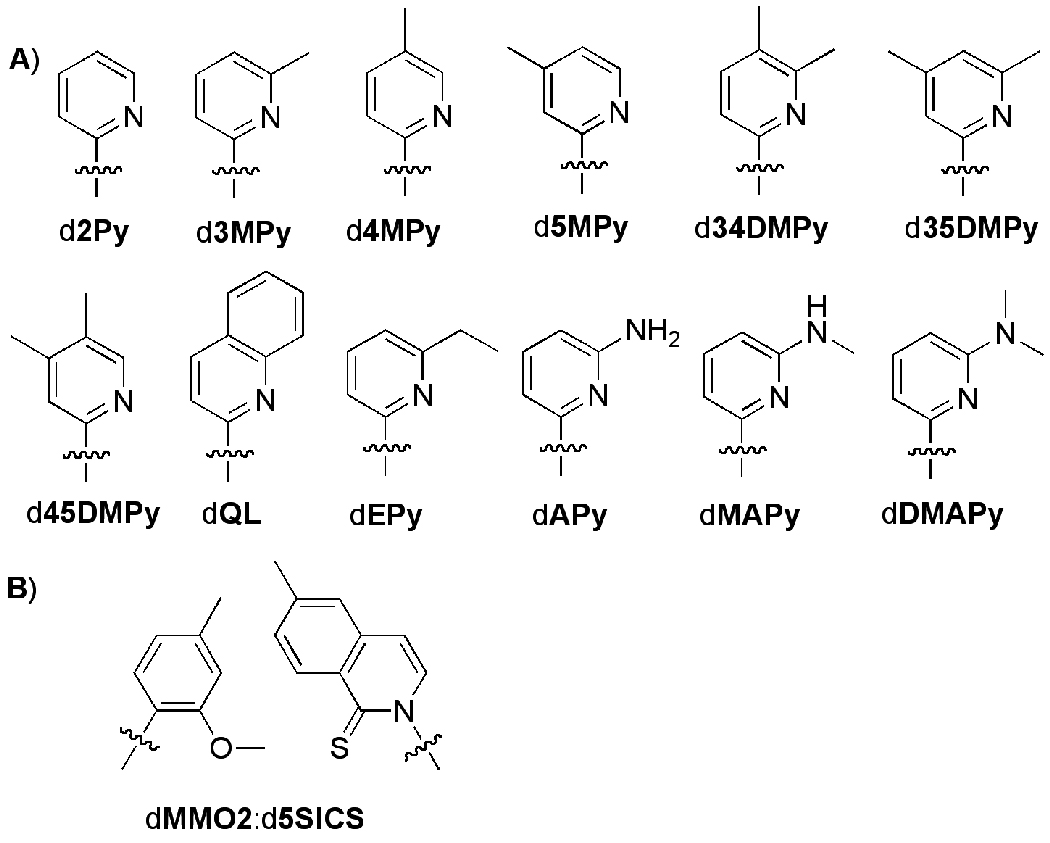
As part of an effort to increase both the biological and biotechnological applications of DNA, we[1–5] and others[6–9] have explored the DNA polymerase-mediated replication of a wide range of unnatural base pairs. In our initial efforts we examined large, aromatic, unnatural nucleotides, both as self pairs of two identical nucleotides and heteropairs of different nucleotides.[1–5,10,11] While several of these unnatural base pairs are efficiently synthesized (i.e. by insertion of the unnatural dNTP opposite its partner in the template) by the exonuclease-deficient Klenow fragment of E. coli DNA polymerase I (Kf), none are efficiently extended (i.e. by continued primer elongation), most likely due to interstrand nucleobase intercalation and distortion of the primer terminus.[10] Thus, a range of nucleotides bearing smaller phenyl-based nucleobases that should be incapable of intercalation were explored, and several modifications that facilitate extension were identified.[1–4] Of these, aza-substitution at the 2 position (2Py, Figure 1) appears to be the only modification that facilitates self pair extension without significantly facilitating mispairing.[3]
Figure 1.
a) 2-Pyridyl nucleotides synthesized and characterized in this study. b) dMMO2:d5SICS base pair. Only the nucleobase analog is shown with the wavy line indicating connection to the sugar and phosphate backbone, which have been omitted for clarity.
