Abstract
We describe here a mass spectrometry (MS)-based analytical platform of RNA, which combines direct nano-flow reversed-phase liquid chromatography (RPLC) on a spray tip column and a high-resolution LTQ-Orbitrap mass spectrometer. Operating RPLC under a very low flow rate with volatile solvents and MS in the negative mode, we could estimate highly accurate mass values sufficient to predict the nucleotide composition of a ∼21-nucleotide small interfering RNA, detect post-transcriptional modifications in yeast tRNA, and perform collision-induced dissociation/tandem MS-based structural analysis of nucleolytic fragments of RNA at a sub-femtomole level. Importantly, the method allowed the identification and chemical analysis of small RNAs in ribonucleoprotein (RNP) complex, such as the pre-spliceosomal RNP complex, which was pulled down from cultured cells with a tagged protein cofactor as bait. We have recently developed a unique genome-oriented database search engine, Ariadne, which allows tandem MS-based identification of RNAs in biological samples. Thus, the method presented here has broad potential for automated analysis of RNA; it complements conventional molecular biology-based techniques and is particularly suited for simultaneous analysis of the composition, structure, interaction, and dynamics of RNA and protein components in various cellular RNP complexes.
INTRODUCTION
RNAs play an essential role during protein biosynthesis by serving as a temporary copy of genes and adaptors for translation of the genetic code. In addition to these classical roles in cell biology, recent genetic and biochemical evidence reveals that diverse types of intronic and small non-coding RNAs play pivotal roles in a variety of cellular processes such as chromatin remodeling, transcriptional regulation, precursor mRNA processing, gene silencing, centromere function and translational regulation (1–5), and participate in the regulation of differentiation, proliferation and programmed cell death (6). Unlike double-stranded genomic DNA, RNA is a single-stranded polynucleotide that folds spontaneously into a variety of secondary and tertiary structures such as hairpins, bulges, pseudoknots and internal loops, which serve as the binding sites for regulatory proteins or directly mediate particular biological processes (7). Most RNAs function as a part of ribonucleoprotein (RNP) complexes and the deregulation of some RNP complexes, such as those containing micro RNA (i.e. naturally occurring short-RNA sequences), leads to severe pathology including tumorigenesis, tumor metastasis, or abnormal morphogenesis (8–10). Thus, isolation and characterization of novel regulatory RNP complexes, such as micro RNPs, small nuclear RNPs (snRNPs), small nucleolar RNPs and heteronuclear RNPs, are crucial to understand normal and aberrant biological processes.
Current mass spectrometry (MS)-based proteomics technology, coupled with various tagging technologies to isolate particular protein complexes, allows large-scale identification and quantitation of protein components in many RNP complexes involved in fundamental cellular processes, such as transcription, precursor mRNA processing and maturation, and translation (11,12). For instance, we isolated a series of pre-ribosomal RNP complexes by a reverse tagging approach using trans-acting protein factors as affinity bait, and characterized hundreds of protein components at various stages of ribosome biogenesis in human cells (13–18). This study provided proteomic snapshots of mammalian ribosome biogenesis and revealed a dynamic aspect of ribosome biogenesis that includes the synthesis, processing, and modification of rRNA directed by hundreds of small nucleolar RNAs and their interactions with trans-acting factors and ribosomal subunits at various stages of the pre-ribosomal RNP complex. Likewise, detailed structural and functional characterization of many cellular RNP complexes, such as those involved in mRNA/miRNA processing or the RNAi-induced gene-silencing complex (19,20), suggest that the assembly of RNP complexes involves a complex series of events performed not only by the components of the final functional complex but also by various additional non-coding RNAs (ncRNAs) and trans-acting protein cofactors that regulate the intermediate processes of biogenesis and ensure the quality of the final products (21–23). Thus, studies of the assembly and function of cellular RNP complexes require detailed characterization of both RNA and protein cofactors.
At present, identification and analysis of RNAs in RNP complexes are mainly carried out using techniques based on genomics and molecular biology, which includes the process of reverse transcription from RNA to cDNA (24). This technique is highly sensitive because of the step of PCR amplification and has proven to be useful for various aspects of RNA research; however, the method has shortcomings of the relatively high error rate of reverse transcriptase—which arises from the presence of both RNA secondary structure and base modifications—and the substrate specificity of reverse transcriptase limits the capacity to obtain quantitative results (25). In addition, the conventional approach does not provide structural information about post-transcriptional modifications of nucleosides (26), which are common in tRNA and rRNA and are essential for their biogenesis and function (22,27). MS offers a sensitive method for the direct chemical analysis of RNA and therefore is ideally suited as a method complementary to conventional techniques.
Numerous attempts have been made to analyze oligonucleotides using both electrospray ionization and matrix-assisted laser desorption/ionization MS (28–31). Although early studies with both techniques were hampered by problems such as cation adduction due to high-affinity binding of Na+, K+ and Mg++ to the polyanionic phosphate backbone or sugar hydroxyl groups of the oligonucleotides, subsequent studies have clarified most of those problems; for example, the addition of a strong organic base such as triethylamine (TEA) or N,N-dimethylbutylamine (DMBA) effectively suppresses adduct formation (32,33). Thus, various MS-based techniques are currently used to analyze synthetic RNA (34) and RNA transcripts, including tRNA, rRNA and ncRNA (35–37). In particular, liquid chromatography (LC)-MS techniques are widely used for chemical analyses of nucleic acids and oligonucleotides by coupling a conventional or capillary reversed-phase column packed with silica-based materials (31,36,37) or a monolithic poly(styrene-divinylbenzene)-based capillary column with an ion-trap, quadrupole, or quadrupole/time-of-flight hybrid mass spectrometer (32,33). Huang et al. have recently studied the ion trap collision-induced dissociation of multiply deprotonated RNA (38) and applied the tandem MS technique for sequencing of a small interfering RNA (39). However, MS-based technology is utilized for RNA analysis much less frequently than proteomics, presumably because of its limited resolution and sensitivity.
We have recently developed a first genome-oriented database searching software, Ariadne, which correlates tandem MS spectra of sample RNA nucleolytic fragments with an RNA nucleotide sequence in a DNA/RNA database, thereby allowing MS/MS-based identification of RNA in biological samples (40). We describe here our continuing effort to develop an MS-based analytical platform for small RNAs. The system reported in this paper is essentially based on the instrumentation that has been developed for the ‘shotgun’ proteomics approach (41–43) and consists of a direct nanoflow LC apparatus equipped with a fritless spray tip column and a high-resolution LTQ-Orbitrap mass spectrometer. Application of the method to the chemical analysis of synthetic siRNA, short oligoribonucleotides generated by ribonuclease digestion of tRNA, and U small nuclear RNA (snRNA) in yeast pre-spliceosomal RNP complexes suggests that it is a highly sensitive and efficient tool for the analysis of small RNAs in biological samples, particularly those associated with cellular RNP complexes.
MATERIALS AND METHODS
Chemicals
Standard laboratory chemicals were obtained from Sigma-Aldrich (St. Louis, MO). RNase T1 was purchased from Worthington (Lakewood, NJ) and further purified by reversed-phase liquid chromatography (RPLC) before use. High-performance liquid chromatography grade methanol and acetonitrile were obtained from Kokusan Chemical Co. (Tokyo, Japan), and lysyl endopeptidase, 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP) and acetic acid were obtained from Wako Pure Chemical Industries (Osaka, Japan). Triethylammonium (TEA) acetate buffer (pH 7.0) was purchased from Glen Research (Sterling, VA). The synthetic oligonucleotides, CACCA-OH (-OH refers to 3′-hydroxyl group), UUUCGUdCdA-OH, CUCAGUdTdT-OH, AAUUCGAdTdT-OH, and the synthetic 21-nucleotide siRNA with the sequence 5′-AGUAGUUGGCAUAGGAGUCdTdT-3′, designed for the mRNA of the human SH3BP1 gene (NM_018957), and its ‘sense’ RNA with the complementary sequence 5′-GACUCCUAUGCCAACUACUdTdT-3′, were obtained from JBioS (Saitama, Japan). The Dynamarker small RNA kit, containing 5-, 8-, 9-, 10-, 20-, 30-, 40-, 50- and 100-nucleotide oligoribonucleotides, was obtained from BioDynamics Laboratory Inc. (Tokyo, Japan).
