Abstract
Background
Previous studies indicate that the endothelin system is involved in hypertension, heart failure, atherosclerosis, chronic kidney disease, and diabetes. To explore the potential genetic effects of copy number variations (CNV) on the endothelin system which underlie these diseases, we studied the association of genome-wide CNVs with gene expression levels of seven genes involved in the endothelin system using independent HapMap subjects including 90 Asians (45 Han Chinese and 45 Japanese), 60 Caucasians, and 60 Africans.
Methods and Results
For each subject, the genome-wide variations were measured using the Affymetrix® 6.0 chip that includes measurements of 906,000 single nucleotide polymorphisms and 946,000 CNV probes. The gene expression profiles of the transformed B-lymphocytes were measured for the same subjects. Among the 210 subjects, we identified 1,529 CNV regions on 22 autosomes. By testing the association between CNVs and the gene expression levels in each racial group using linear regression, we identified four statistically significant CNV associations in all three groups (alpha=0.05). The strongest association was between a 66 kbp CNV region located on chromosome 6 and endothelin-1 (EDN1) expression. The effects of the CNV-EDN1 association in the three racial groups were in the same direction and explained 7%–14% of the variation in EDN1 expression.
Conclusions
Although the biological function of the chromosome 6 CNV is unclear, the significant and consistent association found in three racial groups suggests that CNVs may contribute to variation in underlying risks of common disease through their effects on key molecular signaling pathways.
Keywords: hypertension, heart failure, atherosclerosis, endothelin system
Introduction
A copy number variation (CNV) is a segment of DNA that is present at a variable number of copies compared with a reference genome sequence 1 Initial studies by Redon et. al. used genome-wide single nucleotide polymorphism (SNP) arrays to identify genome wide CNVs using the HapMap population and identified 1,447 CNVs, covering 12% of the human genome 2. Recently, Stranger et al. have demonstrated potentially functional associations between CNV regions and quantitative gene expression levels 3. CNVs represent a unique opportunity to expand genome-wide association (GWA) studies because they quantitate both gene-dosage and the functional implications of SNP variation. Furthermore, CNVs have been reported to be associated with numerous human disorders such as Parkinson’s disease 4, schizophrenia 5–7, autism 8, 9 and Crohn’s disease 10.
While a number of GWA studies to date have identified SNP loci associated with cardiovascular disease (CVD) 11–13, the association between CNVs and CVD has not been investigated widely and is less understood. CNVs are suggested to affect CVD and its clinical phenotypes 14. One approach to assess the role of CNVs in CVD susceptibility is to focus on candidate genes. Endothelin-1 (EDN1) is a well-studied candidate gene for CVD. EDN1 was firstly isolated and cloned as a novel vasoconstrictor in 1988 15. Human EDN1 is located on chromosome 6 and encodes a 21-amino acid peptide. Besides vasoconstriction, EDN1 also contributes to the vasodilation, oxidative stress, inflammation and fibrogenic processes through the endothelin system 16. EDN1 is believed to be an important molecule contributing to the pathogenesis of hypertension, heart failure, atherosclerosis, chronic kidney disease (CKD), and diabetes 16. Importantly, several SNPs in the endothelin system genes have been shown to have functional relevance and association with cardiovascular phenotypes and/or diseases 17.
In this study, we used available data from HapMap subjects to estimate CNV associations with gene expression levels of seven endothelin system genes, including EDN1, endothelin-2 (EDN2), endothelin-3 (EDN3), endothelin converting enzyme 1 (ECE1), endothelin converting enzyme 2 (ECE2), endothelin receptor type A (EDNRA), and endothelin receptor type B (EDNRB) in three racial groups (Asians, Caucasians, and Africans). The goal of the study was to identify associations between CNVs and gene expression levels that were consistent across the three racial groups.
