Abstract
Cyclin dependent kinases (cdks) regulate cell cycle progression and transcription. We report here that the transcriptional co-activator PCAF directly interacts with cdk2. This interaction is mainly produced during S and G2/M phases of the cell cycle. As a consequence of this association, PCAF inhibits the activity of cyclin/cdk2 complexes. This effect is specific for cdk2 because PCAF does not inhibit either cyclin D3/cdk6 or cyclin B/cdk1 activities. The inhibition is neither competitive with ATP, nor with the substrate histone H1 suggesting that somehow PCAF disturbs cyclin/cdk2 complexes. We also demonstrate that overexpression of PCAF in the cells inhibits cdk2 activity and arrests cell cycle progression at S and G2/M. This blockade is dependent on cdk2 because it is rescued by the simultaneous overexpression of this kinase. Moreover, we also observed that PCAF acetylates cdk2 at lysine 33. As this lysine is essential for the interaction with ATP, acetylation of this residue inhibits cdk2 activity. Thus, we report here that PCAF inhibits cyclin/cdk2 activity by two different mechanisms: (i) by somehow affecting cyclin/cdk2 interaction and (ii) by acetylating K33 at the catalytic pocket of cdk2. These findings identify a previously unknown mechanism that regulates cdk2 activity.
INTRODUCTION
Cyclin dependent kinases (cdks) are key enzymes for the regulation of cell cycle progression and transcription (1). Their activities are firstly regulated by their binding to regulatory subunits called cyclins (2). A specific subset of cyclin/cdk complexes participates in the control of cell cycle progression by being activated at different stages of the cell cycle, thus driving the cells through its different phases. It is now clear that cdk1 bound to cyclins A and B governs G2/M transition (3). G1 progression is primarily under the control of cyclin D/cdk4/6 (4). Finally, cyclins E and A paired to cdk2 are required for G1/S transition and progression through S phase (1,5).
Cyclin/cdk complexes are additionally regulated by a number of mechanisms including phosphorylation and binding to inhibitory proteins. Thus, in addition to cyclin binding most cdks require phosphorylation at a conserved residue (Thr 160 in human cdk2) to achieve full kinase activity. The enzyme responsible for this phosphorylation is CAK, that consists in the cdk7/cyclin H/Mat 1 trimer (6). Major cdks can also be inhibited by phosphorylation at a conserved tyrosine (Tyr 15) and at its adjacent threonine (Thr 14). These phosphorylations are carried out by Wee1 and Myt1 in vertebrate cells and can be removed by the phosphatase cdc25 (7). Finally, cdk activity is also regulated by binding to members of two families of inhibitors (CKIs): the Ink4 family (p16ink4a, p15ink4b, p18ink4c and p19ink4d) and the Cip/Kip family (p21Cip1, p27Kip1 and p57Kip2) (8). The members of the Ink4 family only interact with cdk4 and cdk6 inhibiting their activities. In contrast, the Cip/Kip members bind to all known cyclin/cdk complexes. These proteins are potent inhibitors of cyclin/cdk2, but they also inhibit the other cyclin/cdk complexes, although in a less extension.
Apart from participating in cell cycle regulation cyclinA/cdk2 also plays a role in the control of the transcriptional activity of steroid receptors (9). For instance, both the estrogen receptor (ER) and the progesterone receptor (PR) are activated by cyclin A/cdk2. In the first case, this complex directly phosphorylates ER, thus potentiating its transcriptional activity (10). In the second case, cyclin A/cdk2 phosphorylates the co-activator SRC-1, fact that enhances its affinity for PR and thus increases gene expression (11). Thus, in the promoters regulated by these receptors cyclin A/cdk2 participates in multi-protein complexes that also contain transcription factors, co-repressors and co-activators including acetyltransferases.
During the last decade a growing number of evidences indicate that acetylation, a post-translational modification occurring at the Nε-amino-group of lysines, might regulate protein functions in many different ways as, for instance, protein-protein interaction, protein association to DNA and protein stability (12). Recently, it has been shown that cdk9, a member of the cdk family involved in transcriptional regulation, is acetylated by Gcn5 and PCAF at lysines 44 and 48 that are located at the catalytic pocket of the enzyme (13). In particular, K48 is essentially involved in orienting the ATP phosphate residues within the catalytic pocket and thus, acetylation of this lysine residue inactivates the enzyme (13,14). Therefore, acetylation of cdk9 at these specific lysines is a new mechanism involved in transcriptional regulation. Lysine K48 is conserved in all the members of the cdk family and this fact suggests that other cdks may be susceptible to be acetylated at this site. For this reason, we aimed to explore whether acetylases might participate in the regulation of cdk2 activity. Recently, we observed that the acetyltransferase PCAF can acetylate cyclin A at specific lysines, leading to its degradation (15). PCAF is homologous to GCN5 and in vertebrate cells both proteins are subunits of the SAGA-type multiprotein complexes. These complexes are co-activators that stimulate transcription in part via acetylation and modification of nucleosomes, in cooperation with nucleosome remodeling enzymes and by physically recruiting the mediator complex (16,17).
We report here that PCAF directly binds to cdk2, acetylates K33 and as a consequence inhibits its kinase activity. Moreover, our results also revealed that merely the interaction of PCAF with cyclin/cdk2 complexes, in the absence of acetylation, inhibits cdk2 activity. This effect is specific because PCAF does not inhibit the activities of either cyclin B/cdk1 or cyclin D3/cdk6 complexes. Therefore, PCAF can regulate cdk2 activity by two different mechanisms: acetylation of K33 of cdk2 and disturbing cyclin/cdk2 complexes independently of acetylation.
MATERIALS AND METHODS
Plasmids
cDNA of wild-type cdk2 was cloned into pGEX2T and pECFP-C1 vectors. pUHDP1-Flag-cdk2 WT and K33R and pGEX2T-cdk2 K33R were a generous gift from R. Poon (Hong Kong). pECFP-C1-cdk2 K33R, pUHDP1-Flag-cdk2 K33Q and pGEX2T-cdk2 K33Q were generated by site-directed mutagenesis. All the vectors harboring different acetylases or their fragments used in this work were provided by MA. Martínez-Balbás and M. Giacca.
Antibodies and reagents
Antibodies against cyclin A (H-432), PCAF (E-8), cdk2 (M-2) and cdk2 (D-12) were purchased from Santa Cruz Biotechnology. Anti-phospho-histone H3 (Ser 28) (#9713) and anti-acetylated lysines (#9441) were from Cell Signaling. Anti-acetylated cdk9 was from M. Giacca’s laboratory. Antibodies against SPT3 were a kind gift from E. Martinez (California). Antibodies against FLAG (F7425) and PCAF (P7493) were obtained from Sigma. For immunoprecipitation, we used monoclonal anti-FLAG M2 affinity gel from Sigma. For pull-down experiments, we conjugated purified proteins to CNBr-sepharose beads (Pharmacia). Thymidine and Nocodazole used in cell synchronization were from Sigma. [32P]ATP used in kinase assays was purchased from Amersham and [14C]acetylCoA used in the in vitro acetylation experiments was from Perkin Elmer.
Cell culture, transfection and synchronization
Cells were grown in Dulbecco's; modified Eagle's; medium supplemented with 10% fetal calf serum. Transfection experiments were performed using Lipofectamine 2000 from Invitrogen. Transfected synchronized cells were obtained as described (18).
