Abstract
SMYD1 is a heart and muscle specific SET-MYND domain containing protein, which functions as a histone methyltransferase and regulates downstream gene transcription. We demonstrated that the expression of SMYD1 is restricted in the heart and skeletal muscle tissues in human. To reveal the regulatory mechanisms of SMYD1 expression during myogenesis and cardiogenesis, we cloned and characterized the human SMYD1 promoter, which contains highly conserved serum response factor (SRF) and myogenin binding sites. Overexpression of SRF and myogenin significantly increased the endogenous expression level of Smyd1 in C2C12 cells, respectively. Deletion of Srf in the heart of mouse embryos dramatically decreased the expression level of Smyd1 mRNA and the expression of Smyd1 can be rescued by exogenous SRF introduction in SRF null ES cells during differentiation. Furthermore, we demonstrated that SRF binds to the CArG site and myogenin binds to the E-box element on Smyd1 promoter region using EMSA and ChIP assays. Moreover, forced expression of SMYD1 accelerates myoblast differentiation and myotube formation in C2C12 cells. Taken together, these studies demonstrated that SMYD1 is a key regulator of myogenic differentiation and acts as a downstream target of muscle regulatory factors, SRF and myogenin.
INTRODUCTION
Skeletal muscle differentiation is a multistep process, which begins with the commitment of multi-potent mesodermal precursor cells to the skeletal muscle lineage. The committed cells, called the myoblasts, differentiate into myocytes and then fuse into multinucleated myotubes. The final step of muscle differentiation is the maturation of differentiated myotubes into myofibres (1–3). This process is tightly controlled by multiple groups of transcriptional factors, among which the basic helix–loop–helix myogenic regulatory factors (MRFs) and MADS (MCM1, agomous, deficiens, serum response factor) box transcription factors play pivotal roles in regulating muscle-specific gene expression and controlling skeletal muscle lineage determination, differentiation, and myotube formation (4–6).
The MyoD family (also called MRFs) of basic helix–loop–helix proteins includes MyoD, myogenin, Myf5 and MRF4, which binds to E-box (CANNTG) sequences in the promoters and induces downstream muscle specific gene expression (5,7). The MRFs regulate skeletal muscle differentiation through a temporal pattern. MyoD and Myf5 govern myoblast specification and act upstream of myogenin while MRF4 regulates terminal differentiation. Relative normal myogenesis was observed in both MyoD and Myf5 mutant mouse whereas double mutant of these two factors in mouse results in a complete lack of skeletal muscle formation, indicating the functional redundancy of MyoD and Myf5 (8–11). A perinatal lethal phenotype was observed in myogenin-mutant mice, which exhibit no defects in the initiation step of myogenesis but defects in the differentiation of myocytes and myofibers (12,13). Muscle specific transcription requires functional interactions of these muscle-specific bHLH factors with other regulatory proteins that are not restricted to skeletal muscle. The MADS domain transcription factors are important members among these regulatory proteins (14).
Serum response factor (SRF), a MADS box transcription factor related to the MEF2s, regulates skeletal, as well as cardiac and smooth muscle genes by binding to a consensus DNA sequence known as CArG [CC(A/T)6GG] box within the promoter of downstream target genes (15–18). The Myocardin family proteins, including Myocardin, MRTF-A/MKL1 and MRTF-B/MKL2, are powerful SRF coactivators expressed in heart and muscle tissues (19–22). Conditional deletion of the Srf gene in mouse skeletal muscle-lineage leads to perinatal death due to severe skeletal muscle hypoplasia (23). Cardiac-specific deletion of Srf results in embryonic lethality due to cardiac insufficiency during chamber maturation and blocking of the appearance of rhythmic beating myocytes (24,25). Moreover, deletion of Srf in smooth muscle results in embryonic lethality caused by a deficiency of differentiated smooth muscle cells (26). The interactions between MADS-box proteins and MyoD family members are at multiple levels and form a dedicated regulatory network. SRF not only physically interacts with MyoD and myogenin but also regulates the mRNA expression of MyoD family members (27–30). Moreover, SRF and the myogenic bHLH proteins act cooperatively to regulate muscle-specific gene expression through adjacent CArG sites and E-box elements in the target gene promoter (31–34).
SMYD1, also called BOP, is the first identified heart and muscle specific histone methyltransferase which contains a SET domain and is essential for embryogenesis in mouse and fish through regulation of cardiogenesis and myogenesis (35,36). Here we report the characterization of SMYD1 promoter and the identification of the regulation of SMYD1 expression by SRF and myogenin. By northern blot analysis, the mRNA of human SMYD1 is restricted in heart and skeletal muscle tissues. With sequence alignment of SMYD1 promoter across species, we identified myogenin and SRF binding sites which were further characterized by EMSA, ChIP and reporter assays. Over-expression of myogenin and SRF in C2C12 cells stimulates endogenous Smyd1 expression, respectively and co-operatively. In vivo, Smyd1 mRNA level was dramatically decreased in the heart of Srf cardiac-conditional knock-out mouse as demonstrated by in situ hybridization. Finally, over-expression of SMYD1 up-regulates muscle-specific marker genes and promotes myoblasts differentiation and myotube formation in C2C12 cells. Taken together, SMYD1 is a direct downstream target of cardiac and skeletal muscle regulatory factors and acts as a key factor in myogenic differentiation.
MATERIALS AND METHODS
Chemicals, biochemicals and constructs
Luciferase reagent and lysis buffer were obtained from Promega Corp. (Madison, WI). [γ-32P]dCTP (300 Ci/mmol) was obtained from PerkinElmer Life Sciences (Wellesley, MA). Poly (dI–dC) and T4 polynucleotide kinase were purchased from Roche Molecular Biochemicals (Indianapolis, IN). The pCMV-Tag2B mammalian expression vector containing the human SMYD1 cDNA sequence was generated with primers FL-F (5′-AGGATCCACTGAGATGACAATAGGGAGAATG-3′) and FL-R (5′-AGACTCGAGTCCACTGGGCAGTCCTC-3′). The full-length human SMYD1 promoter reporter construct was amplified by PCR with genomic DNA extracted from 293T cells as the templates. PCR products were digested with XhoI and KpnI restriction enzymes followed with ligation into the pGL3-basic vector. All other human SMYD1 truncated promoter fragments were likewise cloned into XhoI and KpnI sites in the pGL3-basic vector, and the primers for subcloning were: SMYD1-R, 5′-CCGCTCGAGCGGACGTTCTCCATTCTCCCTATTG-3′; SMYD1-1120F, 5′-GGGGTACCCCTCCAGCATACCAGCATCACC-3′; SMYD1-620F 5′-GGGGTACCCCTTCTGCTGCTAAACAGTCCAC-3′; SMYD1-519F 5′-GGGGTACCCCACCTTGTTTCTATGGGACC-3′; SMYD1-330F 5′-GGGGTACCCCATGCCATATTAACCCATGGTC-3′; SMYD1-110F 5′-GGGGTACCCCAGCAATGACAAGAGACTTGGC-3′. Expression vectors for SRF, myogenin and E12 were obtained as previously described (37).
Generation of SRF cardiac-conditional null embryos and genotyping
SRF cardiac-conditional knockout mouse strain was generated as described earlier (24,25).