Methods
Sample and Data
Two hundred and seventy subjects in three racial groups were available from the HapMap project for the current study. All 270 subjects were used to identify the specific CNV regions (described below). We used the 210 unrelated HapMap subjects from three racial groups to examine the association between CNVs and the expression level of genes in the endothelin system. There were 60 unrelated subjects from Utah; these individuals represent the United States Caucasian population with Northern and Western European ancestry (parents in 30 trios). There were 60 unrelated subjects collected from the Yoruba people in Nigeria (parents in the 30 trios). Forty-five unrelated Han Chinese in Beijing, China and 45 unrelated Japanese in Tokyo, Japan were collected for the Asian group. All subjects gave specific consent for their inclusion in the HapMap project 18.
The mRNA gene expression data for the HapMap samples were obtained from Wellcome Trust Sanger Institute. Gene expression profiles of the seven endothelin system genes, EDN1, EDN2, EDN3, ECE1, ECE2, EDNRA and EDNRA, were identified using their unique mRNA RefSeq identifiers from the microarray annotation file. The quantification and normalization of the gene expression data were described in a previous report 3.
The genotyping data for the 270 HapMap subjects were produced and distributed by Affymetrix® using the Genome-Wide Human SNP Array 6.0 platform. The CNV genotype data for the 210 HapMap subjects were merged with their gene expression data using the anonymous, unique identifiers. Prior to CNV analysis, the Contract QC (CQC) value was calculated for each chip (Affymetrix 6.0) for each of the 270 HapMap subjects using Affymetrix Genotyping Console™. All of the studied chips passed the CQC threshold of 0.4, which is recommended by Affymetrix for controlling genotyping quality.
Copy Number Variation Analysis
Two different approaches were applied to identify copy number variation. For the first approach, the Copy Number Analysis Module (CNAM) in HelixTree was used to process the Affymetrix® raw intensity files (.CEL) and generated a common reference genome with a normal number of copies, using all 270 HapMap subjects. Then the log2ratio was calculated for each probe (both SNP probes and copy number probes) on each microarray compared to the reference genome. HelixTree implements a dynamic programming based algorithm 19, 20, which exhaustively searches through all possible cutpoint positions to find an optimal segmentation of the log2ratios for each measured subject. A multivariate method implemented in HelixTree segments log2ratios across all subjects simultaneously, finding general copy number regions that may be similar across all subjects (HelixTree manual of copy number analysis module). For a given subject, the mean of the log2ratios within each segment for that subject are used as independent variables to identify the CNV associations with a dependent variable.
The second approach used the Affymetrix® Genotyping Console (GTC) 3.0.1 to generate a reference genome for comparing copy numbers and was generated using all 270 raw intensity files (CEL files) from the human SNP 6.0 array (Affymetrix® Genotyping Console 3.0.1 User Manual). Using the common reference genome for comparisons, the intensity ratio of each probe (both SNP probe and copy number probe) on each array was calculated. The boundaries of the common CNV segments were determined using the predefined CNV regions 21. A hidden Markov model (HMM) was utilized to call the copy number state (i.e. the number of DNA copies, 2 for a diploid genome like human) for each identified CNV. Genotype calls for the common copy number variations (observed in multiple unrelated subjects) were determined using the Canary algorithms 21. The chromosomal boundaries as well as the copy number state were exported and utilized in the association analysis of gene expression levels.
Statistical Analysis
Population genetic parameters for CNVs (copy number states) were calculated, including minor allele frequencies (MAFs), genotype frequencies, and a chi-square test for departures from expectations under Hardy-Weinberg equilibrium (HWE) for the 210 unrelated subjects in three racial groups. Summary statistics for CNVs and linear regression models were generated using the statistical software R. Single CNV associations with gene expression levels were evaluated in each racial group separately using standard linear regression models. For CNV associations with p-value less than 0.05 in all three racial groups, we pooled the 210 HapMap subjects from all racial groups after testing for heterogeneity (described below) and performed the CNV association analysis. To estimate the relationship between the associated CNVs and SNPs, we tested the additive effect of SNPs associated with gene expression levels in the pooled sample.
For the CNV and SNP association tests conducted on the pooled sample of three racial groups, as well as on the sample of each of the three racial groups, we used the principal component analysis (PCA) to adjust for the population stratification 22. The top ten principal components (PCs) of the 752,286 autosomal SNPs (MAF>0.05 and call rate >95%) were calculated within each race and in the pooled sample. The PCs significantly associated with the gene expression levels were used as covariates in the multiple regression model of testing CNV associations. In the CNV association analysis of EDN1 gene expression within each racial group, none of top 10 PCs is significantly associated with the outcome and no PC was used to adjust the linear regression model within each racial group.