Immunocytochemistry and fluorescence microscopy
For intracellular localization analysis of PCAF and cdk2, cells were grown on coverslips and fixed in 4% paraformaldehyde-phosphate-buffered saline (PBS) for 15 min at room temperature. Coverslips were then washed three times (5 min each) in PBS, permeabilized and blocked with 1% bovine serum albumin (BSA) + 0.1% Triton X-100 in PBS for 15 min at room temperature, and then incubated for 1 h at 37°C in a humidified atmosphere with a monoclonal anti-PCAF antibody (E-8, Santa Cruz Biotechnology) at a 1:100 dilution and a polyclonal cdk2 antibody (M2, Santa Cruz Biotechnology), at a 1:200 dilution. Coverslips were then washed three times (5 min each) in PBS and incubated for 45 min at 37°C with an Alexa 594 anti-mouse antibody and an Alexa 488-conjugated anti-rabbit antibody (dilution 1:500 in both cases; Jackson). Coverslips were washed, mounted on glass slides with Mowiol (Calbiochem) and analyzed by fluorescence microscopy. For intracellular localization analysis of CFP and YFP fusion proteins, transfected cells were grown on coverslips and fixed as described above, then washed 3 times in PBS and mounted.
Protein purification, pull-down and immunoprecipitation
Protein expression and purification was performed as described (19). For pull-down experiments, cells were lysed in RIPA buffer (50 mM Tris−HCl pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% Sodium deoxycholate, 0.1% SDS, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, 0.1 mM Na3VO4, 0.5 μg/μl aprotinin, 10 μg/μl leupeptin) for 30 min on ice. Lysates (0.2–2 mg of protein) were incubated with CNBr-sepharose beads conjugated with purified GST, GST-HAT(PCAF) or GST-PCAF (full length) in the case of pull-down, or with anti-FLAG M2 affinity gel in the case of immunoprecipitation, for 2 h at 4°C. After three washes with RIPA buffer, Laemmli buffer was added to the samples and they were subsequently electrophoresed.
Surface plasmon resonance experiments
The Surface plasmon resonance (SPR) analysis is a method that permits to analyze the direct interaction between two proteins. SPR detects binding interactions by monitoring the reflection of a beam of light off the interface between an aqueous solution of potential binding molecules and a biosensor surface carrying an immobilized bait protein. The analysis of the interaction between PCAF and cdk2 was performed at room temperature using a Biacore T100 (Biacore International AB). Purified recombinant proteins GST-PCAF and GST-cdk2 were cleaved with Thrombin protease (Sigma) to remove GST. Then, PCAF was immobilized on a carboxymethylated dextran sensor chip (CM5) using the amine coupling method as described by the manufacturer. A blank immobilization was performed using the same method and was used as the reference surface. Purified full-length cdk2 was diluted in HBS-EP buffer (Biacore International AB) and was injected over the flow cells at a flow rate of 30 µl/min for 60 s. Following a dissociation time of 120 s, final regeneration of the surface was performed with a short pulse of 0.05% (w/v) SDS. The interaction between PCAF and cdk2 was detected and presented as a sensorgram by plotting resonance units against time.
Kinase assays
Purified active cyclin A/cdk2, cyclin E/cdk2, cyclin B/cdk1 and cyclin D3/cdk6 were purchased from Upstate Biotechnology. Purified recombinant proteins were resuspended in a final volume of 30 μl of kinase buffer (50 mM Hepes pH 7.4, 2.5 mM EGTA, 10 mM MgCl2) containing 12.5 μM ATP, 1 μCi of [32P]ATP, 2 mM dithiothreitol and 2 μg of histone H1. Then, they were incubated for 30 min at 30°C. Samples (25 μl) were spotted on the center of a 2 cm × 2 cm P81 paper square. After three washes with 1% phosphoric acid, papers were left to dry, then transferred to vials with 10 ml scintillation cocktail and activity was measured in a liquid scintillation counter (Wallac 1409). In some experiments, reactions were stopped by adding Laemmly buffer and samples were electrophoresed in 12% SDS-polyacrylamide gels and then stained with Coomassie Blue and dried. The radioactivity associated to the gels was detected with a PhosphorImager. Experiments involving immunoprecipitation followed by cdk activity assays were performed as described in (19).
Cell proliferation assays
Cells were transfected with Flag-PCAF or empty vector then counted and seeded in 6 well-plates. Measurements of the number of cells were performed at different times after transfection and represented in a graph. Cell proliferation was also measured using inducible cell lines expressing C-terminus-PCAFΔHAT (CtermΔHAT, 352–832, Δ527–547). This cell line was provided by M. Ventura (Barcelona)
Flow cytometry analysis
Cells were fixed with 70% cold ethanol for 2 h at 4°C, washed with PBS, and finally incubated with 2 μM TOPRO-3 (Invitrogen) and 200 μg/ml RNase for 30 min at room temperature. Analysis of DNA content was carried out in a BD Biosciences FACS Canto II. Data was analysed with WinMDI 2.9 software.
In vitro acetylation
Acetylase assays were performed as described (20). For cdk2 acetylation assays, 1–10 μl of the different acetylases (5000–10 000 c.p.m. activity on histones) were incubated with 6 μM of purified GST or GST-cdk2 and 0.02 μCi [14C]acetylCoA for 30 min at 30°C. Reactions were stopped by addition of Laemmli buffer. Then, samples were electrophoresed and transferred onto a nitrocellulose membrane. After that, the membrane was subjected to autoradiography. For the spot-mapping experiment, the membrane containing the spotted peptides was incubated in 3 ml of HAT buffer (50 mM Tris−HCl pH 8, 500 mM NaCl, 0.1 mM EDTA, 5% glycerol, 0.1% NP-40) in the presence of GST-HAT(PCAF) and [14C]acetylCoA, for 30 min at 30°C. Then the membrane was washed, dried and subjected to autoradiography.
RESULTS
Cdk2 interacts with the acetyltransferase PCAF
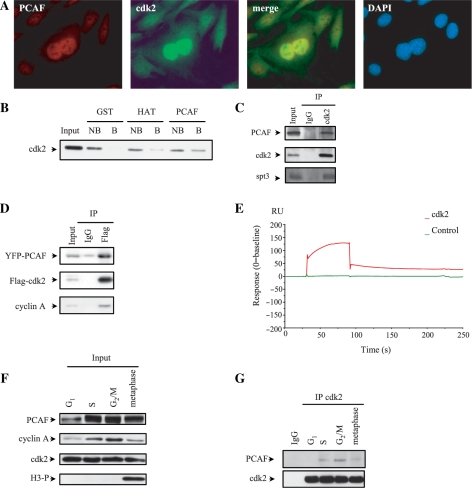
With the aim to analyze the putative interaction between cdk2 and the acetylase PCAF, we first studied the intracellular distribution of both proteins in C2C12 cells by immunofluorescence using specific antibodies against PCAF and cdk2. Figure 1A shows that both proteins co-localized in the nucleus. Supplementary Figure S1A and B show single staining for PCAF or cdk2. Nuclear co-localization of both proteins was also observed by fluorescence confocal microcopy of cells transfected with YFP-PCAF and CFP-cdk2 (Supplementary Figure S1C). Pull-down experiments using the full length GST-PCAF or the catalytic domain of PCAF (HAT domain) revealed that cdk2 binds to both of them (Figure 1B). In contrast, no binding of cdk2 to the control GST-beads was observed.
Figure 1.