LC-MS apparatus for RNA analysis
The LC system used was essentially as described (41). It consisted of a direct nano-flow pump with a pressure limit of ∼300 bars (LC Assist, Tokyo, Japan) which delivers solvent to the fritless spray tip ESI column, a ReNCon gradient device (41) and an injection valve (Cheminart C2-0006, Valco, Houston, TX) for sample loading. The spray tip ESI column was prepared with a fused-silica capillary (150 µm i.d. ×375 µm o.d.) using a laser puller (Sutter Instruments Co., Novato, CA). The column was slurry-packed with reversed-phase material (Develosil C30-UG-3, particle size; 3 µm, Nomura Chemical, Aichi, Japan) to a length of 50 mm and was connected to the LC line with micro-fingertight fittings via a metal union (Valco Instruments). High voltage for ionization in negative mode (−1.4 kV) was applied to the metal union, and the eluate from RPLC was sprayed on-line to an LTQ-Orbitrap hybrid mass spectrometer (model XL, Thermo Fisher Scientific, San Jose, CA). Reverse phase separation of oligoribonucleotides was performed at flow rate of 100 nl/min using a 60-min linear gradient from 5 to 40% methanol in 20 mM TEA acetate (pH 7.0) or in 10 mM N,N-dimethylbutylamine (DMBA) acetate (pH 7.0) with or without 0.4 M HFIP.
The mass spectrometer was operated in a data-dependent mode to automatically switch between Orbitrap-MS and linear ion trap-MS/MS acquisition. Survey full scan MS spectra (from m/z 500 to 1500) were acquired in the Orbitrap with resolution R = 30 000 (after accumulation to a target value of 500 000 ions in the linear ion trap). The most intense ions (up to four, depending on signal intensity) were sequentially isolated for fragmentation in the linear ion trap using CID at a target value of 30 000 ions. An MS scan was accumulated for 2 s and an MS/MS scan for 3 s. The resulting fragment ions were recorded in the linear ion trap with a high-scan rate ‘Enhanced’ mode. Target ions selected for MS/MS were dynamically excluded for 60 s. General mass spectrometric conditions were as follows: electrospray voltage, 1.4 kV; no lock mass option; with sheath and auxiliary gas flow; normalized collision energy, 35% for MS/MS. Ion selection threshold was 10 000 counts for MS/MS. An activation q-value of 0.25 and activation time of 30 ms were applied for MS/MS acquisitions.
Preparation of the yeast Lsm3-associated spliceosome complex
The Lsm3-associated pre-spliceosomal complex was purified from the yeast Saccharomyces cerevisiae strain S288C expressing TAP-tagged Lsm3, essentially as described by Puig et al. (44). Briefly, the yeast cells were grown in 4 l of YPD medium to A600 of 2.0, and suspended in an equal volume of 10 mM HEPES-KOH (pH 7.9), 200 mM NaCl, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol (DTT), 0.5 mM phenylmethylsulfonyl fluoride. The cells were disrupted by passing three times through a French press, and centrifuged at 100 000g for 30 min at 4°C. The extract (∼1.2 g protein) was combined with NP-40 to 0.1%, mixed with 1.8 ml IgG-Sepharose beads (50% slurry, GE Healthcare UK Ltd., Little Chalfont, Buckinghamshire, UK), incubated for 60 min at 4°C, and then poured into a Polyprep column (Bio-Rad Laboratories, Richmond, CA, USA). After the column was washed with 10 mM Tris–HCl (pH 8.0), 150 mM NaCl, 0.1% NP-40, 0.5 mM EDTA and 1 mM DTT, the Lsm3-associated complex was eluted by the same buffer after incubation with 100 U/ml AcTEV protease (Invitrogen, Carlsbad, CA, USA) for 60 min at room temperature. The eluate was then combined with CaCl2 to 3 μM, diluted with 3 volumes of 10 mM Tris–HCl (pH 8.0), 150 mM NaCl, 0.1% NP-40, 1 mM Mg acetate, 1 mM imidazole, 2 mM CaCl2 and 10 mM DTT, and incubated with 200 μl Calmodulin Affinity Resin beads (50% slurry, Stratagene, La Jolla, CA, USA) at 4°C for 60 min. After the column was washed with 10 mM Tris–HCl (pH 8.0), 300 mM NaCl, 0.1% NP-40, 1 mM Mg acetate, 1 mM imidazole, 2 mM CaCl2 and 10 mM DTT, the Lsm3-associated complex was recovered by incubation with 100 μl of 125 mM Tris–HCl (pH 6.8), 4% SDS and 100 mM DTT.
Preparation of RNA and protein from the RNP complex
The RNA and protein components in the purified RNP complex were separated by phenol–chloroform extraction. One hundred microliters of the RNP preparation recovered from Calmodulin affinity beads was added to 10 μl of 3 M sodium acetate (pH 5.2) and 200 μg of glycogen (Roche Applied Science, Mannheim, Germany), and then mixed with an equal volume of phenol:chloroform:3-methyl-1-butanol (25:24:1, v/v). The upper layer was collected as an RNA fraction, precipitated by addition of an equal volume of 2-propanol, and redissolved in RNase-free water. The intermediate layer was collected as the protein fraction, precipitated with 3 volumes of acetone, and redissolved in loading buffer for SDS–PAGE. The RNA and protein solutions were stored frozen at –80°C until use. RNA was separated by denaturing 10% polyacrylamide gel electrophoresis (PAGE) and visualized using SYBR Gold (Invitrogen) as described (45). Protein was separated by SDS–PAGE and visualized by Coomassie Brilliant Blue staining.
Proteomics procedures
SDS–PAGE of protein, in-gel digestion and LC-MS/MS analysis of the resulting peptides were performed as described previously (41,46,47). The LC-MS apparatus used for proteomics analyses was the direct nanoflow LC-MS system equipped with a quadrupole-time-of-flight hybrid mass spectrometer (Q-Tof Ultima, Waters, Bedford, MA, USA) (38). Database search was performed as described (48) using MASCOT software (version 2.2.1., Matrix Science Ltd., London) and the SGD sequence database (release 20060506, http://downloads.yeastgenome.org/) under the following search parameters. The variable modification parameters were pyro-Glu, acetylation (protein N-terminus), oxidation (Met) and phosphorylation (Ser, Thr and Tyr). The maximum missed cleavage was set at 3 with a peptide mass tolerance of ± 500 ppm. Peptide charges from + 2 to + 4 states and MS/MS tolerances of ± 0.5 Da were allowed. We selected the candidate peptides with probability-based Mowse scores (total score) that exceeded its threshold indicating a significant homology (p < 0.05), and referred to them as ‘hits’. The criteria were based on the vendor’s definitions (Matrix Science, Ltd.). Furthermore, we set more strict criteria for protein assignment: (i) any peptide candidate with an MS/MS signal number of<2 was eliminated from the ‘hit’ candidates, regardless of the match score (total score minus threshold); (ii) proteins with match scores exceeding 10 ( p < 0.005) were referred to as ‘identified’; and (iii) if the protein was identified with a single peptide candidate having a match score lower than 10 or with peptides having excessively high mass errors ( >200 ppm), the original MS/MS spectrum was carefully inspected to confirm that the assignment was based on three or more y- or b-series ions.
Preparation of RNase T1 digests
RNase T1 digestion of yeast tRNAPhe-1 (∼5 μg) was performed in 20 μl of 10 mM sodium acetate buffer (pH 5.3) at 37°C for 30 min at an enzyme/substrate ratio of 1/500 (w/w). In-gel digestion of U snRNA was performed as follows: the SYBR gold-stained RNA band was excised from the gel, cut into small pieces and dried under vacuum. The gel pieces were swollen by adding 15 μl of 10 mM sodium acetate buffer (pH 5.3) and added with 2 ng/μl RNaseT1. After the mixture was incubation at 37°C for 1 h, the nucleolytic fragments were extracted from the gel with 100 μl of RNase free water, filtrated through a polyvinylidenfluoride membrane centrifugal filter (Ultrafree-MC, Millipore, Billerica, MA), and finally added with 5 μl of 2 M TEA acetate (pH 7.0). The digests were analyzed immediately by nanoflow LC-MS or stored frozen at –20°C until use. Database search was performed as described (40) using Ariadne software (available through internet, http://ariadne.riken.jp/) and the genome database of S. cerevisiae (release 14 November 2006, http://downloads.yeastgenome.org/).
