Abstract
We have previously reported a natural GTAA deletion within an intronic splicing processing element (ISPE) of the ataxia telangiectasia mutated (ATM) gene that disrupts a non-canonical U1 snRNP interaction and activates the excision of the upstream portion of the intron. The resulting pre-mRNA splicing intermediate is then processed to a cryptic exon, whose aberrant inclusion in the final mRNA is responsible for ataxia telangiectasia. We show here that the last 40 bases of a downstream intronic antisense Alu repeat are required for the activation of the cryptic exon by the ISPE deletion. Evaluation of the pre-mRNA splicing intermediate by a hybrid minigene assay indicates that the identified intronic splicing enhancer represents a novel class of enhancers that facilitates processing of splicing intermediates possibly by recruiting U1 snRNP to defective donor sites. In the absence of this element, the splicing intermediate accumulates and is not further processed to generate the cryptic exon. Our results indicate that Alu-derived sequences can provide intronic splicing regulatory elements that facilitate pre-mRNA processing and potentially affect the severity of disease-causing splicing mutations.
INTRODUCTION
The splicing reaction involves recognition of the exon–intron junction by the spliceosome and excision of the intronic sequences through a two-step transesterification reaction (1). An accurate mRNA biosynthesis requires both the classical splicing signals [the 5′- and 3′-splice sites (ss), the branch-point and polypyrimidine sequences] and a large number of highly degenerate intronic and exonic cis-acting regulatory elements (2–4). The latter are auxillary cis-acting elements recognized by trans-acting regulatory factors, which modulate exon selection and regulate alternative splicing. A large number of exonic splicing regulatory elements have been characterized in detail: in general they stimulate or inhibit spliceosomal assembly on an exon, affecting its definition and favoring or inhibiting the recognition of the adjacent splice sites (4,5). A number of intronic elements are also known (6,7), but less data are available regarding those located at some distance from the splice sites. Some of them have been recently shown to facilitate the formation of pre-mRNA splicing intermediates by acting selectively on the splicing efficiency of upstream or downstream introns (8). Human introns are typically thousands of bases long and abound in both cryptic splice sites and consensus cis-acting regulatory elements. As a consequence, the several potentially cryptic sequences contained in introns have to be distinguished from real exons and skipped in the mature mRNA. In some cases, these sequences can be the origin of non-functional pre-mRNA isoforms through nonsense mediated decay (9).
Genomic variants that affect splicing regulatory elements may change the normal splicing pattern and in consequence cause or modify the severity of human diseases (2,4,10,11). These splicing-affecting mutations can be found either in distant regions of the pre-mRNA or in close proximity of the invariant splice sites. Since the vast majority of genes contain short exons surrounded by introns whose average length can be measured in kilobases, intronic alterations located far away from classical splice sites are quite often considered functionally neutral regarding pre-mRNA processing. Thus, they are mostly excluded from functional studies aimed of mapping and characterizing splicing regulatory elements. Nevertheless, increasing evidence shows that ‘deep’ intronic mutations are indeed implicated in aberrant pre-mRNA processing in a number of genes associated to disease. They frequently act either by creating novel splice sites or by strengthening pre-existing cryptic splice sites located in their proximity (12–15).
Alu repeats are highly conserved primate-specific interspersed repetitive DNA elements ∼300 bp long. They are the most abundant of all mobile elements in the human genome with >1 million copies (16–18). Alu sequences are not uniformly distributed in the human genome but preferentially located within gene-rich regions (19,20) and specifically embedded within introns in both sense and antisense orientation relative to the mRNA (18). Although their pathological incorporation in the protein-encoding portion of a gene has been explored (21), the impact they might have on gene expression upon integration in introns is still to be investigated. Alu sequences contain splicing regulatory elements that contribute to their own exonization, an evolution-related process that generates primate-specific alternatively spliced exons (22,23). In addition, intronic Alu repeats can change the mode of exon splicing from constitutive to alternative during evolution (24). In spite of the fact that the large amounts of transcribed intronic Alu’s are rich in splicing regulatory elements (25,26), their effect on normal and pathological intron processing is largely unexplored.
Ataxia telangiectasia (AT) is an autosomal recessive disease characterized by cerebellar degeneration, immunodeficiency, dilation of blood vessels, hypogonadism, premature aging, genomic instability, radiosensitivity and cancer predispositions (27). The gene whose loss of function is responsible for AT is ataxia telangiectasia mutated (ATM) (28). The ATM gene is composed of 66 exons spanning 150 kb of genomic DNA and results in an mRNA of ∼13 kb in size with an open reading frame of 9.2 kb. Genetic alterations identified in ATM occur throughout the entire gene with no ‘hot spots’ and generally cause protein instability (29). However, analysis of the mutations in ATM gene has revealed that a significant number of them (48%) are splicing-affecting mutations (30). We have previously identified a new disease-causing mechanism that involves an intronic splicing processing element (ISPE) in ATM intron 20 (31,32). The ISPE consists of the CAGGTAAGT sequence, which is fully complementary to U1 snRNA and is located 1870 bp and 570 bp away from neighboring exons 20 and 21. A 4 bp deletion (GTAA) in ISPE disrupts its non-canonical binding to U1 snRNA and leads to the activation of two nearby cryptic donor and acceptor splice sites and the ensuing inclusion of a cryptic 65-bp-long exon. Interestingly, the activation of the cryptic acceptor site results in a stringent order of intron sequence removal around the cryptic exon and is associated with the formation of the splicing precursor (preS1) that retains the intron located downstream but not the one that precedes the cryptic exon (32). On the other hand, the presence of C in position +2 of the weak 5′ cryptic splice site suggests that its subsequent recognition may depend on additional splicing regulatory elements possibly modulated by the RNA secondary structure (33). In this article, we show that the last 40 nucleotides of a downstream intronic antisense Alu repeat are required for the complete activation of the cryptic exon and its final inclusion in mRNA. Our results demonstrate that a new intronic splicing enhancer (ISE) facilitates the processing of the splicing intermediate to generate the cryptic exon, probably by facilitating the recognition of the cryptic weak 5′ss. Therefore, Alu repeats provide splicing intronic regulatory elements, which affect not only the severity of disease-causing mutations but may also facilitate processing of normal intronic sequences.
