Abstract
Bioinformatic analysis of the intergenic regions of Staphylococcus aureus predicted multiple regulatory regions. From this analysis, we characterized 11 novel noncoding RNAs (RsaA‐K) that are expressed in several S. aureus strains under different experimental conditions. Many of them accumulate in the late-exponential phase of growth. All ncRNAs are stable and their expression is Hfq-independent. The transcription of several of them is regulated by the alternative sigma B factor (RsaA, D and F) while the expression of RsaE is agrA-dependent. Six of these ncRNAs are specific to S. aureus, four are conserved in other Staphylococci, and RsaE is also present in Bacillaceae. Transcriptomic and proteomic analysis indicated that RsaE regulates the synthesis of proteins involved in various metabolic pathways. Phylogenetic analysis combined with RNA structure probing, searches for RsaE‐mRNA base pairing, and toeprinting assays indicate that a conserved and unpaired UCCC sequence motif of RsaE binds to target mRNAs and prevents the formation of the ribosomal initiation complex. This study unexpectedly shows that most of the novel ncRNAs carry the conserved C−rich motif, suggesting that they are members of a class of ncRNAs that target mRNAs by a shared mechanism.
INTRODUCTION
RNAs are recognized as major regulators of gene expression, and many of them affect the fate of mRNAs at different levels. In bacteria, systematic searches for functional intergenic regions using a combination of methods ranging from in silico prediction to genome-wide expression studies have led to the discovery of more than 150 small trans-acting non-coding RNAs [generally referred to as small RNAs (sRNAs)]. Most of them have been identified in Escherichia coli and closely related bacteria (1,2). These sRNAs participate in regulatory pathways that allow the bacteria to sense the population density, to modulate their cell surface composition in response to various stresses, and to adjust their metabolism during cell growth. It was thus not surprising to discover that RNA-dependent regulation also enables several pathogenic bacteria to express virulence genes and to adapt their metabolic needs during the infection process (3,4).
Staphylococcus aureus is a Gram-positive pathogen responsible for a wide-range of human diseases, from minor skin infections to life-threatening diseases such as endocarditis, pneumonia, or septic shock. It is also one of the leading causes of hospital-acquired infections, often causing post-surgical wound infections. Moreover, the emergence of multiple antibiotic resistances renders the treatment of the infections difficult (5). The high diversity of disease manifestations caused by S. aureus depends on the expression of numerous virulence factors and stress-response pathways. This requires the coordinated action of multiple regulators including several two-component systems, transcriptional regulatory proteins, the alternative sigma B factor and the regulatory RNAIII (6,7). Remarkably, regulation of the genes encoding virulence factors occurs at multiple levels and in a temporal manner, suggesting that the amount and timing of production of the virulence factors need to be precisely controlled during the course of infection. The complexity of these regulatory networks likely benefits the pathogen by allowing it to integrate multiple external signals. Although many studies have focused on regulatory proteins, few studies have addressed the importance of RNA-dependent regulation in S. aureus (8–12). Only RNAIII, which is so far the largest known regulatory RNA, has been shown to control the switch between the expression of many surface proteins and that of excreted toxins (13). RNAIII plays a key role in the quorum sensing-dependent regulatory circuit and coordinately regulates the synthesis of virulence-associated genes and of a transcriptional regulator at the post-transcriptional level through multiple RNAIII-mRNA interactions (6,9,14,15). Staphylococcus aureus RNAIII belongs to a major class of regulatory RNAs that acts on mRNAs by an antisense mechanism (1,16–18). In Enterobacteriaceae, many sRNAs require the Sm-like protein Hfq to facilitate mRNA-sRNA interaction and to enhance sRNA stability (19). However, S. aureus Hfq is required neither for virulence nor for RNAIII function (10,20) and it is characterized by different RNA-binding properties from that of E. coli Hfq (21). Several stable RNAs have been identified in infectious S. aureus strains (8,12,22). Based on their sequence conservation and GC-rich content, seven sRNAs were found to be encoded by pathogenicity islands, although their functions remain unknown (8). Their localization on mobile genetic elements, which contain clusters of virulence factors and genes encoding antibiotic resistance, suggests that they may contribute to niche adaptation and pathogenicity. Numerous stable RNAs, identified by microarrays, were differently expressed under stringent, cold and heat shock responses, but many of them may be co-transcribed as part of an operon (11,12). More recently, a large-scale computational work predicted numerous ncRNA genes in 932 different bacteria, revealing the potential presence of 47 new ncRNAs in the N315 strain of S. aureus (23).
In the present work, we use computational tools and expression studies to identify sRNA genes in intergenic regions (IGR) of S. aureus and related Gram-positive bacteria. Eleven new sRNAs are described and characterized with respect to transcriptional regulation, Hfq-dependence, stability, their target mRNAs and their likely biological functions. Most notably, one ncRNA regulates metabolism and uses a conserved and unpaired C-rich sequence that binds to the Shine-Dalgarno sequence of mRNA targets. This specific signature was found in many of the identified ncRNAs in S. aureus, suggesting that they regulate gene expression by a shared mechanism.
MATERIALS AND METHODS
Genomes, databases and known sRNA annotation
Genome sequences and ORF files (.gbk, .fna and .ptt extensions) were downloaded from the NCBI ftp database for S. saprophyticus ATCC15305, S. haemolyticus JCSC1435, S. aureus N315 and S. epidermidis RP62A. Currently, there are 110 known annotated RNAs in the database for the S. aureus N315 genome. In this analysis, the coordinates of 78 tRNAs and rRNAs were downloaded from the NCBI. The coordinates of other 32 putative sRNAs and riboswitches were obtained from the RFAM (version 8.1) database. The coordinates of other known sRNAs including RNAIII, sprA, sprB, sprC, sprD, sprE, sprFG and other stable RNAs of S. aureus N315 were obtained from data available in the literature (8,11,12,22,24).
Variable regions resulting from a comparative analysis of available S. aureus strains were obtained from MOSAIC, a relational database developed to compare closely related bacterial genomes (25,26). Tandem repetitive elements were assigned using the Microorganisms Tandem Repeats Database [(27); http://minisatellites.u-psud.fr/GPMS/] for S. aureus N315. Additional sequence comparisons were performed with Blastn (default values) in order to complete the list of repeats. PatScan (28) allowed us to search for the STAR element (29) and we also used it to find σA- and σB-dependent promoters, Fur-boxes (30) and hairpins with lengths greater than 6 bp. Rho-independent transcription terminators were found with TransTermHP with default parameters [(31); http://transterm.cbcb.umd.edu/].
In silico prediction of sRNA genes
The possibility of a biased composition analysis in S. aureus N315 was considered since the G+C content was significantly higher in the set of the 110 annotated ncRNAs than in the rest of the genome (Gaspin et al., unpublished data). This result lead us to use a two state Hidden Markov Model (HMM) to segment the genomic sequence into RNA and other regions. Parameters of the HMM were adapted from the analysis performed on the AT-rich hyperthermophilic Archaea Methanococcus jannaschii and Pyrococcus furiosus (32). From HMM predictions, only putative RNAs of lengths greater than 70 nt located in intergenic regions (not overlapping long ORFs) were kept for further analysis. A comparative analysis of the chromosomes of S. saprophyticus ATCC15305 (NC_007350.1), S. haemolyticus JCSC1435 (NC_0071168.1), S. aureus N315 (NC_002745.2) and S. epidermidis RP62A (NC_002976.3) was performed. A file containing fasta nucleotide sequences of intergenic regions (IGR) greater than 150 nt and ORFs with a size lower than 100 aa was built. For each chromosome, all the RNAs (tRNAs, tmRNA and rRNAs) annotated in Genbank were removed. All the IGR sequences were compared pairwise with the RNAsim software (Supplementary Figures S1 and S2). RNAsim searches for conserved sequences and structural regions between different genomes. It is based on Wu-blast 2.0 pairwise comparisons (this work, W = 11, E < 0.001) of sequences for searching similarities, and it uses QRNA (33) to identify base substitution patterns in pairwise alignments that could correspond to a conserved RNA secondary structure. Finally, in the context of a multi-genome comparison, it combines overlapping predicted loci that are conserved in multiple replicons into a prediction. Only putative RNAs with lengths greater than 150 nt were selected to search for similarities against the NCBI complete bacterial genome database (W = 7 and default parameters).
Integration of data and results of predictions
ApolloRNA (http://www.carlit.toulouse.inra.fr/ApolloRNA) is an extension of the Apollo environment [http://apollo.berkeleybop.org/current/index.html; (34)], which provides information on the orientation of the flanking genes and on all annotated genes. ApolloRNA was used to integrate the HMM and comparative analysis predictions (Supplementary Figure S1). We also assigned riboswitch elements, known ncRNAs (8,11), STAR (30), other repeats (29) and mobile elements (transposons, pathogenicity islands). All these data are reported in Supplementary Tables S1 and S2. ApolloRNA also contains some extensions to Apollo that are devoted to RNA analysis. It includes the integration of RNAfold of the Vienna package to predict the RNA secondary structure of a selected sequence along the genome (35). In addition, ApolloRNA integrates tools to interactively search for possible RNA/RNA interactions, and to visualize the graphs of nucleotide composition and minimum free energy associated with a sliding window all along a genome. In ApolloRNA, all predicted RNAs were analyzed with regard to available data and orientation of the flanking genes. Several were selected for further expression studies (Table 1, Supplementary Figure S1).
Table 1.
List of selected intergenic regions of S. aureus for expression studies
| Rsaa | Start-end (size) | Startb | Flanking genes | Strand | Selection criteriac | Commentsd |
|---|---|---|---|---|---|---|
| RsaA | 637111–637250 (139) | RT, 5′RACE | SA0543/SA0544 | < > < | RNAsim, ΔG, Term | E<<S (BHI, NZM), Staphylococci |
| RsaB | 1778009–1778068 (60) | RT, 5′3′RACE | SA1552/fhs | < > < | RNAsim | E<<S (BHI), S. aureus |
| RsaC | 679909–680452 (544) | 5′3′RACE | SA0586/SA0587 | < > > | ΔG, RNAsim, Term | E<<S (BHI, NZM), S. aureus |
| RsaD | 695867–696043 (177) | RT | SA0600/SA0601 | > < > | RNAsim, Term | E<<S (BHI, NZM), Staphylococci |
| RsaE | 975383–975482 (100) | RT, 5′RACE | SA0859/SA0860 | > > < | RNAsim, Term | E<<S (BHI, NZM), Bacillaceae |
| RsaF | 975461–975564 (104) | 5′RACE | SA0859/SA0860 | > > < | ΔG, RNAsim | E<<S (BHI, NZM), S. aureus |
| RsaG | 254702–254895 (194) | RT, 5′RACE | uhpT/SA0215 | > > < | HMM, ΔG, Term | S (BHI), S. aureus |
| RsaH | 829511–829634 (124) | RT, 5′3′RACE | SA0724/SA0725 | < > > | ΔG, RNAsim, Term | E<<S (BHI), Staphylococci |
| RsaI | 2367918–2368061 (144) | RT, 5′RACE | SA2104/SA2105 | < < > | RNAsim, Term | E<<S (BHI), Staphylococci |
| RsaJ | 2486806–2487092 (287) | nd | bioD/SA2216 | < > < | RNAsim, ΔG, Term | E<<S (BHI), S. aureus |
| RsaK | 216920–217128 (209) | RT | glcA/SA0184 | > > < | RNAsim, Term | Cis-acting leader of glcA |
| RsaX01 | 43000–43600 | SA0035/SA0036 | HMM, RNAsim | – | ||
| RsaX02 | 81706–81855 | kdpC/SA0072 | RNAsim, Term | (23) | ||
| RsaX03 | 95295–95689 | SA0084–SA0085 | RNAsim, Term | (22) | ||
| RsaX04 | 149756–149850 | SA0129–SA0130 | RNAsim, Term | – | ||
| RsaX05 | 470600–470900 | SA0410/ndhF | Prom, Term | Cis-acting leader of ndhF? | ||
| RsaX06 | 659823–659968 | SA0565/SA0566 | RNAsim | – | ||
| RsaX07 | 666122–666257 | SA0575/sarA | RNAsim, Term | – | ||
| RsaX08 | 761087–761230 | SA0667/SA0668 | RNAsim, Term | PreQ1 riboswitch, (22) | ||
| RsaX09 | 905650–905835 | SA0801/SA0802 | Term | Cis-acting leader of SA0802? | ||
| RsaX10 | 1138640–1138848 | SA1005/SA1006 | RNAsim, Term | – | ||
| RsaX11 | 1175489–1175753 | SA1037/lsp | HMM, RNAsim, Term | – | ||
| RsaX12 | 1399044–1399209 | SA1224/lysC | RNAsim, HMM | Upstream of lysine riboswitch | ||
| RsaX13 | 1421983–1422128 | odhA/arlS | Term | UCCC motif | ||
| RsaX14 | 1801062–1801380 | SA1570/SA1571 | RNAsim, Term | – | ||
| RsaX15 | 1839300–1839600 | SA1602/trunc.SA | HMM, RNAsim, Term | – | ||
| RsaX16 | 1914236–1914464 | SA1676/tnp | RNAsim, Term | – | ||
| RsaX17 | 1992090–1992414 | SA1738/SAS056 | HMM, RNAsim, Term | (11) | ||
| RsaX18 | 2436200–2436800 | SA2168–SA2169 | RNAsim, ΔG | (11) | ||
| RsaX19 | 2440668–2440850 | scrA/gltT | HMM, RNAsim, Term | Repeat motifs, STAR | ||
| RsaX20 | 2473450–2473566 | SA2203/SA2204 | RNAsim, ΔG, Term | UCCC motif, sORF upstream | ||
| RsaX21 | 2544796–2545113 | SA2268/SA2269 | Term | – | ||
| RsaX22 | 2586551–2586821 | SA2301/SA2302 | RNAsim | – | ||
| RsaX23 | 2590928–2591046 | fbp/SA2305 | RNAsim, Term | sprA3 (8) | ||
| RsaX24 | 2649194–2649400 | isaA/SA2357 | RNAsim, Term | – | ||
| RsaX25 | 2769241–2769725 | SA2457/icaR | HMM, RNAsim, Term | UCCCC motif |
aThe Rsa sRNAs have been referred in GenBank under the accession numbers: GQ403615 (RsaA), GQ403616 (RsaB), GQ403617 (RsaC), GQ403618 (RsaD), GQ403619 (RsaE), GQ403620 (RsaF), GQ403621 (RsaG), GQ403622 (RsaH), GQ403623 (RsaI), GQ403624 (RsaJ) and GQ403625 (RsaK). RsaD, E, H and I were predicted by Livny et al. (23) and RsaI was also found expressed in the S. aureus N315 strain (22). In RN6390, no significant expression was detected for RsaX01 to RsaX26. sORF is for small open reading frame.