Cdk2 interacts with the acetyltransferase PCAF. (A) C2C12 cells were fixed and stained with antibodies against PCAF and cdk2 and colocalization of both proteins was studied by fluorescence microscopy. (B) CNBr-sepharose beads coupled to GST, GST-HAT (PCAF) or GST-PCAF (full length) were incubated with HCT-116 cell extracts and pull-down experiments were performed. The presence of cdk2 in the precipitates was analysed by WB. NB, not bound; B, bound. (C) C2C12 cell extracts were subjected to IP with IgG as a control and anti-cdk2 in order to immunoprecipitate the endogenous protein. Then, WB was performed to detect endogenous cdk2, PCAF and SPT-3. A sample of cell lysate (input) is shown in the first lane. (D) HeLa cells were transfected with YFP-PCAF and Flag-cdk2. Cell extracts were subjected to IP using anti-Flag or IgG as a control followed by WB with antibodies against Flag, PCAF or cyclin A. A sample of cell lysate (input) was used as a control. (E) The putative direct interaction between PCAF and cdk2 was studied by Surface Plasmon Resonance as described in ‘Materials and Methods’ section. PCAF was fixed on the matrix and cdk2 was left to circulate on the chip. The interaction was represented in the sensorgram. (F) C2C12 cells were synchronized by a double-thymidine block or nocodazole as described in ‘Materials and Methods’ section. Then, the levels of endogenous PCAF, cyclin A and cdk2 were determined by WB. To confirm the time of mitosis a WB with antibodies against phosphorylated histone H3 was performed. (G) Cell extracts from synchronized cells described in (F) were subjected to IP with anti-cdk2 or IgG as a control and the amount of PCAF and cdk2 was analyzed by WB.
To analyze the in vivo interaction between endogenous cdk2 and PCAF, cells were subjected to immunoprecipitation (IP) with anti-cdk2 and the immunoprecipitates were analyzed by western blot (WB). Results showed that PCAF associates with cdk2 in the cells (Figure 1C). Interestingly, these complexes contain SPT3, a protein subunit of the GCN5/PCAF multiprotein complexes (21). A similar interaction was observed with ectopic YFP-PCAF and Flag-cdk2 proteins (Figure 1D). These complexes also contain cyclin A (Figure 1D). To further investigate the putative direct interaction between cdk2 and PCAF, Surface Plasmon Resonance analyses were performed. As observed in Figure 1E results indicate that cdk2 directly interacts with the acetylase.
The interaction between cdk2 and PCAF during cell cycle was subsequently determined. Thus, cells were synchronized at different phases of the cell cycle by a double thymidine block or by a nocodazole treatment as described in the methods section. To check whether synchronization was correct we analyzed the levels of cyclin A, and phosphorylated histone H3 in the cell extracts. It is known that cyclin A levels are high at S and G2/M, whereas they remain low at G1 and mitosis and that phosphorylated histone H3 is considered to be a mitotic marker. As shown in Figure 1F the levels of cyclin A and those of phosphorylated H3 behaved as expected. We also observed that cdk2 remained constant along cell cycle, whereas the levels of PCAF were high at S and G2/M, slightly lower at metaphase and much lower at G1 (Figure 1F). The interaction between cdk2 and PCAF at these different stages of the cell cycle was determined by IP with anti-cdk2 followed by WB with anti-PCAF. Results indicate that cdk2 was preferentially associated with PCAF during G2/M, whereas the association decreased at S phase and metaphase and was almost undetectable at G1 (Figure 1G). Similar results were observed when the interaction between overexpressed proteins was analyzed. However, in this case the association of PCAF with cdk2 at S phase is much higher than that observed in the case of the endogenous proteins (Supplementary Figure S1D).
PCAF inhibits cyclin A/cdk2 activity
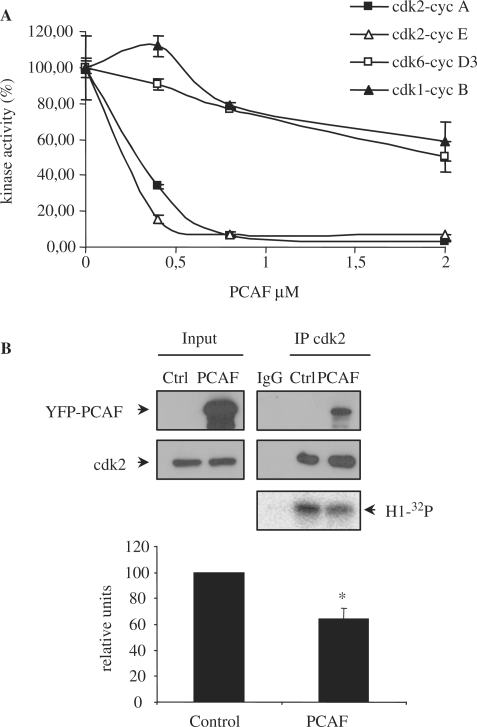
Because of the direct interaction between PCAF and cdk2, we aimed to study whether PCAF might affect cyclin/cdk2 activity. Thus, in vitro cyclin A/cdk2 and cyclin E/cdk2 kinase assays were performed in the presence of different PCAF concentrations. Results showed that PCAF inhibits the activity of both complexes with an IC50 of around 300 nM (Figure 2A). This effect is specific, because PCAF only slightly affected the activities of other cyclin/cdk complexes as cyclin D3/cdk6 or cyclin B/cdk1 determined in similar in vitro experiments (Figure 2A). As a control, we determined the effect of GST on the activity of these different cyclin/cdk complexes and as shown in supplementary Figure S2, GST did not modify the activity of any of them. Therefore, these results indicate that PCAF specifically inhibits with high affinity the in vitro activity of cyclin/cdk2 complexes. We also observed that PCAF is able to inhibit cyclin/cdk2 complexes in vivo. As shown in Figure 2B cdk2-associated kinase activity was significantly decreased in cells transfected with YFP-PCAF.
Figure 2.
PCAF inhibits cyclin/cdk2 complexes in vivo and in vitro. (A) Purified active cyclin A/cdk2, cyclin E/cdk2, cyclin B/cdk1 and cyclin D3/cdk6 complexes were incubated with increasing concentrations of purified recombinant PCAF and kinase assays were performed. (B) HeLa cells were transfected with YFP-PCAF or empty vector as a control. Cell extracts were subjected to IP with anti-cdk2. Kinase assays of the immunoprecipitated endogenous cdk2 were performed and phosphorylation of histone H1 was detected by PhosphorImager. Kinase activity was normalized to the amount of immunoprecipitated cdk2 and represented in the graph. Results shown are the mean ± SE of 3 independent experiments. *P-value <0.05.
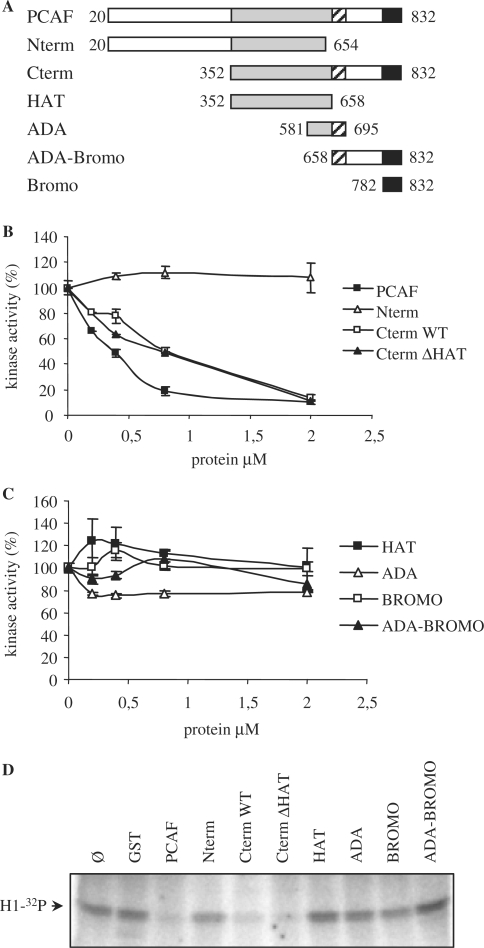
To further identify the domain of PCAF involved in the inhibition of cyclin/cdk2 activity we first generated two different PCAF fragments: the N-terminus (aa 20–654) and the C-terminus (aa 352–832) (Figure 3A). Then, their effect on cyclin A/cdk2 activity was determined. Results indicate that the C-terminus fragment but not the N-terminus was able to significantly inhibit the kinase activity (Figure 3B). An inactive C-terminus fragment of PCAF that lacks a 20 amino acids region (aa 527–547) inside of its catalytic domain (CtΔHAT) (22) also inhibited cyclin A/cdk2 activity as efficiently as Ctwt PCAF did (Figure 3B). These results indicate that the inhibitory effect of PCAF on cyclin A/cdk2 is independent of its acetylase activity.