All regression analyses were performed with the mean log2ratio of each CNV fragment as the independent variable. The null hypothesis of no CNV effect was evaluated by testing whether the regression coefficients were significantly different from zero. The CNV genotypes (copy number states exported from Affymetrix® GTC 3.0.1 using Canary) were also used as independent variables in regression analysis to confirm the findings. In the CNV genotype test, we assumed an additive model of the DNA copy number effect for each CNV. All tests were two-sided and the alpha level was set to 0.05.
For those CNVs that were significant in three racial groups, we applied two methods, Cochran’s Q test 23 and Higgins’s inconsistency test 24, to examine the heterogeneity of the results across the three HapMap racial groups. where wi=1/Si2, Si2 is the an unbiased estimate of the error variance of xi, x̄w = Σwixi/Σwi, k is the number of experiments. Under the null hypothesis, the Q statistic has an approximate chi-square distribution with k−1 degrees of freedom. The I2 inconsistency metric measures the amount of heterogeneity not due to chance 24. I2 = (Q−df)/Q*100%, where Q is Cochran’s heterogeneity statistic and df the degrees of freedom.
906,602 SNPs were genotyped using the Affymetrix® Genome-Wide Human SNP Array 6.0 platform. SNPs were excluded if they had unknown chromosomal location, a call rate less than 95% or a minor allele frequency (MAF) less than 0.05. These quality control filters resulted in 752,286 SNPs available for analysis in 210 independent HapMap subjects. The additive effect of each SNP was estimated by using a linear regression model.
Results
Among the 270 HapMap genotyped subjects, we identified 1,529 CNV regions on 22 autosomes using CNAM implemented in HelixTree 6.2 software package. We identified statistically significant associations between four CNVs and gene expression levels among the endothelin system in each of the three racial groups. One CNV region (chromosome 6: 79,025,772 bp - 79,091,892 bp) was associated with EDN1 expression, one CNV region (chromosome 2: 86,802,346 bp - 86,807,353 bp) was associated with EDN3 expression, one CNV region (chromosome 7: 3,150,183 bp – 3,150,685 bp) was associated with ECE1 expression, and one CNV region (chromosome 20: 35,480,198 bp – 35,488,136 bp) was associated with EDNRA expression. The respective p-values in Asians, Caucasians and Africans were 0.011, 0.009 and 0.003 for the EDN1 association, 0.011, 0.026 and 0.023 for the EDN3 association, 0.016, 0.039 and 0.008 for the ECE1 association, and 0.027, 0.008 and 0.016 for the EDNRA association.
We focused on the CNV-EDN1 associations, as they represented the most statistically significant findings in this study. The CNV-EDN1 associations are summarized in Table 1. The numbers of CNV regions demonstrating statistically significant associations with EDN1 expression were, 77 in Asians, 218 in Caucasians and 106 in Africans. We repeated the CNV association analysis of EDN1 gene expression using the standardized copy number states called by Affymetrix® GTC 3.0.1. Both methods identified the CNV region (CNV_6q14.1) on chromosome 6 with identical boundaries. The p-values are higher when using the discrete CNV states in the linear regression model than those when using the continuous log2ratio. The p-values, however, remain statistically significant within each racial group (Table 1). Pooling the samples from the three racial groups together yielded highly significant p-values for the log2ratio test as well as the copy number test with adjustment of population stratification using principal component analysis 22. Among the top ten PCs, the first and second were associated with EDN1 gene expression. In the pooled CNV association test of EDN1 expression, we used these two PCs to adjust for population stratification. The p-values of CNV_6q14.1 in the multiple regression model were 4.34×10−6 (log2 ratio test) and 1.15×1−5 (copy number test). These p-values were significant after Bonferroni correction for multiple testing, with adjusted p-values of 4.77×1−3 (log2 ratio test) and 1.27×1−2 (copy number test). The results are similar when including the top 10 PCs in the adjustment model (nominal p-values are 1.07×10−5 for log2 ratio test and 2.66×10−5 for copy number test).