bThe 5′ start of Rsa RNAs was determined by primer extension (RT), and Rapid Amplification of cDNA Ends (RACE); 5′3′RACE was done on circularized RNA; nd is for not determined.
cTerm is for Rho-independent terminator.
dThe expression of RsaA to RsaK was analyzed at the exponential (E) and the stationary (S) phase of growth in various S. aureus strains.
Strains, vector and plasmids
Staphylococcus aureus RN6390 derives from the NCTC8325-4 strain (Table 2). In WA400 (ΔrnaIII), the P3 operon is deleted and replaced by the chloramphenicol transacetylase gene (cat86) (36). In the LUG774 and LUG911 strains, the RNase III (rnc) and hfq genes, respectively, have been replaced by the cat86 gene (9,14) (Table 2). In the LUG1430 strain, the rsaE gene has been replaced by aphA-3, which confers kanamycin resistance. These strains are derived from RN6390. For the deletion of the rsaE gene, the deletion/replacement ΔrsaE/aphA-3 mutant of S. aureus RN6390 was obtained by using pMAD, a thermosensitive plasmid (37). Clones were transferred by electroporation into RN4220, a nitrosoguanidine-induced mutant capable of accepting E. coli DNA, before transfer to other strains. Staphylococci were grown on BM agar plates, in brain-heart infusion (BHI) with kanamycin (5 µg/ml) when appropriate, or in NZM medium (10 g/l Casein Tryptone digested, Fluka 95039; 5 g/l NaCl; 2 g/l MgSO4–7H20; pH 7.0). The allelic replacement of rsaE was verified by PCR.
Table 2.
Strains and plasmids
| Name | Revelant characteristics | Reference or source | |
|---|---|---|---|
| Escherichia coli | DH5α, cloning strain | Laboratory stock | |
| Bacillus subtilis | Wild type strain | ATCC6051 | |
| S. aureus strains | |||
| RN4220 | restriction- mutant of 8325‐4 | (105) | |
| RN6390 | derivative of 8325-4, agr positive | (106) | |
| RN6911 | RN6390: Δagr region::tetM | (107) | |
| WA400 | 8325-4: ΔrnaIII region::cat86 | (36) | |
| COL | Methicillin-resistant laboratory strain | (108) | |
| Newman | Wild type strain | (109) | |
| SH1000 | rsbU+ strain derivative of 8325-4 | (56) | |
| RN1HG | rsbU+ strain derivative of RN1 | (57) | |
| LUG774 | RN6390: Δrnc region::cat86 | (14) | |
| LUG911 | RN6390: Δhfq region::cat86 | (9) | |
| LUG1160 | RN6390: Δrot region::cat86 | This work | |
| LUG1397 | RN4220/pLUG766 | This work | |
| LUG1408 | RN6390/pLUG766 | This work | |
| LUG1430 | RN6390: ΔrsaE:: aphA-3 | This work | |
| E. coli plasmids | |||
| pUT7_RsaG | T7 promoter/RsaG | This work | |
| pUC18_T7_RsaA | T7 promoter/RsaA | This work | |
| pUC18_T7_RsaH | T7 promoter/RsaH | This work | |
| pUC18_T7_RsaE | T7 promoter/RsaE | This work | |
| pUC18_T7_oppB | T7 promoter/5′-region of oppB (nt −182 to +182, +1 being the A of AUG codon) | This work | |
| pUC18_T7_SA0873 | T7 promoter/5′-region of SA0873 (nt −22 to +128) | This work | |
| E. coli-staphylococcal shuttle plasmids | |||
| pMAD | Thermosensitive origin of replication, constitutively expressed βgalB gene | (37) | |
| pLUG766 | pMAD derivative for deletion/ replacement of S. aureus rsaE gene | This work | |
Staphylococcus aureus COL strains were submitted to various stress conditions. The cells were grown to mid-exponential phase (OD 600 nm 0.5) at 37°C and stress was induced by the addition of 15% NaCl (osmotic stress), 5 mM hydrogen peroxide (oxidative stress), 0.1 mM paraquat (oxidative stress), 0.25 mM dipyridyl (an iron chelating agent), or HCl to decrease the pH of the medium to 4.5. The cells were submitted to either cold shock (25°C) or heat shock (42°C). After 30 min, the cells were centrifuged and the total RNA was extracted as described previously (9). Total RNAs were prepared from different S. aureus or B. subtilis strains at early (2 h) and late (6 h) exponential phases of growth, or from S. aureus COL strain submitted to stress conditions.
Northern analysis, RACE and primer extension analysis
Electrophoresis of total RNAs was done on 5% polyacrylamide gels containing 8 M urea or 1% agarose gels containing 20 mM guanidine thiocyanate (Sigma, G9277). Hybridizations were carried out with end-labeled specific oligodeoxynucleotides to detect mRNA, Rsa RNAs, RNAIII or 5S rRNA (14).
Primer extension was carried out on 2 µg of total RNA prepared from S. aureus or B. subtilis strains. Total RNA was first denatured for 1 min at 90°C followed by 1 min on ice in 15 µl of RNase-free water in the presence of 2 µl of 5′-end labeled oligodeoxynucleotide complementary to a specific Rsa RNA (Supplementary Table S4). The hybridization was done at 20°C for 15 min in the presence of 3 µl of the commercial AMV 10× buffer (Qbiogen) and 6 µl of dNTP 2.5 mM. The extension was done with 1 µl of AMV reverse transcriptase (20 U/µl, Qbiogen) for 30 min at 45°C. The RNA was then hydrolyzed by RNase A (1 U) for 10 min at 37°C. After the addition of 4 µl of formamide-bromophenol blue, the samples were loaded onto an 8% polyacrylamide gel containing 8 M urea and 1× TBE. The 5′ start and the size of the RNA were evaluated by running sequencing ladders in parallel.
The 5′-end of the Rsa RNAs was also determined by rapid amplification of cDNA ends (RACE) using the First Choice RLM-RACE kit (Ambion). To assign the 5′- and 3′-ends of the RNAs, the RACE approach was done on RNAs previously circularized using T4 RNA ligase according to Redko et al. (38).
Microarray manufacturing and design
The microarray was manufactured by in situ synthesis of 10 807 long oligonucleotide probes (Agilent, Palo Alto, CA, USA), selected as previously described (39). It covers >98% of all ORFs annotated in strains N315 and Mu50 (40), MW2 (41), COL NCTC8325 (42), USA300 (43), MRSA252 and MSSA476 (44) including their respective plasmids.
Preparation of labeled nucleic acids for expression microarrays
Total RNAs were purified from late-exponential (6 h at 37°C) phase cultures grown in BHI. After additional DNase treatment, the absence of remaining DNA traces was evaluated by quantitative PCR (SDS 7700; Applied Biosystems, Framingham, MA) with assays specific for 16S rRNA (45,46). Batches of 8 µg total S. aureus RNA were labeled by Cy-5dCTP (RN6390 strain) or Cy-3dCTP (LUG1430 strain) using SuperScript II (Invitrogen, Basel, Switzerland) following the manufacturer’s instructions. Labeled products were then purified onto Qiaquick columns (Qiagen).
The Cy5-labeled DNA and the Cy3-labeled cDNA mixture (1 µg) were diluted in Agilent hybridization buffer (final volume of 50 µl) and hybridized at 60°C for 17 h in a dedicated hybridization oven (Robbins Scientific, Sunnyvale, CA, USA). Slides were washed, dried under nitrogen flow and scanned with an Agilent scanner (Agilent, Palo Alto, CA, USA) using 100% Photon Multiplier Tube (PMT) power for both wavelengths.
Fluorescence intensities were extracted using the feature extraction software (Agilent, version 9.3). Local background-subtracted signals were corrected for unequal dye incorporation or unequal loads of the labeled product and normalization per gene and per slide was performed using GeneSpring. Data consisting of two independent biological experiments were expressed as log10 ratios and analyzed using GeneSpring (Agilent). The statistical significance of differentially expressed genes was identified by variance analysis (ANOVA, P < 0.05) (45,47), performed using GeneSpring, including the Benjamini and Hochberg false discovery rate correction (5%).
Microarray data accession number
The complete microarray data set has been posted on the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GPL7137 for the platform design and GSE17135 for the original data set.
Proteomic analysis
Strains were grown at 37°C to exponential (2 h) and post-exponential (6 h) phases by inoculating 50 ml of BHI medium with an overnight culture (1:100). The supernatants (1.2 ml) were precipitated by cold acetone, and the pellets were washed with cold ethanol. Crude extracts from the pellets were obtained by Trizol (Sigma) extraction, followed by acetone precipitation. The proteins were dissolved in sample buffer (7 M urea, 2 M thiourea, 4% CHAPS, 20 mM Tris‐HCl pH 8) and 50 µg were labeled with 400 pmol of amine-reactive-Cy2, Cy3 or Cy5 (Amersham) for 30 min in the dark at 4°C. The reaction was stopped by the addition of 20 µl of 10 mM lysine for 10 min. Alkylation then proceeded at 37°C for 15 min in the presence of DTT (up to 50 mM) and 10 µl of 1 M iodoacetamide. The protein extracts were desalted by a G-25 Sephadex chromatography. Ampholytes (Bio-Rad) and bromophenol blue were added, and the samples were used to re-swell pre-cast pH 3–10 or 4–7 IPG strips (Bio-Rad). Isoelectric focusing was performed using an IEF Cell (Bio-Rad) at 75 kV/h. The IPG strips were buffer-exchanged in 1 M Tris‐HCl pH 8.45, 6 M urea, 2% SDS, 30% glycerol. For the second dimension, the strips were loaded onto the top of a 12% polyacrylamide–2% SDS gel (48). After electrophoresis, the gels were scanned for fluorescence detection, stained with colloidal blue and analyzed using the PQuest software (Bio-Rad). The differentially expressed proteins were characterized by mass spectroscopy.
RNA preparation and band shift analysis
Rsa RNAs (RsaA, E, G and H) and mRNA (oppB, SA0873) fragments were transcribed in vitro using T7 RNA polymerase as previously described (24). Before use, the RNAs were renatured by incubation at 90°C for 2 min and 1 min at 4°C in RNase-free water, followed by an incubation step at 20°C for 15 min in TMN buffer (20 mM Tris‐HCl pH 7.5, 10 mM MgCl2, 150 mM NaCl).
Complex formation was done using 5′-end-labeled RsaE (5 nM) and an increasing amount of unlabeled target mRNA (5–400 nM) for 15 min at 37°C in TMN buffer in a total volume of 10 µl. The samples were then separated on a native 6% polyacrylamide gel in 1× Tris‐borate (0.09 M) buffer. Quantification of the complex formation was done using a Bioimager analyzer.
Structure probing and toeprinting
Enzymatic hydrolysis of free or bound RNAs was performed as previously described (14). The formation of a simplified translational initiation complex with mRNA and the extension inhibition conditions were strictly identical to those described by Benito et al. (24).