Production of the anti-SMYD1 antibody
The GST-SMYD1ΔN fusion construct was prepared by amplifying the amino acid residues 279–490 of human SMYD1 into the unique BamHI and XhoI sites of pGEX-4T. GST-SMYD1ΔN recombinant protein was purified with a glutathione–sepharose column (Amersham Biosciences) according to the manufacturer’s directions. Rabbit polyclonal antiserum recognizing Smyd1 was prepared using the GST-SMYD1ΔN recombinant protein. The serum recognizing Smyd1 was filtered through a passage to the column with GST-SMYD1ΔN fusion protein, and that column was eluted with a low-pH buffer to obtain the anti-GST-SMYD1ΔN antibody. It was filtered again through a passage to the column with only GST protein to remove the anti-GST antibody component as best as possible. The antibody specificity was confirmed by western blotting using recombinant Smyd1 protein expressed in bacterial and eukaryotic cells.
Northern blot analysis
An enzyme-digested 633 bp probe binding to the 3′ half of SMYD1 cDNA was prepared by using BamHI and XhoI sites on the pGEX-4T-SMYD1ΔN construct. Multiple Tissue Northern (MTN) blots were prepared from the purified poly(A)+ RNA and blotted onto a positively charged nylon membrane (CLONTECH). The amount of RNA on an MTN blot membrane is adjusted so that the β-actin hybridization signal is of comparable intensity in every lane (CLONTECH).
Cell culture and transfections
C2C12 and 293T cells (American Type Culture Collection, Manassas, VA) were grown in DMEM (Hyclone Laboratories, Inc., Logan, UT) supplemented with 10% fetal bovine serum (FBS) (Hyclone), 4.5 mg/ml glucose, and penicillin/streptomycin, and maintained at 37°C in 5% CO2. Cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s directions. For all experiments other than reporter assays, pools of cells were stably selected with G418 prior to experimental analysis.
Luciferase reporter assays
C2C12 cells were cultured in 24-well plates in DMEM supplemented with 10% FBS. After 18–20 h when cells were ∼60–70% confluent, indicated gene constructs were transfected using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) in serum-free DMEM according to the manufacturer’s protocol. After 6 h, medium was replaced with growth medium (GM) consisting of DMEM supplemented with 10% FBS or differentiation medium consisting of DMEM supplemented with 2% horse serum (DM) as indicated. Thirty-six to forty-eight hours after transfection, cells were harvested, and the luciferase activity was determined using the luciferase assay system (Promega) normalized to β-galactosidase enzyme activity, as previously described (38).
Western blot analysis
Cells were lysed in cell lysis buffer [50 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 µg/ml leupeptin, 10 µg/ml aprotinin, and 1 mM sodium orthovanadate]. The insoluble material was excluded by centrifugation. The resulting supernatant was mixed with sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) loading buffer and was subjected to SDS–PAGE. After electrophoresis, the proteins were transferred to a polyvinylidene difluoride membrane, and the blot was incubated with the appropriate primary antibody. Following incubation with the primary antibody, the membrane was exposed to a horseradish peroxidase-conjugated secondary antimouse, anti-rabbit, or anti-goat antibody, subjected to SuperSignal West Pico Chemiluminescent reagent (Pierce Biotechnology Inc.), and exposed to film. Antibodies were used in the following dilutions: 1:1000 anti-SRF antibody, 1:1000 anti-actin (Santa Cruz Biotechnology); 1:25 anti-myogenin F5D antibody, 1:25 anti-myosin heavy chain (MHC) MF20 antibody (MHC; Developmental Studies Hybridoma Bank).
Electrophoretic mobility shift assays
Cells were cultured in six-well plates in DMEM containing 10% FBS or manipulated as described. The following day, cells were harvested by manual scraping and resuspended in 25 mM HEPES, 1.5 mM EDTA and 1 mM dithiothreitol (pH 7.6), and homogenized with 10% glycerol. Cell lysates were then centrifuged at 4°C, and protein concentration of nuclear extracts was then determined using BCA assay (Pierce, Rockford, IL). Aliquots of nuclear protein were then frozen and stored at −80°C until use. Mouse Smyd1 promoter derived SRF and myogenin binding site oligonucleotides were synthesized and annealed, and 5 pmol were 5′-end-labeled using T4 polynucleotide kinase and [γ32-P]ATP. A 30-µl EMSA reaction containing ∼100 mM KCl, 5 µg crude nuclear extract, 1 µg poly (dI–dC), and 1 µl of 1 mm ZnCl2 with or without unlabeled competitor oligonucleotide, and 10 fmol labeled probe was incubated on ice for 20 min. SRF- and F5D-specific antibodies were then incubated in appropriate reactions for 20 min on ice. DNA–protein complexes were then resolved on 5% PAGE gels at ∼120 V at 4°C for 2.5 h. The gels were dried, and protein DNA complexes were visualized by autoradiography.
Chromatin immunoprecipitation assays
ChIP assays were performed as previously described (38). Briefly, C2C12 cells were grown in 100-mm tissue culture plates and introduced in DM for 2 days. Formaldehyde was then added to the medium to a final concentration of 1%, and the reaction was incubated at room temperature with shaking for 10 min, after which glycine (0.125 M) was added, and the reaction was incubated for another 10 min. The media were then removed, and cells were washed twice with cold PBS and 1 mM phenylmethylsulfonyl fluoride, scraped, collected by centrifugation, and lysed in SDS lysis buffer containing 1 mM phenylmethylsulfonyl fluoride, 1 µg/ml aprotinin and 1 µg/ml pepstatin A. DNA was sheared to fragments of 500–1000 bp by eight 10-s sonications. The chromatin was precleared with salmon sperm DNA/protein A-agarose slurry (Upstate Biotechnology, Waltham, MA) for 1 h at 4°C with gentle agitation. The agarose beads were pelleted, and the precleared supernatant was incubated with or without anti-SRF antibody overnight at 4°C, respectively. The reactions were subsequently washed with low-salt wash buffer [0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris (pH 8.1) and 150 mM NaCl], high-salt wash buffer [0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris (pH 8.1) and 500 mM NaCl], LiCl wash buffer [0.25 m LiCl, 1% IGEPAL CA-630, 1% deoxycholate, 1 mM EDTA, 10 mM Tris (pH 8.1)], and twice in TE buffer [10 mM Tris–HCl (pH 8.0) and 1 mM EDTA]. After reversing the protein/DNA cross-links, DNA was recovered by phenol/chloroform extraction and ethanol precipitation. The region containing SRF binding site on mouse Smyd1 promoter was amplified from the immunoprecipitated chromatin using the following primers: sense, 5′-TCTGGGTGATGAACGAGACC-3′; and antisense, 5′-TCTCCGCAAAGATGACATC-3′. After PCR, the 341 bp products were resolved on a 2.0% agarose gel and stained with ethidium bromide. Samples were visualized under UV light. PCR products were purified and sequenced.
Embryo collection and in situ hybridization
Embryos were collected and fixed in 4% paraformaldehyde overnight at 4°C. For whole mount in situ hybridization, post-fixed embryos were briefly washed in diethylpyrocarbonate (DEPC)-treated standard phosphate-buffered saline, and dehydrated in serial methanol. Whole mount in situ hybridization was performed as previously described (24). For in situ hybridization on sections, fixed embryos were washed in DEPC-treated phosphate-buffered saline and handled as stated above. Sagittal and transverse sections (7 µm) were collected onto PL-100 poly-l-lysine slides (CEL Associates Inc., Pearland, Texas). Two to three sections were placed on each slide and hybridized with cRNA probes. cRNA probes were prepared according to the digoxigenin labeling kit (Roche Applied Science).
Embryonic stem cell culture and lentiviral rescue
Both AB2.2 and Srf−/− ES cell were maintained at optimal condition with lymphocyte inhibitory factor. ES cell infection with SRF-lentivirus and embryoid body (EB) differentiation was previously described (25).
