-phase synthesis. We first tested RNA substrate combinations that have systematic sequence changes outside of the fixed UA↓GG nucleotides at the ligation junction (denoted with the arrowhead). All remaining nucleotides in both RNA substrates were changed by systematic transversions, A↔C and G↔U (the deoxyribozyme binding arm nucleotides were altered to maintain base pairing). Eight of the 7CX clones and five of the 8CW clones had relatively high ligation yields with these alternative substrates. Of these, we then tested four of the 7CX clones and five of the 8CW clones with RNA substrates that included the sequence elements GA↓GU, where the underlined nucleotides one position away from the ligation junction (i.e., the penultimate nucleotides) were altered by transversion from the parent sequence UA↓GG, and all remaining nucleotides were unchanged from the original substrates. None of these nine clones had high ligation yields (<1%; the remaining four 7CX clones were not tested with these substrates). We conclude that whereas the 7CX and 8CW deoxyribozymes have promising RNA substrate sequence generality outside of the ligation site nucleotides UA↓GG, sequence requirements that prohibit very high generality are found closer to the ligation site. The precise nature of these requirements was not investigated.
Discussion
Influence of selection design on direction of site-selectivity and regioselectivity during DNA-catalyzed RNA ligation
One of the primary challenges for any in vitro selection experiment is to devise a selection design that leads to the desired catalytic outcome.2,13 In the case of DNA-catalyzed ligation of a 2′,3′-cyclic phosphate RNA substrate, the principal difficulty revealed by our past experiments has been identifying the best combination of selection design aspects that induce only native 3′–5′ linkages in the RNA products. Here we have found that when Zn2+ is the metal ion cofactor, deoxyribozymes that uniformly have the direction of site-selectivity and regioselectivity necessary to synthesize native 3′–5′ linkages (Scheme 1) can be identified via an appropriate combination of selection design aspects. This is a significant improvement over our previous work, in which native 3′–5′ linkages were formed by only a small subset of the deoxyribozymes.9
In the present work, both the number of overhanging RNA substrate nucleotides (one rather than the previous four) and the identity of the 5′-terminal nucleotide on the right-hand RNA substrate R (switching between 2′-dG and 2′-G during selection, rather than only 2′-dG as used previously) were critical to obtaining the desired selectivity (CX selection; Fig. 1d). In contrast, when only the overhang length was changed (CW selection; Fig. 1c), control over the direction of regioselectivity in 2′,3′-cyclic phosphate opening remained poor, leading to both native 3′–5′ and non-native 2′–5′ linkages. However, control over the direction of site-selectivity was very high, favoring the 5′-OH as the nucleophile.
In the CW and CX selections, the two design changes that were made to the original J/BB selection strategy9 were largely intended to control the direction of site-selectivity. In both cases, reducing the number of overhanging RNA nucleotides (from four in J/BB to one in CW/CX) predictably did result in increased control over site-selectivity, likely due to physically constraining the internal nucleotides of R. In combination with the use of the 2′-deoxy substitution at the 5′-terminal nucleotide of R, which suppresses reactivity of the 2′-OH, this led to complete control over site-selectivity, favoring nucleophilic attack of the 5′-OH.
In contrast to this rational and successful control of the direction of site-selectivity, it is very difficult to envision how to engineer logical control of the direction of regioselectivity during 2′,3′-cyclic phosphate opening. We do not know the mechanistic reason that native 3′–5′ linkages (reflecting control over both site-selectivity and regioselectivity) were dramatically favored in the CX selection but not the CW selection, which differed only in whether the 5′-terminal nucleotide of R was switched between 2′-dG and 2′-G (CX) or always 2′-dG (CW). Similarly, we do not know the mechanistic reason that non-native 2′–5′ linkages were favored in our earlier efforts with Mg2+-dependent deoxyribozymes.3–5 Collectively, these results demonstrate that control over the final outcome of a selection procedure depends on relatively subtle aspects of the selection design.