RESULTS
Computational selection of IGRs using multiple approaches
Based on functional studies performed on S. aureus RNAIII (9,14), we made the assumption that most sRNAs may carry specific structural motifs required for efficient target recognition. A combination of different tools was then used to retain IGRs that present particular sequence/structure features (Supplementary Figure S1). First, the degree of conservation of the IGRs in different microbial genomes was analyzed. Regions conserved either in various firmicutes (Staphylococcus, Bacillus, Listeria and Clostridium), or in four different staphylococcal species (S. aureus N315, S. epidermidis, S. saprophyticus and S. haemolyticus) were analyzed using RNAsim (Supplementary Figure S2 and Table S1). Because unique genes could also contribute to pathogenesis, we searched S. aureus-specific regions (Supplementary Table S1) using data downloaded from MOSAIC, a database developed for comparison of closely related bacterial genomes (25). MOSAIC identified identical sequences constituting the backbone interrupted by strain-specific sequences (loop regions). Finally, in all the conserved and S. aureus-specific IGRs, we searched for orphan Rho-independent terminators located far downstream of the flanking coding genes (49).
As a second approach, a Hidden Markov Model (HMM) derived from Klein et al. (32) was used to exploit the compositional bias between known RNAs (RFAM, GenBank) and the rest of the genome (Supplementary Figure S1). The HMM model was able to distinguish ncRNAs including tRNAs and rRNAs, from mRNAs on the basis of local variations in genomic base composition of AT rich genomes (8,32,50). Although this approach failed to find RNAIII, it allowed the detection of known ncRNAs such as 6S RNA, SRP, tmRNA and sprA RNAs as well as the identification of 28 candidate sRNAs. In addition, this method identified many repeated sequences and structural motifs known as Staphylococcus aureus repeats (STAR) elements (29) (Supplementary Table S2).
The obtained information was assigned to the annotated sequence of the S. aureus N315 genome using ApolloRNA (http://carlit.toulouse.inra.fr/ApolloRNA/) (51). Using this environment, we have selected 36 IGRs based on at least two different criteria (conserved and S. aureus-specific IGR, orphan terminators, promoters, structural motifs and the orientation and nature of the flanking genes), and the expression patterns of these putative transcripts were analyzed (Table 1, Supplementary Figure S1). Home-made arrays were designed using the 36 PCR-amplified IGR-specific dsDNAs which were spotted on nitrocellulose membranes (for experimental details, see Supplementary Data). As positive control, we used dsDNA corresponding to the regulatory RNAIII. Total RNAs were prepared from strain RN6390 grown in BHI (rich) and NZM (stringent) medium and in exponential and stationary phases. Total RNAs were fractionated by a denaturing polyacrylamide gel electrophoresis to remove ribosomal RNAs. The RNAs with a size of 50–600 nt were eluted, labeled at their 3′-end and hybridized to the membrane under stringent conditions (for experimental details, see Supplementary Data). Strong expression signals were obtained for 11 RNA candidates prepared from cultures grown in the stationary phase. The expression of these 11 RNAs was then analyzed by northern blots in various strains and under stress conditions (Figure 1, Supplementary Figure S3). In the rest of the article, these 11 RNAs are denoted Rsa (A–K), for RNA from S. aureus (Table 1). We used the nomenclature of the S. aureus N315 strain for convenience but all of these IGRs are conserved in all other S. aureus strains sequenced to date.
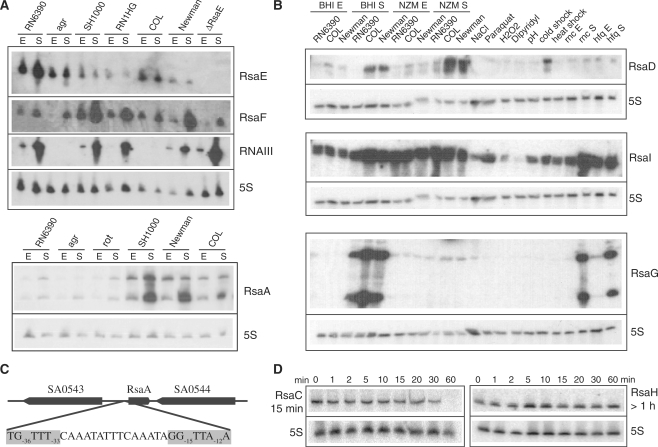
Figure 1.
The expression of Rsa RNAs as monitored under various stresses, culture conditions, and in various S. aureus strains. (A) Northern blot analysis of RsaE, RsaF and RsaA in different S. aureus strains: RN6390 (ΔrbsU, σB−), RN6911 (Δagr, σB−), SH1000, RN1HG, COL and Newman (σB+). The strains LUG1430 (ΔrnaE) and LUG1160 (Δrot) are isogenic to RN6390. Total RNAs were prepared from cells grown in BHI medium and stopped at the exponential (E) or stationary (S) phases of growth. Staphylococcus aureus RNAIII was used as positive control, and 5S rRNA was probed as a quality RNA control. (B) Northern blot analysis of RsaD, RsaI and RsaG in various S. aureus strains (RN6390, COL, Newman, LUG774 (RN6390-Δrnc) and LUG911 (RN6390-Δhfq)) grown in BHI or NZM at exponential (E) and stationary (S) phases of growth. The COL strain was grown under various stress conditions: osmotic stress (NaCl), oxidative stress (H2O2, paraquat), iron chelating agent (dipyridyl), acidic pH, cold shock (25°C) and heat shock (42°C). (C) Genomic organization of the rsaE gene with the σB-binding site; the consensus sequence is highlighted. (D) Northern blot analysis of the half-lives of RsaC and RsaH. The cells were treated with rifampicin and total RNAs were extracted after 1, 2, 5, 10, 20, 30 and 60 min at 37°C in BHI. 5S rRNA was probed to quantify the yield of RNA in each lane.
Expression studies reveal tight transcriptional regulation of Rsa sRNAs
The expression of the 11 Rsa (A‐K) transcripts was further analyzed in several S. aureus reference strains (RN6390, COL and Newman). RN6390, a derivative of NCTC8325-4, has been frequently used for molecular analysis (Table 2). However, this strain carries a mutation in rsbU which results in defective σB activity (52,53). Staphylococcus aureus COL was one of the first methicillin-resistant isolates identified (42). The S. aureus Newman strain was originally isolated from a human infection and has been extensively studied in animal models (54). Based on phylogenetic analysis, these three strains are closely related, while their pathogenicity islands show polymorphisms. The expression of the 11 Rsa RNAs was analyzed in cells grown to exponential and stationary phases. In addition, the COL strain was submitted to stress conditions that might be encountered by the bacteria during infection, such as osmotic and oxidative stress, cold and heat shock, iron limitation and acidity.
Most of the Rsa RNAs (RsaA, C, D, E, F, G, H and I) were highly expressed and showed distinct bands consistent with the length of the predicted IGR (Figure 1, Supplementary Figure S3, Table 1). No significant expression was found for the antisense strands of RsaA, C, H, E, or G (results not shown). The expression of many Rsa RNAs appeared to be tightly regulated, as observed for E. coli and Salmonella typhimurium sRNAs (1,4,55). In rich culture media, RsaB, D, G and H were predominantly expressed in the stationary phase while the expression of RsaA, C, E, F, I and J was observed in the exponential phase, but was strongly enhanced in the stationary phase (Figure 1, Supplementary Figure S3). When cultures were grown in stringent NZM medium, RsaE, F, G and H were not expressed, whereas RsaI expression was significantly enhanced in the exponential phase. Moreover, several Rsa RNAs responded to stress conditions but RsaF and RsaG did not. Indeed, RsaA, E and I were expressed under various stress conditions (osmotic and oxidative stress, acidic pH and temperature variations) while the expression of RsaC and D were significantly induced during cold shock (Figure 1B, Supplementary Figure S3).
RsaA, D, E, F and J were differently expressed in the three S. aureus strains. While RsaA, D and F bands were faint in RN6390, they showed strong expression in the COL and Newman strains (Figure 1A and B, Supplementary Figure S3). An opposite pattern of expression was also observed for RsaE and RsaF (Figure 1A). Since RN6390 has a defect in the alternative sigma B factor due to a mutation in the rsbU gene, we analyzed rsaA, rsaE and rsaF expression in the isogenic rsbU-proficient strain SH1000 (56) and in strain RN1HG (57). The results show that RsaA and RsaF are strongly enhanced in SH1000 and RN1HG, approaching the level observed in the COL and Newman strains (Figure 1A, Supplementary Figure S3). This suggests that transcription of these sRNAs is dependent on rbsU and/or σB. Since the transcription start sites of all sRNAs have been determined either by primer extension and/or RACE (Table 1), we looked for putative promoters directly upstream. We identified a conserved σB promoter sequence [TG−36TTT(N15)GG−15TTA−12A] upstream of the rsaA gene, suggesting a direct role of σB in rsaA transcription (Figure 1C). Moreover, the synthesis of RsaE was significantly repressed in σB+ strains by an unknown mechanism. Interestingly, the partially overlapping RsaE and RsaF transcripts had opposite expression patterns (Figure 1A). RsaF was strongly enhanced in σB+ strains, whereas RsaE was mainly detected in the σB−deficient strain RN6390. The overlap between a σA recognition motif [TTGAAA(N16)TATATT] and a potential σB [AG−36GTT−33GAA(N15)TG−15TTA−12T]-binding site may account for transcriptional interference resulting in the differential expression of rsaE and rsaF (58). RsaE levels were also strongly decreased in an agr mutant strain in the late-exponential phase, suggesting that the synthesis of this RNA is regulated by the quorum sensing system (Figure 1A).
Primary and secondary features of the ncRNAs
The 5′- and 3′-ends of RsaB, C and H were determined by 5′/3′RACE experiments after circularization of total RNAs by T4 RNA ligase (59). The 5′-ends of the other Rsa sRNAs were identified by 5′ RACE (RsaA, E, F, G and I) and primer extension (RsaA, B, D, E, G, H, I and K; Table 1). The transcriptional start sites for most of the sRNAs were consistent with the size of the RNAs as evaluated by northern blots. The 5′-end-mapping of RsaJ failed, probably due to its rather poor expression. Determination of the 5′ starts of the Rsa RNAs helped to predict consensus σA−dependent promoters for RsaB-E/I and a σB-dependent promoter for RsaA (Figure 1C). Most of the Rsa RNAs have at their 3′-ends a typical Rho-independent transcription terminator, except for RsaB and RsaF. The presence of a Rho-independent terminator 136 nt downstream of the 3′-end of RsaF suggests that this RNA might be processed from a longer RNA precursor, which was detectable by Northern blots (Supplementary Figure S3). Noteworthy, the sequences of the terminator structures are identical for RsaJ and SprA, an sRNA expressed from the pathogenicity island (8). The sizes of the sRNAs range from 59 (RsaB) to 544 nt (RsaC) (Table 1).
Their genomic locations indicate that most of the sRNAs are encoded by autonomous genes (Figure 2) and that none of the Rsa RNAs, including the longest one (RsaC, 544 nt), carry a small ORF as has been shown for RNAIII. Four of them (RsaC, H, I and K) are transcribed in the same orientation as their downstream genes, suggesting that they could arise from mRNA leaders, whereas the Rho-independent terminators of RsaC, H and I are located 300, 431, 50 nt upstream of their downstream genes, respectively (Figure 2). The distance and the absence of signals corresponding to longer transcripts suggest that these RNAs are bona fide sRNAs (Figure 1D, Supplementary Figure S3). RsaK, on the other hand, appears to be part of the leader sequence of the glcA mRNA, which encodes an enzyme of the glucose-specific phosphotransferase system (Supplementary Figure S4). RsaK contains a conserved ribonucleic antiterminator sequence (RAT) motif recognized by the transcriptional antiterminator protein GlcT [e.g. (60,61)]. RsaK was detected in several S. aureus strains together with a longer transcript corresponding to the full-length glcA mRNA (Supplementary Figure S4).
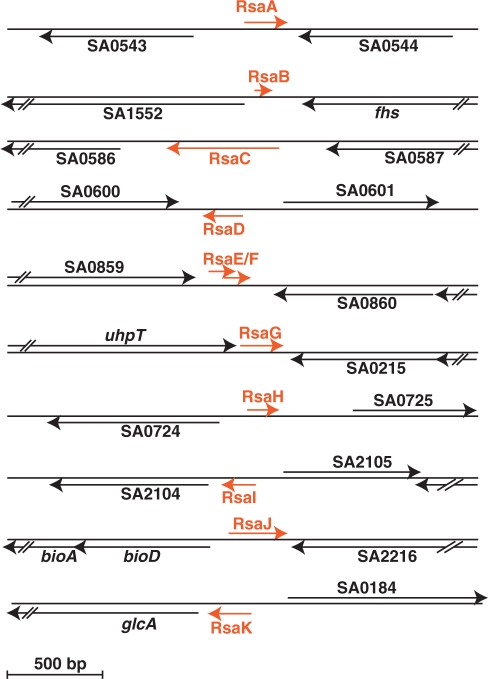
Figure 2.