MATERIALS AND METHODS
Plasmid construction
An ATM cassette that includes the entire 2440 bp ATM intron 20 with part of flanking exons 20 and 21 was amplified in three fragments from normal genomic DNA using primers ATM 185 dir and ATM 1640 rev, ATM 1680 dir and ATM 2250 rev, and primers ATM 2222 dir and ATM BsteII rev, respectively (oligonucleotide sequences are provided in Supplementary Data), and cloned in the BstEII site of the third exon of the α-globin minigene (34), under the control of the α-globin promoter and SV40 enhancer. A unique NdeI-NdeI cassette that spans 263 and 233 bases upstream and downstream the cryptic exon, respectively, was substituted with polymerase chain reaction (PCR)-amplified fragments to generate pATMΔ and pΔSH3. In the latter the sequences downstream cryptic exon from –52 to –263 were deleted. To facilitate subsequent cloning procedure a unique BamHI site was introduced in pATMΔ and pATMWT 40 bp downstream the cryptic exon through a two-step PCR overlap extension method using primers ATM 374 rev and ATM 373 dir. pΔSH5 was created by deleting the 116 bases from position 40 to 233 downstream the cryptic exon between the BamHI and NdeI sites. Intronic-amplified fragments of increasing length were inserted in the unique BamHI site of pΔSH5 to generate pΔ103, pΔ156, pΔ209, pΔSH5-209 and corresponding inv minigenes. pΔ156-209, a derivative of pATMΔ with deletion of intronic sequences from 156 to 209 downstream the cryptic exon substituted with a unique BamHI site, was used to create pΔA, pΔB, pΔC, pΔD pΔE and pΔEmut by direct cloning of paired complementary oligonucleotides.
pATMWT10 was created by overlapping PCRs using ATM 219 dir, ATM spacer10 rev, ATM spacer10 dir and ATM 374 rev primers. pATMWT20 and pATMWT30 were obtained by insertions of corresponding paired oligonucleotide in the unique AccI restriction site of pATMWT10. A similar overlapping PCR strategy was used to change the C to T at the cryptic 5′ss to generate pATMΔST and pATMΔST SH. To prepare U1C2 snRNA, the sequence between BclI and BglII of the parental U1snRNA clone pGEM3U1(WT-U1), a derivative of pHU1, was replaced with mutated oligonucleotides as previously described (31). We inserted a 40 bp spacer derived from pBS in the HindIII site of second α-globin exon to obtain pBglΔ and pBglΔSH5. All minigenes were verified by sequencing.
Analysis of the hybrid minigene expression and splicing precursors
HeLa cells (2 × 106) were grown in standard conditions and transfected with Effectene reagent with 500 ng of each minigene plasmid (35). RNA extraction, reverse transcriptase (RT)–PCR and quantitation of amplified products were done as previously described (35). For the analysis of spliced forms pATM minigenes were amplified with E16 dir and ATM 2550 rev and for the amplification of intermediates with E16 dir and ATM 374 rev. In cotransfection experiments 250 ng of U1C2 were transfected with 250 ng of minigene plasmids.
RESULTS
Identification of an ISE located downstream of the ATM cryptic exon
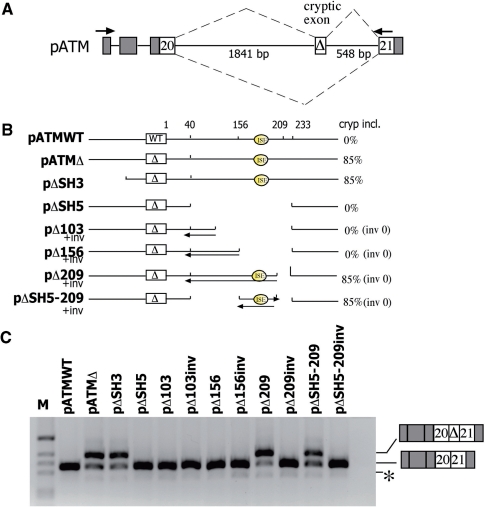
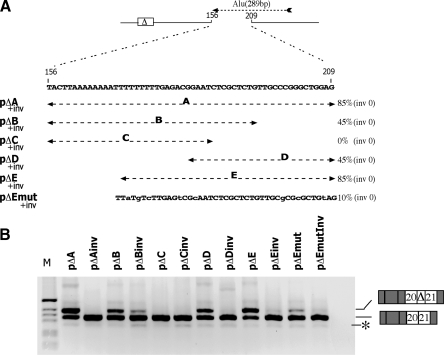
To better understand the processing of the ATM intron 20 and to identify additional splicing regulatory elements involved in cryptic ATM exon activation, we prepared deletion mutants in pATM minigenes. The basic minigene construct was composed of the ATM exons 20 and 21 along with the entire intron 20 embedded in the α-globin context (Figure 1A). As previously reported, the natural disease-causing GTAA deletion within ISPE induces the predominant inclusion of the cryptic exon (∼85% of the total mRNA obtained is aberrantly spliced; Figure 1C, pATMΔ). Furthermore, deletion of intronic sequences located upstream of the cryptic exon has no effect on the splicing pattern (Figure 1C, pΔSH3). On the contrary, the deletion of 116 bp of downstream intronic sequences completely restores normal intron processing, suggesting the presence of an intronic splicing regulatory element necessary for the cryptic exon activation mediated by the natural ISPE mutant (Figure 1C, pΔSH5). We generated a set of mutants in which the large SH5 deletion was progressively restored in order to map this ISE by evaluating the significance of certain intronic portions on the splicing pattern. Functional splicing assay revealed that pΔ209 is the only construct whose splicing pattern shows cryptic exon inclusion in the final transcript, to the same extent as in the pATMΔ. This result indicates that a 53-bp-long region between positions 156 and 209, relative to the cryptic 5′ss, harbors the regulatory element. To rule out a possible spatial effect, we created additional minigenes that contained inverted intronic sequences within SH5 region (Figure 1, ‘inv’). Transfection of these constructs showed normal splicing pattern thus confirming that ISE is indispensable for cryptic exon inclusion. In addition, cloning of the 156–209 stretch in close proximity of the cryptic exon led to aberrant splicing and cryptic exon inclusion (Figure 1, pΔSH5–209). To perform a fine mapping of the ISE element, we introduced different portions of the 53 bp region between positions 156 and 209 in the minigene and analyzed their effect by means of splicing assays. We eventually observed that a 40-bp-long sequence between positions 169 and 209 (pΔE) was sufficient to promote complete cryptic exon activation. To confirm this data, we subjected this element to site-directed mutagenesis, and by introducing 8 bp substitutions we managed to inactivate the ISE function (Figure 2, pΔEmut). Interestingly, the ISE corresponds to the last 40 nucleotides of an inverted Alu Sg repeat situated ∼160 bp downstream of the cryptic exon.
Figure 1.