We have also recently found that another of the phenyl-based nucleotides, dMMO2, forms a heteropair with d5SICS that is synthesized and extended with relatively high efficiency and fidelity by a variety of different DNA polymerases (Figure 1).[5] However, it is unclear whether dMMO2 is the best phenyl-based nucleotide for heteropair formation with d5SICS, or if a derivatized pyridyl analog might optimize heteropair replication.
Here, we report the synthesis and characterization of a series of substituted 2-pyridine derivatives (Figure 1) designed to systematically examine the effect of nucleobase shape, size, and hydrophobicity as well as structural and electronic modifications within the interbase interface. The analogs are examined both as self pairs and as part of a heteropair with d5SICS. All nucleosides, phosphoramidites, oligonucleotides, and triphosphates were synthesized as described in the Supporting Information or in Ref. 12.
The 2Py self pair is largely limited by inefficient self pair synthesis; thus, we first characterized the steady-state rates for Kf-mediated synthesis of the 2-pyridyl-based nucleotide self pairs (Table 1). As with the fully carbocyclic scaffold,[4] methyl group substitution has a significant effect on self pair synthesis. For example, the d4MPy and d45DMPy self pairs are synthesized far more efficiently than the parent d2Py self pair (which is synthesized with an efficiency of 6.2 × 103 M−1min−1),[3] demonstrating that simple methyl substitution can substantially increase the rates of synthesis. The similar rates with which the d4MPy and d45DMPy self pairs are synthesized (1.6 × 104 and 4.1 × 104 M−1min−1, respectively) suggest that substitution at the 4-position is sufficient for the observed increase in efficiency. Additionally, self pairs of dQL, which combines both 3- and 4-position substitution with increased aromatic surface area, are synthesized with an efficiency very similar to those of d4MPy and d45DMPy (Table 1), suggesting that the packing interactions mediated by the 4-position methyl group stabilize the dNTP insertion transition state as much as the intercalative interactions mediated by the larger aromatic group. The rates of synthesis for the self pairs of the remaining analogs are all less than that for d2Py, indicating that substitution at the 3- or 5-positions does not facilitate self pair synthesis.
Table 1.
Incorporation rates of unnatural triphosphates dXTP.[a]
| 5'-dTAATACGACTCACTATAGGGAGA | |||
|---|---|---|---|
| 3'-dATTATGCTGAGTGATATCCCTCTXGCTAGGTTACGGCAGGATCGC | |||
| X | kcat [min−1] | KM [µM] | kcat/KM[M−1 min−1] |
| d3MPy | 0.42 ± 0.14 | 163 ± 23 | 2.6 × 103 |
| d4MPy | 3.2 ± 0.6 | 201 ± 27 | 1.6 × 104 |
| d5MPy | 0.34 ± 0.02 | 117 ± 33 | 2.9 × 103 |
| d34DMPy | 1.2 ± 0.2 | 267 ± 22 | 4.7 × 103 |
| d35DMPy | 0.10 ± 0.02 | 89 ± 19 | 1.2 × 103 |
| d45DMPy | 2.0 ± 0.2 | 49 ± 7 | 4.1 × 104 |
| dQL | 0.74 ± 0.05 | 20 ± 3 | 3.7 × 104 |
| dEPy | nd[b] | nd[b] | <1.0 × 103 |
| dAPy | nd[b] | nd[b] | <1.0 × 103 |
| dMAPy | nd[b] | nd[b] | <1.0 × 103 |
| dDMAPy | 0.26 ± 0.02 | 162 ± 42 | 1.6 × 103 |
See Supporting Information for details.
Reaction was too inefficient for kcat and KM to be determined independently.
We also characterized the efficiency with which Kf inserts the natural dNTPs opposite a pyridyl nucleotide in the template to gauge the fidelity of unnatural base pair synthesis (Table S1). For reference, opposite d2Py, dATP is the most efficiently inserted natural dNTP, and it is actually inserted 32-fold faster than d2PyTP.[3] While substitution at the 5-position has no significant effect, the rate of incorporation of dATP decreases as the steric bulk at the 3- or 4-position increases. In each case dATP remains the most efficiently inserted natural dNTP, followed by dGTP, dTTP, and last by dCTP. The increased rate of self pair synthesis and the reduced rates of mispairing with dA combine so that the d4MPy and d45DMPy self pairs are synthesized only 5- and 2-fold, respectively, slower than dATP is inserted.