Genomic organization of rsa genes in the S. aureus N315 strain. Red arrows denote the rsa transcripts and their orientations, and the blacks arrows are for the flanking open reading frames (ORFs). The genes are represented to scale. The sizes of the rsa genes were determined based on the experimental determination of the 5′ start by RACE mapping and primer extension, estimates from denaturing gel electrophoresis, and the presence of transcription terminator. For RsaJ, the transcription start site was not determined and the length is estimated. RsaB and RsaF do not contain transcription terminator at their 3′-ends. In Table 1, the exact genomic location of rsa genes is given.
Rsa RNA sequences were compared across phylogeny using the RNAsim software (Supplementary Figure S2). Their genes and genomic localizations are well-conserved in all sequenced S. aureus strains although expression of some of Rsa RNAs varies in COL and RN6390. Five rsa genes (rsaB, C, F, G and J) are specific to S. aureus whereas all the others (rsaA, D, E, H, I and K) are conserved in four different staphyloccocal species (S. aureus, S. epidermidis, S. saprophyticus and S. haemolyticus). RsaE is unique since its conservation extends to Macrococcus caseolyticus and Bacillus while the partially overlapping RsaF and its upstream gene are only conserved in S. aureus species (Table 1, Supplementary Figure S5). Interestingly, the rsaE gene is always located downstream of the pepF gene, which encodes the conserved oligoendopeptidase F. These two genes, pepF and rsaE, are not always located in the same genomic environment.
The structures of four RNAs (RsaA, E, G and H) were probed by using single-strand-specific (RNases T1, T2 and A) or double-strand-specific (RNase V1) ribonucleases (Figure 3A). The enzymatic cleavage patterns obtained on these RNAs are well correlated with the existence of several hairpin structures and a well-defined terminator of transcription interconnected by long unpaired regions. Multiple sequence alignment was performed with PARADISE (62) which took into account the secondary structure of S. aureus Rsa RNAs in several staphylococcal species (RsaA, D, E, H and I; Figure 3C and Supplementary Figure S6). This analysis unravels conserved regions that are expected to be functional regulatory elements (4). Although the sequences of the helices vary, the overall secondary structures of RsaA, D, E, H and I remain conserved in all staphylococci (Figure 3C, Supplementary Figure S6). Strikingly, all of the Rsa RNAs, except RsaI and F, contain a C-rich sequence motif (UCCC) located in unpaired regions (Figure 3B, Supplementary Figure S6). RsaG, RsaE and RsaC each carry several copies of this conserved sequence. RsaI is characterized by several conserved stretches of nucleotides: two regions are G-rich, while the third is a long, unpaired region that contains stretches of uridines interrupted by adenines and cytosines (Supplementary Figure S6).
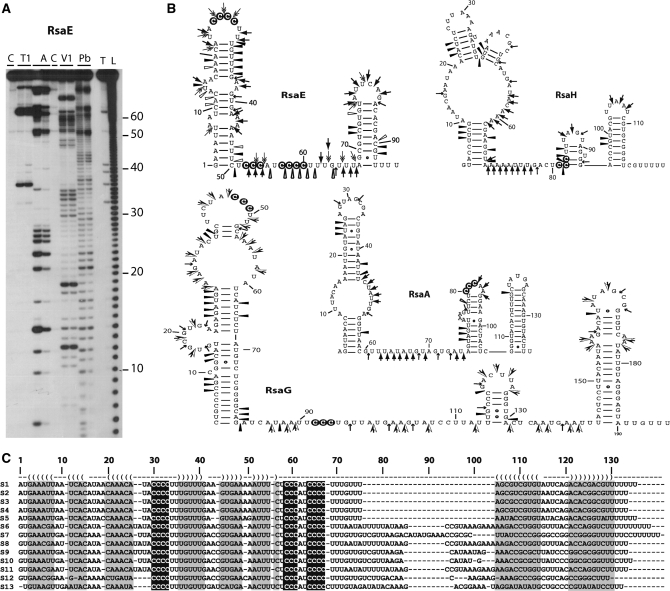
Figure 3.
Secondary structures of various Rsa RNAs. (A) Gel fractionation of enzymatic cleavages and lead-induced cleavages of 5′-end-labeled RsaE: lanes T1: RNase T1, 0.01 and 0.02 U for 10 min at 20°C; lanes A: RNase A, 0.01 µg/ml and 0.001 µg/ml for 10 min at 20°C; lanes V1: RNase V1, 0.05 and 0.1 U for 5 min at 20°C; lanes Pb: lead-induced cleavage, 12.5 and 25 mM for 5 min at 20°C; lane T: RNase T1 under denaturing conditions; lane L: alkaline ladder; lane C: Incubation control. Bars denote the main reactivity changes induced by complex formation. (B) Probing data represented on the secondary structures of RsaE, RsaH, RsaA and RsaG RNAs. Enzymatic cleavages are as follows: RNase T1 ( ), RNase A (
), RNase A ( ), RNase T2 (
), RNase T2 ( ), Lead-cleavages (
), Lead-cleavages ( ) and RNase V1: (
) and RNase V1: ( ) moderate and (
) moderate and ( ) strong cleavages. The cytosines of the C-rich motif are encircled in black. (C) Sequence and structure alignment of RsaE coming from various Staphylococci and Bacillaceae: S1, S. aureus; S2, S. epidermidis; S3, S. saprophyticus; S4, S. haemolyticus; S5, Macrococcus caseolyticus; S6, B. anthracis; S7, B. amyloliquefaciens; S8, B. cereus; S9, B. licheniformis; S10, B. subtilis; S11, B. thuringiensis; S12, Geobacillus thermodenitrificans; S13, Oceanobacillus iheyensis. The helices are shown in grey. The C-rich conserved residues are highlighted in black. The alignment was done with the PARADISE platform (https://simtk.org/home/paradise).
) strong cleavages. The cytosines of the C-rich motif are encircled in black. (C) Sequence and structure alignment of RsaE coming from various Staphylococci and Bacillaceae: S1, S. aureus; S2, S. epidermidis; S3, S. saprophyticus; S4, S. haemolyticus; S5, Macrococcus caseolyticus; S6, B. anthracis; S7, B. amyloliquefaciens; S8, B. cereus; S9, B. licheniformis; S10, B. subtilis; S11, B. thuringiensis; S12, Geobacillus thermodenitrificans; S13, Oceanobacillus iheyensis. The helices are shown in grey. The C-rich conserved residues are highlighted in black. The alignment was done with the PARADISE platform (https://simtk.org/home/paradise).
The ncRNAs are stable
In many enterobacteria, the activity of sRNA acting as antisense RNA depends on the Hfq protein (19,63). Among the multiple functions of this pleiotropic regulator, Hfq stabilizes many sRNAs to provide regulatory activity. Therefore, we compared steady-state levels of the RNAs in RN6390 and its isogenic Δhfq derivative (representative data is shown in Figure 1D and Supplementary Figure S3). Although these data do not exclude that Hfq binds to the sRNAs, deletion of hfq had no major effect on sRNA levels, consistent with previous results for RNAIII (9).
Half-lives of all sRNAs, except for RsaB and RsaF, were determined in BHI medium at 37°C after rifampicin treatment (Figure 1D and Supplementary Figure S3). Most sRNAs are characterized by a long half-life: 15 min for RsaC and RsaI, 20 min and RsaD, 24 min for RsaE, 55 min for RsaG and >60 min for RsaA and RsaH. Among the sRNAs, two distinct transcripts for RsaA and RsaG were visualized in northern experiments. The longer RsaA transcript is most likely generated from a transcriptional readthrough, and transcription stops 140 nt downstream at a second strong terminator. This second transcript is unstable, with a half-life of 2.5 min (Supplementary Figure S3). For RsaG, the shorter fragment most probably results from an endoribonucleolytic cleavage in the unpaired interhelical region of RsaG, generating the stable upstream 5′ hairpin motif. Recent works identified an essential enzyme, RNase J1 in B. subtilis, with dual endonucleolytic and 5′-3′ exonucleolytic activities (64–66). This enzyme contributes significantly to the degradation of mRNAs (67,68) and cleaves small ncRNAs in B. subtilis (69). It remains to be addressed whether RNase J1 is responsible for site-specific cleavage of RsaG.
Functions regulated by RsaE as monitored by transcriptomic and proteomic analysis
The function of RsaE, the only RNA found to be conserved in staphylococcal species and Bacillaceae, was assessed as follows. We constructed a RN6390-derived strain, in which the rsaE gene was deleted and replaced by aphA-3 (aminoglycoside 3′-phosphotransferase, kanamycin resistance). In BHI medium, growth was not affected by the rsaE deletion. We then analyzed the effect of the mutation on gene expression and protein synthesis using microarrays and quantitative proteomics. The experiments were carried out under similar conditions on wild type (RN6390) and ΔrsaE grown in BHI medium to early stationary phase. We have verified that under these conditions, rsaE expression is high in RN6390 at a level similar to S. aureus RNAIII while the expression of RsaE is abolished in the mutant ΔrsaE strain (Figure 1A).
Transcriptional profiles were obtained using extensively validated oligoarrays (39). Statistically significant changes in the levels of gene expression were recorded, and only differences greater than 2-fold were considered. According to these criteria, 86 different mRNAs were differentially expressed in the wild type versus the mutant ΔrsaE strains, representing ∼4% of the transcriptome (Supplementary Table S3). The same proportions of genes were found to be up- and down-regulated by RsaE. Functional classification of these genes according to the COG database showed that many of these mRNAs encoded proteins involved in various metabolic pathways such as amino-acid transport and metabolism, lipid metabolism, inorganic ion transport, coenzyme transport and metabolism, carbohydrate metabolism and energy production. In addition, the synthesis of capsular polysaccharides (Cap5I-O) was significantly repressed (Supplementary Table S3). These cell-surface polysaccharides contribute to virulence by impeding phagocytosis and modulate S. aureus adherence to endothelial surfaces (70).
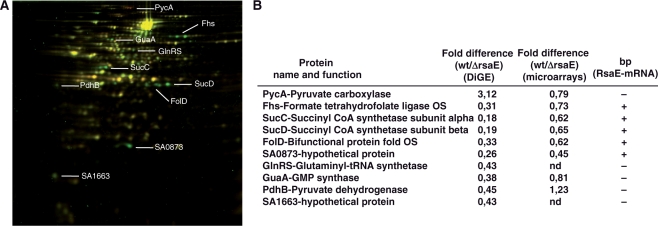
To assess the effect of the rsaE knockout on the staphylococcal proteome, cytosolic and secreted protein extracts were prepared and separated by a two-dimensional difference gel electrophoresis (DiGE), which enables quantitative differential display analysis (Figure 4A). The extracts were prepared from cells grown in BHI in the exponential and post-exponential phases of growth. Protein samples were pre-labeled with two different fluorescent dyes (Cy3 and Cy5). An internal standard, which includes the two samples present in the experiment, was pre-labeled with Cy2 and run on the same multiplexed gel. Quantitative measurements were made for each resolved protein spot relative to the cognate signal from the standard, which was used to normalize the ratio derived from one gel. Four different experiments were carried out to provide confident statistical analysis. We did not observe significant variations of secreted proteins between the two strains suggesting that RsaE does not regulate the synthesis of many of the exotoxins (serine proteases SplA-E and haemolysins α and β), as expected from the transcriptomic analysis (results not shown, Supplementary Table S3). For cytosolic proteins, even though we resolved only a small proportion of all proteins (around 100 proteins), we identified 10 proteins with strongly altered expression patterns (Figure 4). The synthesis of the pyruvate carboxylase PycA was shown up-regulated by RsaE. This enzyme catalyzes the ATP-dependent carboxylation of pyruvate to form oxaloacetate, which is involved in the biosynthesis of aspartate. The synthesis of several proteins (SucC/D, FolD, Fhs and SA0873) was strongly repressed even when the RsaE level was rather low (after 2 h of growth in BHI medium, Figure 1A), suggesting that their RsaE-dependent repression is most likely direct. These proteins are connected to carbohydrate metabolism: succinyl-CoA synthetase subunits α and β (SucC/D) are part of the TCA cycle and the bi-functional protein FolD and the formate-tetrahydrofolate ligase Fhs are involved in glyoxylate and dicarboxylate metabolism and the biosynthesis of folate. The transcriptomic analysis also shows that the levels of sucCD and SA0873 mRNAs are significantly decreased in the RN6390 strain (Figure 4A). We also analyzed the level of SA0873 mRNA in different S. aureus strains in early and late exponential phase (Figure 5A). In RN6390 and WA400 (ΔrnaIII), the expression of the mRNA is very low whereas it significantly accumulates in the late-exponential phase in the mutant ΔrsaE and Δagr strains, consistent with the AgrA-dependent expression of rsaE (Figure 1A).
Figure 4.