Table 1.
Summary of the linear regression analysis of the chromosome 6 CNV associated with EDN1 expression.
| log2 Ratio |
CNV State (0,1,2) |
|||||||
|---|---|---|---|---|---|---|---|---|
| beta | SE | R2 | p-value | beta | SE | R2 | p-value | |
| Asian (N=90) | 0.576 | 0.222 | 0.071 | 1.12×10−2 | 0.381 | 0.164 | 0.058 | 2.23×10−2 |
| Caucasian (N=60) | 0.631 | 0.234 | 0.111 | 9.17×10−3 | 0.427 | 0.166 | 0.102 | 1.27 ×10−2 |
| African (N=60) | 1.210 | 0.391 | 0.142 | 3.02×10−3 | 0.609 | 0.216 | 0.121 | 6.55 ×10−3 |
| Pooled* (N=210) | 0.849 | 0.142 | 0.146 | 4.34×10−6 | 0.571 | 0.097 | 0.143 | 1.15 ×10−5 |
adjusted for the population stratification using principal components computed from 752,286 autosomal SNPs.
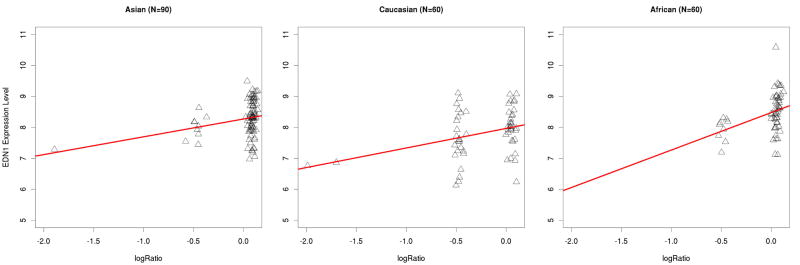
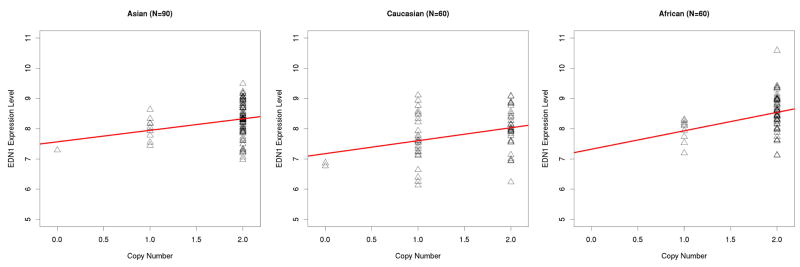
The proportion of variance (R2) explained by this CNV ranges from 0.071 in the Asians to 0.142 in the Africans (log2ratio test), and from 0.058 in the Asians to 0.121 in the Africans (copy number test). The R2 values from the copy number test are slightly smaller than those obtained when using the mean log2ratio as the independent variable. The R2 values between the two pooled tests (mean log2ratio vs. CNV state) are comparable (0.146 vs. 0.143). Previous studies found that R2 values or p-values generated in the association test using the log2 ratios or the CNV genotypes are strongly correlated (Pearson correlation coefficients > 0.9), indicating that log2 ratios can be used directly 3. In our study, we also found that the linear relationship between CNV_6q14.1 and EDN1 expression is very similar using either log2 ratio or CNV genotype (Figure 1), and the R2 values or p-values from the linear regression model are also consistent (Table 1).
Figure 1.
Scatter Plot of copy number measurements and the EDN1 gene expression levels among Asians, Caucasians and Africans. A: median log2Ratio; B: CN State (i.e. genotype)
In addition, we examined the heterogeneity of the estimated betas of the CNV_6q14.1 association with EDN1 gene expression levels in three racial groups, using Cochran’s Q test 23 and Higgins’s inconsistency test 24. The p-value of the Cochran’s Q test was 0.649 and the I2 value was 4.6% in the inconsistency test (possible range 0% to 100%, I2 less than 25% indicates low heterogeneity) 24. Both tests indicate that no heterogeneity was observed across the CNV_6q14.1-EDN1 association results from three racial groups. The beta coefficients for CNV associations are all of the same relative magnitude and direction of association (e.g. 0.381 (Asian), 0.427 (Caucasian), and 0.609 (African) for the CNV genotype association). We recognize, however, that we may have low power to detect heterogeneity with only three groups.