One might consider if we can control the linkage formation using our previously introduced 3′–5′-selective pressure that is based on the 8–17 deoxyribozyme, which only cleaves 3′–5′ linkages.14 In this approach, the 8–17 is used during each selection round immediately after the step depicted in Fig. 2, and only those deoxyribozymes that are inherently attached to a 3′–5′-linked RNA product (which is efficiently cleaved by 8–17) are separated by PAGE and taken into the next round. However, the reversibility of RNA ligation with a 2′,3′-cyclic phosphate L substrate means that the deoxyribozyme attached to the R substrate during a given selection round can cleave the ligation junction that it just formed, regardless of whether this linkage is 3′–5′ or 2′–5′ (or, for that matter, any unnatural linkage) As a consequence, this 3′–5′-selective pressure cannot readily be used with the 2′,3′-cyclic phosphate RNA substrate.
Potential for general utility of deoxyribozymes for RNA ligation involving a 2′,3′-cyclic phosphate
In contrast to the successful control of both site-selectivity and regioselectivity, the generality of the new deoxyribozymes for various RNA substrate sequences was suboptimal, because sequence requirements were found in at least one nucleotide not immediately at the ligation junction. This observation highlights the need to impose a selection pressure specifically in favor of generality, at least for RNA-ligating deoxyribozymes. By reducing the number of overhanging nucleotides on each RNA substrate from four to one, we did successfully limit the sequence requirements to nucleotides near the ligation junction, unlike in our previous efforts where longer single-stranded substrate regions led to more extensive requirements.3,6,9–11 Nevertheless, reducing even further the number of substrate nucleotides that have sequence limitations is important for practical utility.
In other efforts with 5′-triphosphate RNA substrates, we have used a mutant Taq polymerase15 that tolerates mismatches between the template and the 3′-terminus of the PCR primer. This mutant polymerase allows alternation of the penultimate RNA substrate nucleotides near the ligation junction in successive selection rounds, always while maintaining Watson-Crick base pairing between the RNA and the deoxyribozyme (D. A. Baum, S.K.S., and coworkers, data not shown). Such a strategy is required to reliably achieve generality with 5′-triphosphate substrates, because the generality observed in a small number of cases without applying a specific selection pressure8 has not been found in follow-up selection experiments with different RNA substrate sequences (data not shown). We expect that this alternation strategy or a related approach will be necessary to identify deoxyribozymes that ligate 2′,3′-cyclic phosphate RNA substrates with high generality while also creating native 3′–5′ linkages.
The need for a selection pressure specifically to enforce generality during DNA-catalyzed RNA ligation is in sharp contrast to the observations with RNA-cleaving deoxyribozymes such as 10–23, 8–17, and numerous variants,16–19 which require at most one nucleotide on either side of the cleavage site despite no special selection pressure in favor of such generality. This intriguing distinction between the generality of deoxyribozymes that ligate and cleave RNA suggests a fundamental difference in the difficulty of RNA ligation versus cleavage by a DNA catalyst. Understanding the origin of this difference, which may relate to different active-site dynamics for ligation versus cleavage substrates, will require detailed experiments to reveal the mechanisms of catalysis.
Experimental section
Oligonucleotide preparation
DNA oligonucleotides were prepared at IDT (Coralville, IA). The L RNA substrates (bearing a 2′,3′-cyclic phosphate) were synthesized by in vitro transcription using T7 RNA polymerase and a suitable DNA template that encodes a self-cleaving HDV or hammerhead ribozyme at the 3'-terminus of the desired transcript. The R RNA substrates were prepared by solid-phase synthesis at IDT or by in vitro transcription and subsequent CIP dephosphorylation. All DNA and RNA oligonucleotides were purified by denaturing PAGE with running buffer 1× TBE (89 mM each Tris and boric acid and 2 mM EDTA, pH 8.3) as described previously.10 The identities of the RNA transcripts were confirmed by MALDI mass spectrometry. For kinetic assays (Fig. 3), the 5′-32P-radiolabeled L substrate with a 2′,3′-cyclic phosphate was prepared by 5′-32P-radiolabeling (γ-32P-ATP, 3′-phosphatase-free T4 PNK from New England BioLabs) an oligoribo-nucleotide that was previously cleaved with a 10–23 deoxyribozyme to provide the cyclic phosphate terminus. For 60 ligation junction assays (Fig. 4), the L+R ‘product’ strands were prepared by solid-phase synthesis, using a 2′–5′-adenosine phosphoramidite monomer (ChemGenes) for the 2′–5′-linked strand, and then 5′-32P-radiolabeled.