Real-time PCR
Total RNA from ES cell embryoid body was extracted using Trizol (Invitrogen) and treated with DNAseI (Invitrogen) before further analysis. Real-time RT-PCR assay was carried in an ABI Prism 7700 system following manufacture instruction. Gene specific probes were order from ABI: GAPDH (Mm99999915_g1) and Smyd1 (Smyd1: Mm00477663_m1).
Immunofluorescence
Cells growing on glass coverslips were fixed in ice-cold methanol for 10 min, followed by 10 min of permeabilization in 0.1% Triton X-100 in PBS. Reactions were blocked for 30 min with 0.2% bovine serum albumin, followed by 60 min of incubation with a mouse monoclonal MF-20 antibody (1:5 dilution; Iowa Hybridoma Bank). Fluorescein-conjugated goat anti-mouse antibody was added for 30 min (1:500; Molecular Probes). Nuclear staining was observed after 10 min of 4′,6-diamidino-2-phenylindole (DAPI) treatment (1:500 dilution; Molecular Probes). Fluorescence images were captured at 400x on a CCD camera mounted on an inverted research microscope using Ultraview imaging software (Olympus Inc.).
RESULTS
Human SMYD1 is a SET and MYND domain containing protein with specific expression in skeletal muscle and heart tissues
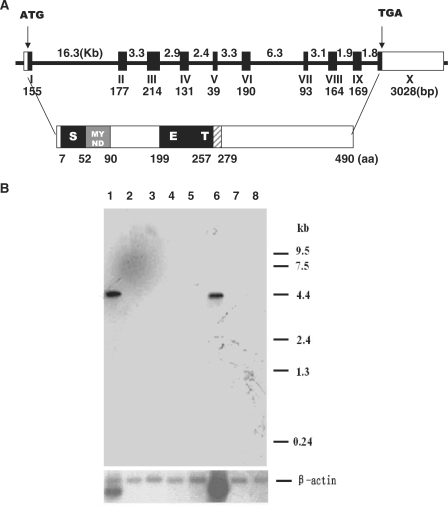
SMYD1 has been identified as a histone H3 lysine 4 methyltransferase, which plays an essential role in myogenesis and cardiogenesis in different species (35,36,39). Since Smyd1 is evolutionary conserved in sequence and function from fish to mouse, we hypothesized that its human orthologue is also a critical gene with important function. As presented in Figure 1A, SMYD1 gene is 4360 bp in length and contains a putative open reading frame of 1473 bp which encodes a 490aa protein with SET domain split by a MYND finger.
Figure 1.
The genomic/protein structure and tissue distribution of human SMYD1. (A) SMYD1 gene spans about 45 kb and contains 10 exons encoding a protein of 490 amino acids. Coding region is depicted as solid black boxes and the 5′- and 3′-untranslated regions of exon 1(I) and 10 (X) are presented as open boxes. The number and size of the exons and introns are indicated. The predicted domain structure of SMYD1 is presented below the genomic structure. (B) Northern blot analysis of SMYD1 expression in different human tissues. Multiple tissue northern (MTN) blots were prepared from the purified poly(A)+ RNA and blotted onto a positively charged nylon membrane (CLONTECH). The amount of RNA on an MTN blot membrane is adjusted so that β-actin hybridization signal is of comparable intensity in each lane (CLONTECH). RNA was isolated from different human tissues: 1, heart; 2, brain; 3, placenta; 4, lung; 5, liver; 6, skeletal muscle; 7, kidney; 8, pancreas. The size of SMYD1 cDNA was estimated to be 4.4 kb.
To analyze the tissue distribution of SMYD1 transcripts, we examined the expression of the mRNA in different human tissues by Northern blot analysis. As shown in Figure 1B, SMYD1 expression is highly restricted in human heart and skeletal muscle tissues. The detected size of the SMYD1 mRNA is ∼4.4 kb, close to the length of the sequence in GenBank database (ACCESSION NM_198274).
SMYD1 promoter has conserved binding sites for myogenic and cardiogenic regulators
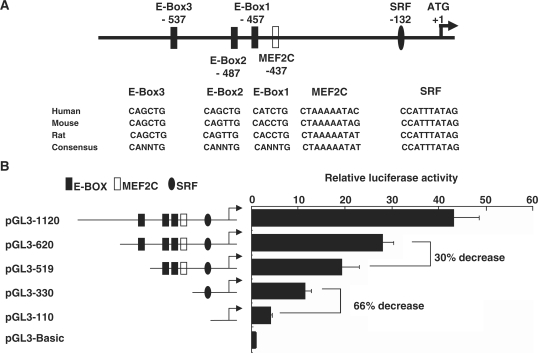
Since the expression of SMYD1 is restricted in muscle and cardiac tissues, it is interesting to investigate the molecular mechanisms through which the tissue-specific expression pattern of SMYD1 is regulated. The genomic sequences from rat and mouse were aligned with the identified corresponding proximal promoter region of human SMYD1, and multiple conserved cis-elements were predicted (Figure 2A). Among these transcription factor binding sites, the MEF2C binding site has been identified as an essential element which directs Smyd1 expression in mouse heart while the E-box1 and E-box2 are responsible for Smyd1 expression in the skeletal muscle lineage (40). In Mef2c null mutant mouse embryos, the expression level of Smyd1 is dramatically reduced but still detectable (40). Therefore, other important factors regulate Smyd1 expression in cardiac muscle. By comparing the promoter regions among human, mouse and rat, we found two important myogenic transcription factor binding sites, the SRF binding element located on −132 bp and the E-box3 located on −537 bp relative to the ATG start codon of SMYD1.
Figure 2.
Sequence analysis of the proximal region of SMYD1 gene promoter and mapping of promoter activity in C2C12 cells. (A) Conserved binding sites of myogenic regulators in SMYD1 gene, which include the basic helix–loop–helix proteins (E-box), MEF2, and serum response factor (SRF) are indicated with relative positions. The sequence of each binding site is listed along with homologous motifs from other species (human, mouse and rat). (B) Deletion analysis of SMYD1 gene promoter fragments with luciferase reporter assays. C2C12 cells were transiently transfected with various SMYD1 promoter deletion constructs and the pGL3-Basic plasmids were transfected as a negative control. Promoter-driven luciferase activity was determined as described in ‘Materials and Methods’ section.
In order to examine the transcriptional activation regulated by SRF and E-box binding sites on SMYD1 proximal promoter, a serial of promoter regions of SMYD1 were cloned into pGL3-basic luciferase reporter vectors. The chimera reporter constructs were transfected into C2C12 cells and luciferase reporter activity assays were performed (Figure 2B). The highest activity was observed in the cells transfected with full-length 1120 bp promoter reporter while the activity dropped with the decrease of the promoter length. The luciferase activity was decreased 66% compared to pGL3-330 construct when the fragment spanning −330 to −120 bp, which contains the SRF binding site, was deleted (Figure 2B). The most proximal 120 bp had basal promoter activity comparing to negative pGL3-Basic control. These data suggest that the proximal SMYD1 promoter contains the putative E-box protein and SRF binding sites and is regulated by certain transcription factors.
