Abstract
CpG methylation within the O6-methylguanine-DNA-methyltransferase (MGMT) promoter is associated with enhanced survival of glioblastoma multiforme (GBM) patients treated with temozolomide (TMZ). Although MGMT promoter is methylated in ~50% of GBM, several studies have reported a lack of correlation between MGMT methylation and protein expression levels and consequently inaccurate discrimination of TMZ sensitive and resistant patients. To understand the limitations of currently used assays, TMZ responsiveness of 13 GBM xenograft lines was correlated with MGMT protein expression and MGMT promoter methylation determined by 1) standard methylation-specific polymerase chain reaction (MS-PCR), 2) quantitative MS-PCR (qMS-PCR) and 3) bisulfite sequencing. For each xenograft line, mice with established intracranial xenografts were treated with vehicle control or TMZ (66 mg/kg × 5 days), and TMZ response was defined as relative prolongation in median survival for TMZ-treated vs. control-treated mice. The relative survival benefit with TMZ was inversely related to MGMT protein expression (r= −0.75; p=0.003) and directly correlated with qMS-PCR (r=0.72; p=0.006). There was a direct correlation between MGMT methylation signal by qMS-PCR and the number of methylated CpG sites within the region amplified by MS-PCR (r =0.78, p=0.002). However, bisulfite sequencing revealed heterogeneity in the extent of CpG methylation in those tumors with a robust qMS-PCR signal. Three of the 4 GBM lines with a qMS-PCR signal greater than 10% had at least 1 unmethylated CpG site, while only one line was fully methylated at all 12 CpG sites. These data highlight one potential limitation of the evaluation of MGMT methylation by MS-PCR assay and suggest that more detailed evaluation of methylation at individual CpG sites relative to TMZ response may be worth pursuing.
Keywords: MGMT, methylation, Glioblastoma, orthotopic xenografts
Introduction
Temozolomide (TMZ) chemotherapy combined with high-dose radiation therapy improves survival for a subset of patients with glioblastoma multiforme (GBM). Cytotoxic TMZ-induced O6-methylguanine lesions are repaired by the O6-methylguanine-DNA-methyltransferase (MGMT) protein, and cells lacking MGMT repair activity are significantly more sensitive to the cytotoxic effects of TMZ than cells with normal levels [1]. MGMT expression is suppressed at the level of transcription by CpG methylation within the MGMT promoter, and almost half of primary GBM tumor samples have evidence of promoter hypermethylation, as determined by methylation-specific PCR (MS-PCR) [2, 3]. In a recent study, tumor MGMT promoter hypermethylation was associated with prolonged survival (46% 2-year survival) in GBM patients treated with TMZ and radiation as compared to those patients without evidence of tumor hypermethylation (14% 2-year survival) [4]. These data suggest that suppression of MGMT expression mediated by promoter hypermethylation may be an important factor influencing TMZ sensitivity.
The predictive accuracy of the MGMT promoter methylation is limited, since approximately 30% of patients experience tumor progression during TMZ therapy regardless of methylation status, and conversely, approximately 15% of patients with an unfavorable unmethylated status have prolonged survival [4]. These apparent inconsistencies may reflect inherent technical limitations of the assay or the role of pathways other than MGMT in mediating TMZ resistance. To better understand the limitations of the standard MS-PCR assay, the influence of TMZ treatment on survival was evaluated in a panel of 13 GBM xenografts using an intracranial therapy evaluation model. TMZ responsiveness was compared with the MGMT protein expression and MGMT promoter methylation status (determined both by 1) standard MS-PCR, 2) quantitative MS-PCR and 3) bisulfite sequencing).
Methods
Xenograft information
The 13 serially passaged xenografts used in this study were derived from individual patients. Molecular alterations and histopathology for 11 xenografts have been previously described [5, 6]: two additional xenografts, GS 22 and GBM 26, diagnosed as gliosarcoma and glioblastoma, respectively, are also included. Prior Institution Review Board approval was obtained for the use of human tissue to establish the xenograft lines.
Orthotopic model
Therapy evaluations were conducted using an orthotopic tumor model as previously described [6, 7]. Athymic nude mice (NCI, Frederick, MD) with established intracranial tumors were randomized into groups of 8 to 10 mice each and treatment was initiated 2 weeks before mice were expected to become moribund. TMZ was purchased from the Mayo Clinic Pharmacy, suspended in Ora-plus (Paddock Laboratories, Minneapolis), and administered by oral gavage at 66 mg/kg for 5 days. This dosing regimen results in a drug exposure in mice that is equivalent to that obtained in humans with the routinely used adjuvant dosing regimen of 200 mg/m2 × 5 days. Mice were observed daily and euthanized when they reached a moribund state. All animal studies were performed with the prior approval of the Mayo Institutional Animal Care and Use Committee.