Table 2 summarizes the population genetics of this EDN1 associated CNV. The minor allele (i.e. deletion allele) frequency (MAF) in Caucasians is 0.242, which is much higher than that in Asian (MAF=0.061) and African (MAF=0.075) populations. None of the three populations have significant p-values from the chi-square test of Hardy-Weinberg equilibrium (HWE), with an alpha level of 0.05 (Table 2). The EDN1 associated CNV spans 66 kbp on 6q14.1 starting at 79,025,772 bp and ending at 79,091,892 bp (NCBI built 36). This CNV has been previously identified and repeatedly reported in several human CNV studies 2, 25, 26.
Table 2.
Population Genetics of the chromosome 6 CNV associated with EDN1 expression.
| Genotype Count |
MAF | HWE p-value | |||
|---|---|---|---|---|---|
| copy number =0 | copy number =1 | copy number =2 | |||
| Asian (n=90) | 1 | 9 | 80 | 0.061 | 0.223 |
| Caucasian (n=60) | 2 | 25 | 33 | 0.242 | 0.289 |
| African (n=60) | 0 | 9 | 51 | 0.075 | 0.530 |
The genome-wide association of 752,277 autosomal SNPs (MAF>5% and call rate > 95%) with EDN1 gene expression level was conducted using all 210 unrelated HapMap subjects with adjustment for population stratification by principal component analysis (Price 2006).The lowest p-value among 752,277 SNP tests for the pooled sample is 1.06×10−6 which is not significant after adjusting for multiple testing (Bonferoni corrected p-value is 0.79). Within the flanking region of CNV_6q14.1 (25 kb upstream and downstream), there are 13 SNPs (5 upstream and 8 downstream) genotyped on the Affymetrix 6.0 array with a minor allele frequency larger than 5% and call rate larger than 95%. Their distances and LD correlations to CNV_6q14.1, and the associations with EDN1 gene expression levels are summarized in supplementary Table 1. The lowest p-value among thirteen SNPs is 0.006 (rs818269). None of these neighboring SNPs have strong LD with CNV_6q14.1.
Discussion
CNVs have been suggested to influence CVD due to their potential biological effects on various CVD candidate genes 14. One plausible mechanism is indicated by the effect of CNV on the gene expression levels of given disease candidate genes 10. Using unrelated Asians, Caucasians, and Africans from HapMap samples, we investigated the associations between genome-wide CNVs with the gene expression levels of the endothelin system, which includes well-studied candidate genes for hypertension, atherosclerosis, heart failure, CKD and diabetes. In this study, a known deletion variation on chromosome 6 was significantly associated with EDN1 expression in each of the three racial groups considered.
Although there is no known gene overlapped with the identified CNV region on chromosome 6, four genes are located within 1 Mbp window from the CNV region, including 5-hydroxytryptamine (serotonin) receptor 1B (HTR1B, 796 Kbp upstream); interleukin-1 receptor-associated kinase 1 binding protein 1 (IRAK1BP1, 608 Kbp downstream), pleckstrin homology domain interacting protein (PHIP, 684 Kbp downstream), and high mobility group nucleosomal binding domain 3 (HMGN3, 942 Kb downstream). Two neighboring blocks within the chromosome 6 CNV, 79033035 bp -79033306 bp and 79033329 bp -79033429 bp, are highly conserved among 10 mammalian species (UCSC genome browser). The log-odds score for the two blocks are 644 and 272 correspondingly computed by PhastCons 27 which is a program for identifying evolutionarily conserved elements in a multiple alignment, given a phylogenetic tree. The log-odds scores range from 0 to 1000 and the higher score indicates more conserved region across species. One the other hand, approximately 60% of conserved bases in the ENCODE regions are assigned at least one molecular function 28. The inter-species conservation within the chromosome 6 CNV region suggests plausible biological functions.