RESULTS AND DISCUSSION
LC-MS apparatus and elution conditions
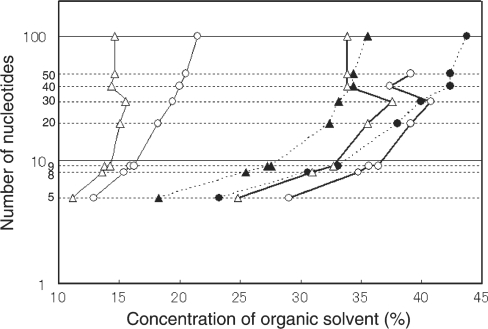
The LC-MS apparatus used in this study was essentially based on the instrumentation for the ‘shotgun’ proteomics approach (41). The direct nanoflow LC apparatus was equipped with a fritless spray tip column and a high-resolution LTQ-Orbitrap mass spectrometer was connected in tandem through an electrospray interface (ESI). To adapt the instrument for oligonucleotide analysis, we operated MS in the negative mode to analyze negatively charged oligonucleotide ions and selected a silica-based C30 packing material (Develosil C30; 3 μm beads) from a number of commercially available packing materials with different chemical characteristics (conventional silica-based material with distinct alkyl chain lengths and polymer-based materials such as alkylated polystyrene divinylbenzene material), mainly because of its strong hydrophobic nature to adsorb hydrophilic ribonucleotides with high affinity. To optimize the elution conditions, we examined several solvents for their capacity to separate 5–100-nucleotide oligonucleotides in a mixture. The plot of the concentration of the organic solvent required to elute each oligonucleotide versus oligonucleotide length (Figure 1) showed that the mobile phase solvents composed of methanol and volatile buffer containing a strong organic base, such as TEA acetate or DMBA acetate, exhibited efficient resolution of oligonucleotides from a few to ∼100 nucleotides in length. We noted that (i) methanol exhibited a much milder effect than the typical RP solvent acetonitrile in the separation of oligonucleotides and was suitable for the oligonucleotide analysis; (ii) organic amines were suitable as a counter ion to bind the negatively charged phosphate moiety of oligonucleotides and improved the retention behavior of oligonucleotides (in particular, oligonucleotides were retarded rather tightly on a reversed-phase column in the presence of DMBA); and (iii) HFIP, a weak organic acid introduced originally by Apffel et al. (49) for oligonucleotide analysis, significantly improved the resolution of oligonucleotides of relatively large size. Thus, the mobile phase solvent could be selected depending on the size of oligonucleotides to be analyzed; however, we used the mobile phase solvents composed of TEA acetate and methanol for most of the subsequent experiments aimed at the analysis of small oligoribonucleotide fragments generated by RNase T1 digestion of RNAs.
Figure 1.
RPLC separation of oligoribonucleotides under different solvent conditions. The oligoribonucleotide mixture, Dynamarker small RNA kit containing 5-, 8-, 9-, 10-, 20-, 30-, 40-, 50- and 100-nucleotide oligoribonucleotides, was applied to a capillary Develosil C30 column (2.1 mm × 150 mm) and eluted by a 60-min gradient of different mobile phase solvents. The concentration of organic solvent required to elute each oligonucleotide was plotted against the nucleotide length (UV detection at 260 nm). The mobile phase solvents used were TEA–acetate (pH 7.0) in acetonitrile/water (solid line with open triangles); DMBA–acetate (pH 7.0) in acetonitrile/water (solid line with open circles); HFIP–TEA–acetate (pH 7.0) in methanol/water (dotted line with closed triangles); HFIP–DMBA–acetate (pH 7.0) in methanol/water (dotted line with closed circles); TEA–acetate (pH 7.0) in methanol/water (bold line with open triangles); DMBA–acetate (pH 7.0) in methanol/water (bold line with open circles).
LC-MS analysis of synthetic oligonucleotides and siRNA
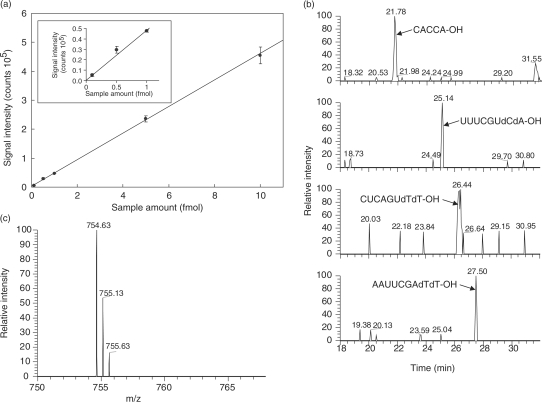
Figure 2a illustrates the sensitivity of the nanoflow LC-LTQ-Orbitrap MS system, as examined by the analysis of the small synthetic oligonucleotides CACCA-OH, UUUCGUdCdA-OH, CUCAGUdTdT-OH and AAUUCGAdTdT-OH. The system was extremely sensitive and exhibited a linear signal response within a range of a few hundred attomoles to 10 fmol of oligonucleotides (Figure 2a). The system provided an extracted ion chromatogram with a sufficient signal-to-noise ratio (Figure 2b) and MS and MS/MS spectra (Figure 2c and Supplementary Figure S1) even with 100 amol oligonucleotide. Note that no adduct ions were observed in the MS spectrum under the solvent conditions employed.
Figure 2.
The sensitivity of the nanoflow LC-LTQ-Orbitrap MS system. (a) Correlation between the signal intensity of [CACCA-OH]2− ion and the amount applied to the LC-MS system. The graph is enlarged in the boxed inset to display below 1 fmol. The system exhibited essentially the same sensitivity for the oligonucleotides UUUCGUdCdA-OH, CUCAGUdTdT-OH, or AAUUCGAdTdT-OH (data not shown). (b and c) Mass chromatogram and spectrum of the synthetic oligonucleotides (100 amol). Chromatography was performed at a flow rate of 100 nl/min as described in the Methods section. (b) Extracted ion monitoring of four oligonucleotide ions extracted from a single LC-MS chromatogram. The mass of each oligonucleotide and mass window used were: CACCA-OH; 754.63 ± 0.5 Da, UUUCGUdCdA-OH; 1206.16 ± 0.5 Da, CUCAGUdTdT-OH; 1220.18 ± 0.5 Da and AAUUCGAdTdT-OH; 1396.71 ± 0.5 Da. (c) the MS spectrum of [CACCA-OH]2− ion.
The nanoflow LC apparatus equipped with this LC-MS system also exhibited excellent peak resolution and reproducibility. Here, the average half-width of four independent peaks was 11.4 s (SD 0.49), and variations in the retention times of these peaks were <0.5 min in three repeated analytical runs (data not shown).
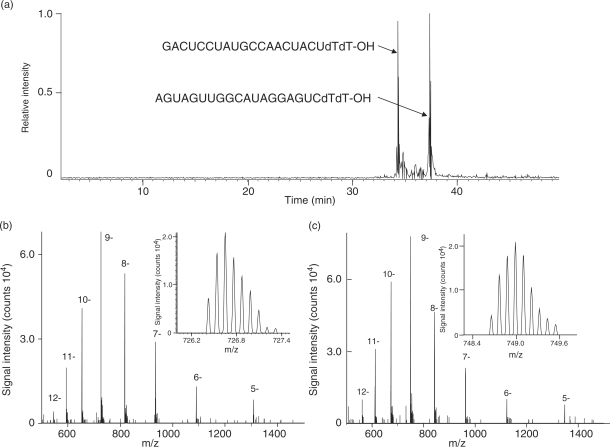
The performance of the LC-MS system was further evaluated by the analysis of a 21-nucleotide synthetic siRNA and its ‘sense’ RNA with the complementary nucleotide sequence. The total ion chromatogram, the raw electrospray negative ion spectra, and the isotopic signals of the [M-9H+]9− ion of the siRNA and sense RNA are shown in Figure 3a–c. The nanoflow RPLC separated siRNA and sense RNA at a sub-femtomole level and the subsequent MS gave rise to a series of multiply charged negative ions ranging from −5H+ to −11H+ or to −12H+ for each RNA species. The LTQ-Orbitrap mass analyzer exhibited extremely high resolution and high mass accuracy sufficient to determine the monoisotopic peak mass, which is the sum of the lightest isotopes from each element in the molecule. Thus, the monoisotopic molecular masses of siRNA and sense RNA were observed to be 6546.940 and 6746.965, respectively, which coincided within 3∼4 ppm of the theoretical mass values. According to our computer-aided statistical analysis, the human genome can generate 21-mer oligonucleotides of ∼4.5-billion distinct sequences having ∼2000 different nucleotide base compositions and, if we assume that those oligonucleotides carry no modified residues and are separated sufficiently without serious overlap of isotopic clusters, the compositions of all potential oligonucleotides can be distinguished by the monoisotopic mass measurement with accuracy of better than 5 ppm. Thus, the LC-MS system reported here should allow for prediction of the nucleotide composition of 21-mer oligonucleotides, such as typical human miRNAs, from the experimentally determined monoisotopic mass values, although the assignment of a particular miRNA will certainly require analysis of the nucleotide sequence.