Western blotting
Flank tumor specimens were processed for western blotting as described previously [5]. Primary antibodies used in this study were MGMT (R&D Biosystems, Minneapolis, MN), β-actin (Sigma, St. Louis, MO) and secondary antibodies were horseradish peroxidase-conjugated rabbit anti-goat and goat anti-mouse (Pierce, Rockford, IL). Blots were developed with Super Signal Chemiluminescence reagent (Pierce).
MGMT Promoter Methylation Assay
DNA was extracted from flank xenograft samples using the Gentra DNA extraction kit (Puregene, Minneapolis, MN). Isolated tumor DNA was bisulfite-treated using the EZ DNA methylation kit (Zymo Research, Orange, CA). The modified DNA was amplified using primers specific for either methylated or unmethylated MGMT promoter sequences as described previously, with the exception that a nested PCR reaction was not used [3, 8]. Thus, the primers used for our standard MS-PCR are identical to those used for a final nested PCR described by Hegi, et. al. PCR products were visualized on ethidium bromide-stained, 3% agarose gels. In this assay, the tumor is methylated when the gel shows a single methylated band or both unmethylated and methylated bands. In contrast, the tumor is unmethylated only when a single unmethylated band is amplified. For quantitative MS-PCR, the custom MGMT assay was synthesized by the Applied Biosystems Inc. (ABI, Foster City, CA). The primers and the probe for this assay were designed to assay the same region of MGMT promoter evaluated by the standard MS-PCR. The sequences were TTCGCGGTGCGTATCGT (forward primer), CACTCTTCCGAAAACGAAACGA (reverse primer), and ACACTCACCAAATCGC (TAqMan® MGB/FAM probe). Similar assay was developed for β-actin as internal control. The primers and probe for β-actin quantitative MS-PCR (qMS-PCR) assay were designed using regions of the DNA devoid of CpG dinucleotides. The sequences for β-actin assay were TTTTATTTAGAGTGTAGGTGTGTGGAGATTTT (forward primer), CAAAAACAAAAACCTAACCCCTAAACCT (reverse primer), and CCCACCCTCTAAAACT (TAqMan® MGB/FAM probe). The reagents for real-time PCR were purchased from ABI. Amplifications were performed in a 50 μl reaction volume in 96-well plates on a 7900 Sequence Detection System (ABI.), at 95°C for 2 min followed by 40 cycles of 95°C for 15 s, and 60°C for 1 min. The universal methylated DNA (Chemicon, Temecula, CA) was used as a positive control and normal human brain DNA was used for negative control. Relative quantification of MGMT promoter methylation status (qMS-PCR signal) was performed using 7000 SDS 1.1 RQ software (ABI).
MGMT bisulfite sequencing
The MGMT CpG island was amplified by nested PCR as described previously using bisulfite-treated template DNA [9]. The resulting PCR amplicon was purified using a PCR purification kit (Qiagen, Valencia, CA) followed by ligation into a pGEM-T Easy cloning vector (Promega, Madison, WI). The ligated PCR product was then transformed into Escherichia coli (Invitrogen, Carlsbad, CA) and plated on agar embedded with ampicillin (50 μg/ml), IPTG (0.5mM) and X-gal (80 μg/ml) (Invitrogen). For each xenograft, direct colony PCR was performed on a minimum of 10 clones using vector based primers (sequences available on request). The final PCR product was incubated with shrimp-alkaline exonuclease (New England Biolab, Ipswich, MA) and submitted for DNA sequencing. In this method, if the cytosine within the CpG site is methylated it will not be deaminated by bisulfite treatment. Therefore, sequencing a methylated CpG will detect a “C”; whereas in unmethylated reaction the cytosine will be deaminated to uracil and eventually replaced by a thymine in the PCR reaction. Consequently, sequencing an unmethylated CpG will detect a “T” instead of “C”. The MGMT promoter region consists of 97 CpG dinucleotides, which we serially numbered as CpG1-97 (Fig. 1). However, in this report bisulfite sequencing is focused on 12 CpG dinucleotides (CpG 78–89), which are located within a region evaluated by the standard MS-PCR assay.
Figure 1.
Map of MGMT promoter region. Vertical lines depict the CpG dinucleotides. Also shown are annealing locations for primers used for MS-PCR assay, the 12 CpG dinucleotide analyzed by bisulfite sequencing for this study, and the location of putative transcription factor Sp1 binding sites. TS; transcription start site.