Figure 3.
The C terminus of PCAF is responsible for the inhibition of cyclin A/cdk2 activity in vitro. (A) Schematic representation of the domains of PCAF used in the experiments. (B) Purified recombinant cyclin A/cdk2 complexes (400 nM) were incubated with increasing concentrations of Nterm, Cterm, Cterm ΔHAT and full-length PCAF and kinase assays were performed. (C) The same as in (B), but using different recombinant fragments of PCAF: HAT, ADA, Bromo and ADA-Bromo. (D) A typical kinase assay using purified recombinant cyclin A/cdk2 complex (400 nM) as a kinase and histone H1 (2 µg) as a substrate. Different reactions were performed by adding GST or different purified fragments of PCAF at a concentration of 800 nM. After incubation, reactions were stopped and samples were electrophoresed. Phosphorylation of histone H1 was detected by autoradiography using a PhosphorImager.
We further analyzed four different domains belonging to the PCAF C-terminus fragment for their ability to inhibit cyclin A/cdk2 kinase activity. These domains were: HAT (aa 352–658), ADA fragment (aa 581–695, involved in the interaction with ADA2 cofactor), bromodomain (aa 782–832, involved in the interaction with acetylated lysines), and ADA-bromodomain (aa 658–832) (Figure 3A). As it can be observed in Figure 3C none of these fragments significantly inhibited the kinase activity of cyclin A/cdk2. Figure 3D shows a typical experiment of the effect of the different PCAF constructs on cyclin A/cdk2 activity. Altogether, these results indicate that an intact C-terminus region is needed to efficiently inhibit cyclin A/cdk2 activity and that the acetylase activity of PCAF is not required for the inhibition.
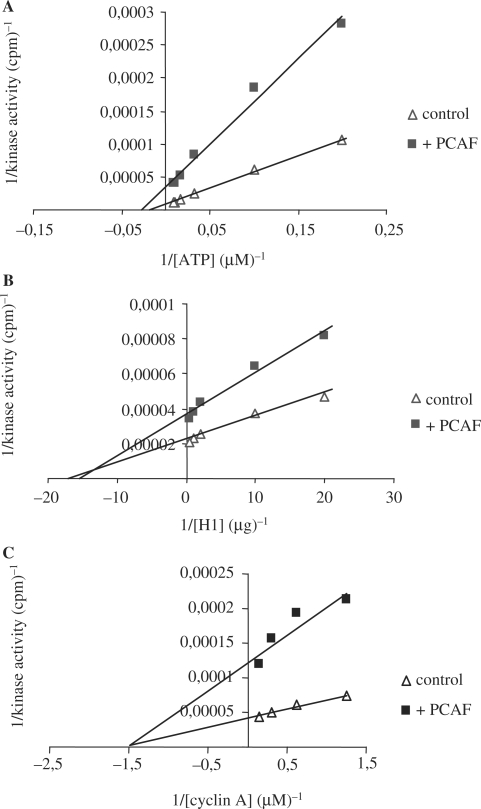
In order to analyze the mechanism of the inhibition of cdk2 activity by PCAF we performed kinetic analysis of cyclin A/cdk2 activity in the presence of increasing amounts of ATP or substrate (histone H1). Results indicate that the inhibition of cyclin A/cdk2 by PCAF follows a non-competitive kinetics in both cases, with ATP (Figure 4A) or with the substrate (Figure 4B). Kinetic analyses of cdk2 activity in the presence of PCAF and growing amounts of cyclin A were also performed. We observed that again, inhibition by PCAF follows a non-competitive kinetics with cyclin A (Figure 4C) meaning that likely PCAF does not separate the components of cyclin A/cdk2 complex, but rather disturbs the complex in some way thereby inducing the inhibition of its kinase activity.
Figure 4.
PCAF inhibition is not competitive with ATP nor with substrate histone H1 or cyclin A. Data from kinetic assays (see ‘Materials and Methods’ section) were analysed by double reciprocal plots. (A) Inhibition of cyclin A/cdk2 activity by PCAF at different ATP concentrations. The assay consisted in the incubation of 2 µg of substrate histone H1 with 400 nM cyclin A/cdk2 in the presence (square) or absence (triangle) of 800 nM PCAF. (B) Inhibition of cyclin A/cdk2 activity by PCAF at different histone H1 concentrations. The assay consisted in the incubation of 12.5 µM ATP with 400 nM cyclin A/cdk2 in the presence (square) or absence (triangle) of 800 nM PCAF. (C) Inhibition of cdk2 activity by PCAF at different cyclin A concentrations. The assay consisted in the incubation of 400 nM cdk2 (control) or 400 nM cdk2 plus 800 nM PCAF (+PCAF) with increasing concentrations of cyclin A in the presence of 12.5 µM ATP and 2 µg of substrate histone H1.
PCAF overexpression blocks cell cycle progression
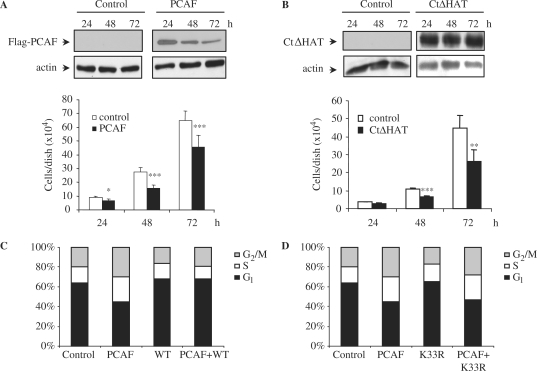
As PCAF inhibits cyclin A/cdk2 activity, that is necessary for cell cycle progression, we aimed to study the effect of PCAF overexpression on cell proliferation. Thus, we first transfected NIH3T3 cells with Flag-PCAF or with an empty vector and the number of cells in the cultures were counted at different times after transfection. As showed in Figure 5A the cell number is significantly lower in PCAF overexpressing cells than in control cells. Similar results were obtained when using a NIH3T3 stable cell line expressing CtΔHAT-PCAF in the absence of tetracycline (Tet-off system). Also in this case, a block of cell proliferation was observed (Figure 5B).
Figure 5.
PCAF impairs cell proliferation by causing S and G2/M cell cycle arrest. (A) NIH3T3 cells were transfected with empty vector or Flag-PCAF, then counted, and equal amounts were seeded in triplicates in 6-well-plates. Cell proliferation was measured by counting the number of cells present in each well 24, 48 and 72 h after transfection. Results shown are the mean of three independent experiments ± SE. *P-value <0.05; **P-value <0.01; ***P-value <0.001. A WB showing the levels of transfected PCAF is shown in the top panel, and a WB with anti-actin was performed as a loading control (bottom panel). (B) Similar experiments as in (A) were performed using a NIH3T3 clone expressing CtΔHAT-PCAF under a Tet-off system generated as described in the ‘Materials and Methods’ section. Equal amounts of cells were seeded in triplicates in 6 well plates. Cell proliferation was measured by counting the number of cells present in each well 24, 48 and 72 h after seeding. CtΔHAT expressing cells were cultured in the absence of tetracyclin. As a control, the same clone was cultured in medium supplemented with tetracyclin. Results shown are the mean of three independent experiments ± SE. *P-value <0.05; **P-value <0.01; ***P-value <0.001. A WB showing the levels of CtΔHAT-PCAF is shown in the top panel, and a WB with anti-actin was performed as a loading control (bottom panel). (C) HCT-116 cells were transfected with YFP-PCAF, CFP-cdk2WT or both. At 48 h after transfection they were fixed, DNA was stained with TOPRO-3 and transfected cells were analysed by FACS. The percentage of cells in each phase of the cell cycle is represented in the graph. (D) The same as in (C), but in this case cells were transfected with YFP-PCAF, CFP-cdk2 K33R, or both.