ATM intron 20 contains sequences that enhance the cryptic exon activation. (A) Schematic representation of the pATM minigene. α-globin and ATM exons are grey and white boxes, respectively, and introns are lines. The cryptic 65 bp ATM exon activated by the GTAA deletion (Δ) is indicated. Splicing pattern is represented with diagonal dashed lines and the arrows indicate location of E16 and 2550 primers used in RT–PCR analysis. (B) Diagram indicating deletions introduced within the intron 20 of the pATM minigenes. The nucleotide distance downstream of the cryptic exon and of the Intronic Splicing Enhancer (ISE) are indicated. On the right it is indicated the percentage of cryptic exon inclusion as deduced form panel C and expressed as mean of three independent experiments done in duplicate. inv refers to sequences inserted in inverted orientation. (C) Splicing assay. The ATM minigenes were transfected in HeLa cells and the pattern of splicing was analyzed with the indicated primers. RT–PCR fragments were resolved on 2% agarose gel. M is the molecular weight marker 1 kb. The cryptic exon inclusion and exclusion forms are indicated and the asterisk corresponds to a minor product in which the hybrid exon made of globin exon 3 and ATM exon 20 is skipped.
Figure 2.
Mapping of the ISE. (A) Diagram indicates in the pΔ minigenes the intron 20 sequence downstream of the cryptic exon (white box) between positions 156 and 209 along with the mutant sequences analyzed. Position, orientation and length of the Alu repeat included in the ISE are indicated with dashed arrow. White box represents the cryptic exon while the introns are lines. Mutant nucleotides in Emut are in lower case. The percentage of cryptic exon inclusion as deduced from panel B is shown on the right and represent the mean of three independent experiments done in duplicate. inv refers to sequences inserted in inverted orientation. (B) Splicing assay. The pΔ minigenes were transfected in HeLa cells and the pattern of splicing analyzed with E16 and 2550 primers. RT–PCR fragments were resolved on 2% agarose gel. M is the molecular weight marker 1 kb. The cryptic exon inclusion and exclusion forms are indicated. The asterisk corresponds to a minor spliced product without the hybrid exon made of globin exon 3 and ATM exon 20.
Effect of the distance between the cryptic 3′ss and the ISPE on intron processing
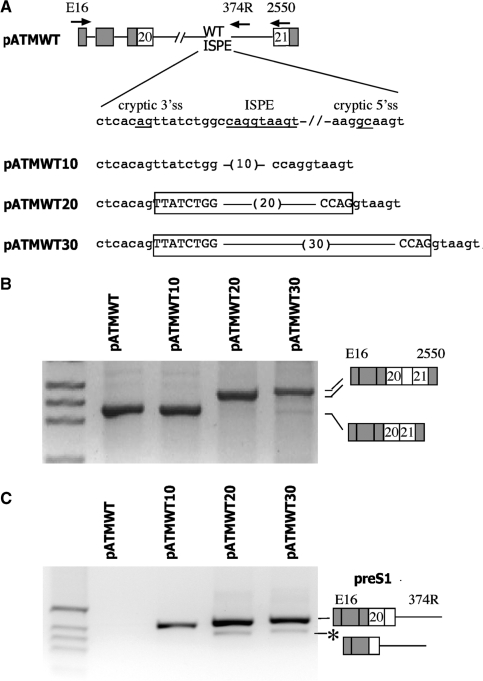
Previous analysis of splicing intermediates showed that the ISPE deletion results in a stringent 5′–3′ order of intron sequence removal around the cryptic exon (32). In fact, both in patient’s lymphoblast cells and in minigene-derived transcripts, the ISPE deletion exclusively activates splicing of the upstream intron leading to the production of a precursor that retains the downstream part of the intron, the preS1 intermediate (32). This preS1 intermediate is then spliced at the cryptic 5′ss with removal of the downstream part of the intron. In addition, the ISPE wild type (WT) sequence, even if it is a perfect consensus of the 5′ and binds to U1 snRNP (31), is not normally used as a donor site. To understand whether the activation of the ATM cryptic exon is due to an interference of the ISPE-U1 snRNP complex with the 12 bp upstream cryptic 3′ss and its relationship with the preS1 formation, we progressively increased the distance between the ISPE and the cryptic 3′ss. Three nucleotide sequences of 10, 30 and 40 bases were inserted between the ISPE and the cryptic 3′ss to generate pATMWT10, pATMWT20 and pATMWT30, respectively (Figure 3A). The resulting minigenes were transfected in HeLa cells and analyzed with specific primers to detect mature mRNA (Figure 3B) and splicing of the upstream part of the intron (i.e. the preS1 intermediate) (Figure 3C). Transfection experiments showed that pATMWT-derived transcripts correspond to a normal processing of the intron with no significant inclusion of cryptic exonic sequences (Figure 2B, lane 1) and absence of the preS1 intermediate (Figure 3C, lane 1). In pATMWT10, the increase in distance between the 3′ss and the ISPE did not result in any significant inclusion of cryptic exonic sequences (Figure 3B, lane 2) but started to produce a low amount of the preS1 intermediate (Figure 3C, lane 2). On the contrary, amplification of pATMWT20 and pATMWT30 showed, in comparison to pATMWT, mature transcripts with higher molecular weight (Figure 3, lanes 3 and 4) leading to the appearance of the corresponding preS1 intermediate. Sequence analysis of these mature transcripts revealed that the higher molecular weight bands include a cryptic exon with activation of the 5′ss contained in the ISPE (Figure 3B). Furthermore, to analyze semi-quantitatively the abundance of splicing intermediates, a cotransfection experiment with pATMWT10 and pATMWT30 constructs was conducted. What we observed is that pATMWT10 produces a substantially lower amount of preS1 when compared to the pATMWT30 construct (Figure S1). Thus in pATMWT10 the cryptic 3′ss is only partially and inefficiently activated and is probably not sufficient to provide enough distance between the splice sites for ‘exon definition’ and subsequent splicing of the downstream part of the intron. These data further reinforce the hypothesis that the natural mutant removes a steric U1 snRNPs interference on the cryptic 3′ss, thus leading to preferential splicing of the upstream part of the intron and activation of the preS1 intermediate.
Figure 3.