Statistical analysis
Survival distributions were estimated using the Kaplan-Meier method. The log rank test was used to compare survival across treatment groups. The two-sample rank sum test was used to compare relative differences in survival relative to MGMT MS-PCR status. Spearman’s correlation coefficient was used to assess the association of relative prolongation in median survival and either MGMT protein expression levels or MGMT qMS-PCR signal relative to β-actin.
Results
Establishment of glioblastoma xenografts
Tumors within the GBM xenograft panel were established by implanting patient tumor samples subcutaneously in the flank of nude mice and then maintained through serial subcutaneous propagation. To evaluate relationships between MGMT and TMZ responsiveness, flank tumors from 13 xenograft lines were used to establish intracranial tumors for TMZ therapy evaluations. Corresponding portions of each flank tumor were examined for MGMT promoter methylation and MGMT protein expression. For 7 xenograft lines, the primary patient tumor sample from which the xenografts were derived also was evaluated for MGMT promoter methylation.
MGMT promoter methylation status in glioblastoma xenografts
MGMT promoter methylation was evaluated by standard MS-PCR for both the xenograft lines and selected primary patient tissues (Fig. 2A). Similar to the observed incidence of methylation in clinical samples, MGMT methylation was detected in 5 of 13 (38%) xenograft lines (GBM8, 12, 16, 36, GS22). No promoter methylation was found in the remaining 8 lines (GBM6, 10, 14, 26, 34, 43, 44, GS28). To investigate the potential for preservation of methylation during the xenografting process, MGMT promoter methylation by standard MS-PCR was analyzed in clinical samples from which the xenografts were derived. In all cases the methylation status was identical between the patient and xenograft samples. There was a good correlation between the gel-based standard MS-PCR and quantitative MS-PCR results (Fig. 2B), although qMS-PCR detected low-levels of MGMT promoter methylation in GBM14 (1.35% relative to positive control) and 34 (0.48% relative to control) that was not seen by the gel-based standard MS-PCR method. Of those tumors defined as MGMT methylated by standard MS-PCR, the extent of MGMT methylation by qMS-PCR in GBM36 (4.16% relative to control) was relatively low compared to other tumor lines (GBM8 – 35.06%, GBM12 – 52.23%, GBM16 – 38.54%, and GS22 – 53.53%). Given the distinct separation in PCR methylation signal between the GBM36 (4.16%) and then next closest methylation signal (GBM8 - 35.06%), GBM36 was classified as being unmethylated on the basis of the qMS-PCR results, while tumors with a PCR signal greater than 10% were classified as being methylated. Consistent with gene silencing via promoter methylation, high-level MGMT methylation signal (>10%) was associated with low to undetectable levels of MGMT protein (Fig. 2C–D) (r=−0.75; p=0.003). Significant variation in MGMT protein expression was evident in tumors with minimal MGMT methylation, with expression ranging from robust (GBM10 and 43) to undetectable (GBM14 and 34). Of importance, the MGMT antibody and PCR primers for these assays were all specific for human MGMT and did not detect murine MGMT in control assays (data not shown).
Figure 2.
MGMT methylation status and protein expression determination. Flank tumor specimens used to establish the orthotopic tumors for TMZ therapy evaluations were analyzed for MGMT promoter methylation status and protein levels. A) Gel-based standard MS-PCR - the lanes corresponding to PCR reactions specific for unmethylated (U) and methylated (M) templates are labeled. For each line, the capitalized letter denotes our interpretation of the methylation status (“U m” depicts unmethylated and “u M” depicts methylated). B) qMS-PCR - the % qMS-PCR signal is the ratio of MGMT methylation qMS-PCR signal in a tumor sample relative to the positive control. C) MGMT protein expression by western blotting. Membrane was stripped and re-probed for β-actin. D) MGMT protein levels were quantitated by film densitometry and plotted relative to the qMS-PCR results.