Figure 3.
Mass chromatogram and spectra of synthetic siRNAs (21-mer). (a) Base peak chromatogram of two complementary siRNAs (50 fmol each). (b and c) The MS spectra of multiply charged (b) GACUCCUAUGCCAACUACUdTdT-OH and (c) AGUAGUUGGCAUAGGAGUCdTdT-OH ions. Insets, the enlarged spectra of the 9− ions.
Nucleotide mapping of yeast tRNAPhe−1
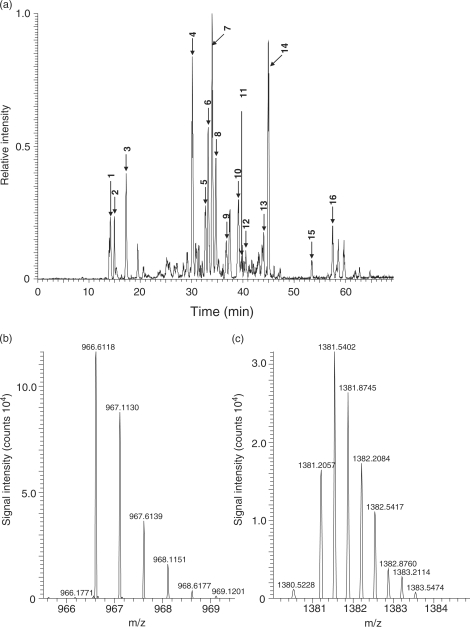
Yeast tRNAPhe−1 contains 76 nucleotides and carries 12 chemically modified nucleotides that result from post-transcriptional modification (50). The tRNAPhe−1 preparation was digested with RNase T1 as described in the ‘Materials and Methods’ section and subjected to the nanoflow LC-LTQ-Orbitrap MS system. Figure 4a shows the base peak chromatogram, and Table 1 lists the retention time and the molecular mass of each nucleotide fragment estimated by MS. Although RNase T1 generated more than 10 fragments of yeast tRNAPhe−1, high-resolution Orbitrap MS estimated monoisotopic mass values for all of the fragments. Figure 4b and c illustrates typical spectra of the oligoribonucleotide ion, [AUUUAm2G > p]2− (m2G refers to N2-methylguanosine, >p; 2′, 3′-cyclic phosphate) and [ACmUGmAAyWAΨUm5CUG > p]3−, (Cm, 2′-O-methylcytidine; Gm, 2′-O-methylguanosine; yW, wybutosine; Ψ, pseudouridine; m5C, 5-methylcytidine), respectively. In each case, Orbitrap MS estimated the monoisotopic mass value, 966.6118, for the theoretical mass 966.6134 of the oligoribonucleotide AUUUAm2G > p (error 1.5 ppm) and 1381.2057 for the theoretical mass 1381.2103 of ACmUGmAAyWAΨUm5CUG > p (error 3.3 ppm). Thus, all fragments were easily assigned to the original tRNAPhe−1 sequence based on the experimentally estimated mass values (Table 1). The assigned fragments covered the total tRNAPhe−1 sequence except for free guanosine monophosphate released by RNase T1 cleavage. All of the mass values estimated were compatible with the post-transcriptional modifications reported previously for yeast tRNAPhe−1 including nine methylated nucleotides, two dihydrouridines and a single wybutosine (27), although we could not distinguish pseudouridine from uridine as the modification is mass-silent. In addition to the oligonucleotide fragments derived from tRNAPhe−1, we detected a number of fragments that were specific to yeast tRNAPhe−2, tRNATyr and tRNALys−2 (indicated in Figure 4a), suggesting that the tRNAPhe−1 preparation used in this study was contaminated with these tRNA species. This was confirmed by the subsequent sequence analysis of these nucleolytic fragments using collision-induced dissociation (CID)-MS/MS (data not shown). Interestingly, the LC-MS analysis also detected a fragment, CACC-OH, which matches the 3′-terminal fragment CACCA-OH without the 3′-A. It is known that the 3′-end CCA of tRNA is added post-transcriptionally by the CCA-adding enzyme without a nucleic acid template (51); however, it was not evident whether the fragment was derived by incomplete biosynthesis or by non-specific nucleolytic cleavage of the 3′-end adenosine during the preparation or the analysis of tRNA.
Figure 4.
LC-MS analysis of RNaseT1 digests of yeast tRNAPhe−1. (a) Base peak chromatogram of the RNase T1 digest of the yeast tRNAPhe−1preparation (200 fmol). Chromatography was performed as described in the ‘Materials and Methods’ section. Sixteen major oligoribonucleotide peaks, indicated by arrows with peak numbers, are assigned to the fragments of yeast tRNAPhe−1, tRNAPhe−2, tRNALys−2, or tRNATyr (see Table 1). (b and c) A typical MS spectrum of the RNase T1 digest of tRNAPhe−1; (b) [AUUUAm2G > p]2− and (c) [ACmUGmAAyWAΨUm5CUG > p]3−.
Table 1.
Assignments of oligonucleotide fragments found in the RNase digest of the yeast tRNAPhe-1 preparation
| Peak number | Observed |
Theoretical |
Δppm | tRNAa | Residue numbersa | Sequencea | ||
|---|---|---|---|---|---|---|---|---|
| m/z | Charge | Molecular mass | Molecular mass | |||||
| 1 | 649.0779 | −1 | 650.0858 | 650.0882 | 3.7 | Phe1, Phe2 | 2–3 | CG>p |
| 2 | 960.1170 | −1 | 961.1249 | 961.1284 | 3.6 | Phe1, Phe2 | 16–18 | DDG>p |
| 3 | 650.0623 | −1 | 651.0702 | 651.0722 | 3.1 | Phe1, Phe2 | 52–53 | UG>p |
| 4 | 590.1003 | −2 | 1182.2164 | 1182.2195 | 2.6 | Phe1, Phe2 | 72–75 | CACC-OH |
| 5 | 673.0939 | −1 | 674.1018 | 674.0995 | −3.4 | Phe1, Phe2 | 44–45 | AG>p |
| 6 | 794.0939 | −2 | 1590.2036 | 1590.2065 | 1.8 | Phe1, Phe2 | 11–15 | CΨCAG>p |
| 7 | 637.0674 | −2 | 1276.1506 | 1276.1538 | 2.5 | Phe1, Phe2 | 54–57 | TΨCG>p |
| 8 | 754.6255 | −2 | 1511.2668 | 1511.2718 | 3.3 | Phe1, Phe2 | 72–76 | CACCA-OH |
| 9 | 649.0736 | −2 | 1300.1630 | 1300.1651 | 1.6 | Lys2 | 7–10 | UUAm2G>p |
| 10 | 969.1186 | −2 | 1940.2530 | 1940.2576 | 2.4 | Phe1, Phe2 | 46–51 | m7GUCm5CUG>p |
| 11 | 980.1395 | −2 | 1962.2948 | 1962.3009 | 3.1 | Phe1, Phe2 | 25–30 | Cm2,2GCCAG>p |
| 12 | 1282.6711 | −2 | 2567.3580 | 2567.3677 | 3.8 | Phe1, Phe2 | 58–65 | m1AUCCACAG>p |
| 13 | 966.1183 | −2 | 1934.2524 | 1934.2584 | 3.1 | Phe2 | 5–10 | ACUUAm2G>p |
| 14 | 966.6118 | −2 | 1935.2394 | 1935.2424 | 1.6 | Phe1 | 5–10 | AUUUAm2G>p |
| 14 | 959.1121 | −2 | 1920.2400 | 1920.2428 | 1.5 | Phe1 | 66–71 | AAUUCG>p |
| 15 | 1463.6897 | −2 | 2929.3952 | 2929.4075 | 4.2 | Tyr | 37–45 | ΨA(i6A)AΨCUUG>p |
| 16 | 1381.2057 | −3 | 4146.6408 | 4146.6543 | 3.3 | Phe1, Phe2 | 31–42 | ACmUGmAAyWAΨm5CUG>p |
aAccording to the MODMICS database27 (http://genesilico.pl/modomics).