Effect of the distance between the cryptic 3′ss and the ISPE on the splicing pattern. (A) A schematic representation of the pATMWT minigenes. α-globin and ATM exons are grey and white boxes, respectively, and introns are lines. A sequence of 10, 20 and 30 nucleotides (in brackets) was inserted between the cryptic 3′ss and the ISPE (both underlined). The exonic sequences activated by the spacer insertions are boxed and the introns are indicated in lower letters. The arrows indicate location of the primers used in RT–PCR analysis. (B) To analyze the mature transcript the hybrid minigenes were transfected in HeLa cells and analyzed with E16 and 2550 primers. RT–PCR results of the transfection experiments were resolved on 2% agarose gel; the resulting bands were analyzed by direct sequencing and their identity is schematically represented. M is the molecular weight marker 1 kb. (C) To analyze the preS1 transcript the hybrid minigenes were transfected in HeLa cells and analyzed with E16 and 374R primers. RT–PCR results of the transfection experiments were run on 2% agarose gel and the resulting bands were analyzed by direct sequencing. M is the molecular weight marker 1 kb. The asterisk corresponds to a minor spliced product without the hybrid exon made of globin exon 3 and ATM exon 20.
Turnover of pre-mRNA splicing intermediates from the ISE minigenes
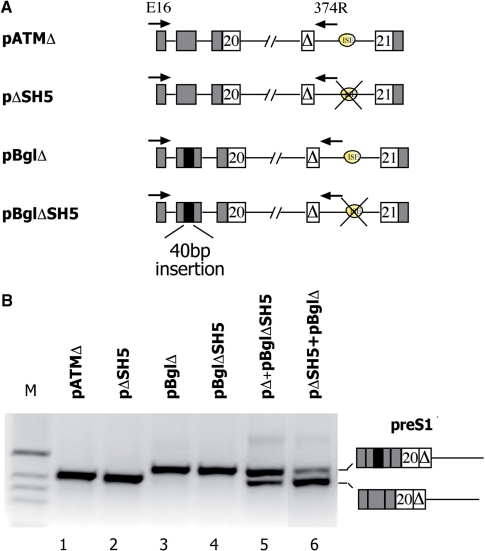
Since the generation of the cryptic exon through the ISPE deletion (32) or spacer insertions (Figure 3) led to the unique formation of the preS1 splicing precursor, we asked whether this intermediate appears in cells transfected with the ISE-deletion mutant pΔSH5. To address this question, we performed RT–PCR using a pair of primers that exclusively amplify the preS1 precursor. Strikingly, we detected the preS1 intermediate (Figure 4, pΔSH5, lane 2) even though the cryptic exon was not included in mature mRNA (Figure 1C). To quantify the relative amount of preS1 RNA produced, we set up a cotransfection experiment using pBgl–globin minigenes (Figure 4A). These minigenes contain a 40-bp-long insertion within the second exon of α-globin and RT–PCR amplification results in band at a slightly higher position, thus allowing us to distinguish preS1 intermediates deriving from different constructs. In fact, the primers used to co-amplify preS1 precursors were designed to recognize the first α-globin exon and intronic sequence just upstream the ISE, which means that the 40-bp-long insertion represented the only difference between analyzed amplicons. When equal amounts of the pΔ and pΔBglSH5 were cotransfected in HeLa cells, the intensity of the higher band deriving from the pΔBglSH5 intermediate was more pronounced in comparison with the lower pΔ precursor band (Figure 4, lane 5). Similarly, cotransfection of the same amount of pΔSH5 and pΔBgl constructs led to a significantly increased intensity of the pΔSH5 preS1 band in comparison with that deriving from the pΔBgl minigene (Figure 4, lane 6). Altogether, these data indicate that all the constructs allow reaching the preS1 precursor stage regardless of the presence of the regulatory element. Successively, the intermediates without the ISE are not further processed into a mature mRNA that contains a cryptic exon and eventually accumulate in cells. Hence, the effect of the ISE may be to facilitate processing of the nascent transcript.
Figure 4.
ISE affects turnover of pre-mRNA splicing intermediates. (A) Schematic representation of the hybrid minigenes used to evaluate the splicing intermediates. α-globin and ATM exons are grey and white boxes, respectively, the black box is the 40 bp insertion and introns are lines. The cryptic 65 bp ATM exon (Δ) is indicated. The arrows indicate location of primers used in RT–PCR analysis. (B) Analysis of preS1 intermediate abundance. The indicated minigenes were transfected in HeLa cells (250 ng each) and preS1 intermediate amplified with the primers. In cotransfection experiments equal amount of plasmids (250 ng) were transfected. The splicing products are shown and their identity was verified by direct sequencing.
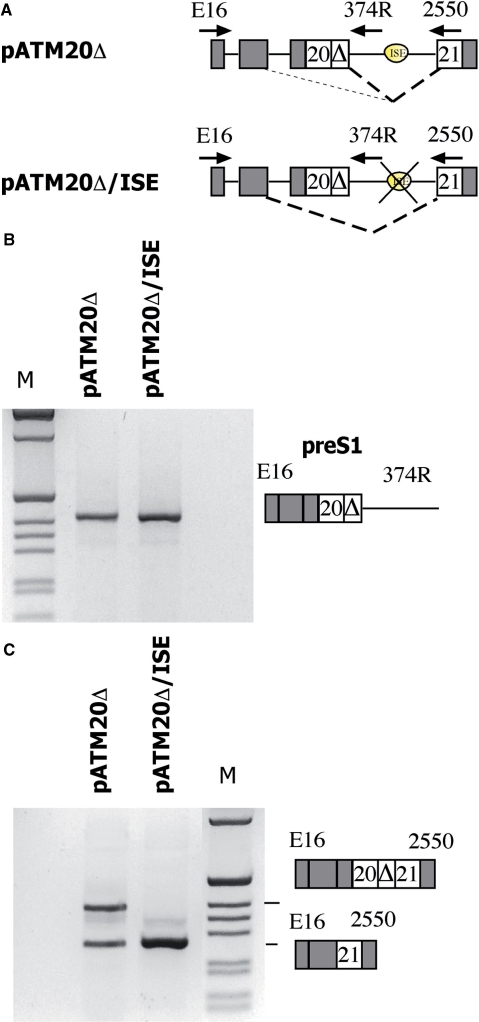
To further explore this hypothesis, we studied the effect of the ISE deletion in two partially spliced intermediate minigenes, pATM20Δ and pATM20Δ/ISE, in which the section of the intron 20 located upstream of the cryptic exon was completely deleted (Figure 5A). The splicing precursor was present in both minigenes (Figure 5B) but the final mRNA was substantially different. More precisely, whereas pATMΔ20 preferentially generated a mature mRNA that contained exon 20 along with the cryptic exon, pATM20Δ/ISE produced a mature mRNA with complete exclusion of these exons (Figure 5C). Thus, we conclude that the ISE facilitates the turnover of the intermediate. In its absence, the intermediate cannot be efficiently processed into a mature mRNA and accumulates in cells.
Figure 5.