MGMT promoter methylation in relation to TMZ response
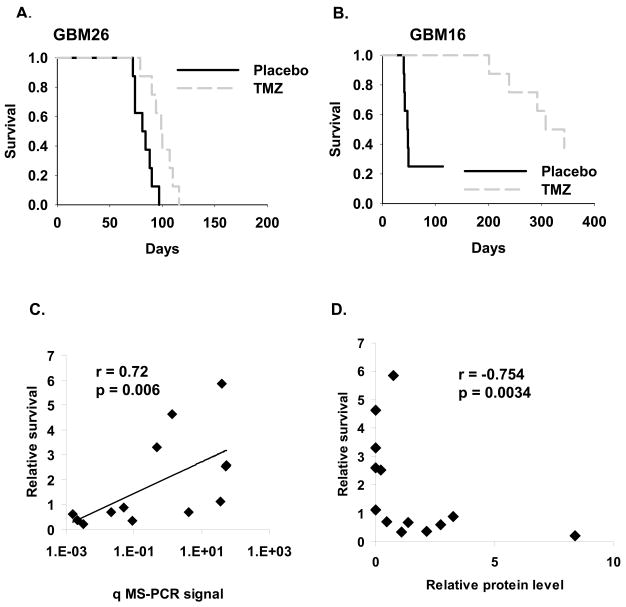
TMZ sensitivity was evaluated using an orthotopic model. Mice with established intracranial tumors were randomized to placebo or TMZ treatment for 5 days, and then mice were followed for survival (Table 1 and Supplemental Fig. 1). There was a broad range of TMZ responsiveness, with treatment extending the median survival of mice from 21% (GBM26) to 585% (GBM16) relative to placebo (Fig. 3A–B). Consistent with previously published clinical results, there was a good correlation between MGMT promoter methylation and survival prolongation with TMZ therapy (Fig. 3C; r = 0.72, p =0.006). However, 2 tumors lacking robust MGMT methylation signal were highly sensitive to TMZ (GBM14 – survival prolongation of 464%; GBM34 – survival prolongation of 331%) and a single MGMT methylated tumor, GBM8 was relatively resistant to TMZ (111% prolongation in survival with TMZ therapy). In a similar analysis, MGMT protein expression levels from Figure 2C were measured by densitometry, and normalized values were plotted relative to survival benefit (Fig. 3D). This demonstrated an inverse relationship between MGMT protein levels and TMZ sensitivity (r= −0.75; p=0.003), with high level MGMT expression invariably associated with TMZ resistance. For tumors with a low but detectable MGMT expression (GBM8, 12, 16, 36, GS28) there was considerable variability in survival benefit ranging from 68% to 585% prolongation in median survival. Thus, while there was good correlations between TMZ sensitivity and the MGMT assays, neither low MGMT expression nor MGMT promoter methylation status were entirely reliable in predicting TMZ responsiveness in this orthotopic model.
TABLE 1.
Survival summary of temozolomide treated glioblastoma xenografts lines
| Placebo | Temozolomide | ||||||
|---|---|---|---|---|---|---|---|
| Tumor | n | median survival | n | median survival | Survival Prolongationa | Log rank p-value | qMS-PCR |
| GBM6 | 8 | 41 | 8 | 56 | 37% | 0.019 | U |
| GBM8 | 8 | 59 | 8 | 124 | 111% | <0.001 | M |
| GBM10 | 8 | 41 | 9 | 55 | 34% | 0.249 | U |
| GBM12 | 10 | 15 | 10 | 53 | 253% | <0.001 | M |
| GBM14 | 10 | 33 | 10 | 186 | 464% | <0.001 | U |
| GBM16 | 8 | 48 | 8 | 326 | 585% | 0.021 | M |
| GS22 | 8 | 43 | 8 | 155 | 259% | <0.001 | M |
| GBM26 | 8 | 83 | 8 | 100 | 21% | 0.003 | U |
| GS28 | 8 | 33 | 8 | 56 | 68% | 0.013 | U |
| GBM34 | 8 | 85 | 8 | 367 | 331% | 0.001 | U |
| GBM36 | 8 | 126 | 7 | 214 | 71% | 0.156 | U |
| GBM43 | 8 | 23 | 8 | 43 | 89% | <0.001 | U |
| GBM44 | 9 | 35 | 10 | 56 | 60% | 0.336 | U |
Determined as a ratio of the median survival of TMZ-treated to the median survival of placebo-treated GBM xenografts.
Figure 3.
TMZ survival evaluation relative to MGMT methylation and protein expression. Mice with established orthotopic xenografts were randomized and then treated with placebo or 66 mg/kg TMZ orally for 5 days. Survival curves for the A) least and B) most responsive tumors are shown. For each tumor line, the ratio of median survival for TMZ-treated vs. placebo-treated mice (relative survival) is plotted relative to C) the qMS-PCR signal and D) MGMT protein levels.
Determination of MGMT methylation status by standard MS-PCR relies on annealing of primers across 9 CpG sites (CpG78-82 and CpG86-89, Fig. 1). To address possible heterogeneity of methylation across these sites, bisulfite sequencing was performed and the results for CpG sites 78 to 89 are reported here. For this assay, PCR products amplifying the MGMT promoter were cloned and at least 10 clones were individually sequenced. The sequencing results were tabulated and compared with the standard MS-PCR results (Fig. 4A–B). There was a direct correlation between MGMT methylation signal by qMS-PCR and the number of methylated CpG sites (r =0.78, p=0.002), although there was significant heterogeneity in the extent of CpG methylation in those tumors with a robust qMS-PCR signal. Three of the 4 GBM lines with a qMS-PCR signal greater than 10% had at least 1 unmethylated CpG site, while only GBM16 was fully methylated at all 12 CpG sites. Thus, there was a heterogenous methylation of CpGs within this region relative to the MGMT methylation status measured by qMS-PCR.