Tandem MS of RNase T1 fragments of yeast tRNAPhe−1
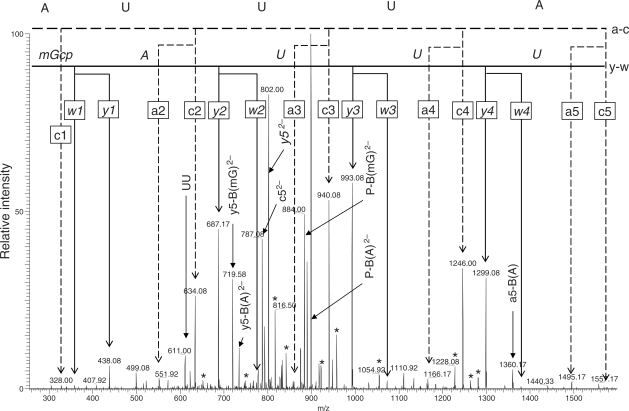
The fragmentation profiles of oligodeoxyribonucleotides and/or oligoribonucleotides upon low-energy CID tandem MS have been studied extensively using various types of mass analyzers under a variety of conditions (33,52–54). These studies have shown that CID of oligoribonucleotides most frequently generates the c and y series ions and less frequently generates the a, a-B [an ion losing [a] nucleotide base; refer to the nomenclature in reference (55)], and w ions regardless of the mass analyzer types, whereas oligodeoxyribonucleotides tends to decompose more frequently into the a, a-B and w ions under similar CID conditions (54). In our present study using a nanoflow LC-LTQ-Orbitrap MS system, all of the oligoribonucleotide fragments derived by RNase T1 cleavage of yeast tRNAPhe−1 (Figure 4) could be assigned at a sequence level by the analysis of MS/MS spectra collected automatically by data-dependent CID. Although the RNA fragments generated a complex series of product ions in the CID-MS/MS analysis, most of the ions could be assigned to the original sequence. Thus, the product ions observed included the a/w and c/y series ions and their derivatives (hydrated or dehydrated ions and those ions that lost nucleotide bases), and internal ions (such as UU). In most cases, however, the a/w and c/y ion series were the major product ions as reported earlier (33,52–54) and thereby allowing the mass ladder assignments of the nucleotide sequence. Figure 5 illustrates a typical tandem MS spectrum of an oligoribonucleotide ion [AUUUAm2G > p]2−. In this particular case, the ladder from the a and c series indicated the sequence 5′-AUUUA … with OH at the 5′-end, and the ladder from the w and y series indicated the sequence … UUAm2G with 2′, 3′-cyclic phosphate at the 3′-end. We could not distinguish whether the methyl group was attached to the base or sugar of guanosine in the CID spectrum; however, given that RNase T1 does not cleave a phosphodiester bond if the ribose is 2′-O-methylated (56) or the guanine base is N7-methylated (56,57), the sequence of this oligoribonucleotide was deduced as 5′-HO-AUUUAm2G > p-3′. Likewise, most of the tRNAPhe−1 fragments shown in Figure 5 were confirmed at a sequence level by data-dependent LC-CID-MS/MS analyses.
Figure 5.
CID-MS/MS spectrum of [AUUUAm2G]2− derived from RNase T1 digestion of yeast tRNAPhe−1. The doubly charged ion with m/z = 966.6 was analyzed by CID. The nucleotide sequence was verified by manual interpretation of the a- and c-type (normal text) and w- and y-type (italic text) product ion series as indicated in the figure. The parent ion losing methyl guanine [P−B(mG)2−], the parent ion losing adenine [P−B(A)2−], the y5 ion losing methyl guanine [y5−B(mG)2−], the y5 ion losing adenine [y5−B(A)2−], y52− and c52− were doubly charged products. All other assigned signals were singly charged products, unless indicated otherwise. The asterisks indicate hydrated or dehydrated ions of a-, c-, w- or y-type products.
Analysis of U snRNA in the yeast pre-spliceosomal RNP complex
To examine whether the MS-based technology reported here could be used to analyze RNA components in the RNP complexes isolated from cells, we prepared the Lsm-associated RNP complex from yeast cells using tandem affinity purification (TAP)-tagged Lsm3 as affinity bait. The Lsm-associated RNP complex is a multifunctional complex that participates in the processing and/or turnover of various RNAs (58,59). The yeast S. cerevisiae has eight Lsm proteins (Lsm1–8), parts of which generate two types of ring-shaped heteroheptameric complexes (59). One of the complexes consisting of Lsm1–7 has a role in mRNA decapping and decay in the cytoplasm (58,60,61), whereas the other heteroheptameric complex consisting of Lsm2–8 binds to the 3′-end of U6 snRNA, increases its stability (62–65), and accelerates nuclear accumulation (66) in the yeast cell. In addition, the Lsm2–8 complex facilitates incorporation of U6 snRNPs into U4/U6 di-snRNPs and U4/U6.U5 tri-snRNPs, and thereby has a chaperone-like function in remodeling RNP particles (67). Recent advances in proteomics technologies have provided a dynamic aspect to the analysis of molecular interactions that regulate the complex series of events required for RNP assembly and RNA processing of this gigantic molecular machine (21,59).
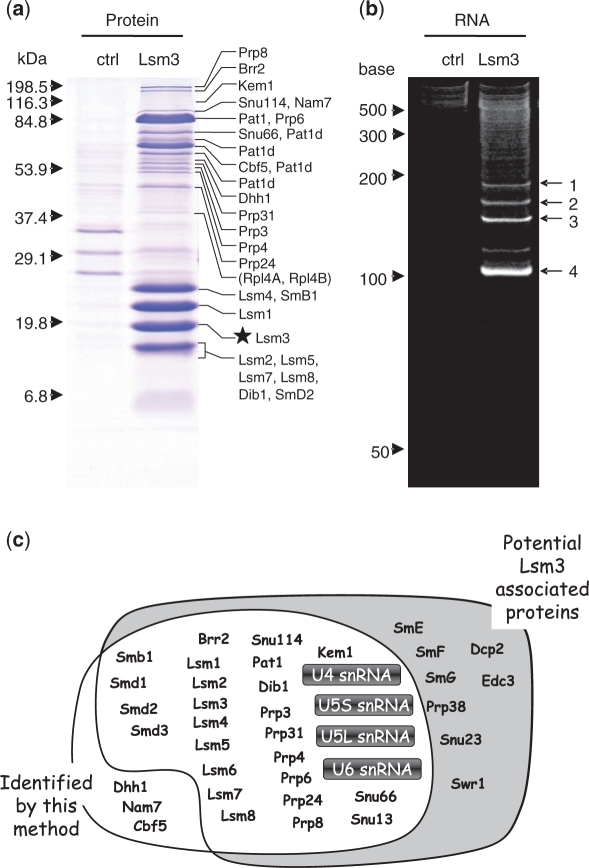
After the two-step affinity purification using the TAP-tag (see ‘Materials and Methods’ section), the Lsm3-associated RNP complex contained many proteins as examined by SDS–PAGE (Figure 6a). To characterize the purified RNP complex, we analyzed the protein composition by the proteomic LC-MS/MS method (41) after in-gel digestion of the individual bands excised from the SDS gel (Figure 6a and Supplementary Table S1) as well as by the LC-MS/MS shotgun method (42,43) after direct lysylendopeptidase digestion of the RNP preparation without gel separation (Supplementary Tables S1 and S2 and Supplementary Figure S2). In total, these analyses identified 25 of 33 potential components of the yeast Lsm3-associated RNP complex (58,68,69) (Figure 6c and Supplementary Table S1), suggesting that our preparation contained a typical Lsm3-associated RNP complex. Interestingly, we found additional proteins—Cbf5, Nam7 and Dhh1—in the Lsm3-associated RNP complex; Cbf5 is a pseudouridine synthase catalytic subunit found in box H/ACA small nucleolar RNP particles (70), Nam7 is an ATP-dependent RNA helicase of the SFI superfamily required for nonsense-mediated mRNA decay and for efficient translation termination at nonsense codons (71), and Dhh1 is a highly conserved DEAD-box RNA helicase that stimulates mRNA decapping and deadenylation (72) and is found associated with Lsm-3 by means of a protein-fragment complementation assay (73). However, whether these proteins are endogenous cofactors of this RNP complex and have roles in the RNA metabolism awaits further investigation.
Figure 6.