RsaE regulates the synthesis of several metabolic enzymes. (A) 2D fluorescence difference gel electrophoresis (DiGE) performed on the RN6390 and ΔrsaE strains. Total protein extracts were prepared from cultures stopped at the stationary phase of growth and labeled with different cyanine-dyes. Yellow spots represent unchanged proteins; green spots, protein synthesis repressed in the strain expressing RsaE; red spots, protein synthesis increased in the strain expressing RsaE. The proteins were identified by mass spectroscopy analysis. (B) Summary of the DiGE analysis, and comparative analysis with the microarray data. Ratios correspond to the quantification obtained for RN6390 versus the ΔrsaE strain (data from four different experiments). (bp) for base pairing between RsaE and mRNA targets: (+) potential bp, (−) no bp.
Figure 5.
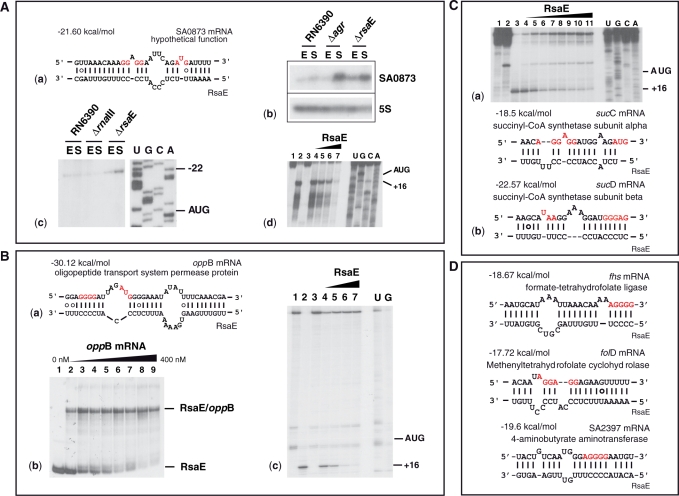
RsaE regulates gene expression by direct interaction with target mRNAs. (A) RsaE binds to the RBS of SA0873 mRNA: (a) Base pairings predicted between SA0873 and RsaE. The Shine‐Dalgarno sequence (SD) and the initiation codon AUG are shown in red. (b) Northern analysis of SA0873 mRNA prepared from various S. aureus strains (RN6390, RN6911-Δagr and RN6390-ΔrsaE). The 5S rRNA was probed as an internal control. (c) The 5′ start of the SA0873 mRNA was determined by primer extension with reverse transcriptase. The mRNA was only observed in the ΔrsaE strain. Lanes U, G, C and A: sequencing ladders. (d) RsaE binding to SA0873 mRNA prevents the formation of the ribosomal initiation complex. Lanes 1 and 3: Incubation controls of mRNA alone and with RsaE, respectively; lane 2: formation of the ternary complex formed from the S. aureus 30S subunit, the initiator tRNA, and the mRNA; lanes 4–7: Formation of the ternary ribosomal mRNA‐30S‐tRNA complex in the presence of increasing RsaE concentrations of 50, 100, 150 and 200 nM, respectively. (B) RsaE binds to oppB mRNA and prevents the ribosome binding: (a) Base pairings between oppB and RsaE. Same legend as above. (b) Band shift experiment showing a stable RsaE-oppB mRNA complex. Lane 1, incubation control of RsaE alone; lanes 2–9, complex formation between 5′-end-labeled RsaE with increasing oppB mRNA concentrations of 10, 50, 100, 150, 200, 250, 300 and 400 nM, respectively. (c) RsaE binds to oppB mRNA and prevents the formation of the 30S initiation complex. Same legend as above. (C) The RsaE-sucD mRNA complex prevents the formation of the ribosomal initiation complex. (a) Toeprinting assays: lanes 1, 2, incubation controls on the mRNA alone and with 200 nM of RsaE; lane 3, ternary mRNA‐30S‐tRNA complex; lanes 4–11, ternary complex with increasing RsaE concentrations 50, 100, 200, 300, 400, 500, 600 and 800 nM, respectively. (b) RsaE-sucC/D mRNA base pairing interaction. The hybrid formed between RsaE and sucD mRNA includes the stop codon of the upstream sucC mRNA. (D) Predicted base pairing interactions between RsaE and other mRNAs repressed by RsaE.
Taken together, these data show that RsaE altered mRNA and/or protein levels for enzymes involved in several metabolic pathways that are necessary to synthesize biosynthetic intermediates. This may contribute to facilitate the transition from the exponential to the stationary phase of growth (71).
Staphylococcus aureus RsaE regulates gene expression through mRNA pairing
Although the transcriptomic and proteomic analyses give insights into the putative functions of RsaE, they do not permit us to distinguish between primary and secondary effects. As mentioned above, two conserved unpaired regions within RsaE contain a C-rich sequence motif. We postulated that these regions might interact with ribosome-binding sites (RBS) of target mRNAs, as does RNAIII (9,14). We searched for potential base pairing that involves the conserved unpaired regions of RsaE (nt 19–65) and the RBSs of mRNAs (30 nt downstream and upstream the AUG start codon) using the program RNAup (72). This program takes the structures of both RNAs into account and assesses the energy cost of melting local secondary structures for the formation of intermolecular base pairs. We first restricted the search to the genes whose mRNA and/or protein levels changed in an RsaE-dependent fashion. The mRNA–RsaE hybrids that were considered had a minimum free energy of ≤−18 kcal/mol (Figure 5C and D). Significant base pairing was predicted between RsaE and mRNAs encoding OppB (oligopeptide transport system, opp-3 operon), SucC and SucD, SA0873 (hypothetical protein), Fhs (formate tetrahydrofolate synthase), SA2397 (4-aminobutyrate aminotransferase) and FolD (bi-functional protein).
We used two other ab initio analyses to predict mRNA targets. The sequences of each RBS for all of the S. aureus mRNAs were concatenated to the sequence of RsaE by a linker and submitted to RNAfold for predicting base pairing (9). This approach estimates the hybridization of two RNA sequences and calculates the minimum free energy of interaction. Prediction of targets was also carried out with the algorithm developed for Listeria monocytogenes (73). This program quantifies the strength of RNA duplexes and takes into account positive contributions due to pairing nucleotides and negative contributions due to bulged nucleotides and internal loops. In addition, statistical significance of the pairings is assessed with respect to an ensemble of random sequences. This program was successfully trained on several regulatory L. monocytogenes sRNAs and S. aureus RNAIII (73). By comparing the two approaches, stable base pairings were predicted between RsaE and the hutH (encoding histidine ammonia-lyase), oppB and sucD mRNAs.
Thus, the in silico data predict that the oppB, sucD and SA0873 mRNAs are among the best candidates to form stable duplexes with RsaE (Figure 5). Indeed, band shift experiments show that RsaE forms a stable complex with an oppB mRNA fragment (370 nt) encompassing the entire 5′ untranslated region and part of the coding sequence (Figure 5B). A time-course analysis suggests that this complex is rapidly formed, as was shown for S. aureus RNAIII and its mRNA targets (data not shown) (9). We further analyzed the effect of RsaE on the formation of initiation ribosomal complexes using toeprinting assays. The ternary initiation complex formed by S. aureus 30S subunits, the initiator tRNAMet and the three mRNAs (oppB, sucD and SA0873) blocked primer extension 16 nt downstream of the initiation codon. Binding of RsaE to any of the three mRNAs significantly decreased the toeprint signal (Figure 5A–C) indicating that the formation of active ribosomal initiation complexes is prevented. Therefore, we hypothesized that RsaE inhibits the initiation of translation of the oppB, sucD and SA0873 mRNAs by masking the RBS.
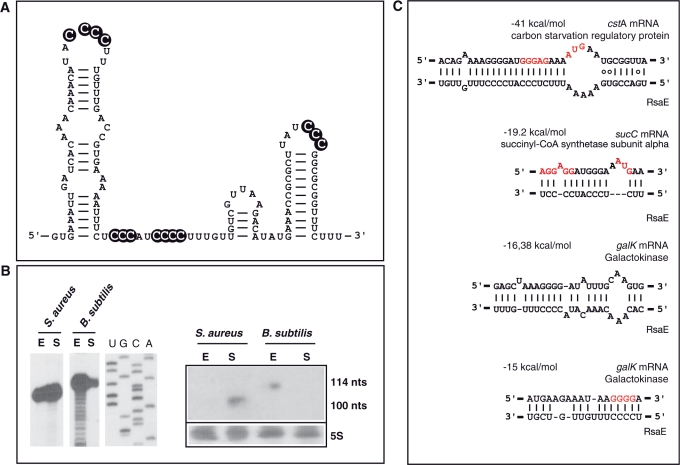
RsaE is expressed in Bacillus subtilis
Since RsaE is conserved in Bacillaceae, we have analyzed the expression of rsaE in B. subtilis grown in BHI medium. Northern experiments indicated a homogenous RNA transcript that was preferentially expressed in the exponential phase (Figure 6). Determination of the 5′-end by primer extension revealed a RNA 114-nt long, consistent with the predicted length (Figure 6B). As in S. aureus, its secondary structure is characterized by a 5′ hairpin motif and a stable terminator structure at the 3′-end, and it contains several highlighted C-rich sequences (Figure 6A). Interactions between the RBS of B. subtilis mRNAs and RsaE were predicted using the method applied to S. aureus RNAIII (9). This analysis revealed several mRNA candidates that encode proteins involved in carbohydrate and sugar utilization (Figure 6C). Indeed the most stable duplex was predicted between RsaE and the cstA mRNA, encoding the carbon starvation protein (CstA). Interestingly, pairings between RsaE and sucC mRNA appear to be conserved in B. subtilis and S. aureus. We also found that the two C-rich motifs of B. subtilis RsaE could interact with the RBS and part of the coding sequence of the galK mRNA, encoding galactokinase, respectively (Figure 6C).
Figure 6.
RsaE ortholog is expressed in B. subtilis. (A) Secondary structure of B. subtilis RsaE derived from the structural alignment shown in Figure 2. The C-rich conserved boxes are shown in black. (B) Expression of B. subtilis RsaE: (left) Primer extension was used to map the 5′ start of S. aureus and B. subtilis RsaE. The coordinates of the RNA are 1232743–1232852 corresponding to Bacillus subtilis subsp. subtilis strain 168 (NC_000964.2). The RNA has been referred in GenBank with the accession number GQ403626. Lanes U, G, C and A: sequencing ladders. (Right) Northern experiment performed on S. aureus and B. subtilis RsaE. Both experiments were carried out on total RNA extracts prepared at the exponential (E) and stationary (S) phases. (C) Potential base pairings between B. subtilis RsaE and mRNAs encoding proteins involved in carbohydrate metabolism and sugar utilization. The Shine‐Dalgarno (SD) sequence is in red.
DISCUSSION
A plethora of RNA-dependent regulatory events in Staphylococcus aureus
In recent years, new in silico and experimental strategies have been developed to identify functional regulatory intergenic regions in a variety of model bacteria (74,75). Here, we used a combination of several bioinformatic tools to predict more than 500 potential functional IGRs from S. aureus in addition to the highly conserved bacterial ncRNAs (tmRNA, 6S RNA, 4.5S RNA and RNase P). These IGRs were selected based on the presence of orphan terminators, on their GC content and their sequence and structure conservation among S. aureus, staphylococcal species and firmicutes (Supplementary Figure S1). The visualization of the data with ApolloRNA provided an easy tool for allowing the characterization of three main families of IGRs: repeated sequences, conserved leader regions of mRNAs, and bona fide ncRNAs. In this study, we did not search specifically for antisense RNAs (73,76).
The first class of non-coding regions includes Staphylococcus aureus repeats (STARs) (29). Some of them carry features reminiscent of the clustered regularly interspaced short palindromic repeats (Gaspin et al., unpublished data). The expression of one of these elements failed to be significant in RN6390 grown in BHI and NZM medium (RsaX19, Table 1). However, we do not exclude that STAR elements may have evolved regulatory functions under defined stress conditions. These repeats were also present in the leader region of mRNA (22) as well as in the S. aureus sprA3 ncRNA (Supplementary Table S1) (8).