Figure 4.
Correlation between methylation of individual CpG sites and MGMT promoter methylation A) The methylation status of 12 CpG sites (78–89) determined by bisulfite sequencing is shown relative to the corresponding qMS-PCR results for each xenograft line. The percent of PCR clones that were methylated at each site is indicated. Also, shown is the relative MGMT protein level and the survival benefit from TMZ therapy for each xenograft line B) The extent of CpG methylation is plotted relative to MGMT methylation signal by qMS-PCR.
Discussion
TMZ is an important component of therapy for GBM, and developing robust predictive models for TMZ responsiveness is an important step towards customizing therapies for this highly malignant disease. In this study, we used a panel of GBM xenograft lines to correlate TMZ response with MGMT protein expression and MGMT promoter methylation status. In contrast to previous studies using flank xenografts to address this question [10–12], the efficacy of TMZ therapy was assessed using an intracranial tumor model with prolongation in survival as the primary endpoint. TMZ therapy was initiated in mice with established intracranial tumors, and to maximize clinical relevance, TMZ was delivered using a dosing regimen (66 mg/kg daily × 5 days) that provides a similar drug exposure in mice as a routinely used clinical dosing regimen for GBM patients (200 mg/mg2 daily × 5 days). In contrast to previous studies using established glioma cell lines [10, 12, 13], we have previously described that our method of serial transplantation in mice stably maintains genetic features commonly associated with gliomagenesis and represents a more clinically relevant model system than traditional tumor lines [5, 7, 14]. The current analysis in 13 GBM xenograft lines represents the most extensive analysis of TMZ therapy response in an animal model of GBM.
Evaluation of MGMT methylation by standard MS-PCR has been suggested as a predictive assay that could be used to select those patients who would benefit from TMZ therapy. In the current study, promoter hypermethylation was significantly associated with greater TMZ responsiveness, although methylation status was not an absolute predictor for TMZ responsiveness. Similar to clinical observations [2, 4, 15], one of the xenografts with robust MGMT promoter hypermethylation was relatively resistant to TMZ therapy (GBM8), and conversely, 2 xenografts lacking MGMT hypermethylation were highly sensitive to TMZ therapy (GBM14 and 34). These data would support the idea that early clinical progression during TMZ therapy in some patients with ‘favorable’ tumor MGMT promoter hypermethylation may be due to unspecified, intrinsic TMZ resistance. Conversely, the sensitivity observed in the MGMT non-methylated GBM14 and 34 tumors is reminiscent of patients with an ‘unfavorable’ tumor methylation status but a prolonged survival. Thus, the range of TMZ responses in the current study closely parallels clinical observations and suggests that this panel may be a clinically relevant platform for testing TMZ-based therapeutics.
MGMT promoter methylation suppresses gene transcription and should significantly reduce MGMT protein expression. Consistent with this model, MGMT methylated tumors expressed low levels of MGMT protein (Fig. 2). However, a subset of tumors lacking MGMT methylation also had low MGMT protein levels (GBM14, GS28 and GBM34) similar to those seen in some MGMT methylated tumors (GBM12, GBM16 and GBM36). In this subset of low MGMT-expressing tumors, there was a broad range in TMZ responsiveness with prolongation in survival ranging from 68% to 464%. Similarly, clinical studies have demonstrated a range in TMZ responsiveness in tumors with low MGMT expression levels as assessed by immunohistochemistry [16], and our group has observed similar discrepancies between MGMT promoter methylation status and MGMT protein expression levels in patient samples [17]. Collectively, these results suggest that the predictive accuracy of MGMT methylation by standard MS-PCR or MGMT protein expression is insufficient to confidently predict tumor TMZ responsiveness in individual patients.