Overexpression of SRF and myogenin regulates Smyd1 expression in C2C12 cells
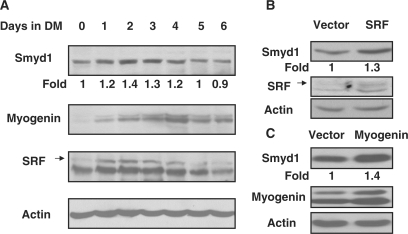
Since the expression of SMYD1 was only detected in heart and skeletal muscle, we investigated its expression during myogenic differentiation. C2C12 cell line is a well-studied multipotent mesenchymal progenitor cell model which is capable of differentiating into myotube when 10% FCS is replaced by 2% horse serum in culture medium (37). As evidenced by western blot with specific antibodies, the protein level of SMYD1 was significantly elevated upon switching the cells in differentiation conditions (Figure 3A). The highest expression level of Smyd1 was observed in Day 2 of differentiation, and the expression was decreased from Day 4. The endogenous expression pattern of myogenin and SRF in C2C12 cells was similar to that of Smyd1, but the induction was much more dramatic in the first 2 days (Figure 3A). This result is consistent with the report by other group (39). In order to investigate whether Smyd1 was the down stream target of SRF and myogenin, the mammalian expression constructs of these two genes were transfected into C2C12 cells independently. The exogenous SRF and myogenin could not significantly induce Smyd1 expression in growth medium (data not shown), but the protein level of SMYD1 was obviously increased in SRF and myogenin over-expressed cells after 2 days of differentiation (Figure 2B and C). These results indicate that Smyd1 expression is regulated by SRF and myogenin.
Figure 3.
Regulation of Smyd1 expression by SRF and myogenin. (A) Smyd1 expression during C2C12 cell differentiation. C2C12 myoblasts were cultured in proliferation medium to full confluence and induced to differentiate for indicated days by culturing in differentiation medium. Endogenous protein levels of Smyd1, myogenin and SRF were detected by western blot analysis (actin as a protein loading control). (B) C2C12 cells were transfected with pcDNA3.1 and pcDNA3.1-SRF, respectively. After 7 days of G418 selection, cells were pooled and cultured in differentiation medium for 2 days. Protein extracts were subjected to immunoblot analysis using anti-Smyd1 and anti-SRF antibodies, respectively. (C) C2C12 cells were transfected with pEMSV and pEMSV-myogenin respectively. After 7 days of G418 selection, cells were pooled and cultured in differentiation medium for 2 days. The protein levels of Smyd1 and myogenin were detected by western blotting with specific antibodies, respectively.
Decreased expression of Smyd1 in Srf cardiac knockout embryos
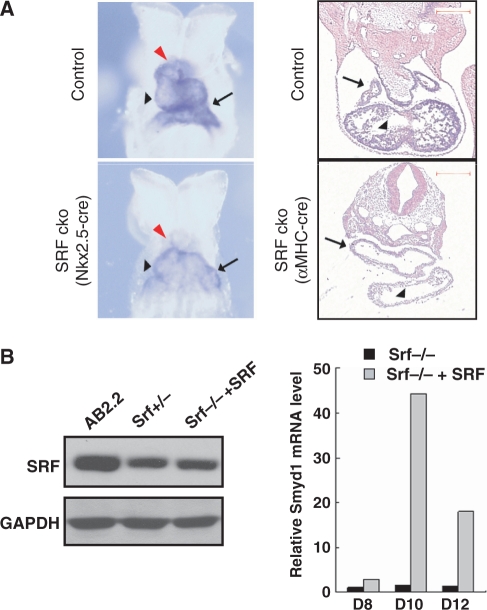
During embryonic development, expression of Smdy1 is detected as early as 8.0 dpc in the developing heart tube and outflow tract (35). To examine Smyd1 expression in Srf cardiac knockout mutants, we performed in situ hybridization to determine Smyd1 expression in two Srf cardiac specific knockout mutants (Figure 4A). In one mutant, Srf gene is disrupted at the cardiac progenitor stage by Nkx2.5-cre and the expression of Smyd1 is severely decreased in the developing heart tube including outflow tract. In the second mutant, Srf gene is ablated in myocyte of heart chambers by αMHC-cre and the expression of Smyd1 was significantly decreased in both atrial and ventricular myocyte. These findings strongly support that Smyd1 is specifically expressed in heart tissue and a downstream target of SRF in vivo.
Figure 4.
Expression of Smyd1 in SRF null ES cells and the SRF cardiac-conditional knockout embryo. (A) Reduced expression of Smyd1 was revealed by in situ hybridization in the SRF Nkx2.5-cre cko embryo (left panel, 8.5 dpc) and SRF αMHC-cre cko (right panel, 10.5 dpc) embryo compared to controls. The arrow: atrium, black arrow head: ventricle; red arrow head: outflow tract. Scale bar: 100 μm. (B) Rescue of Smyd1 expression in Srf-null ES cells with recombinant wild-type SRF. Immunoblot showing the expression of SRF protein in Srf-null ES cells infected with SRF-lentiviruses comparing with wild type and Srf+/− ES cells (left panel). Smyd1 expression was induced by SRF expression during Srf−/− ES cell differentiation measured by qRT-PCR. Significant induction of Smyd1 by SRF expression was observed by Days 8, 10, 12 of embryoid body differentiation (right panel).
Srf induces Smyd1 expression in ES cell myogenic differentiation
To further validate that SRF is responsible for Smyd1 induction in myogenic differentiation, we studied Smyd1 expression in Srf−/− ES cell. The Srf−/− ES cell failed to differentiate into beating myocyte in vitro (24). However, when wild-type SRF was introduced into the Srf−/− ES cell, dramatic induction of Smyd1 was observed along with the formation of beating myocytes. Highest level of Smyd1 induction by SRF was observed in Day 10 as measured with qRT–PCR assays (Figure 4B, right panel). Although the protein level of SRF was comparable in Srf +/− and SRF reconstituted ES cells (Figure 4B, left panel), the induction of Smyd1 mRNA was slightly higher (1.1-fold examined by realtime RT-PCR) in reconstituted cells at the same time points (data not shown).
SRF directly binds to the CArG site in Smyd1 promoter
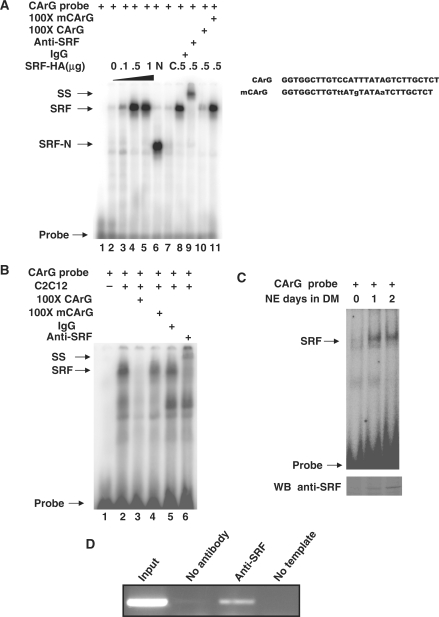
SRF activates Smyd1 expression when ectopically expressed in C2C12 cells and Smyd1 expression is dramatically down-regulated in SRF null mutant embryos. However, whether the regulation is directly through DNA-binding of SRF to the promoter region of Smyd1 remains unclear. To answer this question, electrophoretic mobility shift assays (EMSA) were performed using synthetic probes identical to CArG site found in SMYD1 promoter. Different concentrations of full-length or truncated forms of SRF constructs were transfected into 293T cells. Nuclear extracts from the transfected cells were harvested and EMSAs were performed. As shown in Figure 5A, the intensity of the binding complex band was increased with the increasing amount of SRF-HA plasmid transfected (lane 2–5). The binding complex band intensity was affected by wild type cold probes but not mutant cold probes. Moreover, the complex was shifted with addition of SRF specific antibody but not non-specific IgG (Figure 5A, lanes 8–11). A fast-migrating band was detected when the hot probes were incubated with the nuclear extracts from SRF-N terminus domain-transfected cells but no band was detected from SRF-C terminus domain-transfected cells (Figure 5A, lanes 6–7). Since the DNA binding domain of SRF was located in the N-terminus of SRF, our data demonstrated the direct binding affinity of exogenous SRF and CArG element in Smyd1 promoter.