Protein and RNA composition of the purified yeast Lsm3-associated snRNP complex. (a) SDS–PAGE profile of the control (ctrl) and the Lsm3-associated RNP complex (Lsm3) pulled down with TAP-tagged Lsm3 as affinity bait (visualized by Coomassie brilliant blue staining). The control experiment was performed using TAP-tagged Ist3, which forms a pre-spliceosomal RNP complex that shares no protein/RNA components with the Lsm3-associated complex (78). The molecular mass markers are indicated on the left and the proteins identified by LC-MS/MS analysis of individual bands excised from the gel are shown on the right. The star indicates the position of TAP-tagged Lsm3 protein. Note that the gel-based proteomic analysis identified 25 known protein cofactors of the yeast Lsm3 complex and several Pat1-related proteins with apparently different molecular size (designated as Pat1d; Supplementary Table S1), as well as other yeast proteins, Rpl4A and B, that were also detected in the control Ist3 complex and thereby considered as contaminants in our Lsm3 complex preparation (indicated in parentheses). (b) PAGE profile of RNA components in the control (ctrl) and the Lsm3-associated complex (Lsm3). The bands 1–4, visualized by SYBR Gold staining containing 4–10 ng RNA (100–300 fmol), were subjected to LC-MS analysis. The RNA size markers are indicated on the left. (c) A schematic representation of the yeast Lsm3-associated proteins and RNAs identified in this study (white background) and their overlap with the known components reported in previous studies (grey background). Note that the MS-based technology reported here identified most protein cofactors and four U snRNAs (highlighted) of this pre-spliceosomal RNP complex. Detailed data for the protein and RNA analyses are given in Supplementary Figure S2 and Supplementary Tables S1–6.
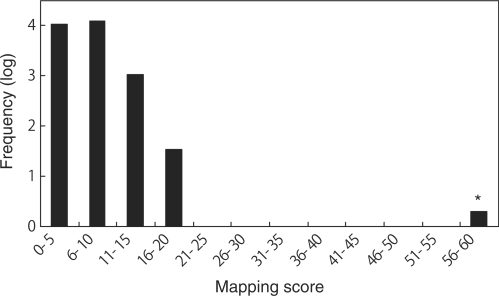
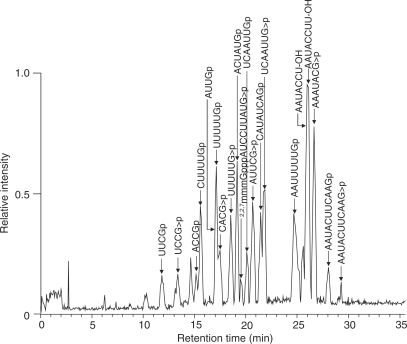
We analyzed the same Lsm3-associated RNP complex by 8 M urea–10% PAGE and using SYBR Gold staining, we detected four major RNA bands with approximate sizes of 100–200 nucleotides (Figure 6b). These bands were excised from the gel and in-gel digested with RNase T1. The digests were then analyzed by the nanoflow LC-LTQ-Orbitrap MS system, respectively, and the resulting MS/MS data were used to Ariadne search (40) against the genome database of S. cerevisiae for the identification of RNA species. Figure 7 illustrates a typical result obtained with one of the RNAs (band 3 in Figure 6b), where Ariadne assigned U4 snRNA for the band 3 with a significantly high score (cf. a false-positive rate of ∼1/1020). Figure 8 shows a base peak chromatogram of the nucleolytic fragments derived from band 3, together with the assignments of each fragment determined by the tandem MS analyses (Supplementary Table S3). The sequences of all the fragments coincided with those expected from the sequence of yeast U4 snRNA (65% sequence coverage), confirming that band 3 is U4 snRNA. Likewise, bands 1, 2 and 4 in Figure 6b were identified as U5S, U5L and U6 snRNA, respectively, by Ariadne search (data not shown) as well as by detailed MS and MS/MS analysis of the RNase T1 fragments (Supplementary Tables S4–6). Thus, the MS-based analysis clearly shows that the Lsm3-associated RNP complex isolated in this study is the core of the yeast spliceosome, which contains the major RNA components U4, U5 and U6 and the protein cofactors associated with U4/U6.U5 tri-snRNP (69). We note that the present MS analysis identified the 5′-terminal fragment AUCCUUAUG with a 5′-trimethylguanosine cap of U4 snRNA (Figure 8), as well as the equivalent trimethylguanosine-capped 5′ fragments of U5S and U5L, all of which are transcribed by RNA polymerase II. However, we failed to detect the 5′-terminal fragment of U6 snRNA, a transcript of RNA polymerase III, probably because the corresponding RNase T1 fragment, mpppGp, was not sufficiently hydrophobic to be retained on the C30 RP column used for LC-MS analysis. On the other hand, we found that the yeast U4 snRNA had multiple 3′-terminal fragments containing one, two and three uridines at the 3′-end (AAUACCU1–3-OH, Figure 8 and Supplementary Table S3). Although the biological significance of this heterogeneity is obscure, it is known that a primary transcript of U4 snRNA is digested by Rnt1 RNase III and a subsequent exonuclease catalyzes 3′-trimming during the biogenesis of spliceosomal U snRNPs (74). Likewise, analysis of U6 identified multiple 3′-terminal fragments consisting of a stretch of four, five, six and seven uridines (Supplementary Table S6). It is known that the 3′-uridine stretch of U6 is incorporated post-transcriptionally by a unique terminal uridylate transferase (75,76) and forms the binding site for a distinct heteroheptameric ring of Lsm2–8 proteins (23,77).
Figure 7.
Mapping score histogram of the Ariadne search results for band 3 in Figure 6 (b). Scores for all entries in the yeast genome database are summarized in the histogram. Frequencies of entries within a 10-point scoring range were counted, converted to common logarithm of frequency +1 and plotted. Note that the histogram shows a ‘hit’ (designated by an asterisk) for the query, as demonstrated by a distinctly high score, which indicates that the band 3 is U4 snRNA.
Figure 8.
Base peak chromatogram of the RNaseT1 digest of yeast U4 snRNA isolated from the Lsm3-associated RNP complex. The gel piece containing U4 snRNA was in-gel digested with RNaseT1 and subjected to the LC-MS analysis. Major oligoribonucleotide peaks assigned as RNase T1 fragments of yeast U4 snRNA are indicated by arrows with the corresponding sequence. Detailed data for MS/MS-based assignment of each fragment are given in Supplementary Table S3.
CONCLUSIONS
We described here a MS-based technology for RNA analysis, which combines direct nano-flow LC on a spray tip column and a high-resolution LTQ-Orbitrap mass spectrometer. The LC-MS system exhibited considerably high resolution that is sufficient for nucleotide mapping of small RNAs such as tRNA and U snRNA (Figures 4 and 8), and permits highly sensitive RNA analysis at a sub-femtomole level that is compatible with the current MS-based proteomics technology (Figure 2). Thus, the method reported here, coupled with the unique genome-oriented database search engine Ariadne, should provide a powerful tool for the analysis of short oligonucleotides such as siRNA and miRNA, as well as for the simultaneous identification and chemical analysis of small RNAs in RNP complexes such as those purified from cultured cells using affinity tags. In particular, the method should be useful to analyze the posttranscriptional nucleolytic processing and chemical modification of RNA, as exemplified here by its application to the identification and nucleotide mapping of U4 snRNA in the yeast Lsm3-associated pre-spliceosomal RNP complex (Figure 8). Thus, the LC-MS technology described here should permit, in combination with Ariadne, the development of MS-based technology similar to that used in ‘shotgun proteomics’, which allows simultaneous analysis of multiple RNA species in biological mixtures. This will open up the possibility to analyze the composition, chemical structure, and dynamics of RNA and protein components in the intermediate and functional cellular RNP complexes using a common MS-based technology platform.
In the course of this study, however, we noticed that the development of ‘shotgun ribonucleomics’ would still require surmounting several technical challenges. First, spurious RNA can be generated through an unknown mechanism from yet undefined genomic loci that make up ∼40% of the entire genome (77). Furthermore, the chemical characteristics of RNA are much less variable than protein with respect to the number of constituents (4 nucleotides versus 20 amino acids). Thus, RNA fragments often have similar compositions, making them inherently difficult to distinguish. However, we assume that the accumulation of our knowledge of non-coding RNA and the capability of LC-MS technology will rapidly solve most of these problems and accelerate the use of automated MS-based RNA analyses that are complementary to conventional techniques based on genomics and molecular biology.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Core Research for Evolutionary Science and Technology (CREST), Japan Science and Technology Agency. Funding for open access charge: Core Research for Evolutionary Science and Technology (CREST), Japan Science and Technology Agency.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Takashi Ito at University of Tokyo for kind donation of the yeast Saccharomyces cerevisiae strain S288C.