Standard MS-PCR assay for MGMT relies on annealing of PCR primers across 9 CpG sites and does not delineate the methylation status of individual CpG sites. To address whether this inherent limitation affects the predictive accuracy, the standard MS-PCR results were compared to methylation-specific sequence analysis of 12 CpG sites across this region. Although there was general agreement between the assays, the methylation status of one or more CpG sites was discordant with the qMS-PCR results in 6 of 13 xenograft lines. Consistent with these results, others have demonstrated heterogeneity in the methylation status of discrete CpG sites in tumor cell lines and human tumor tissues [18–20]. In the relatively resistant GBM36 tumor, bisulfite-sequencing revealed lack of methylation at all CpG sites, while standard MS-PCR demonstrated amplification of both unmethylated and methylated products. These discordant results likely reflect heterogeneous methylation of the MGMT promoter and highlight the inherent limitation of non-quantitative MS-PCR, which may detect a small fraction of cells with MGMT promoter methylation [21–23].
CpG methylation within promoter regions provides docking sites for a family of methyl-CpG binding proteins, which in turn recruit chromatin modifying complexes that ultimately regulate gene expression [24]. Most of these methyl-CpG binding proteins recognize and bind to a single methylated CpG site (reviewed in [25]), within a particular sequence context [26, 27]. Several studies have demonstrated discrete methylation of individual CpG sites in association with silencing of the BRCA1 gene in breast cancer and TCEAL7 in ovarian cancer [28, 29]. The MGMT promoter contains multiple putative Sp1 transcription factor binding sites (Fig. 1), and DNA binding and transcriptional activity of Sp1 and some related family members can be modulated by discrete methylation within DNA promoter elements [30]. From these data, we hypothesize that methylation of individual CpG sites within the MGMT promoter may be key elements that modulate MGMT expression and that evaluation of methylation at these discrete CpG sites may provide a more robust predictor of TMZ responsiveness than the conventionally used MGMT standard MS-PCR assay. While the labor intensive nature of bisulfite sequencing precludes routine use in patient samples, other methylation detection platforms could be adapted to evaluate discrete CpG methylation sites. Towards this end, we are expanding our analysis to involve all CpG sites within the MGMT promoter region (Fig. 1) and including additional xenograft lines as well as patient tumor samples to fully delineate the most important CpG sites within the promoter region for regulation of MGMT-dependent TMZ resistance.
In conclusion, the data presented suggest that MGMT promoter methylation (determined by standard MS-PCR and/or qMS-PCR) is an important but not absolute predictor of TMZ response in an orthotopic GBM xenograft model. The heterogeneity in CpG methylation within the region of MGMT promoter evaluated by the standard MS-PCR assay highlight one potential limitation of this assay.
Supplementary Material
Acknowledgments
Grant Support: National Institutes of Health CA108961 (JNS/CG), CA127716 (JNS), NS49720 (CDJ), CA097257 (CDJ); American Cancer Society Research Scholar Grant (JNS); Brain Tumor Funders Consortia (JNS).
References
- 1.Liu L, Gerson SL. Targeted Modulation of MGMT: Clinical Implications. Clin Cancer Res. 2006;12:328–331. doi: 10.1158/1078-0432.CCR-05-2543. [DOI] [PubMed] [Google Scholar]
- 2.Paz MF, Yaya-Tur R, Rojas-Marcos I, Reynes G, Pollan M, Aguirre-Cruz L, Garcia-Lopez JL, Piquer J, Safont MJ, Balana C, Sanchez-Cespedes M, Garcia-Villanueva M, Arribas L, Esteller M. CpG island hypermethylation of the DNA repair enzyme methyltransferase predicts response to temozolomide in primary gliomas. Clin Cancer Res. 2004;10:4933–4938. doi: 10.1158/1078-0432.CCR-04-0392. [DOI] [PubMed] [Google Scholar]