Three other CNVs were also associated with gene expression levels of EDN3, ECE1 and EDNRA. Taken together, these associations demonstrate a possible impact of CNVs on CVD, CKD, and diabetes through the endothelin system, although further epidemiological studies are needed to confirm the potential relationships between the CNVs and these complex common diseases.
In this study, the gene expression profile was measured in transformed B-lymphocytes. Therefore the CNV association may not be generalized to other cell-types or tissues due to potential cell-type/tissue specific gene expression. Because of the high heritability of many gene expression levels, studying CNV association with gene expression in transformed B-lymphocytes can be an essential approach to understanding the molecular mechanisms between CNV and disease traits through examining the intermediate gene expression traits.
For CNV analysis, data quality needs to be carefully controlled to guard against false CNV calls. In Affymetrix Genotyping Console™, the Contract QC (CQC) value was calculated for each chip (Affymetrix 6.0) for each of the 270 HapMap subjects. The CQC values of the chips from the 210 unrelated HapMap subjects range from 1.0 to 3.7 with a mean of 2.6. All of the chips passed the CQC threshold of 0.4, which is recommended by Affymetrix for controlling genotyping quality. These high-quality chip results were used in the CNV analysis.
CNV association studies show promise as a complement to the current GWA studies using SNPs to identify disease loci on the human genome 10. Because the CNVs have large base-pair coverage on the human genome 2, their functional roles in human disease development are substantial. At least one thousand human CNVs have been characterized and validated in recent years 2, 25, 29 and more will be discovered with improved technology 30. Unlike the SNPs, CNVs tend to have differential boundaries across individuals 30. This property of CNV limits the current population studies with the more conserved CNVs with common boundaries among individuals. The advancement of technology in both CNV measurements and CNV calling algorithms 21 will improve our understanding of the boundaries, origins and distributions of human CNVs, as well as our knowledge of their functional roles in human diseases. The ongoing GWA studies provide a unique opportunity to study the genome-wide CNV association with CVD and other common human diseases. Along with the genome-wide SNP data, the genome-wide CNV data will help us to better understand the genetic architecture of common diseases.
Supplementary Material
Acknowledgments
We would like to thank Michael Todd Greene, University of Michigan, Ann Arbor, Michigan, and Greta M. Linse, Christophe G. Lambert, Golden Helix Inc., Bozeman, Montana, for their insightful comments.
Funding Sources
This work was supported by National Institute of Health grant HL087660 and HL086694.
Footnotes
Conflict of Interest Disclosures
None.
References
- 1.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 2.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, Gonzalez JR, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Zhang J, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, de Grassi A, Lee C, Tyler-Smith C, Carter N, Scherer SW, Tavare S, Deloukas P, Hurles ME, Dermitzakis ET. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, Lincoln S, Crawley A, Hanson M, Maraganore D, Adler C, Cookson MR, Muenter M, Baptista M, Miller D, Blancato J, Hardy J, Gwinn-Hardy K. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 5.The International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–41. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, Stray SM, Rippey CF, Roccanova P, Makarov V, Lakshmi B, Findling RL, Sikich L, Stromberg T, Merriman B, Gogtay N, Butler P, Eckstrand K, Noory L, Gochman P, Long R, Chen Z, Davis S, Baker C, Eichler EE, Meltzer PS, Nelson SF, Singleton AB, Lee MK, Rapoport JL, King MC, Sebat J. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 7.Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of denovo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 8.