REFERENCES
- 1.Hirota K, Miyoshi T, Kugou K, Hoffman CS, Shibata T, Ohta K. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature. 2008;456:130–134. doi: 10.1038/nature07348. [DOI] [PubMed] [Google Scholar]
- 2.Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fischer SE, Butler MD, Pan Q, Ruvkun G. Trans-splicing in C. elegans generates the negative RNAi regulator ERI-6/7. Nature. 2008;455:491–496. doi: 10.1038/nature07274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 5.Cullen BR. Transcription and processing of human microRNA precursors. Mol. Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27:5959–5974. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- 7.Serganov A, Patel DJ. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat. Rev. Genet. 2007;8:776–790. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132:945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 10.Chien KR. Molecular medicine: microRNAs and the tell-tale heart. Nature. 2007;447:389–390. doi: 10.1038/447389a. [DOI] [PubMed] [Google Scholar]
- 11.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi N, Isobe T. Proteomic Biology using LC/MS: Large Scale Analysis of Cellular Dynamics and Function. NJ, USA: Wiley Interscience, John Wiley & Sons Inc; 2007. [Google Scholar]
- 13.Fujiyama S, Yanagida M, Hayano T, Miura Y, Isobe T, Fujimori F, Uchida T, Takahashi N. Isolation and proteomic characterization of human Parvulin-associating preribosomal ribonucleoprotein complexes. J. Biol. Chem. 2002;277:23773–23780. doi: 10.1074/jbc.M201181200. [DOI] [PubMed] [Google Scholar]
- 14.Hayano T, Yanagida M, Yamauchi Y, Shinkawa T, Isobe T, Takahashi N. Proteomic analysis of human Nop56p-associated pre-ribosomal ribonucleoprotein complexes. Possible link between Nop56p and the nucleolar protein treacle responsible for Treacher Collins syndrome. J. Biol. Chem. 2003;278:34309–34319. doi: 10.1074/jbc.M304304200. [DOI] [PubMed] [Google Scholar]
- 15.Stavreva DA, Kawasaki M, Dundr M, Koberna K, Muller WG, Tsujimura-Takahashi T, Komatsu W, Hayano T, Isobe T, Raska I, et al. Potential roles for ubiquitin and the proteasome during ribosome biogenesis. Mol. Cell Biol. 2006;26:5131–5145. doi: 10.1128/MCB.02227-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi N, Yanagida M, Fujiyama S, Hayano T, Isobe T. Proteomic snapshot analyses of preribosomal ribonucleoprotein complexes formed at various stages of ribosome biogenesis in yeast and mammalian cells. Mass Spectrom Rev. 2003;22:287–317. doi: 10.1002/mas.10057. [DOI] [PubMed] [Google Scholar]
- 17.Yanagida M, Hayano T, Yamauchi Y, Shinkawa T, Natsume T, Isobe T, Takahashi N. Human fibrillarin forms a sub-complex with splicing factor 2-associated p32, protein arginine methyltransferases, and tubulins alpha 3 and beta 1 that is independent of its association with preribosomal ribonucleoprotein complexes. J. Biol. Chem. 2004;279:1607–1614. doi: 10.1074/jbc.M305604200. [DOI] [PubMed] [Google Scholar]
- 18.Yanagida M, Shimamoto A, Nishikawa K, Furuichi Y, Isobe T, Takahashi N. Isolation and proteomic characterization of the major proteins of the nucleolin-binding ribonucleoprotein complexes. Proteomics. 2001;1:1390–1404. doi: 10.1002/1615-9861(200111)1:11<1390::AID-PROT1390>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 19.Wu L, Belasco JG. Let me count the ways: mechanisms of gene regulation by miRNAs and siRNAs. Mol. Cell. 2008;29:1–7. doi: 10.1016/j.molcel.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 20.Engels BM, Hutvagner G. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene. 2006;25:6163–6169. doi: 10.1038/sj.onc.1209909. [DOI] [PubMed] [Google Scholar]
- 21.Valadkhan S. The spliceosome: caught in a web of shifting interactions. Curr. Opin. Struct. Biol. 2007;17:310–315. doi: 10.1016/j.sbi.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 22.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 23.Neuenkirchen N, Chari A, Fischer U. Deciphering the assembly pathway of Sm-class U snRNPs. FEBS Lett. 2008;582:1997–2003. doi: 10.1016/j.febslet.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 24.Lu C, Meyers BC, Green PJ. Construction of small RNA cDNA libraries for deep sequencing. Methods. 2007;43:110–117. doi: 10.1016/j.ymeth.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 25.Brooks EM, Sheflin LG, Spaulding SW. Secondary structure in the 3′ UTR of EGF and the choice of reverse transcriptases affect the detection of message diversity by RT-PCR. Biotechniques. 1995;19:806–812, 814–805. [PubMed] [Google Scholar]
- 26.Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat. Struct. Mol. Biol. 2007;14:349–350. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- 27.Czerwoniec A, Dunin-Horkawicz S, Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, Grosjean H, Rother K. MODOMICS: a database of RNA modification pathways. 2008 update. Nucleic Acids Res. 2009;37:D118–D121. doi: 10.1093/nar/gkn710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Douthwaite S, Kirpekar F. Identifying modifications in RNA by MALDI mass spectrometry. Methods Enzymol. 2007;425:1–20. doi: 10.1016/S0076-6879(07)25001-3. [DOI] [PubMed] [Google Scholar]
- 29.Tost J, Gut IG. DNA analysis by mass spectrometry-past, present and future. J. Mass Spectrom. 2006;41:981–995. doi: 10.1002/jms.1096. [DOI] [PubMed] [Google Scholar]
- 30.Lin ZJ, Li W, Dai G. Application of LC-MS for quantitative analysis and metabolite identification of therapeutic oligonucleotides. J. Pharm. Biomed. Anal. 2007;44:330–341. doi: 10.1016/j.jpba.2007.01.042. [DOI] [PubMed] [Google Scholar]
- 31.Emmerechts G, Barbe S, Herdewijn P, Anne J, Rozenski J. Post-transcriptional modification mapping in the Clostridium acetobutylicum 16S rRNA by mass spectrometry and reverse transcriptase assays. Nucleic Acids Res. 2007;35:3494–3503. doi: 10.1093/nar/gkm248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Premstaller A, Oberacher H, Huber CG. High-performance liquid chromatography-electrospray ionization mass spectrometry of single- and double-stranded nucleic acids using monolithic capillary columns. Anal. Chem. 2000;72:4386–4393. doi: 10.1021/ac000283d. [DOI] [PubMed] [Google Scholar]
- 33.Holzl G, Oberacher H, Pitsch S, Stutz A, Huber CG. Analysis of biological and synthetic ribonucleic acids by liquid chromatography-mass spectrometry using monolithic capillary columns. Anal. Chem. 2005;77:673–680. doi: 10.1021/ac0487395. [DOI] [PubMed] [Google Scholar]
- 34.Zou Y, Tiller P, Chen IW, Beverly M, Hochman J. Metabolite identification of small interfering RNA duplex by high-resolution accurate mass spectrometry. Rapid Commun. Mass Spectrom. 2008;22:1871–1881. doi: 10.1002/rcm.3561. [DOI] [PubMed] [Google Scholar]
- 35.Wagner TM, Nair V, Guymon R, Pomerantz SC, Crain PF, Davis DR, McCloskey JA. A novel method for sequence placement of modified nucleotides in mixtures of transfer RNA. Nucleic Acids Symp. Ser. (Oxf) 2004;48:263–264. doi: 10.1093/nass/48.1.263. [DOI] [PubMed] [Google Scholar]
- 36.Guymon R, Pomerantz SC, Crain PF, McCloskey JA. Influence of phylogeny on posttranscriptional modification of rRNA in thermophilic prokaryotes: the complete modification map of 16S rRNA of Thermus thermophilus. Biochemistry. 2006;45:4888–4899. doi: 10.1021/bi052579p. [DOI] [PubMed] [Google Scholar]
- 37.Rozhdestvensky TS, Crain PF, Brosius J. Isolation and posttranscriptional modification analysis of native BC1 RNA from mouse brain. RNA Biol. 2007;4:11–15. doi: 10.4161/rna.4.1.4306. [DOI] [PubMed] [Google Scholar]
- 38.Huang TY, Kharlamova A, Liu J, McLuckey SA. Ion trap collision-induced dissociation of multiply deprotonated RNA: c/y-ions versus (a-B)/w-ions. J. Am. Soc. Mass Spectrom. 2008;19:1832–1840. doi: 10.1016/j.jasms.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 39.Huang TY, Liu J, Liang X, Hodges BD, McLuckey SA. Collision-induced dissociation of intact duplex and single-stranded siRNA anions. Anal. Chem. 2008;80:8501–8508. doi: 10.1021/ac801331h. [DOI] [PubMed] [Google Scholar]