To better determine, at which cell cycle stage PCAF blocks cell proliferation FACS analysis were carried out in YFP-PCAF transfected cells. Supplementary Figure S3A shows the expression of YPF-PCAF in these transfected cells. Results indicate that in cells overexpressing PCAF a block in S and G2/M was produced (Figure 5C). This blockade is mediated by cdk2 because it is reversed by the simultaneous overexpression of this kinase (Figure 5C). Interestingly, the overexpression of the kinase dead mutant cdk2K33R did not reverse this cell cycle block (Figure 5D and supplementary Figure S3B), indicating that cdk2 activity is essential to overcome the cell cycle arrest induced by PCAF. FACS images of these studies are shown in supplementary Figure S3C.
PCAF acetylates cdk2
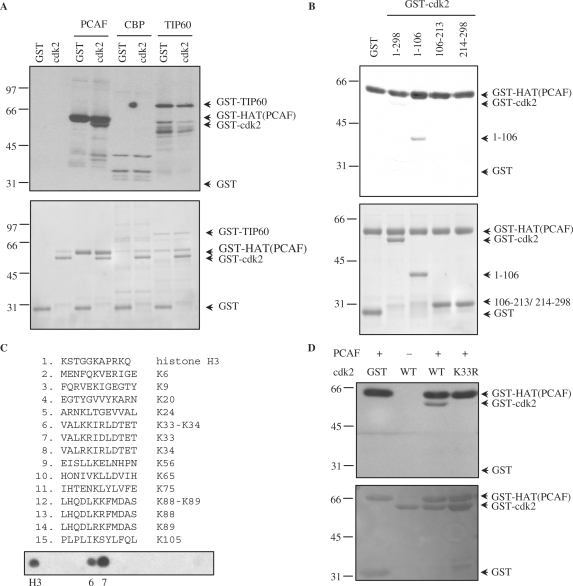
The association of cdk2 with the catalytic domain of PCAF suggested that cdk2 might be a substrate of the acetylase. Thus, an in vitro acetyltransferase assay using purified recombinant GST-cdk2 as a substrate and purified GST-HAT as enzyme was performed. Results revealed that cdk2 was acetylated by PCAF. In contrast, other acetyltransferases as CBP or Tip60 were unable to acetylate this kinase (Figure 6A and Supplementary Figure S4A). To identify the acetylation sites of cdk2 we first performed in vitro acetylation assays using three GST-cdk2 fragments (including aa 1–106, 106–213 and 214–298) as substrates and GST-HAT as acetylase. These fragments show different electrophoretic mobility in SDS-acrylamide gels, as shown in Figure 6B, bottom panel. Results indicated that only the cdk2 fragment including aa 1–106 was acetylated in vitro by PCAF (Figure 6B, upper panel).
Figure 6.
Cdk2 is a substrate acetylated by PCAF in vitro. (A) Purified GST-cdk2 was subjected to in vitro acetylation assays using the catalytic domain of PCAF [GST-HAT(PCAF)], GST-CBP or GST-Tip60 in the presence of [14C]acetylCoA. Purified GST was used as a negative control substrate. In the assays with PCAF or Tip60 their autoacetylation was used as a positive control, whereas in the case of CBP, histones were used as a positive control substrate (see Supplementary Data Figure S4A). Acetylated proteins were visualized by autoradiography (top panel). A loading control gel was stained with coomassie blue (bottom panel). (B) Purified recombinant cdk2 fragments were subjected to in vitro acetylation assays as in (A) using GST-HAT (PCAF) as acetylase. Acetylated proteins were visualized by autoradiography (top panel). A loading control gel was stained with coomassie blue (bottom panel). (C) Fourteen peptides including one or two consecutive lysines from the cdk2 fragment including aa 1–106, were spotted on a membrane. As a positive control a peptide from histone H3 was added. The membrane was subjected to in vitro acetylation assays with GST-HAT(PCAF) and [14C]acetylCoA. Acetylation was visualized by autoradiography. (D) Purified GST-cdk2 WT and GST-cdk2 K33R were subjected to in vitro acetylation assays with GST-HAT(PCAF). Acetylation was visualized by autoradiography (top panel). A loading control gel was stained with red ponceau (bottom panel).
This cdk2 fragment contains 12 lysine residues at positions K6, K9, K20, K24, K33, K34, K56, K65, K75, K88, K89 and K105. To identify the lysine/es that might be acetylated by PCAF, in vitro ‘spot mapping’ experiments were performed. Thus, 14 peptides, each one containing one or two of the lysines present in this fragment, were synthesized and spotted on a nitrocellulose membrane (Figure 6C, upper panel). A peptide from histone H3 was used as a positive control (peptide 1). Then, this membrane was subjected to an in vitro acetylation assay using GST-HAT as acetylase. Results indicated that the control peptide and peptides containing K33 (spots 6 and 7) were clearly acetylated by PCAF (Figure 6C, bottom panel). To analyze whether K33 was the acetylation site in the full length cdk2 protein, a mutational analysis was performed. Thus, a cdk2 protein harboring the K33R substitution was used for an in vitro acetylation assay. Results revealed that the cdk2 K33R mutant was not acetylated by PCAF (Figure 6D). These results indicate that K33 of cdk2 is a specific acetylation site for PCAF.
We further aimed to study the in vivo acetylation of cdk2. Thus, cells were transfected with Flag cdk2 alone or together with Flag PCAF, then cell extracts were subjected to IP with anti-Flag and the immunoprecipitates analyzed for the acetylation of cdk2 by WB with anti-acetyl K. Results revealed that in cells non-transfected with the acetylase cdk2 acetylation is not observed. In contrast, in PCAF-transfected cells cdk2 acetylation is clearly seen (Figure 7A). Similar experiments carried out after transfection of cells with GCN5, an acetylase homologous to PCAF, revealed that this acetylase is also able to acetylate cdk2 (Figure 7A). Experiments performed with the mutant cdk2 K33R, revealed that lysine 33 is also the in vivo acetylation site of cdk2, because it was not acetylated (Figure 7B). Finally, we aimed to determine whether PCAF immunoprecipitated from cell extracts was able to acetylate purified cdk2. Results indicated that cdk2 is acetylated by immunoprecipitated PCAF (Figure 7C). Interestingly, the immunoprecipitated PCAF forms part of specific multiprotein complexes that, as shown in Figure 7D, contain the protein spt3 (21).
Figure 7.
Cdk2 is acetylated by PCAF and GCN5 in vivo. (A) Cells were transfected with Flag-cdk2 alone or together with three different acetylases (p300, PCAF or GCN5). Cell extracts were subjected to IP with anti-Flag and WBs were performed with anti-Flag and anti-Acetyl-K. (B) Cells were transfected with Flag-cdk2 or Flag-cdk2K33R alone or together with HA-GCN5. Then, cell extracts were subjected to IP with anti-Flag followed by WB with anti-Flag and anti-Acetyl-K. (C) C2C12 cell extracts were subjected to IP with anti-PCAF. The obtained immunoprecipitates were used as a source of active PCAF and were used for in vitro cdk2 acetylation experiments. Autoradiography indicates cdk2 acetylation (top panel). A WB anti-cdk2 of the membrane used for autoradiography is shown on the bottom panel. Asterisks indicates non-specific bands. (D) The immunoprecipitates obtained in (C) were checked for the content of PCAF and SPT3 by WB.