We next examined the rate at which each derivatized self pair is extended by insertion of dCTP opposite a dG in the template (Table 2). Substitution at the 3-position has widely varying effects. While methyl substitution has no effect, ethyl substitution (dEPy) increases the rate of extension by 2-fold, relative to the d2Py self pair, and the amino substituents (dAPy, dMAPy, and dDMAPy) decrease efficiency of extension to an extent that is correlated with substituent size. Thus, it appears that while the interface between these pyridyl nucleobase analogs may be optimized by increased packing, it is not tolerant of altered electrostatics. The effects at the 4- and 5-position are much more promising with a 6- to 12-fold increase in the rate of self pair extension with methyl substitution. The effects are roughly additive, with extension of the d45DMPy self pair increased 70-fold relative to the unmodified self pair to a rate that is only 70-fold slower than that for a natural base pair. In fact, the d45DMPy self pair is the most efficiently extended self pair identified to date. This data suggests that increased steric bulk in the nucleobase interface induces a structure that is less efficiently extended, perhaps by causing a widening or distortion of the base pair, while modification at the 4- and 5-positions apparently favors extension, perhaps due to better interbase packing within the major groove or optimized packing with flanking nucleobases. Considering all steps, methyl substitution at the 4-position is the most beneficial as it increases both the efficiency and fidelity of both unnatural base pair synthesis and extension. Substitution at the 5-position, which does not significantly affect synthesis, but does favor extension, also facilitates replication.
Table 2.
Extension rates of unnatural self pairs.[a]
| 5'-dTAATACGACTCACTATAGGGAGAX | |||
|---|---|---|---|
| 3'-dATTATGCTGAGTGATATCCCTCTXGCTAGGTTACGGCAGGATCGC | |||
| X | kcat [min−1] | KM [µM] | kcat/KM [M−1 min−1] |
| d3MPy | 2.2 ± 0.1 | 100 ± 3 | 2.2 × 104 |
| d4MPy | 7.5 ± 1.6 | 36 ± 6 | 2.1 × 105 |
| d5MPy | 6.5 ± 1.1 | 16 ± 2 | 4.1 × 105 |
| d34DMPy | 1.3 ± 0.08 | 132 ± 23 | 9.9 × 103 |
| d35DMPy | 6.8 ± 0.9 | 68 ± 15 | 1.0 × 105 |
| d45DMPy | 15 ± 2 | 6.2 ± 0.3 | 2.4 × 106 |
| dQL | 0.073 ± 0.007 | 57 ± 5 | 1.3 × 103 |
| dEPy | 8.6 ± 1.6 | 102 ± 36 | 8.4 × 104 |
| dAPy | 1.0 ± 0.1 | 41 ± 2 | 2.5 × 104 |
| dMAPy | 0.77 ± 0.04 | 123 ± 17 | 6.3 × 103 |
| dDMAPy | nd[b] | nd[b] | <1.0 × 103 |
See Supporting Information for details.
Reaction was too inefficient for kcat and KM to be determined independently.
We next examined the potential of the pyridyl nucleotides as dMMO2 analogs by screening them for their ability to pair with d5SICS. Examination of primer extension by gel electrophoresis revealed that d5MPy and d34DMPy were most efficiently paired with d5SICS (Figure S1). Thus, we characterized the d5MPy:d5SICS and d34DMPy:d5SICS heteropairs in greater detail (Table 3). The triphosphates of d5MPy and d34DMPy are inserted opposite d5SICS with second order rate constants that are approximately 10-fold slower than that for insertion of dMMO2TP. Likewise, d5SICSTP is inserted opposite either 2-pyridyl analog in the template with rates that are ~10-fold less efficient than insertion opposite dMMO2. Thus, at least with these analogs, methyl substitution at the 3-, 4-, and 5-positions appears to have similar effects on synthesis, and the aza nitrogen atom appears to be slightly less beneficial than the methoxy substituent at the 2-position.
Table 3.