The second class of S. aureus IGRs comprises numerous cis-acting regulatory elements located in the 5′ untranslated regions of mRNAs, which regulate enzymes and proteins involved in the translational machinery and in various metabolic pathways. These regions are highly conserved among firmicutes, and in B. subtilis many of them provide specific binding sites for trans-acting regulators (metabolites, tRNAs or proteins) (77–80). RNAsim has identified all the assigned riboswitches with sizes of more than 150 nt that are regulated by small metabolites such as thiamine pyrophosphate, flavin mononucleotide, S-adenosyl methionine, glucosamine 6-phosphate, lysine, purine and preQ1 (77,78) (Supplementary Table S1). The program also revealed conserved leaders that include T-box antitermination signals. These cis-acting elements are sensed by uncharged tRNA to regulate the expression of genes involved in tRNA aminoacylation and the transport and biosynthesis of amino acids (79,80). Other conserved leaders recognized by regulatory proteins have been identified in the S. aureus genome. A typical example is RsaK which is derived from the 5′ leader of glcA mRNA. This region carries a long terminator structure overlapping the conserved RAT motif (Supplementary Figure S4). This regulatory region is also present upstream of the S. aureus ptsG gene, which encodes the glucose-specific phosphotransferase system. In B. subtilis and S. carnosus, the RAT motif is recognized by the transcriptional antiterminator protein GlcT, which senses the intracellular concentration of glucose (60,81). The RAT–GlcT complex prevents the formation of the terminator structure, allowing transcription of the downstream gene. Hence, we propose that S. aureus glcA mRNA is under the control of GlcT and that the detection of RsaK would be a consequence of a premature termination event. Similarly, RNAsim detected two conserved 5′UTRs with a terminator just upstream of the pyrR and pyrP genes. These two leader regions are characterized by an AGAGAG hairpin loop motif, proposed to be the main recognition element for the regulatory protein PyrR in B. subtilis (82). PyrR, bound to UMP, binds to target mRNAs and induces premature termination of transcription (82) (Supplementary Figure S5). RNAsim pointed out several highly conserved leaders of operons encoding ribosomal proteins and the three initiation factors, which are regulated by a feedback mechanism (Supplementary Table S1). For instance, the S. aureus infC leader carries a typical binding site for the L20 ribosomal protein (83). In B. subtilis, binding of L20 to the infC leader region stabilizes the formation of a terminator structure to induce premature arrest of transcription (83). We have identified typical Rho-independent terminators located upstream of several genes (i.e. SA1002, SA1108, SA1841, splA and uppS) suggesting that these genes might be regulated by a premature transcription termination event (Supplementary Table S1). In agreement with this observation, cis-acting RNAs in low-GC Gram-positive bacteria were shown to predominantly regulate gene expression through a transcription termination/antitermination mechanism (77,80).
The third class of IGRs reported here are the bona fide sRNAs. The expression from 36 IGRs was analyzed in S. aureus RN6390 grown in two different media. From this analysis, 10 new ncRNAs (RsaA to J) of sizes ranging from 59 to 544 nt were identified (Table 1, Figure 2). However, this fraction of ncRNAs is probably an underestimate since many growth conditions and stress protocols were not investigated. None of the Rsa RNAs carries a small ORF, and the RNA genes are dispersed in the genome. The location of the rsaC gene is of interest since its 5′ start site is located close to a repeat element and the downstream gene encodes a transposase. Many genes encoding sRNAs from various bacteria have also been found to be close to mobile elements such as transposons, prophages and pathogenicity islands (8,84,85). Based on their genomic location, it was proposed that sRNAs, like structured tRNAs and tmRNA, might play a role in genome plasticity by serving as integration sites for foreign genes (85). We have predicted most of the spr RNA expressed from the pathogenicity islands (8), except sprE (Supplementary Tables S1 and S2). The conserved region of sprE comprises only 70 nt, which is below the cut off used by RNAsim. A recent high throughput computational tool identified Rho-independent terminators downstream of conserved intergenic regions (23). This powerful analysis predicted 49 new ncRNA genes from S. aureus, 35 of which were also predicted by our approach (Supplementary Table S1). We show above that four of them are highly expressed (RsaH, RsaD, RsaE and RsaI), while we assigned other 14 IGRs to STAR elements (Table 1, Supplementary Tables S1 and S2). Recently, a phylogenetic profiling study has been performed on all available bacterial genomes. Based on specific signatures, this method allows clustering of intergenic and coding regions (22). From this analysis, six RNAs were shown to be expressed in the S. aureus N315 strain. These RNAs were also predicted by RNAsim (Supplementary Table S1), but were not further experimentally analyzed, as many of them resemble riboswitches or leaders of mRNAs. However, our analysis confirms the high expression of rsaI in RN6390 and other strains. All in all, our study revealed ten new ncRNAs in addition to the pathogenicity island spr RNAs (8).
The sRNAs are endowed with a common signature and different expression patterns
Many of the Rsa RNAs are expressed under particular conditions. Some conditions, such as acidic pH, oxidative stress and osmotic stress, could reflect environments that the bacteria encounter within a host. The synthesis of RsaA, RsaH and RsaE is induced under osmotic stress, oxidative stress and at acidic pH, while RsaC and RsaD are cold shock-induced RNAs. Five of the RNAs (RsaA, RsaD, RsaE, RsaF and RsaJ) show significant variation in expression among S. aureus strains although their genes are conserved in all S. aureus strains. RsaA, D and F synthesis is strongly enhanced in strains expressing an active σB factor while the expression of RsaE is negatively controlled by σB via an unknown mechanism (Figure 1A and C). Interestingly, we found a typical σB consensus binding site upstream of rsaA (Figure 1C). Several σB-dependent ncRNAs were recently found in L. monocytogenes (86,87), but their functions remain to be identified. Staphylococcus aureus encodes three sigma factors: the housekeeping σA (a homolog of E. coli σ70), two alternative factors σB (88) and σH (89). While σH is required for transcriptional regulation of the com operon encoding competence factors (89), σB positively controls the synthesis of proteins involved in cell envelope biosynthesis, metabolism, virulence and signaling pathways (52,90,91). A significant number of genes that are upregulated by σB do not contain a promoter consensus, and several genes are negatively influenced by σB (90). These data suggest that σB indirectly regulates the expression of these genes most likely through the induction of transcription of regulatory proteins or ncRNAs. Of interest, we found that RsaA could potentially form stable duplexes with σB-repressed mRNAs (52,90). These mRNAs encode the magnesium-citrate transporter (CitM), an APC amino-acid permease, and the pyruvate carboxylase PycA (Supplementary Figure S7). Although further experiments will be required to validate these targets, we propose that RsaA is part of the σB regulon and regulates gene expression via an antisense mechanism.
Probing of various Rsa RNA structures combined with RNA sequence and structure alignments revealed similar common features (Figure 3, Supplementary Figure S6). Most of them carry a hairpin structure at the 5′-end and a classical terminator at the 3′-end. The 5′ hairpin most probably confers the high stability of the Rsa RNAs, as has been shown for mRNAs in B. subtilis (92,93) and S. aureus (9,11). Intriguingly, most of the Rsa RNAs, except RsaF and RsaI, contain an unpaired UCCC motif which is located in the most conserved regions of the RNAs (Figure 7). Staphylococcus aureus RNAIII also carries UCCC motifs in three hairpin loops; all are known to interact with the Shine-Dalgarno (SD) sequences of target mRNAs (9). In RsaA, this motif is predicted to interact with σB-repressed mRNAs (Supplementary Figure S7). From these sequence characteristics, the ncRNAs may have resulted from a convergent evolution which selected efficient repressors. Similarly, a conserved C-rich hairpin loop of Salmonella typhimurium CyaR sRNA binds to the SD sequence of ompX mRNA to repress its translation (94). This sequence motif is also found in other ncRNAs recently identified in L. monocytogenes (73,86,87) and B. subtilis (95), suggesting that these RNAs act at the mRNA level. Even though the majority of low-GC Gram-positive bacterial mRNAs carry a strong SD sequence (96) specificity could be attained by the surrounding nucleotides and the structure of both interacting RNAs. Thus we postulate that the C-rich box might be a signature for regulatory RNAs that repress translation initiation of target mRNAs.
Figure 7.
The C-rich box conserved motif in S. aureus regulatory RNAs. The alignment of the C-rich sequence motif in S. aureus RNAIII (9) and Rsa RNAs was done with the help of the PARADISE platform (https://simtk.org/home/paradise).
RsaE regulates several metabolic pathways
The above rule can be applied to S. aureus RsaE, the only sRNA which is conserved in all staphylococcal species studied and in Bacillaceae (Figure 3, Supplementary Figure S5). The expression pattern of RsaE is rather complex since a high level of RsaE is dependent on the quorum sensing agr system, whereas its expression drops considerably in strains expressing a fully active σB factor. A stable σB-dependent RNA (RsaF) was also identified although its function remains to be determined. Both RNAs are differently expressed, suggesting that transcriptional interference regulates their synthesis. Alternatively, transcription may occur at the same start site of rsaE but termination readthrough could lead to the synthesis of a larger RNA precursor, which would subsequently be processed (Supplementary Figure S5). The functions of RsaE were then addressed by a combination of approaches, including a comparative analysis of the transcriptome and of the proteome performed on RN6390 and the isogenic ΔrsaE mutant strain. Transcriptomic analysis revealed many genes that were up- and down-regulated by RsaE, suggesting additional indirect regulation (Supplementary Table S3). In this scenario, RsaE may regulate the expression of a regulatory protein by direct binding to the mRNA, as does RNAIII, or alternatively, it may sequester a regulatory protein. Even though further experiments are required to assign the entire set of direct RsaE targets, these data demonstrate a link between RsaE and the regulation of various metabolic pathways (including amino acid and peptide transport, cofactor synthesis, lipid metabolism, carbohydrate metabolism and the TCA cycle). Our data strongly support that RsaE directly represses the synthesis of OppB, the two subunits of the succinyl-CoA synthetase involved in the TCA cycle, two proteins (FhS and FolD) involved in the folate biosynthesis, and SA0873 of unknown function. Direct interactions are strengthened by in vitro experiments showing that RsaE binds efficiently to oppB, sucD and SA0873 mRNAs and inhibits the formation of ribosomal initiation complexes (Figure 5A–C). One of the main roles of bacterial Opp systems is to supply bacteria with essential amino acids and peptides. Staphylococcus aureus genomes carry four polycistronic opp-1–4 operons and one monocistronic opp5A gene (97). Of the four putative Opp systems, only opp-3 can supply S. aureus with peptides as nutritional source and it is the only one whose expression is modulated by amino acids (97). These proteins thus play a key role in nitrogen and carbon metabolism. Our data further suggest that RsaE directly regulates several proteins involved in carbohydrate metabolism and in the TCA cycle. TCA is an essential source of energy and biosynthetic intermediates for many organisms (71). In Gram-positive bacteria, the TCA cycle is repressed in the presence of glucose, and conversely, the derepression coincides with the depletion of the catabolized carbon source. Staphylococcus aureus uses the pentose phosphate and glycolytic pathways to catabolize glucose to pyruvate. Under aerobic conditions, pyruvate undergoes oxidative decarboxylation to produce acetyl-coenzyme A, which in turn leads to the accumulation of acetate in the extracellular medium during the exponential phase of growth. In stationary phase, when glucose has been consumed, acetate and amino acids are used as alternate carbon sources and are oxidized via the TCA cycle. A higher level of enzymes involved in glycolysis was indeed identified in the exponentially growing S. aureus COL strain while enzymes of the TCA cycle and gluconeogenesis were enhanced in the stationary phase (98). Thus, it is quite surprising that RsaE strongly represses the synthesis of SucCD enzymes in the stationary phase. RN6390 which is σB-deficient, has an impaired TCA cycle-mediated acetate catabolism (99). The isogenic strain SH1000, with restored σB activity, had significantly increased acetate catabolism (99). Since RsaE is strongly repressed by σB (Figure 1A), the ncRNA might be partly responsible for this deficiency in acetate catabolism in the stationary phase. The loss of acetate catabolism neither alters stationary-phase survival nor virulence factor expression (99). Interestingly enough, RsaE is highly conserved in most of the Bacillaceae. The rsaE gene of B. subtilis is mainly expressed in the exponential phase and decreases in the stationary phase (Figure 5B). The presence of RsaE in S. aureus, M. caseolyticus and Bacillus, but not in other firmicutes, raises evolutionary considerations (Supplementary Figure S5). The M. caseolyticus genome is closely related to S. aureus and B. anthracis (100). Indeed, the essential biological pathways of M. caseolyticus are similar to those of Staphylococci, although it lacks several sugar and amino-acid metabolism pathways and a plethora of virulence genes that are present in S. aureus. In addition, the genome possesses oxidative phosphorylation machineries that are closely related to those present in Bacillaceae. This phylogenetic analysis suggests that the Bacillus, Macrococcus and Staphylococcus derive from a common Gram-positive ancestor, and that the speciation of Macrococcus into Staphylococcus was accompanied by the acquisition of arrays of virulence genes for host adaptation (100). RsaE, which is present in all three genera, probably arose from the common ancestor genome before the divergence of Bacillus and Macrococcus. Therefore it is not so surprising that RsaE regulates common metabolic pathways. Consistent with this hypothesis, the B. subtilis RsaE ortholog is predicted to regulate the translation of sucC mRNA in the TCA cycle, as in S. aureus, as well as the translation of galK and cstA mRNAs, which are both involved in carbon source utilization (Figure 6C). In E. coli, Spot42 sRNA inhibits the initiation of translation of galK mRNA in response to glucose availability (101). Spot42 is also predicted to form a stable duplex with the 5′-end of the sucC mRNA (101). Hence, Spot42 could be the functional equivalent of RsaE. These studies strongly suggest that, as in Gram-negative bacteria, S. aureus and B. subtilis bacteria use RsaE to control the use of carbon sources and sugar metabolism (102). Further experimental works will characterize how far the regulatory functions of RsaE have evolved in Bacillus, and Staphylococcaceae.