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, Leotta A, Pai D, Zhang R, Lee YH, Hicks J, Spence SJ, Lee AT, Puura K, Lehtimaki T, Ledbetter D, Gregersen PK, Bregman J, Sutcliffe JS, Jobanputra V, Chung W, Warburton D, King MC, Skuse D, Geschwind DH, Gilliam TC, Ye K, Wigler M. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, Platt OS, Ruderfer DM, Walsh CA, Altshuler D, Chakravarti A, Tanzi RE, Stefansson K, Santangelo SL, Gusella JF, Sklar P, Wu BL, Daly MJ Autism Consortium. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 10.McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, Zody MC, Hall JL, Brant SR, Cho JH, Duerr RH, Silverberg MS, Taylor KD, Rioux JD, Altshuler D, Daly MJ, Xavier RJ. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, Masson G, Gudbjartsson DF, Magnusson KP, Andersen K, Levey AI, Backman VM, Matthiasdottir S, Jonsdottir T, Palsson S, Einarsdottir H, Gunnarsdottir S, Gylfason A, Vaccarino V, Hooper WC, Reilly MP, Granger CB, Austin H, Rader DJ, Shah SH, Quyyumi AA, Gulcher JR, Thorgeirsson G, Thorsteinsdottir U, Kong A, Stefansson K. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 12.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, Boerwinkle E, Hobbs HH, Cohen JC. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, Konig IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H WTCCC and the Cardiogenics Consortium. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pollex RL, Hegele RA. Copy number variation in the human genome and its implications for cardiovascular disease. Circulation. 2007;115:3130–3138. doi: 10.1161/CIRCULATIONAHA.106.677591. [DOI] [PubMed] [Google Scholar]
- 15.Yanagisawa M, Kurihara H, Kimura S, Tomobe Y, Kobayashi M, Mitsui Y, Yazaki Y, Goto K, Masaki T. A novel potent vasoconstrictor peptide produced by vascular endothelial cells. Nature. 1988;332:411–415. doi: 10.1038/332411a0. [DOI] [PubMed] [Google Scholar]
- 16.Re RN. Molecular Mechanisms in Hypertension. London, UK: Taylor & Francis; 2006. [Google Scholar]
- 17.Rossi GP, Pitter G. Genetic variation in the endothelin system: do polymorphisms affect the therapeutic strategies? Ann N Y Acad Sci. 2006;1069:34–50. doi: 10.1196/annals.1351.004. [DOI] [PubMed] [Google Scholar]
- 18.The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 19.Hawkins DM. On the Choice of Segments in Piecewise Approximation. IMA Journal of Applied Mathematics. 1972;9:250–256. [Google Scholar]
- 20.Hawkins DM. Fitting multiple change-point models to data. Computational Statistics and Data Analysis. 2001;37:323–341. [Google Scholar]
- 21.Korn JM, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, Cawley S, Hubbell E, Veitch J, Collins PJ, Darvishi K, Lee C, Nizzari MM, Gabriel SB, Purcell S, Daly MJ, Altshuler D. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 23.Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10:101–129. [Google Scholar]
- 24.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H, Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, Shapero MH, de Bakker PI, Maller JB, Kirby A, Elliott AL, Parkin M, Hubbell E, Webster T, Mei R, Veitch J, Collins PJ, Handsaker R, Lincoln S, Nizzari M, Blume J, Jones KW, Rava R, Daly MJ, Gabriel SB, Altshuler D. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 27.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, Weinstock GM, Wilson RK, Gibbs RA, Kent WJ, Miller W, Haussler D. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perry GH, Ben-Dor A, Tsalenko A, Sampas N, Rodriguez-Revenga L, Tran CW, Scheffer A, Steinfeld I, Tsang P, Yamada NA, Park HS, Kim JI, Seo JS, Yakhini Z, Laderman S, Bruhn L, Lee C. The fine-scale and complex architecture of human copy-number variation. Am J Hum Genet. 2008;82:685–695. doi: 10.1016/j.ajhg.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, Haugen E, Zerr T, Yamada NA, Tsang P, Newman TL, Tuzun E, Cheng Z, Ebling HM, Tusneem N, David R, Gillett W, Phelps KA, Weaver M, Saranga D, Brand A, Tao W, Gustafson E, McKernan K, Chen L, Malig M, Smith JD, Korn JM, McCarroll SA, Altshuler DA, Peiffer DA, Dorschner M, Stamatoyannopoulos J, Schwartz D, Nickerson DA, Mullikin JC, Wilson RK, Bruhn L, Olson MV, Kaul R, Smith DR, Eichler EE. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.