In vitro selection procedure
We previously described the generic in vitro selection strategy that is based on separation of catalytically active deoxyribozymes by PAGE.3 For both of the selections performed here (CW and CX), the initial deoxyribozyme pool was 200 pmol (ca. 1014 molecules). In the key selection step, the DNA pool—previously attached to the R substrate using T4 RNA ligase—was heated to 95°C for 3 min in 5 mM Tris, pH 7.5, 15 mM NaCl and 0.1 mM EDTA, then placed on ice for 5 min. The sample was adjusted to 70 mM Tris, pH 7.5, 150 mM NaCl, 2 mM KCl, and 1 mM ZnCl2, then incubated for 3 h at 23 °C. The metal Zn2+ was added from a 10× stock solution containing 10 mM ZnCl2, 20 mM HNO3, and 200 mM Tris at pH 7.5; this stock solution was freshly prepared from a 100× stock of 100 mM ZnCl2 in 200 mM HNO3. After incubation with 1 mM Zn2+, the samples were separated by 8% denaturing PAGE.
Cloning and initial screening of individual deoxyribozyme clones was performed using DNA strands prepared by PCR from miniprep DNA derived from individual E. coli colonies. The miniprep DNA samples were first assayed by digestion with EcoRI to ascertain the presence of the expected insert. The concentration of each PAGE-purified deoxyribozyme strand was estimated from the intensity (UV shadowing) relative to suitable standards. Each screening assay used ca. 0.1 pmol of 5′-32P-radiolabeled L substrate, 2 pmol of the deoxyribozyme strand, and 20 pmol of R substrate. Samples were incubated at 23 °C with timepoints at 0, 1, and 7–15 h, using the procedure described below for kinetic assays.
Ligation activity assays (Fig. 3)
Individual deoxyribozymes prepared by solid-phase synthesis were assayed for ligation activity as follows. A sample was prepared in 7 µL containing 0.5 pmol 5′-32P-radiolabeled L substrate, 2 pmol deoxyribozyme, and 10 pmol R substrate in 5 mM Tris, pH 7.5, 15 mM NaCl, 0.1 mM EDTA. (Alternatively. some of the generality assays used 0.5 pmol 3′-32P-radiolabeled R substrate [32P-pCp, T4 RNA ligase], 2 pmol deoxyribozyme, and 10 pmol L substrate.) The sample was annealed by heating at 95 °C for 3 min and cooling on ice for 5 min. Stock solutions were then added to provide a total volume of 10 µL containing 70 mM Tris, pH 7.5, 150 mM NaCl, 2 mM KCl, and 1 mM ZnCl2 (the ZnCl2 stock solution was as described above for the in vitro selection procedure). The sample was incubated at 23 °C, with timepoints taken between 0–300 min and separated by 20% denaturing PAGE.
Ligation junction assays (Fig. 4)
The ligation junctions created by each deoxyribozyme were assayed by testing for cleavage of the 3′–5′-linked and 2′–5′-linked ‘product’ strands. The 5′-32P-radiolabeled product strand (0.5 pmol) was incubated with the deoxyribozyme (2 pmol) using the procedure described above for ligation activity assays, with timepoints taken at 0, 1, and 7–20 h and separated by 20% denaturing PAGE. The deoxyribozyme strand was prepared either by solid-phase synthesis (some 7CX clones) or by PCR from miniprep DNA followed by PAGE purification (remaining 7CX clones; all 8CW clones; and all new 12BB clones).
Supplementary Material
Acknowledgements
This research was supported by the US National Institutes of Health (GM-65966 to S.K.S.), the David and Lucile Packard Foundation (fellowship to S.K.S.), and Sigma Xi (Grant-in-Aid of Research to J.P.G.).