Acetylation of cdk2 inhibits its kinase activity
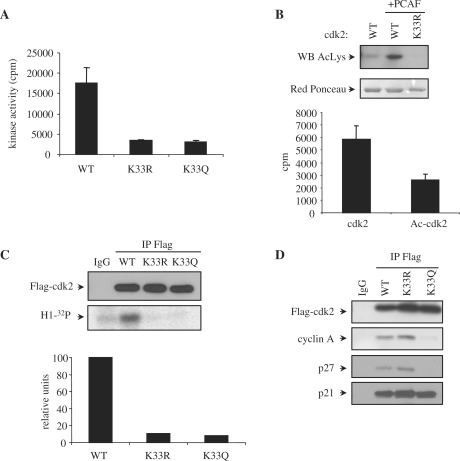
Lysine 33 is located in the catalytic pocket of cdk2 and it is involved in the binding of ATP. This lysine residue is conserved in all the members of the cdk family (Supplementary Figure S4B). It is well known that this lysine is essential for the kinase activity of cdk2 and the mutated form cdk2 K33R is fully inactive (23). To confirm that acetylation at this specific lysine of cdk2 inactivates the kinase we generated purified recombinant cdk2, in which K33 was substituted by glutamine (K33Q mutant) in order to mimic acetylation of this residue (24,25). Thus, the activities of cdk2 K33Q and cdk2 K33R associated with cyclin A were determined in vitro. Results indicate that whereas cyclin A/cdk2 WT was active, the cyclin A complexes with the non-acetylatable mutant cdk2 K33R or the pseudoacetylated mutant cdk2 K33Q were inactive (Figure 8A).
Figure 8.
Acetylation of cdk2 impairs its kinase activity both in vivo and in vitro. (A) In vitro kinase assays were performed using 400 nM of purified recombinant GST-cdk2WT, K33R or K33Q together with 400 nM of cyclin A. Kinase activity ± SE was represented in the graph. (B) GST-cdk2 WT and K33R were expressed in bacteria co-expressing 6His-PCAF. After purification of GST-cdk2 WT and K33R, they were analysed by WB with anti-Acetyl-K (top panel). Red Ponceau staining of the proteins is shown in the bottom panel as a loading control. GST-cdk2 WT purified in the absence or presence (Ac-cdk2) of PCAF were tested for in vitro kinase activity. 400 nM of the proteins were incubated with 400 nM of purified GST-cyclin A in the presence of histone H1 as a substrate and [32P]ATP as a cofactor. Kinase activity ± SE was represented in the graph. (C) HeLa cells were transfected with Flag-cdk2 WT, K33R or K33Q. Cell extracts were subjected to IP with anti-Flag or IgG as a control. A WB performed with anti-Flag is shown in the top panel. Kinase assays were also performed with the immunoprecipitates and their kinase activity was quantitated with a PhosphorImager (bottom panel). Normalization of cdk activity with respect to the amount of immunoprecipitated cdk2 is shown in the graph. (D) 293-T cells were transfected with Flag-cdk2WT, K33R or K33Q. Cell extracts were subjected to IP with anti-Flag or IgG as a control. A WB performed with anti-Flag is shown in the upper panel. Interaction of the different cdk2 forms with cyclin A, p21 and p27 was analyzed by WB.
To further confirm that acetylation at lysine 33 of cdk2 inactivates its kinase activity we generated acetylated GST-cdk2 in bacteria. To this aim, bacteria were transformed with GST-cdk2WT alone, GST-cdk2WT plus PCAF or with the mutant GST-cdk2 K33R plus PCAF. Then, these different cdk2s were purified and subsequently analyzed for their acetylation status by WB using an anti-acetyl-K antibody. We observed that GST-cdk2WT in the absence of PCAF was only slightly acetylated, GST-cdk2WT in the presence of PCAF was highly acetylated and GST-cdk2 K33R even in the presence of PCAF was not acetylated at all (Figure 8B, upper panel). In such a way, we obtained cdk2 with different levels of acetylation at K33. These purified cdk2 fractions did not contain PCAF as checked by WB (data not shown). These fractions were subsequently used for the determination of their activity, when associated to cyclin A. Results clearly indicated that the activity of acetylated cdk2 was much lower than non-acetylated cdk2 (Figure 8B, bottom panel). Finally, we also analyzed the in vivo activity of the cdk2 mutants K33R and K33Q. Cells were transfected with Flag-cdk2 WT, Flag- performed. The kinase activity of the immunoprecipitates was analyzed and results showed that in contrast to cdk2 WT, the cdk2 mutants K33R and K33Q were inactive in vivo (Figure 8C).
To analyze the functional relevance of cdk2 acetylation, we analyzed whether in addition to its inactivation, cdk2 acetylation could affect its interaction with other cell cycle regulatory proteins. To this aim, cells transfected with cdk2WT, cdk2K33R or cdk2K33Q were subsequently subjected to IP and then analyzed for its interaction with different cyclins and CKIs. Interestingly, it was observed that the pseudoacetylated mutant cdk2 K33Q lost its interaction with cyclin A and p27 but not with p21 (Figure 8D). These results indicate that cdk2 acetylation also modulates its interaction with some other cell cycle regulatory proteins.
DISCUSSION
Cdks are serine/threonine kinases that play a key role in the regulation of cell cycle progression and transcription. Those members of the family that participate in the regulation of the G1 phase of the cell cycle act as integrators of the extracellular mitogenic stimuli, that include growth factors, interaction with the extracellular matrix and cell−cell interactions (26,27). These external factors, when associate with specific plasma membrane receptors trigger different signaling pathways that converge in the regulation of the activity of the G1-operating cdks. In addition to that, cdks also integrate the signaling emanated from cell cycle checkpoints (28). Therefore, these kinases have multiple regulatory mechanisms that allow them to coordinate these different pathways and qualify cdks for doing this complex integrative activity. As mentioned above, the now classical general mechanisms that regulate cdk activity include the interaction with cyclins, that stimulates cdk activity, and with CKIs that, on the contrary, inhibit cdk activity. Phosphorylation also regulates positively and negatively the activity of cdks. However, more recently, new mechanisms that specifically regulate the activity of some cdks are emerging. For example, it should be mentioned that the oncogene and chromatin remodeling protein SET specifically inhibits cyclin B/cdk1 activity by associating to this complex (29). Other examples are that acetylation of K48 of cdk9 by PCAF and Gcn5 inactivates this enzyme (13) and that acetylation of cyclin A at specific lysines promotes its degradation (15).
We report here that the acetyltransferase PCAF inhibits cyclin/cdk2 activity by two different mechanisms: (i) by interacting with cyclin/cdk2 complexes in experimental conditions that do not allow protein acetylation (absence of the acetyl donor Acetyl-CoA) and (ii) by acetylating K33 of the cdk2 sequence, similarly to that observed for cdk9.
PCAF is a histone acetyltransferase homologous to Gcn5, both belonging to the GNAT family (Gcn5 related N-acetyltransferases), which are important for transcriptional initiation (30). PCAF participates in the reversible acetylation of various transcriptional regulators as the general transcription factors TFIIEβ and TFIIF (31) and the sequence-specific transcription factors E2F1 (20), c-myc (32), myo D (33) and p53 (34,35) among others (36). In the cell, PCAF is a subunit of multiprotein complexes that posses global histone acetylation activity and locus-specific co-activator functions together with acetyl transferase activity on non-histone susbtrates (21,37). Interestingly, PCAF possesses a domain with E3 ubiquitin ligase activity in its N-terminal region (38).