Steady-state rate constants of d5MPy:d5SICS and d34DMPy:d5SICS heteropairs.[a]
| 5'-dTAATACGACTCACTATAGGGAGA | ||||
|---|---|---|---|---|
| 3'-dATTATGCTGAGTGATATCCCTCTYGCTAGGTTACGGCAGGATCGC | ||||
| dXTP | Y | kcat [min−1] | KM [µM] | kcat/KM [M−1 min−1] |
| d5MPy | d5SICS | 4.1 ± 0.1 | 152 ± 12 | 2.7 × 104 |
| d34DMPy | d5SICS | 5.3 ± 0.1 | 132 ± 4 | 4.0 × 104 |
| d5SICS | d5MPy | 4.5 ± 0.5 | 0.69 ± 0.03 | 6.6 × 106 |
| d5SICS | d34DMPy | 4.1 ± 0.7 | 2.5 ± 0.2 | 1.6 × 106 |
| 5'-dTAATACGACTCACTATAGGGAGAX | ||||
| 3'-dATTATGCTGAGTGATATCCCTCTYGCTAGGTTACGGCAGGATCGC | ||||
| X | Y | kcat [min−1] | KM [µM] | kcat/KM [M−1 min−1] |
| d5MPy | d5SICS | 1.5 ± 0.1 | 0.056 ± 0.015 | 2.7 × 107 |
| d34DMPy | d5SICS | 2.1 ± 0.1 | 7.9 ± 0.5 | 2.6 × 105 |
| d5SICS | d5MPy | 3.2 ± 0.4 | 13 ± 1 | 2.4 × 105 |
| d5SICS | d34DMPy | 0.47 ± 0.06 | 106 ± 3 | 4.4 × 103 |
See Supporting Information for details.
Interestingly, the extension of the two pyridyl heteropairs is very different. While the d34DMPy:d5SICS (primer:template) and d5SICS:d34DMPy heteropairs are extended 10- to 100-fold slower than the dMMO2 heteropair, the d5MPy:d5SICS heteropair is extended with an efficiency of 2.7 × 107 M−1min−1, which is actually more efficient than the corresponding dMMO2 heteropair, and remarkably, only 6-fold slower than extension of a natural base pair in the same sequence context. Moreover, the d5SICS:d5MPy heteropair is extended with an efficiency that is only marginally reduced relative to the heteropair with dMMO2. Thus, derivatization, particularly methyl substitution at the 5-position, has a significant and beneficial effect on the pairing of the 2-pyridyl analogs with d5SICS, and at least for extension, can actually optimize the heteropair so that it is better recognized than dMMO2:d5SICS.
While these data will be helpful for the design of optimized base pairs, it is apparent that the d45DMPy self pair is the most promising of the unnatural base pairs examined in the current study. To further explore the utility of this self pair, we determined the rates at which all possible mispairs are extended (Table S2), which along with the rates at which the mispairs are synthesized (see above), allow for a determination of the overall fidelity. The most efficiently synthesized mispair, dA:d45DMPy, is extended approximately 20-fold less efficiently than the correct pair. Thus, the overall fidelity for self pair replication (synthesis and extension) relative to the mispair with dA is 11. The most efficiently extended mispair is that with dT, which is extended with a rate of 6.4 × 105 M−1min−1; however, due to the mispair’s inefficient synthesis, the overall fidelity of the self pair relative to the mispair with dT is 33. The mispairs with dC and dG are extended with rates of 3.9 × 103 and 2.1 × 103 M−1min−1, respectively, resulting in overall fidelity of 1.1 × 104 for dC and 6.0 × 103 for dG.