Footnotes
Electronic Supplementary Information (ESI) available: deoxyribozyme sequences. See http://dx.doi.org/10.1039/b000000x/
In a small number of cases for which the assays of Fig. 4 were not entirely conclusive, the ligation product was separately synthesized, and the 8–17 deoxyribozyme,3 complement/100 mM Mg2+/pH 9.0,3 and partial alkaline hydrolysis9 assays were used to clarify the nature of the linkage (data not shown).
Notes and references
- 1.Peracchi A. ChemBioChem. 2005;6:1316–1322. doi: 10.1002/cbic.200500098. [DOI] [PubMed] [Google Scholar]
- 2.Silverman SK. Chem. Commun. 2008:3467–3485. doi: 10.1039/b807292m. [DOI] [PubMed] [Google Scholar]
- 3.Flynn-Charlebois A, Wang Y, Prior TK, Rashid I, Hoadley KA, Coppins RL, Wolf AC, Silverman SK. J. Am. Chem. Soc. 2003;125:2444–2454. doi: 10.1021/ja028774y. [DOI] [PubMed] [Google Scholar]
- 4.Flynn-Charlebois A, Prior TK, Hoadley KA, Silverman SK. J. Am. Chem. Soc. 2003;125:5346–5350. doi: 10.1021/ja0340331. [DOI] [PubMed] [Google Scholar]
- 5.Semlow DR, Silverman SK. J. Mol. Evol. 2005;61:207–215. doi: 10.1007/s00239-004-0326-y. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Silverman SK. J. Am. Chem. Soc. 2003;125:6880–6881. doi: 10.1021/ja035150z. [DOI] [PubMed] [Google Scholar]
- 7.Coppins RL, Silverman SK. Nat. Struct. Mol. Biol. 2004;11:270–274. doi: 10.1038/nsmb727. [DOI] [PubMed] [Google Scholar]
- 8.Purtha WE, Coppins RL, Smalley MK, Silverman SK. J. Am. Chem. Soc. 2005;127:13124–13125. doi: 10.1021/ja0533702. [DOI] [PubMed] [Google Scholar]
- 9.Hoadley KA, Purtha WE, Wolf AC, Flynn-Charlebois A, Silverman SK. Biochemistry. 2005;44:9217–9231. doi: 10.1021/bi050146g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Silverman SK. Biochemistry. 2003;42:15252–15263. doi: 10.1021/bi0355847. [DOI] [PubMed] [Google Scholar]
- 11.Ricca BL, Wolf AC, Silverman SK. J. Mol. Biol. 2003;330:1015–1025. doi: 10.1016/s0022-2836(03)00654-5. [DOI] [PubMed] [Google Scholar]
- 12.Prior TK, Semlow DR, Flynn-Charlebois A, Rashid I, Silverman SK. Nucleic Acids Res. 2004;32:1075–1082. doi: 10.1093/nar/gkh263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joyce GF. Annu. Rev. Biochem. 2004;73:791–836. doi: 10.1146/annurev.biochem.73.011303.073717. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Silverman SK. Biochemistry. 2005;44:3017–3023. doi: 10.1021/bi0478291. [DOI] [PubMed] [Google Scholar]
- 15.Ghadessy FJ, Ramsay N, Boudsocq F, Loakes D, Brown A, Iwai S, Vaisman A, Woodgate R, Holliger P. Nat. Biotechnol. 2004;22:755–759. doi: 10.1038/nbt974. [DOI] [PubMed] [Google Scholar]
- 16.Santoro SW, Joyce GF. Proc. Natl. Acad. Sci. USA. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cruz RPG, Withers JB, Li Y. Chem. Biol. 2004;11:57–67. doi: 10.1016/j.chembiol.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 18.Schlosser K, Gu J, Sule L, Li Y. Nucleic Acids Res. 2008;36:1472–1481. doi: 10.1093/nar/gkm1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silverman SK. Nucleic Acids Res. 2005;33:6151–6163. doi: 10.1093/nar/gki930. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.