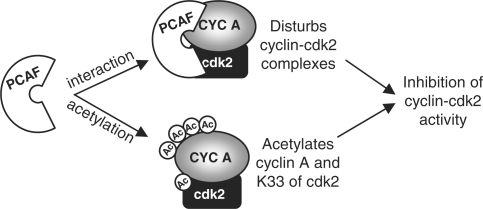
We have recently observed that PCAF directly interacts with and acetylates cyclin A (15). This acetylation is produced at early mitosis and stimulates cyclin A degradation at prometaphase with the consequent inactivation of cyclin A/cdk2 complexes. These results together with those reported here indicate that the mechanism involved in the inhibition of cyclin A/cdk2 activity by PCAF is quite complex and include at least three different actions: alteration of cyclin A/cdk2 association, acetylation of cdk2 and acetylation of cyclin A (Figure 9). Likely, these three actions of PCAF on cyclin A/cdk2 proceed in a specific sequence in order to block the activity of this complex during a given period of time. We propose that after binding of PCAF to cyclin A/cdk2 the activity of this complex is inhibited simply by the affectation of the interaction between both subunits. Then, cdk2 could be acetylated on K33 at the catalytic pocket, a fact that would additionally inactivate the enzyme but also, and probably most important, would provoke its separation from cyclin A. This is suggested by the data reported here revealing that the pseudoacetylated cdk2K33Q mutant does not interact with cyclin A. Subsequently, acetylated cdk2 would remain inactive and also protected from the putative activation by other type of cyclins.
Figure 9.
An outline representing the two different mechanisms, by which PCAF inhibits the activity of cyclin A/cdk2 complexes is shown.
Kinetic analyses of cyclin A/cdk2 activity in the presence or absence of PCAF reveal that it does not compete either with ATP or with the substrate. As PCAF directly interacts with both cyclin A and cdk2 it is likely that this double interaction disturbs somehow cyclin A/cdk2 association, thus reducing its enzymatic efficiency. This is supported by the evidence that, to effectively inhibit the activity of cyclin A/cdk2 complexes, the integrity of a long fragment of PCAF is needed (C-terminus including aa 352–382). Specific domains within this C-terminus fragment of PCAF, as HAT, ADA and Bromodomain, when tested alone, do not inhibit the activity of the complex. Likely, this long C-terminus fragment should contain specific domains responsible for the specific interaction with cyclin A and cdk2. However, the identity of these putative interacting domains still remains unknown.
Overexpression of PCAF blocks cell cycle progression at S and G2/M as observed by FACS analysis. This is consistent with the evidence that at these stages of the cell cycle the interaction of PCAF with cdk2 and also with cyclin A (15) is high. Interestingly, this cell cycle blockade is reversed, when cells are simultaneously co-transfected with PCAF and cdk2. These results indicate that overexpression of PCAF is affecting the cellular availability of cdk2 and that by increasing the amount of cdk2 in the cells, the cell cycle arrest is overcome. We still do not know the role of PCAF on the regulation of S phase but a recent report indicates that GCN5 modulates S phase by regulating cdc6 phosphorylation by cyclin A/cdk2 complexes (39). In this article authors describe that overexpression of GCN5 generates a block in S phase similarly to that we observed in cells transfected with PCAF. Because these two acetylases are highly homologous it can be postulated that PCAF can also play a role in regulating DNA synthesis. The putative role of PCAF on G2/M could be related to the inactivation of cyclin A/cdk2 complexes at the G2 phase. However, to demonstrate this hypothesis it is necessary to determine the endogenous acetylation of cdk2 along the cell cycle. Unfortunately, detection of endogenous acetylated cdk2 in vivo has been widely elusive until now.
PCAF acetylation of K33 of cdk2 is a similar event to that occurring in cdk9. It has been previously reported that this kinase can also be acetylated by PCAF and Gcn5 at K48, which is the equivalent to K33 in cdk2 (13). As cdk9 is involved in transcription (40), authors demonstrated that acetylation of this kinase specifically inhibits cdk9 dependent transcriptional activity. As this lysine is located in the catalytic pocket of all cdks but also of many other kinases, one can speculate that acetylation of this specific residue could be a more general mechanism of the regulation of kinase activity.
PCAF also plays an important role in the DNA damage checkpoint response by acetylating p53 and E2F1. After DNA damage, PCAF acetylates and stabilizes E2F1 and as a consequence complexes containing both proteins are formed. These complexes re-localize from cell cycle progression-genes to the promoters of pro-apoptotic genes (41). Moreover, PCAF acetylates p53 at lysine 320 and induces the expression of selected p53 target genes as PIG3 and NOXA under DNA damage conditions (42,43). Results reported here allow us to speculate about the possibility that PCAF could also participate in the DNA damage response by inhibiting cyclin/cdk2 complexes in order to arrest cell cycle progression. Thus, PCAF activation after DNA damage would induce the acetylation of both cyclin A and cdk2 leading to the inhibition of cdk2 activity and cell cycle arrest.
As cyclin A/cdk2 complexes are involved in the regulation of steroid hormone mediated transcription, it appears that the inhibition of these complexes by PCAF could affect the activity of SR-mediated gene expression. However, this possibility still remains to be explored.
As a summary, our results reveal a new mechanism for the regulation of cell cycle progression by PCAF. This acetylase controls cdk2 activity by a mechanism that includes the direct binding to cyclin/cdk2 complexes that might compromise the interaction between both proteins and by acetylating an essential lysine located in the catalytic pocket of the kinase.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants SAF2006-05212 and SAF2007-60491 from the Ministerio de Ciencia e Innovación of Spain and Rticc RD06/0020/0010 from the Instituto de Salud Carlos III. Funding for open access charge: Grant from the Spanish Government.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Morgan DO. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 2.Sherr CJ. Growth factor-regulated G1 cyclins. Stem. Cells. 1994;12(Suppl. 1):47–55. [PubMed] [Google Scholar]
- 3.Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- 4.Sherr CJ, Kato J, Quelle DE, Matsuoka M, Roussel MF. D-type cyclins and their cyclin-dependent kinases: G1 phase integrators of the mitogenic response. Cold Spring Harb. Symp. Quant. Biol. 1994;59:11–19. doi: 10.1101/sqb.1994.059.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Malumbres M, Barbacid M. Mammalian cyclin-dependent kinases. Trends in Biochemical Sciences. 2005;30:630–641. doi: 10.1016/j.tibs.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Nigg EA. Cyclin-dependent kinase 7: at the cross-roads of transcription, DNA repair and cell cycle control? Curr. Opin. Cell Biol. 1996;8:312–317. doi: 10.1016/s0955-0674(96)80003-2. [DOI] [PubMed] [Google Scholar]
- 7.Coleman TR, Dunphy WG. Cdc2 regulatory factors. Curr. Opin. Cell Biol. 1994;6:877–882. doi: 10.1016/0955-0674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 8.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 9.Weigel NL, Moore NL. Cyclins, cyclin dependent kinases, and regulation of steroid receptor action. Mol. Cell Endocrinol. 2007;265–266:157–161. doi: 10.1016/j.mce.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rogatsky I, Trowbridge JM, Garabedian MJ. Potentiation of human estrogen receptor alpha transcriptional activation through phosphorylation of serines 104 and 106 by the cyclin A/cdk2 complex. J. Biol. Chem. 1999;274:22296–22302. doi: 10.1074/jbc.274.32.22296. [DOI] [PubMed] [Google Scholar]