- 3.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA Repair Gene O6-Methylguanine-DNA Methyltransferase by Promoter Hypermethylation is a Common Event in Primary Human Neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- 4.Hegi ME, Diserens A-C, Gorlia T, Hamou M-F, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, Bromberg JEC, Hau P, Mirimanoff RO, Cairncross JG, Janzer RC, Stupp R. MGMT Gene Silencing and Benefit from Temozolomide in Glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 5.Sarkaria JN, Yang L, Grogan PT, Kitange GJ, Carlson BL, Schroeder MA, Galanis E, Giannini C, Wu W, Dinca EB, James CD. Identification of molecular characteristics correlated with glioblastoma sensitivity to EGFR kinase inhibition through use of an intracranial xenograft test panel. Mol Cancer Ther. 2007;6:1167–1174. doi: 10.1158/1535-7163.MCT-06-0691. [DOI] [PubMed] [Google Scholar]
- 6.Sarkaria JN, Carlson BL, Schroeder MA, Grogan P, Brown PD, Giannini C, Ballman KV, Kitange GJ, Guha A, Pandita A, James CD. Use of an Orthotopic Xenograft Model for Assessing the Effect of Epidermal Growth Factor Receptor Amplification on Glioblastoma Radiation Response. Clin Cancer Res. 2006;12:2264–2271. doi: 10.1158/1078-0432.CCR-05-2510. [DOI] [PubMed] [Google Scholar]
- 7.Giannini C, Sarkaria J, Saito A, Uhm J, Galanis E, Carlson B, Schroeder M, James C. Patient Tumor EGFR and PDGFRA Gene Amplifications Retained in an Invasive Intracranial Xenograft Model of GBM. Neuro-Oncology. 2005;7:164–176. doi: 10.1215/S1152851704000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB, Herman JG. Inactivation of the DNA-Repair Gene MGMT and the Clinical Response of Gliomas to Alkylating Agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 9.Matsukura S, Soejima H, Nakagawachi T, Yakushiji H, Ogawa A, Fukuhara M, Miyazaki K, Nakabeppu Y, Sekiguchi M, Mukai T. CpG methylation of MGMT and hMLH1 promoter in hepatocellular carcinoma associated with hepatitis viral infection. Br J Cancer. 2003;88:521–529. doi: 10.1038/sj.bjc.6600743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kokkinakis DM, Bocangel DB, Schold SC, Moschel RC, Pegg AE. Thresholds of O6-alkylguanine-DNA alkyltransferase which confer significant resistance of human glial tumor xenografts to treatment with 1,3-bis(2-chloroethyl)-1-nitrosourea or temozolomide. Clin Cancer Res. 2001;7:421–428. [PubMed] [Google Scholar]
- 11.Leuraud P, Taillandier L, Medioni J, Aguirre-Cruz L, Criniere E, Marie Y, Kujas M, Golmard J-L, Duprez A, Delattre J-Y, Sanson M, Poupon M-F. Distinct Responses of Xenografted Gliomas to Different Alkylating Agents Are Related to Histology and Genetic Alterations. Cancer Res. 2004;64:4648–4653. doi: 10.1158/0008-5472.CAN-03-3429. [DOI] [PubMed] [Google Scholar]
- 12.Middlemas DS, Stewart CF, Kirstein MN, Poquette C, Friedman HS, Houghton PJ, Brent TP. Biochemical correlates of temozolomide sensitivity in pediatric solid tumor xenograft models. Clin Cancer Res. 2000;6:998–1007. [PubMed] [Google Scholar]
- 13.Friedman HS, Dolan ME, Pegg AE, Marcelli S, Keir S, Catino JJ, Bigner DD, Schold SC., Jr Activity of temozolomide in the treatment of central nervous system tumor xenografts. Cancer Research. 1995;55:2853–2857. [PubMed] [Google Scholar]
- 14.Pandita A, Aldape KD, Zadeh G, Guha A, James CD. Contrasting in vivo and in vitro fates of glioblastoma cell subpopulations with amplified EGFR. Genes Chromosomes & Cancer. 2003;39:29–36. doi: 10.1002/gcc.10300. [DOI] [PubMed] [Google Scholar]
- 15.Hegi ME, Diserens A-C, Godard S, Dietrich P-Y, Regli L, Ostermann S, Otten P, Van Melle G, de Tribolet N, Stupp R. Clinical Trial Substantiates the Predictive Value of O-6-Methylguanine-DNA Methyltransferase Promoter Methylation in Glioblastoma Patients Treated with Temozolomide. Clin Cancer Res. 2004;10:1871–1874. doi: 10.1158/1078-0432.ccr-03-0384. [DOI] [PubMed] [Google Scholar]
- 16.Friedman HS, McLendon RE, Kerby T, Dugan M, Bigner SH, Henry AJ, Ashley DM, Krischer J, Lovell S, Rasheed K, Marchev F, Seman AJ, Cokgor I, Rich J, Stewart E, Colvin OM, Provenzale JM, Bigner DD, Haglund MM, Friedman AH, Modrich PL. DNA mismatch repair and O6-alkylguanine-DNA alkyltransferase analysis and response to Temodal in newly diagnosed malignant glioma. J Clin Oncol. 1998;16:3851–3857. doi: 10.1200/JCO.1998.16.12.3851. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez FJ, Thibodeau SN, Jenkins RB, Schowalter KV, Caron BL, O’Neill BP, James CD, Passe S, Slezak J, Giannini C. MGMT immunohistochemical expression and promoter methylation in human glioblastoma. Applied Immunohistochemistry & Molecular Morphology. 2007 doi: 10.1097/PAI.0b013e31802fac2f. in press. [DOI] [PubMed] [Google Scholar]