Effect of the ISE on the prespliced minigene. (A) Schematic representation of the prespliced minigenes with and without the ISE. α-globin and ATM exons are grey and white boxes, respectively and introns are lines. Major and minor splicing patterns are represented with strong and weak diagonal dashed lines, respectively and the arrows indicate location of primers used in RT–PCR analysis. (B) Analysis of preS1 intermediates. The minigenes were transfected in HeLa cells and the preS1 intermediate amplified with the E16 and 374R primers. (C) Analysis of mature transcripts; the transfected minigenes were analyzed with the E16 and 2250 primers and RT–PCR fragments resolved on 2% agarose gel. M is the molecular weight marker 1 kb. Identity of the different was verified by direct sequencing.
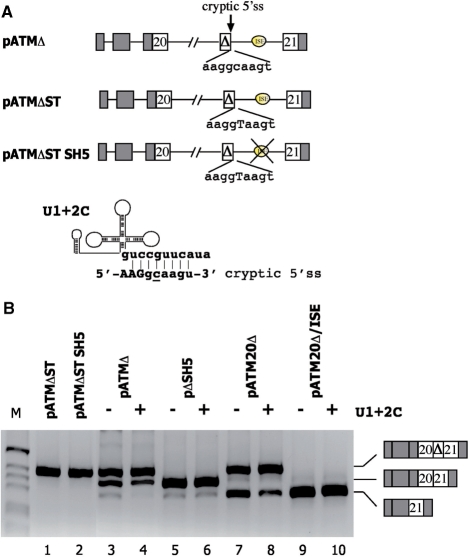
ISE-dependent splicing enhancement is dependent on the weak cryptic 5′ss
To further understand the ISE-dependent mechanism of splicing regulation, we focused on the cryptic 5′ss. This downstream site, activated by the natural ISPE deletion, is intrinsically weak, with a non-canonical C in position +2 (CAGGCAAGT) (Figure 6A). To test if this weak donor site is involved in ISE-dependent intron processing, we improved its strength by replacing the C in position +2 with a T. The resulting donor site is fully complementary to the WT U1 snRNA. The resulting minigenes with or without the ISE, pATMΔST and pATMΔST SH5, respectively, were tested in the splicing assay. The C to T mutation induced complete cryptic exon inclusion and the pattern was not affected by the ISE deletion (Figure 6C, lanes 1 and 2). To further evaluate the relationship between the ISE and the non-canonical cryptic donor site, we prepared a modified version of U1 snRNA complementary to the 5′ cryptic splice site (U1 + 2C) (Figure 6B). Cotransfection of U1 + 2C significantly increased the cryptic exon inclusion in minigenes that contain the ISE: pATMΔ and the corresponding pATM20Δ splicing intermediate minigene (Figure 6). On the contrary, cotransfection of U1 + 2C in minigenes without the ISE (pΔSH5 and pATM20Δ/ISE) did not affect the splicing pattern, suggesting that the ISE-dependent turnover of the splicing intermediate is related to U1 snRNP recruitment to the cryptic 5′ss.
Figure 6.
Relationship between the ISE and the strength of the cryptic 5′ss. (A) Schematic representation of the pATM minigenes with improved cryptic 5′ss. α-globin and ATM exons are grey and white boxes, respectively, introns are lines and the ISE is indicated as oval. (B) Schematic representation of the complementarity between the modified U1 + 2C RNA and the cryptic 5′ss. The non-canonical C in position +2 of the cryptic 5′ss is underlined. (C) Splicing assay. The ATM minigenes (250 ng) were transfected in HeLa cells alone (–) or with the U1 + 2C (+) vector (250 ng) and the splicing pattern was analyzed with the E16 and 2550 primes. RT–PCR fragments were resolved on 2% agarose gel. M is the molecular weight marker 1 kb. The resulting splicing products are indicated. Cotransfection with WT U1 did not affect the splicing pattern (not shown).
DISCUSSION
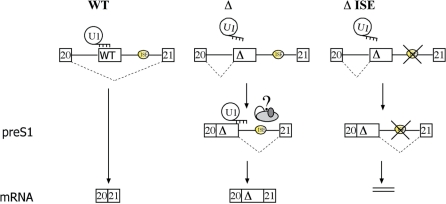
Long introns contain several potential splicing regulatory sequences, including cryptic splice sites and splicing enhancers or silencers (36,37) that, when activated, can be involved in aberrant processing of pre-mRNA. In the majority of cases, deep intronic disease-causing mutations can affect intron processing directly by creating or strengthening a splice site with subsequent inclusion of a cryptic exon in the final transcript. The mutation in the intronic ISPE element in ATM differs from the majority of described intronic variants in that it is not directly concerned with changes at splice sites. In fact, the deletion within the ISPE element, which normally binds to U1 snRNP, activates two nearby cryptic splice sites (31). In this article, we report that the activation of the pre-existing cryptic splice sites by the ISPE deletion requires an ISE embedded in an Alu repeat, which is located downstream of the cryptic exon. Thus, the generation of the final aberrant transcript in ATM intron 20 is due to a complex mechanism that affects sequentially intron processing (Figure 7). Spatially restricted events occurring close to the ISPE deletion initiate a defective turnover of the intron. In the normal situation, the non-canonical interaction between U1 snRNP and ISPE inhibits the upstream cryptic 3′ss. The ISPE deletion removes the steric interference of U1 snRNP on the cryptic splice site and results in the preferential splicing of the upstream section of the intron, with generation of the preS1 5′ splicing precursor. Further processing of the precursor depends on the presence of a downstream ISE embedded in an antisense Alu, which probably facilitates recognition of the weak cryptic 5′ss. In the absence of the ISE, the splicing intermediate activated by the ISPE deletion accumulates and is not efficiently processed. Analogous U1 snRNP-mediated repression during RNA processing has been observed in other gene systems. In Drosophila, inactivation of the genuine 5′ss by shifting U1 snRNA binding to the pseudo splice site modulates P-element pre-mRNA splicing (38,39), in Saccharomyces cerevisiae stable association of U1 snRNP inhibits spliceosomal formation of the RPL30 transcript (40) and aberrant binding of high mobility group A1 protein adjacent to the 5′ss lead to splicing defects in presenilin 2 pre-mRNA (41,42). Similarly, hyper-stabilizing U1 snRNP binding to 5′ss leads to defects in polyadenylation (43,44). These examples of defective or regulated mRNA processing have been associated to aberrant U1 snRNP complexes formation. It would be interesting to clarify the composition of the U1 snRNP complex formed on the ISPE.
Figure 7.