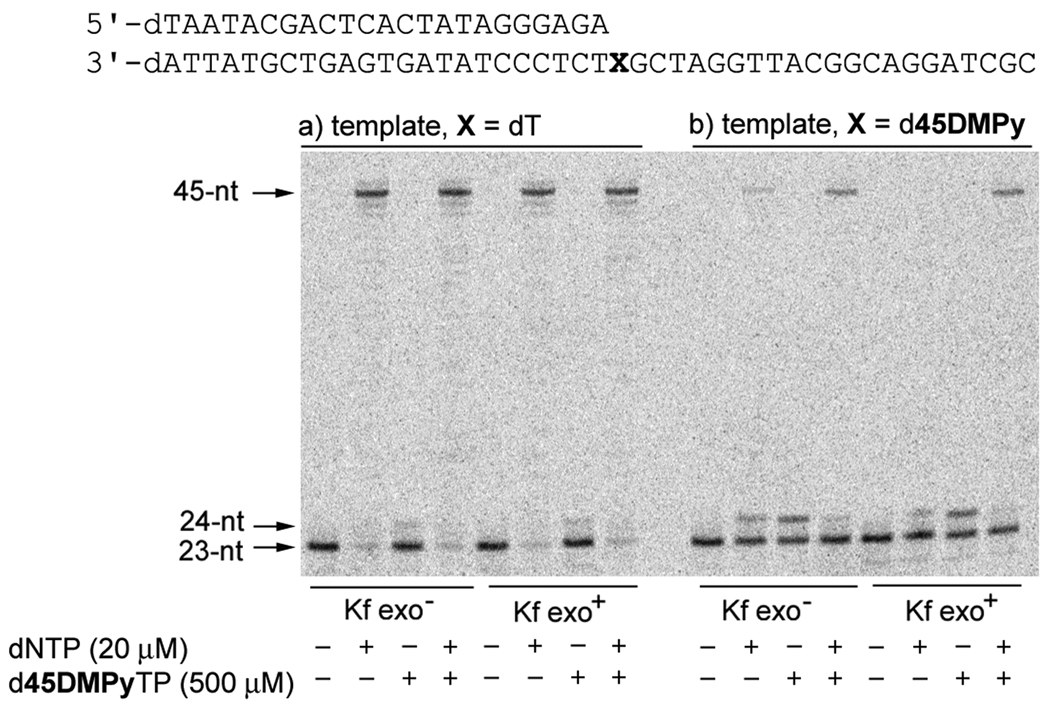
To explore the potential elimination of mispairs that are efficiently synthesized but not extended, we characterized full length DNA synthesis with Kf or exonuclease-proficient Kf (Kf exo+) (Figure 2). As expected, full-length synthesis was observed with a natural template (X = dT) with both Kf and Kf exo+. With d45DMPy in the template, Kf exo+, and only natural triphosphates present, an equilibrium was observed between the primer and the n+1 extension product, presumably resulting from dATP insertion and excision. However, when d45DMPyTP was added to the reaction, full length product is observed in good yield. Thus, while further optimization is clearly desired, the d45DMPy self pair might have immediate use in a variety of different in vitro applications.[13]
Figure 2.
Extension of 23-nt primer with 45-nt templates (X = dT or d45DMPy). Reactions contain either Kf exo+ or Kf exo− and the reaction time was 3 min.
Cumulatively, these results demonstrate that every step of replication may be optimized by derivatization of the 2-pyridyl nucleobase analogs. Judicious placement of methyl groups alone, yielding d45DMPy, results in a self pair that is extended with a natural-like rate and can be used to synthesize site-specifically modified DNA in good yield. Moreover, while none of the pyridyl analogs is actually a better heteropair partner for d5SICS than dMMO2 itself, the results demonstrate that at least in some contexts, heteropair extension is better facilitated by a 2-pyridyl nitrogen atom than a methoxy group. Thus, the 2-pyridyl scaffold remains among the most promising nucleobase analogs and further optimization should result in self pairs or heteropairs for in vitro, and possibly even in vivo, applications.[13,14]
Supplementary Material
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Footnotes
Acknowledgement for financial support is given to the National Institutes of Health (GM60005 to F.E.R.). Support was also provided by the Uehara Memorial Foundation (Y.H.), the Czech Academy of Sciences (Z04055905 to M.H.) and MSMT (LC512 to N.J. and M.H.).
Contributor Information
Dr. Yoshiyuki Hari, Department of Chemistry, The Scripps Research Institute, La Jolla, CA 92037 (USA).
Dr. Gil Tae Hwang, Department of Chemistry, The Scripps Research Institute, La Jolla, CA 92037 (USA).