- 18.Qian X, von Wronski MA, Brent TP. Localization of methylation sites in the human O6-methylguanine-DNA methyltransferase promoter: correlation with gene suppression. Carcinogenesis. 1995;16:1385–1390. doi: 10.1093/carcin/16.6.1385. [DOI] [PubMed] [Google Scholar]
- 19.Watts GS, Pieper RO, Costello JF, Peng YM, Dalton WS, Futscher BW. Methylation of discrete regions of the O6-methylguanine DNA methyltransferase (MGMT) CpG island is associated with heterochromatinization of the MGMT transcription start site and silencing of the gene. Molecular & Cellular Biology. 1997;17:5612–5619. doi: 10.1128/mcb.17.9.5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herfarth KK, Brent TP, Danam RP, Remack JS, Kodner IJ, Wells SA, Jr, Goodfellow PJ. A specific CpG methylation pattern of the MGMT promoter region associated with reduced MGMT expression in primary colorectal cancers. Molecular Carcinogenesis. 1999;24:90–98. doi: 10.1002/(sici)1098-2744(199902)24:2<90::aid-mc3>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 21.Yamada H, Vijayachandra K, Penner C, Glick A. Increased sensitivity of transforming growth factor (TGF) beta 1 null cells to alkylating agents reveals a novel link between TGFbeta signaling and O(6)-methylguanine methyltransferase promoter hypermethylation. J Biol Chem. 2001;276:19052–19058. doi: 10.1074/jbc.M100615200. [DOI] [PubMed] [Google Scholar]
- 22.Danam RP, Howell SR, Remack JS, Brent TP. Heterogeneous methylation of the O(6)-methylguanine-DNA methyltransferase promoter in immortalized IMR90 cell lines. International Journal of Oncology. 2001;18:1187–1193. doi: 10.3892/ijo.18.6.1187. [DOI] [PubMed] [Google Scholar]
- 23.Vlassenbroeck I, Califice S, Diserens AC, Migliavacca E, Straub J, Di Stefano I, Moreau F, Hamou MF, Renard I, Delorenzi M, Flamion B, Diguiseppi J, Bierau K, Hegi ME. Validation of Real-Time Methylation-Specific PCR to Determine O6-Methylguanine-DNA Methyltransferase Gene Promoter Methylation in Glioma. J Mol Diagn. 2008;10:332–337. doi: 10.2353/jmoldx.2008.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Danam RP, Howell SR, Brent TP, Harris LC. Epigenetic regulation of O6-methylguanine-DNA methyltransferase gene expression by histone acetylation and methyl-CpG binding proteins. Mol Cancer Ther. 2005;4:61–69. [PubMed] [Google Scholar]
- 25.Ballestar E, Esteller M, Ballestar E, Esteller M. Methyl-CpG-binding proteins in cancer: blaming the DNA methylation messenger. Biochemistry & Cell Biology. 2005;83:374–384. doi: 10.1139/o05-035. [DOI] [PubMed] [Google Scholar]
- 26.Fraga MF, Ballestar E, Montoya G, Taysavang P, Wade PA, Esteller M, Fraga MF, Ballestar E, Montoya G, Taysavang P, Wade PA, Esteller M. The affinity of different MBD proteins for a specific methylated locus depends on their intrinsic binding properties. Nucleic Acids Res. 2003;31:1765–1774. doi: 10.1093/nar/gkg249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sekimata M, Homma Y, Sekimata M, Homma Y. Sequence-specific transcriptional repression by an MBD2-interacting zinc finger protein MIZF. Nucleic Acids Res. 2004;32:590–597. doi: 10.1093/nar/gkh249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mancini DN, Rodenhiser DI, Ainsworth PJ, O’Malley FP, Singh SM, Xing W, Archer TK. CpG methylation within the 5′ regulatory region of the BRCA1 gene is tumor specific and includes a putative CREB binding site. Oncogene. 1998;16:1161–1169. doi: 10.1038/sj.onc.1201630. [DOI] [PubMed] [Google Scholar]
- 29.Chien J, Staub J, Avula R, Zhang H, Liu W, Hartmann LC, Kaufmann SH, Smith DI, Shridhar V. Epigenetic silencing of TCEAL7 (Bex4) in ovarian cancer. Oncogene. 2005;24:5089–5100. doi: 10.1038/sj.onc.1208700. [DOI] [PubMed] [Google Scholar]
- 30.Costello JF, Futscher BW, Kroes RA, Pieper RO. Methylation-related chromatin structure is associated with exclusion of transcription factors from and suppressed expression of the O-6-methylguanine DNA methyltransferase gene in human glioma cell lines. Molecular & Cellular Biology. 1994;14:6515–6521. doi: 10.1128/mcb.14.10.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.