The present study shows that the S. aureus genome likely encodes a high diversity of regulatory RNAs, including mRNA leaders that affect expression in cis, the pleiotropic regulator RNAIII (6,103), and the small noncoding RNAs [(8,11,12,22); this work]. Deep sequencing efforts and tiling arrays would certainly help to provide a more complete view of the RNA-dependent regulatory circuits in this pathogenic bacterium (86,104). A particular challenge will now concern their roles in response to the host environment and during the infection process of this major pathogen.
ACCESSION NUMBERS
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National de la Recherche Scientifique (CNRS; to P.R.); Institut National pour la Recherche Médicale (INSERM; to F.V.); Agence Nationale pour la Recherche (ANR05-MIME: to C.G., F.V. and P.R.; ANR09-BLAN: to F.V. and P.R.); European Community (BacRNA, FP6-018618; to P.R.); Swiss National Science Foundation PP00B-103002/1 (to J.S.) and 3100A0-116075 (to P.F.). Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are thankful to Anne-Catherine Helfer for providing technical help for the DiGE experiments, Philippe Hammann and Philippe Wolf for the identification of proteins by mass spectroscopy analysis, and Elodie Vogel for the expression analysis of several ncRNAs. They are grateful to Gerhart Wagner, Joerg Vogel and Eric Westhof for helpful comments and critical reading of the manuscript, and to Efthimia Lioliou and Gérard Lina for discussions. This project used equipment facilities provided by Toulouse Midi-Pyrénées bioinformatic platform.
REFERENCES
- 1.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogel J, Wagner EGH. Target identification of small noncoding RNAs in bacteria. Curr. Opin. Microbiol. 2007;10:262–270. doi: 10.1016/j.mib.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Toledo-Arana A, Repoila F, Cossart P. Small noncoding RNAs controlling pathogenesis. Curr. Opin. Microbiol. 2007;10:182–188. doi: 10.1016/j.mib.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Vogel J. A rough guide to the non-coding RNA world of Salmonella. Mol. Microbiol. 2009;71:1–11. doi: 10.1111/j.1365-2958.2008.06505.x. [DOI] [PubMed] [Google Scholar]
- 5.Taubes G. The bacteria fight back. Science. 2008;321:356–361. doi: 10.1126/science.321.5887.356. [DOI] [PubMed] [Google Scholar]
- 6.Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu. Rev. Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 7.van Schaik W, Abee T. The role of sigmaB in the stress response of Gram-positive bacteria – targets for food preservation and safety. Curr. Opin. Biotechnol. 2005;16:218–224. doi: 10.1016/j.copbio.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 8.Pichon C, Felden B. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc. Natl Acad. Sci. USA. 2005;102:14249–14254. doi: 10.1073/pnas.0503838102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007;21:1353–1366. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geisinger E, Adhikari RP, Jin R, Ross HF, Novick RP. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol. Microbiol. 2006;61:1038–1048. doi: 10.1111/j.1365-2958.2006.05292.x. [DOI] [PubMed] [Google Scholar]
- 11.Roberts C, Anderson KL, Murphy E, Projan SJ, Mounts W, Hurlburt B, Smeltzer M, Overbeek R, Disz T, Dunman PM. Characterizing the effect of the Staphylococcus aureus virulence factor regulator, SarA, on log-phase mRNA half-lives. J. Bacteriol. 2006;188:2593–2603. doi: 10.1128/JB.188.7.2593-2603.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson KL, Roberts C, Disz T, Vonstein V, Hwang K, Overbeek R, Olson PD, Projan SJ, Dunman PM. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J. Bacteriol. 2006;188:6739–6756. doi: 10.1128/JB.00609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huntzinger E, Boisset S, Saveanu C, Benito Y, Geissmann T, Namane A, Lina G, Etienne J, Ehresmann B, Ehresmann C, et al. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 2005;24:824–835. doi: 10.1038/sj.emboj.7600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morfeldt E, Taylor D, von Gabain A, Arvidson S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995;14:4569–4577. doi: 10.1002/j.1460-2075.1995.tb00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altuvia S. Identification of bacterial small non-coding RNAs: experimental approaches. Curr. Opin. Microbiol. 2007;10:257–261. doi: 10.1016/j.mib.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Wassarman KM. 6S RNA: a small RNA regulator of transcription. Curr. Opin. Microbiol. 2007;10:164–168. doi: 10.1016/j.mib.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 18.Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 19.Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 20.Bohn C, Rigoulay C, Bouloc P. No detectable effect of RNA-binding protein Hfq absence in Staphylococcus aureus. BMC Microbiol. 2007;7:10. doi: 10.1186/1471-2180-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Večerek B, Rajkowitsch L, Sonnleitner E, Schroeder R, Bläsi U. The C-terminal domain of Escherichia coli Hfq is required for regulation. Nucleic Acids Res. 2008;36:133–143. doi: 10.1093/nar/gkm985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchais A, Naville M, Bohn C, Bouloc P, Gautheret D. Single-pass classification of all noncoding sequences in a bacterial genome using phylogenetic profiles. Genome Res. 2009;19:1084–1092. doi: 10.1101/gr.089714.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livny J, Teonadi H, Livny M, Waldor MK. High-throughput, kingdom-wide prediction and annotation of bacterial non-coding RNAs. PLoS ONE. 2008;3:e3197. doi: 10.1371/journal.pone.0003197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benito Y, Kolb FA, Romby P, Lina G, Etienne J, Vandenesch F. Probing the structure of RNAIII, the Staphylococcus aureus agr regulatory RNA, and identification of the RNA domain involved in repression of protein A expression. RNA. 2000;6:668–679. doi: 10.1017/s1355838200992550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiapello H, Bourgait I, Sourivong F, Heuclin G, Gendrault-Jacquemard A, Petit MA, El Karoui M. Systematic determination of the mosaic structure of bacterial genomes: species backbone versus strain-specific loops. BMC Bioinformatics. 2005;6:171. doi: 10.1186/1471-2105-6-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiapello H, Gendrault A, Caron C, Blum J, Petit MA, El Karoui M. MOSAIC: an online database dedicated to the comparative genomics of bacterial strains at the intra-species level. BMC Bioinformatics. 2008;9:498. doi: 10.1186/1471-2105-9-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Flèche P, Hauck Y, Onteniente L, Prieur A, Denoeud F, Ramisse V, Sylvestre P, Benson G, Ramisse F, Vergnaud G. A tandem repeats database for bacterial genomes: application to the genotyping of Yersinia pestis and Bacillus anthracis. BMC Microbiol. 2001;1:2. doi: 10.1186/1471-2180-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dsouza M, Larsen N, Overbeek R. Searching for patterns in genomic data. Trends Genet. 1997;13:497–498. doi: 10.1016/s0168-9525(97)01347-4. [DOI] [PubMed] [Google Scholar]
- 29.Cramton SE, Schnell NF, Götz F, Brückner R. Identification of a new repetitive element in Staphylococcus aureus. Infect. Immun. 2000;68:2344–2348. doi: 10.1128/iai.68.4.2344-2348.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petersohn A, Bernhardt J, Gerth U, Hoper D, Koburger T, Volker U, Hecker M. Identification of sigma(B)-dependent genes in Bacillus subtilis using a promoter consensus-directed search and oligonucleotide hybridization. J. Bacteriol. 1999;181:5718–5724. doi: 10.1128/jb.181.18.5718-5724.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kingsford CL, Ayanbule K, Salzberg SL. Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biol. 2007;8:R22. doi: 10.1186/gb-2007-8-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klein RJ, Misulovin Z, Eddy SR. Noncoding RNA genes identified in AT-rich hyperthermophiles. Proc. Natl Acad. Sci. USA. 2002;99:7542–7547. doi: 10.1073/pnas.112063799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rivas E, Eddy SR. Noncoding RNA gene detection using comparative sequence analysis. BMC Bioinformatics. 2001;2:8. doi: 10.1186/1471-2105-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis SE, Searle SM, Harris N, Gibson M, Lyer V, Richter J, Wiel C, Bayraktaroglir L, Birney E, Crosby MA, et al. Apollo: a sequence annotation editor. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0082. RESEARCH0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janzon L, Arvidson S. The role of the ™-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 1990;9:1391–1399. doi: 10.1002/j.1460-2075.1990.tb08254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arnaud M, Chastanet A, Debarbouille M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 2004;70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Redko Y, Bechhofer DH, Condon C. Mini-III, an unusual member of the RNase III family of enzymes, catalyses 23S ribosomal RNA maturation in B. subtilis. Mol. Microbiol. 2008;68:1096–1106. doi: 10.1111/j.1365-2958.2008.06207.x. [DOI] [PubMed] [Google Scholar]
- 39.Charbonnier Y, Gettler B, François P, Bento M, Renzoni A, Vaudaux P, Schlegel W, Schrenzel J. A generic approach for the design of whole-genome oligoarrays, validated for genomotyping, deletion mapping and gene expression analysis on Staphylococcus aureus. BMC Genomics. 2005;6:95. doi: 10.1186/1471-2164-6-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, et al. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357:1225–1240. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 41.Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, Oguchi A, Nagai Y, Iwama N, Asano K, Naimi T, et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–1827. doi: 10.1016/s0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 42.Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan M, et al. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 2005;187:2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 44.Holden MT, Feil EJ, Lindsay JA, Peacock SJ, Day NP, Enright MC, Foster TJ, Moore CE, Hurst L, Atkin R, et al. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl Acad. Sci. USA. 2004;101:9786–9791. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scherl A, Francois P, Charbonnier Y, Deshusses JM, Koessler T, Huyghe A, Bento M, Stahl-Zeng J, Fischer A, Masselot A, et al. Exploring glycopeptide-resistance in Staphylococcus aureus: a combined proteomics and transcriptomics approach for the identification of resistance-related markers. BMC Genomics. 2006;7:296. doi: 10.1186/1471-2164-7-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Renzoni A, Barras C, Francois P, Charbonnier Y, Huggler E, Garzoni C, Kelley WL, Majcherczyk P, Schrenzel J, Lew DP, et al. Transcriptomic and functional analysis of an autolysis-deficient, teicoplanin-resistant derivative of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2006;50:3048–3061. doi: 10.1128/AAC.00113-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Churchill GA. Using ANOVA to analyze microarray data. BioTechniques. 2004;37:173–175, 177. doi: 10.2144/04372TE01. [DOI] [PubMed] [Google Scholar]
- 48.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 49.de Hoon MJ, Makita Y, Nakai K, Miyano S. Prediction of transcriptional terminators in Bacillus subtilis and related species. PLoS Comput. Biol. 2005;1:e25. doi: 10.1371/journal.pcbi.0010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schattner P. Searching for RNA genes using base-composition statistics. Nucleic Acids Res. 2002;30:2076–2082. doi: 10.1093/nar/30.9.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cros MJ, Moisan A, Cierco-Ayrolles C, Gaspin C. Visualizing and exploring genomic information for non-protein-coding RNA identification using ApolloRNA. Nature Protocols. doi: 10.1036/nprot.20.07.285. [Google Scholar]