Model of ISE function: the ISE facilitates processing of the splicing intermediate. In WT context binding of U1 snRNP to the ISPE element interferes with the recognition of the nearby upstream cryptic 3′ss and no preS1 intermediate is produced leading to normal processing of the intron. In the affected patient, the natural deletion (Δ) relieves the steric interference of the U1 snRNP and activates the cryptic 3′ss resulting in the generation of the preS1 intermediate. In the presence of the ISE this intermediate is processed to the mature mRNA that contains the cryptic exon whereas in the absence of it (ΔISE) the preS1 is not further processed and gets accumulated. At the preS1 level, ISE could interact with several splicing factors that assist U1snRNP recognition at the cryptic 5′ss.
Few studies have evaluated the effect of splicing-affecting mutations on the accumulation of splicing intermediates in PolII-transcribed genes. The majority of studies that evaluated splicing intermediates in human pathology focused on in vitro splicing assays, a system that does not allow the analysis of splicing intermediates derived from the co-transcriptional processing of large intronic sequences. Some novel in vitro co-transcriptionally coupled splicing systems have been developed (45,46) but never applied to study pathological splicing. To study the abundance of splicing intermediates in a more physiological context we performed cotransfection experiments with minigenes, from which the amount of splicing intermediates can be easily evaluated in a semi-quantitative manner (Figure 4). The same approach was also applied to study the influence of Friedreich ataxia GAA intronic expansions on pre-mRNA processing, showing that these repeats induced the accumulation of an upstream splicing intermediate, which is not converted into mature mRNA (47). Thus, these two pathological events can share a similar intron-processing mechanism. Changes in splicing intermediate kinetics in vivo have been observed in alternative splicing regulation mediated by the neuronal specific splicing factor NOVA. Binding of NOVA to exonic or intronic sequences was shown in vivo to induce preferentially the activation of one pre-mRNA splicing intermediate (i.e. of one upstream or downstream intron), in this manner resulting in different splicing isoforms (8). In the ATM intron 20, the ISE facilitating the processing of the 5′ precursor intermediate can operate similarly. It is interesting to note that intronic clusters of NOVA-target sequences enhanced spliceosomal assembly and exon inclusion, promoting U1 snRNP binding to the alternative spliced donor site. Similarly, we observed that the ISE-dependent splicing enhancement of the upstream cryptic exon is dependent on a weak cryptic 5′ss. In fact, in minigenes with optimal cryptic donor sites, the ISE is dispensable and cotransfection experiments with a modified U1 snRNA complementary to the defective cryptic splice site demonstrates activation of intron processing only if the ISE is present (Figure 6C). This suggests that the ISE-dependent turnover of the splicing intermediate in ATM intron 20 is related to U1 snRNP recruitment to the cryptic 5′ss. Trans-acting factors binding and/or associated to RNA secondary structures of the Alu ISE might be involved in the facilitated processing of the precursor intermediate and the associated U1 snRNP recruitment. Potential splicing factors that facilitate recruitment of U1 snRNP to the donor site include TIA-1 and the related TIAR, which show a preference for U-rich sequences (48). In some cases, intronic TIA-1 interaction occurs at relatively short distance from the 5′ss (49–51). Future studies will try to identify the splicing factor(s) that, interacting with this highly abundant Alu-derived intronic regulatory sequence, is involved in splicing enhancement.
The effect of intronic Alu repeats on pre-mRNA splicing has implications both in human pathology and in primate-specific evolution. The intronic insertion of these repeats has been associated with pathological skipping of adjacent exons in several human diseases (52–56). These events can be due to an Alu-mediated disruption of pre-existing intronic splicing regulatory elements or to a gain of function provided by the repeat itself. In this article we show for the first time that a portion of an intronic Alu can affect the severity of the effect of a disease-causing splicing mutation. Probably, without the evolution-related insertion of the Alu repeat, the activation of the cryptic 3′ss by the GTAA deletion in ATM intron 20 would not be sufficient to induce aberrant intron processing. Thus, the effect of the ISE is detrimental to the disease phenotype, as in the absence of this intronic regulatory element ISPE deletion by itself would not induce the activation of the cryptic exon and its inclusion in the final mRNA transcript. This provides a unique example of how an apparently innocuous Alu-derived sequence may be pathogenic by enhancing the splicing defect.
Processing of intronic sequences can be influenced by Alu’s (24,25,57,58). Recently, a genome-wide analysis showed that many Alu elements preferentially flank alternatively spliced exons rather than constitutively spliced ones (24). This is particularly significant for exons whose mode of splicing has been modified during evolution. A RABL5 primate-specific transcript, due to exon 5 alternative splicing, has been shown to be activated by two Alu insertions upstream of the regulated exon. The suggested mechanism relies on the potential formation of inter Alu’s secondary structures that subsequently undergo RNA editing by adenosine deamination (24,59). Although the role of editing in Alu-mediated splicing regulation is unclear, the formation of inter Alu secondary structures is not possible in our case since the ATM intron 20 contains only one Alu repeat.
The large amount of intronic antisense Alu’s with ISE sequences that facilitate pre-mRNA processing could be of relevance for the spreading of Alu elements throughout the primate genome during evolution. It is tempting to speculate that the primate-specific insertion of these intronic Alu’s can be tolerated since their ISE-like sequences might facilitate intron processing and clearance of normal splicing intermediates of the host gene. This will not affect pre-mRNA processing of the original gene but will provide optimal sequences for exonization, which will shape primate-specific alternative splicing events.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant from the Associazione Italiana Ricerca sul Cancro and Telethon Onlus Foundation Italy (grant number GGP09183). Funding for open access charge: Institutional funding.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Cristiana Stuani for technical assistance and Rodolfo Garcia for helpful discussion.