- 11.Moore NL, Narayanan R, Weigel NL. Cyclin dependent kinase 2 and the regulation of human progesterone receptor activity. Steroids. 2007;72:202–209. doi: 10.1016/j.steroids.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spange S, Wagner T, Heinzel T, Kramer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int. J. Biochem. Cell Biol. 2009;41:185–198. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 13.Sabo A, Lusic M, Cereseto A, Giacca M. Acetylation of conserved lysines in the catalytic core of cyclin-dependent kinase 9 inhibits kinase activity and regulates transcription. Mol. Cell Biol. 2008;28:2201–2212. doi: 10.1128/MCB.01557-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Bondt HL, Rosenblatt J, Jancarik J, Jones HD, Morgan DO, Kim SH. Crystal structure of cyclin-dependent kinase 2. Nature. 1993;363:595–602. doi: 10.1038/363595a0. [DOI] [PubMed] [Google Scholar]
- 15.Mateo F, Vidal-Laliena M, Canela N, Busino L, Martinez-Balbas MA, Pagano M, Agell N, Bachs O. Degradation of cyclin A is regulated by acetylation. Oncogene. 2009;28:2654–2666. doi: 10.1038/onc.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizuguchi G, Vassilev A, Tsukiyama T, Nakatani Y, Wu C. ATP-dependent nucleosome remodeling and histone hyperacetylation synergistically facilitate transcription of chromatin. J. Biol. Chem. 2001;276:14773–14783. doi: 10.1074/jbc.M100125200. [DOI] [PubMed] [Google Scholar]
- 17.Liu X, Vorontchikhina M, Wang YL, Faiola F, Martinez E. STAGA recruits Mediator to the MYC oncoprotein to stimulate transcription and cell proliferation. Mol. Cell Biol. 2008;28:108–121. doi: 10.1128/MCB.01402-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donzelli M, Squatrito M, Ganoth D, Hershko A, Pagano M, Draetta GF. Dual mode of degradation of Cdc25 A phosphatase. EMBO J. 2002;21:4875–4884. doi: 10.1093/emboj/cdf491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Canela N, Orzaez M, Fucho R, Mateo F, Gutierrez R, Pineda-Lucena A, Bachs O, Perez-Paya E. Identification of an Hexapeptide That Binds to a Surface Pocket in Cyclin A and Inhibits the Catalytic Activity of the Complex Cyclin-dependent Kinase 2-Cyclin A. J. Biol. Chem. 2006;281:35942–35953. doi: 10.1074/jbc.M603511200. [DOI] [PubMed] [Google Scholar]
- 20.Martinez-Balbas MA, Bauer UM, Nielsen SJ, Brehm A, Kouzarides T. Regulation of E2F1 activity by acetylation. EMBO J. 2000;19:662–671. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagy Z, Tora L. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene. 2007;26:5341–5357. doi: 10.1038/sj.onc.1210604. [DOI] [PubMed] [Google Scholar]
- 22.Santos-Rosa H, Valls E, Kouzarides T, Martinez-Balbas M. Mechanisms of P/CAF auto-acetylation. Nucleic Acids Res. 2003;31:4285–4292. doi: 10.1093/nar/gkg655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreau JL, Marques F, Barakat A, Schatt P, Lozano JC, Peaucellier G, Picard A, Geneviere AM. Cdk2 activity is dispensable for the onset of DNA replication during the first mitotic cycles of the sea urchin early embryo. Dev. Biol. 1998;200:182–197. doi: 10.1006/dbio.1998.8961. [DOI] [PubMed] [Google Scholar]
- 24.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 25.Li M, Luo J, Brooks CL, Gu W. Acetylation of p53 inhibits its ubiquitination by Mdm2. J. Biol. Chem. 2002;277:50607–50611. doi: 10.1074/jbc.C200578200. [DOI] [PubMed] [Google Scholar]
- 26.Walker JL, Assoian RK. Integrin-dependent signal transduction regulating cyclin D1 expression and G1 phase cell cycle progression. Cancer Metastasis Rev. 2005;24:383–393. doi: 10.1007/s10555-005-5130-7. [DOI] [PubMed] [Google Scholar]
- 27.Takai Y, Miyoshi J, Ikeda W, Ogita H. Nectins and nectin-like molecules: roles in contact inhibition of cell movement and proliferation. Nat. Rev. Mol. Cell Biol. 2008;9:603–615. doi: 10.1038/nrm2457. [DOI] [PubMed] [Google Scholar]
- 28.Lobrich M, Jeggo PA. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat. Rev. Cancer. 2007;7:861–869. doi: 10.1038/nrc2248. [DOI] [PubMed] [Google Scholar]
- 29.Canela N, Rodriguez-Vilarrupla A, Estanyol JM, Diaz C, Pujol MJ, Agell N, Bachs O. The SET protein regulates G2/M transition by modulating cyclin B-cyclin-dependent kinase 1 activity. J. Biol. Chem. 2003;278:1158–1164. doi: 10.1074/jbc.M207497200. [DOI] [PubMed] [Google Scholar]
- 30.Vetting MW, Carvalho LP, S.d, Yu M, Hegde SS, Magnet S, Roderick SL, Blanchard JS. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 2005;433:212–226. doi: 10.1016/j.abb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 31.Imhof A, Yang XJ, Ogryzko VV, Nakatani Y, Wolffe AP, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr. Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 32.Patel JH, Du Y, Ard PG, Phillips C, Carella B, Chen CJ, Rakowski C, Chatterjee C, Lieberman PM, Lane WS, et al. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol. Cell Biol. 2004;24:10826–10834. doi: 10.1128/MCB.24.24.10826-10834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puri PL, Sartorelli V, Yang XJ, Hamamori Y, Ogryzko VV, Howard BH, Kedes L, Wang JY, Graessmann A, Nakatani Y, et al. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol. Cell. 1997;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 34.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 35.Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A, Anderson CW, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schiltz RL, Nakatani Y. The PCAF acetylase complex as a potential tumor suppressor. Biochim. Biophys. Acta. 2000;1470:M37–M53. doi: 10.1016/s0304-419x(99)00037-2. [DOI] [PubMed] [Google Scholar]
- 37.Li X, Wong J, Tsai SY, Tsai MJ, O'M;alley BW. Progesterone and glucocorticoid receptors recruit distinct coactivator complexes and promote distinct patterns of local chromatin modification. Mol. Cell. Biol. 2003;23:3763–3773. doi: 10.1128/MCB.23.11.3763-3773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linares LK, Kiernan R, Triboulet R, Chable-Bessia C, Latreille D, Cuvier O, Lacroix M, Le Cam L, Coux O, Benkirane M. Intrinsic ubiquitination activity of PCAF controls the stability of the oncoprotein Hdm2. Nat. Cell Biol. 2007;9:331–338. doi: 10.1038/ncb1545. [DOI] [PubMed] [Google Scholar]
- 39.Paolinelli R, Mendoza-Maldonado R, Cereseto A, Giacca M. Acetylation by GCN5 regulates CDC6 phosphorylation in the S phase of the cell cycle. Nat. Struct. Mol. Biol. 2009;16:412–420. doi: 10.1038/nsmb.1583. [DOI] [PubMed] [Google Scholar]
- 40.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 41.Ianari A, Gallo R, Palma M, Alesse E, Gulino A. Specific role for p300/CREB-binding protein-associated factor activity in E2F1 stabilization in response to DNA damage. J. Biol. Chem. 2004;279:30830–30835. doi: 10.1074/jbc.M402403200. [DOI] [PubMed] [Google Scholar]
- 42.Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, Berger SL. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Terui T, Murakami K, Takimoto R, Takahashi M, Takada K, Murakami T, Minami S, Matsunaga T, Takayama T, Kato J, et al. Induction of PIG3 and NOXA through acetylation of p53 at 320 and 373 lysine residues as a mechanism for apoptotic cell death by histone deacetylase inhibitors. Cancer Res. 2003;63:8948–8954. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.