Aaron M. Leconte, Department of Chemistry, The Scripps Research Institute, La Jolla, CA 92037 (USA)
Nicolas Joubert, Institute of Organic Chemistry and Biochemistry, Academy of Sciences of the Czech Republic, Gilead Sciences & IOCB Research Center, CZ-16610, Prague 6 (Czech Republic)
Dr. Michal Hocek, Institute of Organic Chemistry and Biochemistry, Academy of Sciences of the Czech Republic, Gilead Sciences & IOCB Research Center, CZ-16610, Prague 6 (Czech Republic)
Dr. Floyd E. Romesberg, Department of Chemistry, The Scripps Research Institute, La Jolla, CA 92037 (USA).
References
- 1.Henry AA, Olsen AG, Matsuda S, Yu C, Geierstanger BH, Romesberg FE. J. Am. Chem. Soc. 2004;126:6923–6931. doi: 10.1021/ja049961u. [DOI] [PubMed] [Google Scholar]
- 2.Leconte AM, Matsuda S, Hwang GT, Romesberg FE. Angew. Chem. Int. Ed. 2006;45:4326–4329. doi: 10.1002/anie.200601272. [DOI] [PubMed] [Google Scholar]
- 3.Kim Y, Leconte AM, Hari Y, Romesberg FE. Angew. Chem. Int. Ed. 2006;45:7809–7812. doi: 10.1002/anie.200602579. [DOI] [PubMed] [Google Scholar]
- 4.Matsuda S, Henry AA, Romesberg FE. J. Am. Chem. Soc. 2006;128:6369–6375. doi: 10.1021/ja057575m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leconte AM, Hwang GT, Matsuda S, Capek P, Hari Y, Romesberg FE. J. Am. Chem. Soc. 2008;130:2336–2343. doi: 10.1021/ja078223d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Z, Sismour AM, Sheng P, Puskar NL, Benner SA. Nucleic Acids Res. 2007;35:4238–4249. doi: 10.1093/nar/gkm395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krueger AT, Lu H, Lee AHF, Kool ET. Acc. Chem. Res. 2007;40:141–150. doi: 10.1021/ar068200o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirao I, Mitsui T, Kimoto M, Yokoyama S. J. Am. Chem. Soc. 2007;129:15549–15555. doi: 10.1021/ja073830m. [DOI] [PubMed] [Google Scholar]
- 9.a) Chairamonte M, Moore CL, Kincaid K, Kuchta RD. Biochemistry. 2003;42:10472–10481. doi: 10.1021/bi034763l. [DOI] [PubMed] [Google Scholar]; b) Kincaid K, Beckman J, Zivkovic A, Halcomb RL, Engels JW, Kuchta RD. Nucleic Acids Res. 2005;33:2620–2628. doi: 10.1093/nar/gki563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuda S, Fillo JD, Henry AA, Rai P, Wilkens SJ, Dwyer TJ, Geierstanger BH, Wemmer DE, Schultz PG, Spraggon G, Romesberg FE. J. Am. Chem. Soc. 2007;129:10466–10473. doi: 10.1021/ja072276d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ogawa A, Wu Y, McMinn DL, Liu J, Schultz PG, Romesberg FE. J. Am. Chem. Soc. 2000;122:3274–3287. [Google Scholar]
- 12.Urban M, Pohl R, Klepetarova B, Hocek M. J. Org. Chem. 2006;71:7322–7328. doi: 10.1021/jo061080d. [DOI] [PubMed] [Google Scholar]
- 13.a) Ieven MJ. Clin. Virol. 2007;40:259–276. doi: 10.1016/j.jcv.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Cropp TA, Chin JW. Curr. Opin. Chem. Biol. 2006;10:601–606. doi: 10.1016/j.cbpa.2006.10.008. [DOI] [PubMed] [Google Scholar]; c) Bittker JA, Phillips KJ, Liu DR. Curr. Opin. Chem. Biol. 2002;6:367–374. doi: 10.1016/s1367-5931(02)00321-6. [DOI] [PubMed] [Google Scholar]
- 14.a) Wang L, Xie J, Schultz PG. Annu. Rev. Biophys. Biomol. Struct. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]; b) Xie J, Schultz PG. Curr. Opin. Chem. Biol. 2005;9:548–554. doi: 10.1016/j.cbpa.2005.10.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.