REFERENCES
- 1.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 2.Pagani F, Baralle FE. Genomic variants in exons and introns: identifying the splicing spoilers. Nat. Rev. Genet. 2004;5:389–396. doi: 10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- 3.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 4.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 5.Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ladd AN, Cooper TA. Finding signals that regulate alternative splicing in the post-genomic era. Genome Biol. 2002;3:0008. doi: 10.1186/gb-2002-3-11-reviews0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 8.Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 9.Grellscheid SN, Smith CW. An apparent pseudo-exon acts both as an alternative exon that leads to nonsense-mediated decay and as a zero-length exon. Mol. Cell Biol. 2006;26:2237–2246. doi: 10.1128/MCB.26.6.2237-2246.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Blanco MA, Baraniak AP, Lasda EL. Alternative splicing in disease and therapy. Nat. Biotechnol. 2004;22:535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- 11.Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 12.Christie PT, Harding B, Nesbit MA, Whyte MP, Thakker RV. X-linked hypophosphatemia attributable to pseudoexons of the PHEX gene. J. Clin. Endocrinol. Metab. 2001;86:3840–3844. doi: 10.1210/jcem.86.8.7730. [DOI] [PubMed] [Google Scholar]
- 13.Metherell LA, Akker SA, Munroe PB, Rose SJ, Caulfield M, Savage MO, Chew SL, Clark AJ. Pseudoexon activation as a novel mechanism for disease resulting in atypical growth-hormone insensitivity. Am. J. Hum. Genet. 2001;69:641–646. doi: 10.1086/323266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chillon M, Dork T, Casals T, Gimenez J, Fonknechten N, Will K, Ramos D, Nunes V, Estivill X. A novel donor splice site in intron 11 of the CFTR gene, created by mutation 1811 + 1.6 kbA–>G, produces a new exon: high frequency in Spanish cystic fibrosis chromosomes and association with severe phenotype. Am. J. Hum. Genet. 1995;56:623–629. [PMC free article] [PubMed] [Google Scholar]
- 15.Vervoort R, Gitzelmann R, Lissens W, Liebaers I. A mutation (IVS8 + 0.6kbdelTC) creating a new donor splice site activates a cryptic exon in an Alu-element in intron 8 of the human beta-glucuronidase gene. Hum. Genet. 1998;103:686–693. doi: 10.1007/pl00008709. [DOI] [PubMed] [Google Scholar]
- 16.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 17.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 18.Sela N, Mersch B, Gal-Mark N, Lev-Maor G, Hotz-Wagenblatt A, Ast G. Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu's; unique role in shaping the human transcriptome. Genome Biol. 2007;8:R127. doi: 10.1186/gb-2007-8-6-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korenberg JR, Rykowski MC. Human genome organization: Alu, lines, and the molecular structure of metaphase chromosome bands. Cell. 1988;53:391–400. doi: 10.1016/0092-8674(88)90159-6. [DOI] [PubMed] [Google Scholar]
- 20.Chen C, Gentles AJ, Jurka J, Karlin S. Genes, pseudogenes, and Alu sequence organization across human chromosomes 21 and 22. Proc. Natl Acad. Sci. USA. 2002;99:2930–2935. doi: 10.1073/pnas.052692099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Britten RJ. Mobile elements inserted in the distant past have taken on important functions. Gene. 1997;205:177–182. doi: 10.1016/s0378-1119(97)00399-5. [DOI] [PubMed] [Google Scholar]
- 22.Sorek R, Lev-Maor G, Reznik M, Dagan T, Belinky F, Graur D, Ast G. Minimal conditions for exonization of intronic sequences: 5′ splice site formation in alu exons. Mol. Cell. 2004;14:221–231. doi: 10.1016/s1097-2765(04)00181-9. [DOI] [PubMed] [Google Scholar]
- 23.Lev-Maor G, Sorek R, Shomron N, Ast G. The birth of an alternatively spliced exon: 37′ splice-site selection in Alu exons. Science. 2003;300:1288–1291. doi: 10.1126/science.1082588. [DOI] [PubMed] [Google Scholar]
- 24.Lev-Maor G, Ram O, Kim E, Sela N, Goren A, Levanon EY, Ast G. Intronic Alus influence alternative splicing. PLoS Genet. 2008;4:e1000204. doi: 10.1371/journal.pgen.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartz S, Gal-Mark N, Kfir N, Oren R, Kim E, Ast G. Alu exonization events reveal features required for precise recognition of exons by the splicing machinery. PLoS Comput. Biol. 2009;5:e1000300. doi: 10.1371/journal.pcbi.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lei H, Day IN, Vorechovsky I. Exonization of AluYa5 in the human ACE gene requires mutations in both 3′ and 5′ splice sites and is facilitated by a conserved splicing enhancer. Nucleic Acids Res. 2005;33:3897–3906. doi: 10.1093/nar/gki707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavin MF, Shiloh Y. The genetic defect in ataxia-telangiectasia. Annu. Rev. Immunol. 1997;15:177–202. doi: 10.1146/annurev.immunol.15.1.177. [DOI] [PubMed] [Google Scholar]
- 28.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, Smith S, Uziel T, Sfez S, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 29.Sandoval N, Platzer M, Rosenthal A, Dork T, Bendix R, Skawran B, Stuhrmann M, Wegner RD, Sperling K, Banin S, et al. Characterization of ATM gene mutations in 66 ataxia telangiectasia families. Hum. Mol. Genet. 1999;8:69–79. doi: 10.1093/hmg/8.1.69. [DOI] [PubMed] [Google Scholar]
- 30.Teraoka SN, Telatar M, Becker-Catania S, Liang T, Onengut S, Tolun A, Chessa L, Sanal O, Bernatowska E, Gatti RA, et al. Splicing defects in the ataxia-telangiectasia gene, ATM: underlying mutations and consequences. Am. J. Hum. Genet. 1999;64:1617–1631. doi: 10.1086/302418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pagani F, Buratti E, Stuani C, Bendix R, Dork T, Baralle FE. A new type of mutation causes a splicing defect in ATM. Nat. Genet. 2002;30:426–429. doi: 10.1038/ng858. [DOI] [PubMed] [Google Scholar]
- 32.Lewandowska MA, Stuani C, Parvizpur A, Baralle FE, Pagani F. Functional studies on the ATM intronic splicing processing element. Nucleic Acids Res. 2005;33:4007–4015. doi: 10.1093/nar/gki710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buratti E, Dhir A, Lewandowska MA, Baralle FE. RNA structure is a key regulatory element in pathological ATM and CFTR pseudoexon inclusion events. Nucleic Acids Res. 2007;35:4369–4383. doi: 10.1093/nar/gkm447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goina E, Skoko N, Pagani F. Binding of DAZAP1 and hnRNPA1/A2 to an exonic splicing silencer in a natural BRCA1 exon 18 mutant. Mol. Cell Biol. 2008;28:3850–3860. doi: 10.1128/MCB.02253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pagani F, Stuani C, Tzetis M, Kanavakis E, Efthymiadou A, Doudounakis S, Casals T, Baralle FE. New type of disease causing mutations: the example of the composite exonic regulatory elements of splicing in CFTR exon 12. Hum. Mol. Genet. 2003;12:1111–1120. doi: 10.1093/hmg/ddg131. [DOI] [PubMed] [Google Scholar]