Figure 5.
Identification of SRF binding to the CArG site of SMYD1 promoter in vitro and in vivo. (A) Nuclear extracts from 293T cells transfected with SRF-HA (lanes 2–5 and 8–11), SRF-N (lane 6) or SRF-C (lane 7) constructs were incubated with [32P] radiolabeled CArG oligonucleotides and competed with 100× unlabeled cold or mutant competitor (lane 10 and 11, respectively), non-specific IgG (lane 8), anti-SRF antibody (lane 9) as described in ‘Materials and Methods’ section. Oligo sequence used for EMSA is listed on the right side. (B) Nuclear extracts from C2C12 myotubes were incubated with [32P] radiolabeled CArG oligonucleotides (lane 2) and competed with 100-fold unlabeled cold wild type or mutant competitor (lanes 3 and 4, respectively), non-specific IgG (lane 5), anti-SRF antibody (lane 6) as described in ‘Materials and Methods’ section. (C) Nuclear extracts from C2C12 cells in different time point of differentiation were incubated with [32P] radiolabeled CArG oligonucleotides as described in ‘Materials and Methods’ section. The protein level of SRF was demonstrated with Western blots with SRF specific antibody. (D) ChIP analysis of SRF with Smyd1 promoter region. C2C12 cells were differentiated for 2 days, harvested, and processed. After IP of the cross-linked complexes, the chromatin was analyzed by PCR. PCR amplified a 341-bp region containing the SRF-binding site, as described in ‘Materials and Methods’ section. Single PCR product was obtained and the identity of the band was confirmed by sequencing (data not shown). Results in these assays are from triplicate experiments. NE, nuclear extract; SS, super-shift; DM, differentiation medium; WB, western blot.
To further confirm the ability of endogenous SRF binding to the CArG motif in Smyd1 promoter, we performed similar assays using C2C12 cells. As shown in Figure 5B, nuclear extracts from differentiated C2C12 cells bound to a [γ32-P]-labeled probes and the complex was further confirmed by both wild type, mutant competitor, and super-shift assays with SRF specific antibody (Figure 5B), suggesting SRF directly binds to the CArG motif of Smyd1 promoter. Since the expression of Smyd1 gene is elevated during myogenesis, we analyzed whether formation of the shifted complex was differentially regulated in C2C12 myoblasts and myotubes. Our results indicated that the association of SRF and CArG element was dramatically increased when the myoblasts differentiated into myotubes due to the increase of SRF protein level (Figure 5C). To investigate whether SRF binds to Smyd1 gene promoter in vivo, chromatin immunoprecipitation (ChIP) experiments were performed using differentiated C2C12 cells. The protein–DNA complexes were precipitated with antibody specific to SRF. The DNA fragments containing the CArG site in the Smyd1 promoter were then amplified. As shown in Figure 5D, PCR with primers flanking the proximal promoter region produced a band from DNA co-precipitated with SRF antibody but not with non-specific antibody control. Taken together, these results suggest that SRF directly binds to the CArG motif located in the proximal region of Smyd1 promoter both in vitro and in vivo.
Myogenin binds to the E-box element in Smyd1 promoter
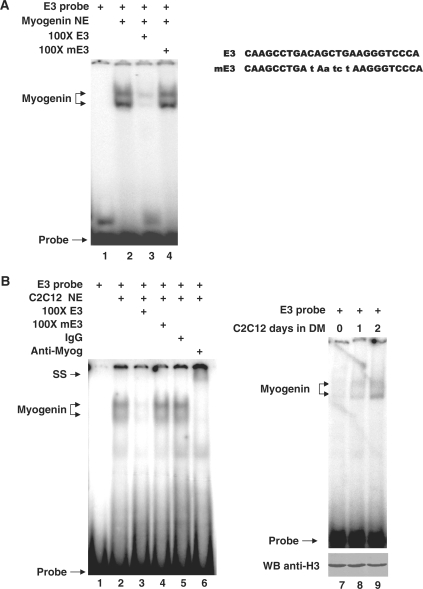
Members of bHLH myogenic regulators, which bind to the consensus E-box elements (CANNTG), play critical roles during skeletal myoblast specification, differentiation and maturation (5). As shown in Figure 2A, there are three E-box elements located in the proximal region of SMYD1 promoter. Among them, E-box1 and E-box2 sites (Figure 2A) are essential for skeletal muscle specific expression of SMYD1 (40). Since the E-box3 motif is more conserve than the other two, we hypothesized that this E-box element also plays an important role in controlling of SMYD1 expression and in myogenesis. To study the role of this E-box element (named E3) in SMYD1 transcriptional regulation, nuclear extracts from myogenin transfected 293T cells were performed gel shift assays with radioactive labeled E3 probes. E3 probes formed complex with nuclear extracts and the signal was decreased with cold E3 probes but not mutant cold probes (Figure 6A). In Figure 6B, nuclear extracts from differentiated C2C12 cells were subjected to gel mobility shift assays. The hot E3 probes formed slow migrating complexes in the gel (Figure 6B, lane 2). The complex band was dramatically decreased by 100-fold excessive cold probes (Figure 6B, lane 3), but not mutant cold probes (Figure 6B, lane 4). The DNA–protein complexes were super shifted in the presence of specific antibody against myogenin protein but not non-specific IgG (Figure 6, lanes 5 and 6). In order to examine the binding ability of myogenin to the E3 DNA motif during myogenesis, the nuclear extracts from myoblasts and myotubes were subjected to EMSA experiments (Figure 6B, lanes 7–9). The results demonstrated that the binding signal of myogenin and E3 probes was stronger in myotubes than in myoblasts, suggesting that myogenin binds to E-box3 element and the DNA–protein association is enriched during myogenic differentiation of C2C12 cells.
Figure 6.
Identification of myogenin binding to the E-box site of SMYD1 promoter with EMSA. (A) Nuclear extracts from Myogenin transfected 293T cells were incubated with [32P] radiolabeled E3 oligonucleotides and competed with 100-fold unlabeled cold wild type or mutant competitor (lanes 3 and 4, respectively) as described in ‘Materials and Methods’ section. H3 protein was presented as loading control of nuclear proteins. (B) Nuclear extracts from C2C12 myotubes were incubated with [32P] radiolabeled E3 oligonucleotides (lanes 2) and competed with 100-fold unlabeled cold wild type or mutant competitor (lane 3 and 4, respectively), non-specific IgG (lane 5), anti-Myogenin (F5D) antibody (lane 6) as described in ‘Materials and Methods’ section. Oligo sequences used for EMSA were listed on top. Similar results were observed in at least three replicate experiments. NE, nuclear extract; SS, super-shift; DM, differentiation medium; WB, western blot.