- 52.Pané-Farré J, Jonas B, Forstner K, Engelmann S, Hecker M. The σB regulon in Staphylococcus aureus and its regulation. Int. J. Med. Microbiol. 2006;296:237–258. doi: 10.1016/j.ijmm.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 53.Pané-Farré J, Jonas B, Hardwick SW, Gronau K, Lewis RJ, Hecker M, Engelmann S. Role of RsbU in controlling SigB activity in Staphylococcus aureus following alkaline stress. J. Bacteriol. 2009;191:2561–2573. doi: 10.1128/JB.01514-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J. Bacteriol. 2008;190:300–310. doi: 10.1128/JB.01000-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Padalon-Brauch G, Hershberg R, Elgrably-Weiss M, Baruch K, Rosenshine I, Margalit H, Altuvia S. Small RNAs encoded within genetic islands of Salmonella typhimurium show host-induced expression and role in virulence. Nucleic Acids Res. 2008;36:1913–1927. doi: 10.1093/nar/gkn050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horsburgh MJ, Aish JL, White IJ, Shaw L, Lithgow JK, Foster SJ. B modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 2002;184:5457–5467. doi: 10.1128/JB.184.19.5457-5467.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pohl K, François P, Stenz L, Schlink F, Geiger T, Herbert S, Goerke C, Schrenzel J, Wolz C. CodY in Staphylococcus aureus: a regulatory link between metabolism and virulence gene expression. J. Bacteriol. 2009;191:2953–2963. doi: 10.1128/JB.01492-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shearwin KE, Callen BP, Egan JB. Transcriptional interference–a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Britton RA, Wen T, Schaefer L, Pellegrini O, Uicker WC, Mathy N, Tobin C, Daou R, Szyk J, Condon C. Maturation of the 5' end of Bacillus subtilis 16S rRNA by the essential ribonuclease YkqC/RNase J1. Mol. Microbiol. 2007;63:127–138. doi: 10.1111/j.1365-2958.2006.05499.x. [DOI] [PubMed] [Google Scholar]
- 60.Langbein I, Bachem S, Stulke J. Specific interaction of the RNA-binding domain of the Bacillus subtilis transcriptional antiterminator GlcT with its RNA target, RAT. J. Mol. Biol. 1999;293:795–805. doi: 10.1006/jmbi.1999.3176. [DOI] [PubMed] [Google Scholar]
- 61.Aymerich S, Steinmetz M. Specificity determinants and structural features in the RNA target of the bacterial antiterminator proteins of the BglG/SacY family. Proc. Natl Acad. Sci. USA. 1992;89:10410–10414. doi: 10.1073/pnas.89.21.10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jossinet F, Westhof E. Sequence to Structure (S2S): display, manipulate and interconnect RNA data from sequence to structure. Bioinformatics. 2005;21:3320–3321. doi: 10.1093/bioinformatics/bti504. [DOI] [PubMed] [Google Scholar]
- 63.Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 2007;10:134–139. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 64.Mathy N, Benard L, Pellegrini O, Daou R, Wen T, Condon C. 5'-to-3' exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5' stability of mRNA. Cell. 2007;129:681–692. doi: 10.1016/j.cell.2007.02.051. [DOI] [PubMed] [Google Scholar]
- 65.Even S, Pellegrini O, Zig L, Labas V, Vinh J, Brechemmier-Baey D, Putzer H. Ribonucleases J1 and J2: two novel endoribonucleases in B subtilis with functional homology to E. coli RNase E. Nucleic Acids Res. 2005;33:2141–2152. doi: 10.1093/nar/gki505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de la Sierra-Gallay IL, Zig L, Jamalli A, Putzer H. Structural insights into the dual activity of RNase J. Nat. Struct. Mol. Biol. 2008;15:206–212. doi: 10.1038/nsmb.1376. [DOI] [PubMed] [Google Scholar]
- 67.Mader U, Zig L, Kretschmer J, Homuth G, Putzer H. mRNA processing by RNases J1 and J2 affects Bacillus subtilis gene expression on a global scale. Mol. Microbiol. 2008;70:183–196. doi: 10.1111/j.1365-2958.2008.06400.x. [DOI] [PubMed] [Google Scholar]
- 68.Deikus G, Bechhofer DH. Initiation of decay of Bacillus subtilis trp leader RNA. J. Biol. Chem. 2007;282:20238–20244. doi: 10.1074/jbc.M702747200. [DOI] [PubMed] [Google Scholar]
- 69.Yao S, Blaustein JB, Bechhofer DH. Processing of Bacillus subtilis small cytoplasmic RNA: evidence for an additional endonuclease cleavage site. Nucleic Acids Res. 2007;35:4464–4473. doi: 10.1093/nar/gkm460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.O'R;iordan K, Lee JC. Staphylococcus aureus capsular polysaccharides. Clinical Microbiol. Rev. 2004;17:218–234. doi: 10.1128/CMR.17.1.218-234.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Somerville GA, Proctor RA. At the crossroads of bacterial metabolism and virulence synthesis in Staphylococci. Microbiol. Mol. Biol. Rev. 2009;73:233–248. doi: 10.1128/MMBR.00005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tafer H, Hofacker IL. RNAplex: a fast tool for RNA-RNA interaction search. Bioinformatics. 2008;24:2657–2663. doi: 10.1093/bioinformatics/btn193. [DOI] [PubMed] [Google Scholar]
- 73.Mandin P, Repoila F, Vergassola M, Geissmann T, Cossart P. Identification of new noncoding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res. 2007;35:962–974. doi: 10.1093/nar/gkl1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hüttenhofer A, Vogel J. Experimental approaches to identify non-coding RNAs. Nucleic Acids Res. 2006;34:635–646. doi: 10.1093/nar/gkj469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eddy SR. Computational genomics of noncoding RNA genes. Cell. 2002;109:137–140. doi: 10.1016/s0092-8674(02)00727-4. [DOI] [PubMed] [Google Scholar]
- 76.Fozo EM, Hemm MR, Storz G. Small toxic proteins and the antisense RNAs that repress them. Microbiol. Mol. Biol. Rev. 2008;72:579–589. doi: 10.1128/MMBR.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yao Z, Barrick J, Weinberg Z, Neph S, Breaker R, Tompa M, Ruzzo WL. A computational pipeline for high- throughput discovery of cis-regulatory noncoding RNA in prokaryotes. PLoS Comput. Biol. 2007;3:e126. doi: 10.1371/journal.pcbi.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vitreschak AG, Mironov AA, Lyubetsky VA, Gelfand MS. Comparative genomic analysis of T-box regulatory systems in bacteria. RNA. 2008;14:717–735. doi: 10.1261/rna.819308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gutierrez-Preciado A, Henkin TM, Grundy FJ, Yanofsky C, Merino E. Biochemical features and functional implications of the RNA-based T-box regulatory mechanism. Microbiol. Mol. Biol. Rev. 2009;73:36–61. doi: 10.1128/MMBR.00026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Knezevic I, Bachem S, Sickmann A, Meyer HE, Stulke J, Hengstenberg W. Regulation of the glucose-specific phosphotransferase system (PTS) of Staphylococcus carnosus by the antiterminator protein GlcT. Microbiology. 2000;146:2333–2342. doi: 10.1099/00221287-146-9-2333. [DOI] [PubMed] [Google Scholar]
- 82.Lu Y, Turner RJ, Switzer RL. Function of RNA secondary structures in transcriptional attenuation of the Bacillus subtilis pyr operon. Proc. Natl Acad. Sci. USA. 1996;93:14462–14467. doi: 10.1073/pnas.93.25.14462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Choonee N, Even S, Zig L, Putzer H. Ribosomal protein L20 controls expression of the Bacillus subtilis infC operon via a transcription attenuation mechanism. Nucleic Acids Res. 2007;35:1578–1588. doi: 10.1093/nar/gkm011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Balbontin R, Figueroa-Bossi N, Casadesus J, Bossi L. Insertion hot spot for horizontally acquired DNA within a bidirectional small-RNA locus in Salmonella enterica. J. Bacteriol. 2008;190:4075–4078. doi: 10.1128/JB.00220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sridhar J, Rafi ZA. Identification of novel genomic islands associated with small RNAs. In Silico Biol. 2007;7:601–611. [PubMed] [Google Scholar]
- 86.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 87.Nielsen JS, Olsen AS, Bonde M, Valentin-Hansen P, Kallipolitis BH. Identification of a sigma B-dependent small noncoding RNA in Listeria monocytogenes. J. Bacteriol. 2008;190:6264–6270. doi: 10.1128/JB.00740-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kullik II, Giachino P. The alternative sigma factor sigmaB in Staphylococcus aureus: regulation of the sigB operon in response to growth phase and heat shock Arch. Microbiol. 1997;167:151–159. doi: 10.1007/s002030050428. [DOI] [PubMed] [Google Scholar]
- 89.Morikawa K, Inose Y, Okamura H, Maruyama A, Hayashi H, Takeyasu K, Ohta T. A new staphylococcal sigma factor in the conserved gene cassette: functional significance and implication for the evolutionary processes. Genes Cells. 2003;8:699–712. doi: 10.1046/j.1365-2443.2003.00668.x. [DOI] [PubMed] [Google Scholar]
- 90.Bischoff M, Dunman P, Kormanec J, Macapagal D, Murphy E, Mounts W, Berger-Bachi B, Projan S. Microarray-based analysis of the Staphylococcus aureus σB regulon. J. Bacteriol. 2004;186:4085–4099. doi: 10.1128/JB.186.13.4085-4099.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ziebandt AK, Becher D, Ohlsen K, Hacker J, Hecker M, Engelmann S. The influence of agr and σB in growth phase dependent regulation of virulence factors in Staphylococcus aureus. Proteomics. 2004;4:3034–3047. doi: 10.1002/pmic.200400937. [DOI] [PubMed] [Google Scholar]
- 92.Sharp JS, Bechhofer DH. Effect of 5′-proximal elements on decay of a model mRNA in Bacillus subtilis. Mol. Microbiol. 2005;57:484–495. doi: 10.1111/j.1365-2958.2005.04683.x. [DOI] [PubMed] [Google Scholar]
- 93.Condon C. Maturation and degradation of RNA in bacteria. Curr. Opin. Microbiol. 2007;10:271–278. doi: 10.1016/j.mib.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 94.Papenfort K, Pfeiffer V, Lucchini S, Sonawane A, Hinton JC, Vogel J. Systematic deletion of Salmonella small RNA genes identifies CyaR, a conserved CRP-dependent riboregulator of OmpX synthesis. Mol. Microbiol. 2008;68:890–906. doi: 10.1111/j.1365-2958.2008.06189.x. [DOI] [PubMed] [Google Scholar]
- 95.Gaballa A, Antelmann H, Aguilar C, Khakh SK, Song KB, Smaldone GT, Helmann JD. The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc. Natl Acad. Sci. USA. 2008;105:11927–11932. doi: 10.1073/pnas.0711752105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chang B, Halgamuge S, Tang SL. Analysis of SD sequences in completed microbial genomes: non-SD-led genes are as common as SD-led genes. Gene. 2006;373:90–99. doi: 10.1016/j.gene.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 97.Hiron A, Borezee-Durant E, Piard JC, Juillard V. Only one of four oligopeptide transport systems mediates nitrogen nutrition in Staphylococcus aureus. J. Bacteriol. 2007;189:5119–5129. doi: 10.1128/JB.00274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kohler C, Wolff S, Albrecht D, Fuchs S, Becher D, Buttner K, Engelmann S, Hecker M. Proteome analyses of Staphylococcus aureus in growing and non-growing cells: a physiological approach. Int. J. Med. Microbiol. 2005;295:547–565. doi: 10.1016/j.ijmm.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 99.Somerville GA, Said-Salim B, Wickman JM, Raffel SJ, Kreiswirth BN, Musser JM. Correlation of acetate catabolism and growth yield in Staphylococcus aureus: implications for host-pathogen interactions. Infect. Immun. 2003;71:4724–4732. doi: 10.1128/IAI.71.8.4724-4732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Baba T, Kuwahara-Arai K, Uchiyama I, Takeuchi F, Ito T, Hiramatsu K. Complete genome sequence of Macrococcus caseolyticus strain JSCS5402, reflecting the ancestral genome of the human-pathogenic staphylococci J. Bacteriol. 2009;191:1180–1190. doi: 10.1128/JB.01058-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Møller T, Franch T, Hojrup P, Keene DR, Bachinger HP, Brennan RG, Valentin-Hansen P. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 102.Goerke C, Fluckiger U, Steinhuber A, Bisanzio V, Ulrich M, Bischoff M, Patti JM, Wolz C. Role of Staphylococcus aureus global regulators sae and sigmaB in virulence gene expression during device-related infection. Infect. Immun. 2005;73:3415–3421. doi: 10.1128/IAI.73.6.3415-3421.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Marzi S, Fechter P, Chevalier C, Romby P, Geissmann T. RNA switches regulate initiation of translation in bacteria. Biol. Chem. 2008;389:585–598. doi: 10.1515/bc.2008.055. [DOI] [PubMed] [Google Scholar]
- 104.Sittka A, Sharma CM, Rolle K, Vogel J. Deep sequencing of Salmonella RNA associated with heterologous Hfq proteins in vivo reveals small RNAs as a major target class and identifies RNA processing phenotypes. RNA Biol. 2009;6:266–275. doi: 10.4161/rna.6.3.8332. [DOI] [PubMed] [Google Scholar]
- 105.Kreiswirth BN, Lofdahl S, Betley MJ, O'R;eilly M, Schlievert PM, Bergdoll MS, Novick RP. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 106.Peng HL, Novick RP, Kreiswirth B, Kornblum J, Schlievert P. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 1988;170:4365–4372. doi: 10.1128/jb.170.9.4365-4372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kornblum J, Kreiswirth B, Projan SJ, Ross H, Novick R. Agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus. In: Novick R, editor. Molecular Biology of the staphylococci. New York: Wiley-VCH; 1990. pp. 373–401. [Google Scholar]
- 108.Dyke KG, Jevons MP, Parker MT. Penicillinase production and intrinsic resistance to penicillins in Staphylococcus aures. Lancet. 1966;1:835–838. doi: 10.1016/s0140-6736(66)90182-6. [DOI] [PubMed] [Google Scholar]
- 109.Duthie ES, Lorenz LL. Staphylococcal coagulase; mode of action and antigenicity. J. Gen. Microbiol. 1952;6:95–107. doi: 10.1099/00221287-6-1-2-95. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.