- 36.Sironi M, Menozzi G, Riva L, Cagliani R, Comi GP, Bresolin N, Giorda R, Pozzoli U. Silencer elements as possible inhibitors of pseudoexon splicing. Nucleic Acids Res. 2004;32:1783–1791. doi: 10.1093/nar/gkh341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun H, Chasin LA. Multiple splicing defect in an intronic false exon. Mol. Cell Biol. 2000;20:6414–6425. doi: 10.1128/mcb.20.17.6414-6425.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Labourier E, Adams MD, Rio DC. Modulation of P-Element pre-mRNA splicing by a direct interaction between PSI and U1 snRNP 70 K protein. Mol. Cell. 2001;8:363–373. doi: 10.1016/s1097-2765(01)00311-2. [DOI] [PubMed] [Google Scholar]
- 39.Ignjatovic T, Yang JC, Butler J, Neuhaus D, Nagai K. Structural basis of the interaction between P-element somatic inhibitor and U1-70 k essential for the alternative splicing of P-element transposase. J. Mol. Biol. 2005;351:52–65. doi: 10.1016/j.jmb.2005.04.077. [DOI] [PubMed] [Google Scholar]
- 40.Vilardell J, Chartrand P, Singer RH, Warner JR. The odyssey of a regulated transcript. RNA. 2000;6:1773–1780. doi: 10.1017/s135583820000145x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manabe T, Katayama T, Sato N, Gomi F, Hitomi J, Yanagita T, Kudo T, Honda A, Mori Y, Matsuzaki S, et al. Induced HMGA1a expression causes aberrant splicing of Presenilin-2 pre-mRNA in sporadic Alzheimer's; disease. Cell Death Differ. 2003;10:698–708. doi: 10.1038/sj.cdd.4401221. [DOI] [PubMed] [Google Scholar]
- 42.Manabe T, Ohe K, Katayama T, Matsuzaki S, Yanagita T, Okuda H, Bando Y, Imaizumi K, Reeves R, Tohyama M, et al. HMGA1a: sequence-specific RNA-binding factor causing sporadic Alzheimer's; disease-linked exon skipping of presenilin-2 pre-mRNA. Genes Cells. 2007;12:1179–1191. doi: 10.1111/j.1365-2443.2007.01123.x. [DOI] [PubMed] [Google Scholar]
- 43.Gunderson SI, Beyer K, Martin G, Keller W, Boelens WC, Mattaj LW. The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell. 1994;76:531–541. doi: 10.1016/0092-8674(94)90116-3. [DOI] [PubMed] [Google Scholar]
- 44.Guan F, Caratozzolo RM, Goraczniak R, Ho ES, Gunderson SI. A bipartite U1 site represses U1A expression by synergizing with PIE to inhibit nuclear polyadenylation. RNA. 2007;13:2129–2140. doi: 10.1261/rna.756707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Das R, Dufu K, Romney B, Feldt M, Elenko M, Reed R. Functional coupling of RNAP II transcription to spliceosome assembly. Genes Dev. 2006;20:1100–1109. doi: 10.1101/gad.1397406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghosh S, Garcia-Blanco MA. Coupled in-vitro synthesis and splicing of RNA polymerase II transcripts. RNA. 2000;6:1325–1334. doi: 10.1017/s1355838200992537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baralle M, Pastor T, Bussani E, Pagani F. Influence of Friedreich ataxia GAA noncoding repeat expansions on pre-mRNA processing. Am. J. Hum. Genet. 2008;83:77–88. doi: 10.1016/j.ajhg.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dember LM, Kim ND, Liu KQ, Anderson P. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J. Biol. Chem. 1996;271:2783–2788. doi: 10.1074/jbc.271.5.2783. [DOI] [PubMed] [Google Scholar]
- 49.Forch P, Puig O, Kedersha N, Martinez C, Granneman S, Seraphin B, Anderson P, Valcarcel J. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell. 2000;6:1089–1098. doi: 10.1016/s1097-2765(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 50.Zuccato E, Buratti E, Stuani C, Baralle FE, Pagani F. An intronic polypyrimidine-rich element downstream of the donor site modulates cystic fibrosis transmembrane conductance regulator exon 9 alternative splicing. J. Biol. Chem. 2004;279:16980–16988. doi: 10.1074/jbc.M313439200. [DOI] [PubMed] [Google Scholar]
- 51.Choi EY, Pintel D. Splicing of the large intron present in the nonstructural gene of minute virus of mice is governed by TIA-1/TIAR binding downstream of the nonconsensus donor. J. Virol. 2009;83:6306–6311. doi: 10.1128/JVI.00213-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y, Dipple KM, Vilain E, Huang BL, Finlayson G, Therrell BL, Worley K, Deininger P, McCabe ER. AluY insertion (IVS4-52ins316alu) in the glycerol kinase gene from an individual with benign glycerol kinase deficiency. Hum. Mutat. 2000;15:316–323. doi: 10.1002/(SICI)1098-1004(200004)15:4<316::AID-HUMU3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 53.Wallace MR, Andersen LB, Saulino AM, Gregory PE, Glover TW, Collins FS. A de novo Alu insertion results in neurofibromatosis type 1. Nature. 1991;353:864–866. doi: 10.1038/353864a0. [DOI] [PubMed] [Google Scholar]
- 54.Tighe PJ, Stevens SE, Dempsey S, Le Deist F, Rieux-Laucat F, Edgar JD. Inactivation of the Fas gene by Alu insertion: retrotransposition in an intron causing splicing variation and autoimmune lymphoproliferative syndrome. Genes Immun. 2002;3:S66–S70. doi: 10.1038/sj.gene.6363864. [DOI] [PubMed] [Google Scholar]
- 55.Oldridge M, Zackai EH, McDonald-McGinn DM, Iseki S, Morriss-Kay GM, Twigg SR, Johnson D, Wall SA, Jiang W, Theda C, et al. De novo alu-element insertions in FGFR2 identify a distinct pathological basis for Apert syndrome. Am. J. Hum. Genet. 1999;64:446–461. doi: 10.1086/302245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ganguly A, Dunbar T, Chen P, Godmilow L, Ganguly T. Exon skipping caused by an intronic insertion of a young Alu Yb9 element leads to severe hemophilia A. Hum. Genet. 2003;113:348–352. doi: 10.1007/s00439-003-0986-5. [DOI] [PubMed] [Google Scholar]
- 57.Ram O, Schwartz S, Ast G. Multifactorial interplay controls the splicing profile of Alu-derived exons. Mol. Cell Biol. 2008;28:3513–3525. doi: 10.1128/MCB.02279-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gal-Mark N, Schwartz S, Ast G. Alternative splicing of Alu exons—two arms are better than one. Nucleic Acids Res. 2008;36:2012–2023. doi: 10.1093/nar/gkn024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.