SRF and myogenin synergistically activate Smyd1 promoter activity
The results of EMSA and ChIP assays described above indicate that SRF and myogenin bind to the proximal Smyd1 promoter, but what is the function of the binding is still not determined. To examine the roles of these elements in the regulation of Smyd1 gene, 293T cells were transfected with various lengths of Smyd1 promoter reporter constructs, together with expression plasmids encoding myogenin, E12, and SRF, respectively (Figure 7). The luciferase reporter, pGL3-620 promoter construct of Smyd1 that contains three E-box elements, one MEF2-, and one SRF-binding site, was slightly activated by myogenin and dramatically stimulated by SRF but not truncated SRF fragments (Figure 7A). The promoter was not strongly activated by E12 alone but significant activation was observed when myogenin were co-transfected with E12 or SRF. The reporter activity of pGL3-330 construct, in which the E-box and MEF2 binding sites were deleted, was less than half of the activity of pGL3-620 (Figure 7B). The activity of pGL3-330 was activated ∼6-fold by SRF but not by SRF truncated fragments and E-box proteins (Figure 7B). Co-transfection of myogenin and E12 failed to significantly activate pGl3-330 promoter activity. However, myogenin and SRF synergistically activated the promoter activity about 10-fold (Figure 7B). To examine the role of the CArG motif in activation of SMYD1 by SRF, the CArG site was mutated to the sequence shown in Figure 5A (mCArG) in the Smyd1 promoter constructs. No SRF binding activity of the mutated sequence was detected by EMSA experiments (data not shown). SRF induced the activity of wild type promoter about 5.5-fold, but both the basal promoter activity and the SRF induction were dramatically decreased when the CArG motif was mutated (Figure 7C). These results suggest that myogenin and SRF synergistically stimulate Smyd1 promoter activity and the CArG site in Smyd1 promoter is required for SRF induction.
Figure 7.
Activation of SMYD1 promoter by SRF and myogenin. (A) Responsiveness of the 620 bp proximal region of SMYD1 promoter to SRF. 293T cells were transfected with a pGL3-Luc reporter containing the region from −620 to +1 bp of Smyd1 gene and the expression vector of myogenic factors (Myogenin, SRF, SRF-N, and SRF-C) as indicated. (B) Responsiveness of the 330 bp proximal region of Smyd1 promoter to SRF. 293T cells were transfected with a luciferase reporter containing the region from −330 to +1 bp of the Smyd1 gene and the expression vector of myogenic factors (myogenin, SRF, SRF-N, SRF-C) as indicated. (C) SRF binding site is essential for basal promoter activity and the responsiveness to SRF. 293T cells were cotransfected with or without SRF and serious of Smyd1 mutant promoter constructs. Luciferase activity was determined as described in ‘Materials and Methods’ section. Results in these assays are from triplicate experiments.
SMYD1 promotes C2C12 differentiation
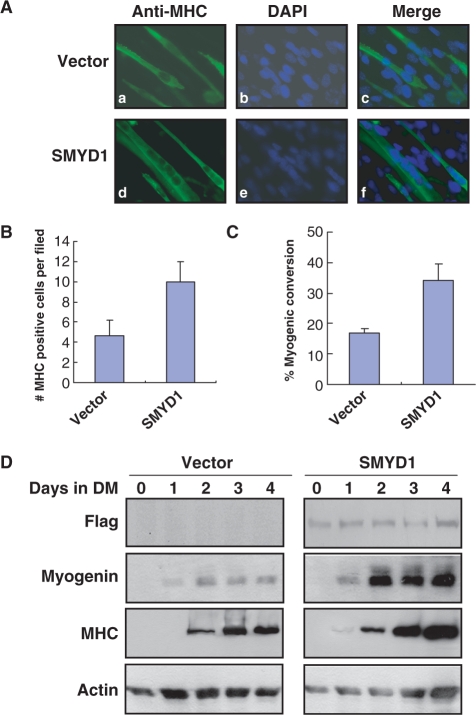
We have demonstrated that SMYD1 is the direct downstream target of myogenic regulators such as SRF and myogenin in this study. To understand the function of Smyd1 during myogenesis, we have examined how Smyd1 regulated myogenesis in C2C12 cells. C2C12 cells are capable of proliferative growth when subjected to serum-rich, subconfluent growth conditions (GM). However, upon 100% confluence and serum deprivation, these cells will withdraw from the cell cycle, polarize with respect to each other, and fuse to form multinucleated myotubes. The time course of Smyd1 expression during C2C12 myogenic differentiation strongly implicates its function in myogenic regulation. In order to test the overall effect of SMYD1 on myogenesis, we analyzed the SMYD1-mediated effects on the above-mentioned myogenesis processes. SMYD1-overexpressed C2C12 cells have been subjected to differentiation for 4 days and the cells were fixed for immunofluorescence staining to detect myogenesis using the MHC immunoreactive signals. The results showed that both the number of MHC positive myotubes and the number of nucleuses detected in single myotube were significantly higher than that of the vector-transfected cells (Figure 8A–C). Several molecular markers of myogenesis are commonly used to judge the degree of differentiation in C2C12 cells. Myogenin is considered as the marker of early differentiation and MHC is a marker of the later ones. C2C12 cells stably transfected with either vector control or Smyd1 were induced to differentiation, and cell lysates were harvested at different time points (Days 0–4). Subsequent western blot analyses were performed to determine the expression levels of different myogenic markers. Myogenin expression was first detected on Day 1 of differentiation, and the signal was much stronger in Smyd1-transfected cells than that of the control (Figure 8D). The weak expression of MHC was detected in Day 1 of differentiation in SMYD1-transfected cells but not in the control cells. Moreover, on matched time point, the protein level of MHC in Smyd1-transfected cells was significantly elevated when compared with the control ones. These results indicate that over-expression of human SMYD1 promotes myogenic differentiation of C2C12 cells, suggesting that SMYD1 is a positive regulator for myogenesis.
Figure 8.
Smyd1 promotes myogenesis in C2C12 cell. (A) C2C12 cells, stably transfected with either vector (a–c) or Smyd1 (d–f) gene, were subjected to differentiation conditions. Immunofluorescent staining with the MHC-specific MF-20 antibody on transfected C2C12 cells at Day 4 of differentiation. Photos were taken at Day 4 of differentiation, showing a significant increase of myotubes in Smyd1-expressing cells. Myosin heavy chain positive cells appear green. (B) Histogram illustrating the mean number of myotubes per vision field in vector- and Smyd1-transfected cells on Day 4 of differentiation. (C) Percentage of converted myotubes (both multinucleated converted and mononucleated converted myocytes) per vision field in vector- and Smyd1-transfected cells on Day 4 of differentiation as described in ‘Materials and Methods’ section. (D) SMYD1 enhances myogenesis in C2C12 cells. C2C12 cells transfected with either vector control or Flag-SMYD1 were subjected to differentiation. Cell lysates were collected at different time points (Days 0–4) of differentiation as indicated and subsequent western blot analysis was performed to determine the level of different myogenic markers. Results in these assays are from triplicate experiments.
DISCUSSION
SMYD1 is an evolutionary conserved histone methyltransferase which contains a SET and a MYND zinc finger domain. The expression of this protein is mainly restricted in muscle tissues in fish, frog, chicken and mouse (35,41,42), but human SMYD1 gene has not been reported yet. Using mouse Smyd1 cDNA sequence as a reference, we have cloned human orthologue of Smyd1 from human heart cDNA library and report here that it is expressed specifically in heart and skeletal muscle tissues. Smyd1 has been demonstrated to play an essential role in heart and skeletal muscle development (35,36), however, it is not fully understood how specific expression pattern of Smyd1 in the muscle lineage is regulated. Dillon Phan and colleagues have reported that mouse Smyd1 proximal promoter can drive LacZ reporter gene to express specifically throughout the developing cardiac tissue and skeletal muscle cells within the somite myotomes and the restricted expression is dependent on MEF2-response element and E-boxes in the promoter, respectively (40). However, lower expression of Smyd1 was still detected in the developing heart of Mef2c mutant embryos, suggesting the existence of other important transcriptional regulatory mechanisms. In this study, we demonstrated that overexpression of SRF increased endogenous Smyd1 expression during C2C12 cell and embryonic stem cell differentiation. Moreover, in SRF ablated mouse embryonic heart, Smyd1 expression was dramatically down-regulated. Different from MEF2 binding site, we demonstrated that SRF induced Smyd1 expression not only in the heart but also in skeletal muscle. The sequence of CArG site in Smyd1 promoter is identical in mouse, rat and human, but it [CC(A/T)6AG] diverges slightly at the second last nucleotide from known SRF consensus sequence [CC(A/T)6GG] which has been reported as a hybrid SRF and MEF2-binding element in MyoD distal promoter (29). The hybrid-binding element in MyoD promoter was recognized by both SRF and MEF2 in vitro. SRF binds to this element in myoblasts and MEF2 complexes can compete out binding of SRF at the onset of differentiation (29). Although the sequence of CArG site of SMYD1 is identical to that of MyoD, we failed to demonstrate the binding of MEF2 complexes to SMYD1 CArG probes in our experiments (data not shown). We hypothesized that this SRF binding site is only response to SRF activation before and after the onset of muscle differentiation. It is also possible that the sequence is not a hybrid binding element and the flanking sequence of the CArG site may affect the association of MEF2 to the target DNA.