- 40.Nakayama H, Akiyama M, Taoka M, Yamauchi Y, Nobe Y, Ishikawa H, Takahashi N, Isobe T. Ariadne: a database search engine for identification and chemical analysis of RNA using tandem mass spectrometry data. Nucleic Acids Res. 2009;37:e47. doi: 10.1093/nar/gkp099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Natsume T, Yamauchi Y, Nakayama H, Shinkawa T, Yanagida M, Takahashi N, Isobe T. A direct nanoflow liquid chromatography-tandem mass spectrometry system for interaction proteomics. Anal. Chem. 2002;74:4725–4733. doi: 10.1021/ac020018n. [DOI] [PubMed] [Google Scholar]
- 42.Kaji H, Saito H, Yamauchi Y, Shinkawa T, Taoka M, Hirabayashi J, Kasai K, Takahashi N, Isobe T. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat. Biotechnol. 2003;21:667–672. doi: 10.1038/nbt829. [DOI] [PubMed] [Google Scholar]
- 43.Taoka M, Yamauchi Y, Shinkawa T, Kaji H, Motohashi W, Nakayama H, Takahashi N, Isobe T. Only a small subset of the horizontally transferred chromosomal genes in Escherichia coli are translated into proteins. Mol. Cell Proteom. 2004;3:780–787. doi: 10.1074/mcp.M400030-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 46.Ichimura T, Wakamiya-Tsuruta A, Itagaki C, Taoka M, Hayano T, Natsume T, Isobe T. Phosphorylation-dependent interaction of kinesin light chain 2 and the 14-3-3 protein. Biochemistry. 2002;41:5566–5572. doi: 10.1021/bi015946f. [DOI] [PubMed] [Google Scholar]
- 47.Taoka M, Ichimura T, Wakamiya-Tsuruta A, Kubota Y, Araki T, Obinata T, Isobe T. V-1, a protein expressed transiently during murine cerebellar development, regulates actin polymerization via interaction with capping protein. J. Biol. Chem. 2003;278:5864–5870. doi: 10.1074/jbc.M211509200. [DOI] [PubMed] [Google Scholar]
- 48.Mawuenyega KG, Kaji H, Yamuchi Y, Shinkawa T, Saito H, Taoka M, Takahashi N, Isobe T. Large-scale identification of Caenorhabditis elegans proteins by multidimensional liquid chromatography-tandem mass spectrometry. J. Proteome Res. 2003;2:23–35. doi: 10.1021/pr025551y. [DOI] [PubMed] [Google Scholar]
- 49.Apffel A, Chake JA, Fischer S, Lichtenwalter K, Hancock WS. Analysis of oligonucleotides by HPLC-electrospray ionization mass spectrometry. Anal. Chem. 1997;69:1320–1325. doi: 10.1021/ac960916h. [DOI] [PubMed] [Google Scholar]
- 50.RajBhandary UL, Chang SH. Studies on polynucleotides. LXXXII. Yeast phenylalanine transfer ribonucleic acid: partial digestion with ribonuclease T-1 and derivation of the total primary structure. J. Biol. Chem. 1968;243:598–608. [PubMed] [Google Scholar]
- 51.Martin G, Keller W. RNA-specific ribonucleotidyl transferases. RNA. 2007;13:1834–1849. doi: 10.1261/rna.652807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu J, McLuckey SA. Gas-phase fragmentation of oligonucleotide ions. Int. J. Mass Spectrom. 2004;237:197–241. [Google Scholar]
- 53.Ni J, Pomerantz C, Rozenski J, Zhang Y, McCloskey JA. Interpretation of oligonucleotide mass spectra for determination of sequence using electrospray ionization and tandem mass spectrometry. Anal. Chem. 1996;68:1989–1999. doi: 10.1021/ac960270t. [DOI] [PubMed] [Google Scholar]
- 54.Tromp JM, Schurch S. Gas-phase dissociation of oligoribonucleotides and their analogs studied by electrospray ionization tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 2005;16:1262–1268. doi: 10.1016/j.jasms.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 55.McLuckey SA, Van Berkel GJ, Glish GL. Tandem mass spectrometry of small, multiply charged oligonucleotide. J. Am. Soc. Mass Spectrom. 1992;3:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
- 56.Osterman HL, Walz FG., Jr Subsites and catalytic mechanism of ribonuclease T1: kinetic studies using GpA, GpC, GpG, and GpU as substrates. Biochemistry. 1978;17:4124–4130. doi: 10.1021/bi00613a003. [DOI] [PubMed] [Google Scholar]
- 57.Brimacombe RLC, Griffin BE, Haines JA, Haslam WJ, Reese CB. An approach to the methylation of polynucleotides. Biochemistry. 1965;4:2452–2458. [Google Scholar]
- 58.Bouveret E, Rigaut G, Shevchenko A, Wilm M, Seraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beggs JD. Lsm proteins and RNA processing. Biochem. Soc. Trans. 2005;33:433–438. doi: 10.1042/BST0330433. [DOI] [PubMed] [Google Scholar]
- 60.Boeck R, Lapeyre B, Brown CE, Sachs AB. Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol. Cell Biol. 1998;18:5062–5072. doi: 10.1128/mcb.18.9.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- 62.Ingelfinger D, Arndt-Jovin DJ, Luhrmann R, Achsel T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- 63.Fromont-Racine M, Mayes AE, Brunet-Simon A, Rain JC, Colley A, Dix I, Decourty L, Joly N, Ricard F, Beggs JD, et al. Genome-wide protein interaction screens reveal functional networks involving Sm-like proteins. Yeast. 2000;17:95–110. doi: 10.1002/1097-0061(20000630)17:2<95::AID-YEA16>3.0.CO;2-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pannone BK, Xue D, Wolin SL. A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 1998;17:7442–7453. doi: 10.1093/emboj/17.24.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Salgado-Garrido J, Bragado-Nilsson E, Kandels-Lewis S, Seraphin B. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. 1999;18:3451–3462. doi: 10.1093/emboj/18.12.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spiller MP, Boon KL, Reijns MA, Beggs JD. The Lsm2-8 complex determines nuclear localization of the spliceosomal U6 snRNA. Nucleic Acids Res. 2007;35:923–929. doi: 10.1093/nar/gkl1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Verdone L, Galardi S, Page D, Beggs JD. Lsm proteins promote regeneration of pre-mRNA splicing activity. Curr. Biol. 2004;14:1487–1491. doi: 10.1016/j.cub.2004.08.032. [DOI] [PubMed] [Google Scholar]
- 68.Nash R, Weng S, Hitz B, Balakrishnan R, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, et al. Expanded protein information at SGD: new pages and proteome browser. Nucleic Acids Res. 2007;35:D468–D471. doi: 10.1093/nar/gkl931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hacker I, Sander B, Golas MM, Wolf E, Karagoz E, Kastner B, Stark H, Fabrizio P, Luhrmann R. Localization of Prp8, Brr2, Snu114 and U4/U6 proteins in the yeast tri-snRNP by electron microscopy. Nat. Struct. Mol. Biol. 2008;15:1206–1212. doi: 10.1038/nsmb.1506. [DOI] [PubMed] [Google Scholar]
- 70.Lafontaine DL, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.de la Cruz J, Kressler D, Linder P. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci. 1999;24:192–198. doi: 10.1016/s0968-0004(99)01376-6. [DOI] [PubMed] [Google Scholar]
- 72.Fischer N, Weis K. The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J. 2002;21:2788–2797. doi: 10.1093/emboj/21.11.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tarassov K, Messier V, Landry CR, Radinovic S, Serna Molina MM, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- 74.van Hoof A, Lennertz P, Parker R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell Biol. 2000;20:441–452. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wickens M, Kwak JE. Molecular biology. A tail tale for U. Science. 2008;319:1344–1345. doi: 10.1126/science.1154946. [DOI] [PubMed] [Google Scholar]
- 76.Trippe R, Guschina E, Hossbach M, Urlaub H, Luhrmann R, Benecke BJ. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA. 2006;12:1494–1504. doi: 10.1261/rna.87706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 78.Dziembowski A, Ventura AP, Rutz B, Caspary F, Faux C, Halgand F, Laprevote O, Seraphin B. Proteomic analysis identifies a new complex required for nuclear pre-mRNA retention and splicing. EMBO J. 2004;23:4847–4856. doi: 10.1038/sj.emboj.7600482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.