The existence of SRF binding site in SMYD1 promoter suggests that SMYD1 is the direct target of SRF and acts in the same regulatory cascade involved in SRF. SRF, the master regulator of the contractile apparatus, plays a crucial role in cellular migration and normal actin cytoskeleton and contractile biology (43). Our recent findings revealed that cardiac specific deletion of SRF blocked the appearance of rhythmic beating myocytes (25). The non-beating phenotype was also observed in Smyd1 knockdown zebrafish, which could not swim and had no heartbeat due to defects of myofibril organization and muscle contraction (36). All the interesting findings suggest that SMYD1 is the direct target of SRF and functionally acts as a downstream regulator of SRF.
We identified an E-box element in SMYD1 promoter and demonstrated that the element was recognized by an early myogenic regulator myogenin. There are multiple E-box elements have been reported, seven in zebrafish and three in mouse (40,42). In this report, our data is the first direct evidence to show myogenin can bind to the SMYD1 promoter. Myogenin can activate SMYD1 promoter reporter in which all the E-boxes are deleted when SRF is co-transfected. It suggests that the SRF binding site not only directs SMYD1 expression in cardiac tissues but also can be recruited by bHLH myogenic regulators. Why so many E-boxes locate in SMYD1 promoter and what is the exact function of each E-box is still under investigation. Over-expression of human SMYD1 in C2C12 cells promotes mouse myogenic differentiation, suggesting that SMYD1 is a functionally conserved myogenic activator. Since histone methyltransferases usually act as co-factors for core transcriptional factors, what is (are) the core transcriptional factor(s) of SMYD1 is still not determined and is under active investigation.
FUNDING
Partially supported by the Research Platform of Cell Signaling Networks from the Science and Technology Commission of Shanghai Municipality (06DZ22923), the National Natural Science Foundation of China (no. 30800627 and 30871340) and a grant from the State Key Development Programs of China (2010CB945403). Funding for open access charge: 06DZ22923.
Conflict of interest statement. None declared.
REFERENCES
- 1.Buckingham M, Bajard L, Chang T, Daubas P, Hadchouel J, Meilhac S, Montarras D, Rocancourt D, Relaix F. The formation of skeletal muscle: from somite to limb. J. Anat. 2003;202:59–68. doi: 10.1046/j.1469-7580.2003.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McKinsey TA, Zhang CL, Olson EN. Signaling chromatin to make muscle. Curr. Opin. Cell Biol. 2002;14:763–772. doi: 10.1016/s0955-0674(02)00389-7. [DOI] [PubMed] [Google Scholar]
- 3.Christ B, Brand-Saberi B. Limb muscle development. Int. J. Dev. Biol. 2002;46:905–914. [PubMed] [Google Scholar]
- 4.Duprey P, Lesens C. Control of skeletal muscle-specific transcription: involvement of paired homeodomain and MADS domain transcription factors. Int. J. Dev. Biol. 1994;38:591–604. [PubMed] [Google Scholar]
- 5.Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Brand-Saberi B. Genetic and epigenetic control of skeletal muscle development. Ann. Anat. 2005;187:199–207. doi: 10.1016/j.aanat.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 7.Aurade F, Pinset C, Chafey P, Gros F, Montarras D. Myf5, MyoD, myogenin and MRF4 myogenic derivatives of the embryonic mesenchymal cell line C3H10T1/2 exhibit the same adult muscle phenotype. Differentiation. 1994;55:185–192. doi: 10.1046/j.1432-0436.1994.5530185.x. [DOI] [PubMed] [Google Scholar]
- 8.Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- 9.Rudnicki MA, Braun T, Hinuma S, Jaenisch R. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- 10.Rawls A, Morris JH, Rudnicki M, Braun T, Arnold HH, Klein WH, Olson EN. Myogenin's; functions do not overlap with those of MyoD or Myf-5 during mouse embryogenesis. Dev. Biol. 1995;172:37–50. doi: 10.1006/dbio.1995.0004. [DOI] [PubMed] [Google Scholar]
- 11.Kablar B, Asakura A, Krastel K, Ying C, May LL, Goldhamer DJ, Rudnicki MA. MyoD and Myf-5 define the specification of musculature of distinct embryonic origin. Biochem. Cell Biol. 1998;76:1079–1091. [PubMed] [Google Scholar]
- 12.Hasty P, Bradley A, Morris JH, Edmondson DG, Venuti JM, Olson EN, Klein WH. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- 13.Nabeshima Y, Hanaoka K, Hayasaka M, Esumi E, Li S, Nonaka I. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature. 1993;364:532–535. doi: 10.1038/364532a0. [DOI] [PubMed] [Google Scholar]
- 14.Molkentin JD, Olson EN. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc. Natl Acad. Sci. USA. 1996;93:9366–9373. doi: 10.1073/pnas.93.18.9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohun TJ, Chambers AE, Towers N, Taylor MV. Expression of genes encoding the transcription factor SRF during early development of Xenopus laevis: identification of a CArG box-binding activity as SRF. EMBO J. 1991;10:933–940. doi: 10.1002/j.1460-2075.1991.tb08027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papadopoulos N, Crow MT. Transcriptional control of the chicken cardiac myosin light-chain gene is mediated by two AT-rich cis-acting DNA elements and binding of serum response factor. Mol. Cell Biol. 1993;13:6907–6918. doi: 10.1128/mcb.13.11.6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mack CP, Thompson MM, Lawrenz-Smith S, Owens GK. Smooth muscle alpha-actin CArG elements coordinate formation of a smooth muscle cell-selective, serum response factor-containing activation complex. Circ. Res. 2000;86:221–232. doi: 10.1161/01.res.86.2.221. [DOI] [PubMed] [Google Scholar]
- 18.McDonald OG, Wamhoff BR, Hoofnagle MH, Owens GK. Control of SRF binding to CArG box chromatin regulates smooth muscle gene expression in vivo. J. Clin. Invest. 2006;116:36–48. doi: 10.1172/JCI26505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hauschka SD. Myocardin. a novel potentiator of SRF-mediated transcription in cardiac muscle. Mol. Cell. 2001;8:1–2. doi: 10.1016/s1097-2765(01)00297-0. [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Kitchen CM, Streb JW, Miano JM. Myocardin: a component of a molecular switch for smooth muscle differentiation. J. Mol. Cell Cardiol. 2002;34:1345–1356. doi: 10.1006/jmcc.2002.2086. [DOI] [PubMed] [Google Scholar]
- 21.Selvaraj A, Prywes R. Megakaryoblastic leukemia-1/2, a transcriptional co-activator of serum response factor, is required for skeletal myogenic differentiation. J. Biol. Chem. 2003;278:41977–41987. doi: 10.1074/jbc.M305679200. [DOI] [PubMed] [Google Scholar]
- 22.Cen B, Selvaraj A, Prywes R. Myocardin/MKL family of SRF coactivators: key regulators of immediate early and muscle specific gene expression. J. Cell Biochem. 2004;93:74–82. doi: 10.1002/jcb.20199. [DOI] [PubMed] [Google Scholar]
- 23.Li S, Czubryt MP, McAnally J, Bassel-Duby R, Richardson JA, Wiebel FF, Nordheim A, Olson EN. Requirement for serum response factor for skeletal muscle growth and maturation revealed by tissue-specific gene deletion in mice. Proc. Natl Acad. Sci. USA. 2005;102:1082–1087. doi: 10.1073/pnas.0409103102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niu Z, Yu W, Zhang SX, Barron M, Belaguli NS, Schneider MD, Parmacek M, Nordheim A, Schwartz RJ. Conditional mutagenesis of the murine serum response factor gene blocks cardiogenesis and the transcription of downstream gene targets. J. Biol. Chem. 2005;280:32531–32538. doi: 10.1074/jbc.M501372200. [DOI] [PubMed] [Google Scholar]
- 25.Niu Z, Iyer D, Conway SJ, Martin JF, Ivey K, Srivastava D, Nordheim A, Schwartz RJ. Serum response factor orchestrates nascent sarcomerogenesis and silences the biomineralization gene program in the heart. Proc. Natl Acad. Sci. USA. 2008;105:17824–17829. doi: 10.1073/pnas.0805491105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miano JM, Ramanan N, Georger MA, de Mesy Bentley KL, Emerson RL, Balza RO, Jr, Xiao Q, Weiler H, Ginty DD, Misra RP. Restricted inactivation of serum response factor to the cardiovascular system. Proc. Natl Acad. Sci. USA. 2004;101:17132–17137. doi: 10.1073/pnas.0406041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.L'H;onore A, Lamb NJ, Vandromme M, Turowski P, Carnac G, Fernandez A. MyoD distal regulatory region contains an SRF binding CArG element required for MyoD expression in skeletal myoblasts and during muscle regeneration. Mol. Biol. Cell. 2003;14:2151–2162. doi: 10.1091/mbc.E02-07-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Groisman R, Masutani H, Leibovitch MP, Robin P, Soudant I, Trouche D, Harel-Bellan A. Physical interaction between the mitogen-responsive serum response factor and myogenic basic-helix-loop-helix proteins. J. Biol. Chem. 1996;271:5258–5264. doi: 10.1074/jbc.271.9.5258. [DOI] [PubMed] [Google Scholar]
- 29.L'H;onore A, Rana V, Arsic N, Franckhauser C, Lamb NJ, Fernandez A. Identification of a new hybrid serum response factor and myocyte enhancer factor 2-binding element in MyoD enhancer required for MyoD expression during myogenesis. Mol. Biol. Cell. 2007;18:1992–2001. doi: 10.1091/mbc.E06-09-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gauthier-Rouviere C, Vandromme M, Tuil D, Lautredou N, Morris M, Soulez M, Kahn A, Fernandez A, Lamb N. Expression and activity of serum response factor is required for expression of the muscle-determining factor MyoD in both dividing and differentiating mouse C2C12 myoblasts. Mol. Biol. Cell. 1996;7:719–729. doi: 10.1091/mbc.7.5.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biesiada E, Hamamori Y, Kedes L, Sartorelli V. Myogenic basic helix-loop-helix proteins and Sp1 interact as components of a multiprotein transcriptional complex required for activity of the human cardiac alpha-actin promoter. Mol. Cell Biol. 1999;19:2577–2584. doi: 10.1128/mcb.19.4.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sartorelli V, Webster KA, Kedes L. Muscle-specific expression of the cardiac alpha-actin gene requires MyoD1, CArG-box binding factor, and Sp1. Genes Dev. 1990;4:1811–1822. doi: 10.1101/gad.4.10.1811. [DOI] [PubMed] [Google Scholar]
- 33.Catala F, Wanner R, Barton P, Cohen A, Wright W, Buckingham M. A skeletal muscle-specific enhancer regulated by factors binding to E and CArG boxes is present in the promoter of the mouse myosin light-chain 1A gene. Mol. Cell Biol. 1995;15:4585–4596. doi: 10.1128/mcb.15.8.4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng G, Hagen TP, Dawson ML, Barnes KV, Menick DR. The role of GATA, CArG, E-box, and a novel element in the regulation of cardiac expression of the Na+-Ca2+ exchanger gene. J. Biol. Chem. 1999;274:12819–12826. doi: 10.1074/jbc.274.18.12819. [DOI] [PubMed] [Google Scholar]
- 35.Gottlieb PD, Pierce SA, Sims RJ, Yamagishi H, Weihe EK, Harriss JV, Maika SD, Kuziel WA, King HL, Olson EN, et al. Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat. Genet. 2002;31:25–32. doi: 10.1038/ng866. [DOI] [PubMed] [Google Scholar]
- 36.Tan X, Rotllant J, Li H, De Deyne P, Du SJ. SmyD1, a histone methyltransferase, is required for myofibril organization and muscle contraction in zebrafish embryos. Proc. Natl Acad. Sci. USA. 2006;103:2713–2718. doi: 10.1073/pnas.0509503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bryan BA, Mitchell DC, Zhao L, Ma W, Stafford LJ, Teng BB, Liu M. Modulation of muscle regeneration, myogenesis, and adipogenesis by the Rho family guanine nucleotide exchange factor GEFT. Mol. Cell Biol. 2005;25:11089–11101. doi: 10.1128/MCB.25.24.11089-11101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li D, Mitchell D, Luo J, Yi Z, Cho SG, Guo J, Li X, Ning G, Wu X, Liu M. Estrogen regulates KiSS1 gene expression through estrogen receptor alpha and SP protein complexes. Endocrinology. 2007;148:4821–4828. doi: 10.1210/en.2007-0154. [DOI] [PubMed] [Google Scholar]
- 39.Sims RJ, 3rd, Weihe EK, Zhu L, O'M;alley S, Harriss JV, Gottlieb PD. m-Bop, a repressor protein essential for cardiogenesis, interacts with skNAC, a heart- and muscle-specific transcription factor. J. Biol. Chem. 2002;277:26524–26529. doi: 10.1074/jbc.M204121200. [DOI] [PubMed] [Google Scholar]
- 40.Phan D, Rasmussen TL, Nakagawa O, McAnally J, Gottlieb PD, Tucker PW, Richardson JA, Bassel-Duby R, Olson EN. BOP, a regulator of right ventricular heart development, is a direct transcriptional target of MEF2C in the developing heart. Development. 2005;132:2669–2678. doi: 10.1242/dev.01849. [DOI] [PubMed] [Google Scholar]
- 41.Kawamura S, Yoshigai E, Kuhara S, Tashiro K. smyd1 and smyd2 are expressed in muscle tissue in Xenopus laevis. Cytotechnology. 2008;57:161–168. doi: 10.1007/s10616-008-9128-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Du SJ, Rotllant J, Tan X. Muscle-specific expression of the smyd1 gene is controlled by its 5.3-kb promoter and 5′-flanking sequence in zebrafish embryos. Dev. Dyn. 2006;235:3306–3315. doi: 10.1002/dvdy.20984. [DOI] [PubMed] [Google Scholar]
- 43.Miano JM, Long X, Fujiwara K. Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am. J. Physiol. Cell Physiol. 2007;292:C70–C81. doi: 10.1152/ajpcell.00386.2006. [DOI] [PubMed] [Google Scholar]