Abstract
Cellular retinoic acid binding protein 1 (Crabp1) gene is biphasically (proliferation versus differentiation) regulated by thyroid hormone (T3) in 3T3-L1 cells. This study examines T3-repression of Crabp1 gene during adipocyte differentiation. T3 repression of Crabp1 requires receptor interacting protein 140 (RIP140). During differentiation, the juxtaposed chromatin configuration of Crabp1 promoter with its upstream region is maintained, but the 6-nucleosomes spanning thyroid hormone response element to transcription initiation site slide bi-directionally, with the third nucleosome remaining at the same position throughout differentiation. On the basal promoter, RIP140 replaces coactivators GRIP1 and PCAF and forms a repressive complex with CtBP1, HDAC3 and G9a. Initially active chromatin marks on this promoter, histone modifications H3-Ac and H3K4-me3, are weakened whereas repressive chromatin marks, H3K9-me3 and H3K27-me3 modification and recruitment of G9a, HP1α, HP1γ and H1, are intensified. This is the first study to examine chromatin remodeling, during the phase of hormone repression, of a bi-directionally regulated hormone target gene, and provides evidence for a functional role of RIP140 in chromatin remodeling to repress hormone target gene expression.
INTRODUCTION
To understand hormone-regulated gene expression, a dogma has centered on the principle of gene activation by hormones that induce recruitment of coactivators to holo-nuclear receptors, and gene repression/silencing under conditions when hormones are absent and corepressors such as N-CoR (1), SMRT (2) and Alien (3) are recruited to apo-nuclear receptors (4,5). Since several hormone-dependent corepressors such as receptor interacting protein 140 (RIP140) (6), LCoR (7), PRAME (8) and REA (9) were reported, the notion of direct repression of genes by hormones and holo-receptors has begun to attract attention. This possibility was supported by studies of genes directly repressed by hormones, such as thyrotropin beta gene by thyroid hormones (T3/T4) (10,11), Oct4 gene by retinoic acid (RA) (12,13) and gonadotropin releasing hormone gene by estrogen (14,15), etc. However, it was less clear whether and how the same gene could be subjected to opposing (activating and repressing) regulations by the same hormone, and in what context this might occur.
The mouse cellular retinoic acid binding protein I (Crabp1) gene encodes a cytoplasmic protein that specifically binds RA to modulate intracellular free RA concentrations (16). Transcriptional regulation of Crabp1 involves numerous players such as RA, T3/T4, sphinganine and DNA methylation, etc. (17–26) as demonstrated in various cellular backgrounds. Of most relevance to the topic of hormonal regulation is the interesting response of this gene to T3/T4. In proliferative mouse embryonic fibroblast (MEF) and certain commonly used preadipocyte cell line models such as 3T3-L1 in the pre-differentiation stage, Crabp1 gene is activated by T3/T4 through holo-thyroid hormone receptors/retinoid receptors binding to a thyroid response element (TRE) located ∼1 kb upstream of its basal promoter that contains five GC boxes to which Sp1 can bind. Using this model system, we previously reported chromatin remodeling underlying T3 activation of Crabp1 gene in the pre-differentiation stage of these cells. This occurred through chromatin juxtaposition between TRE and GC boxes, an event requiring MED1/TRAP220-containing Mediator complex, downward sliding of the nucleosome array and disassembly of the nucleosome covering the transcription initiation site (TIS) (24). The current study was designed to demonstrate, using the T3-biphasically regulated Crabp1 as an example, chromatin remodeling in the hormone-repressive phase.
Studies of the action of transcription factors, their coregulators, Mediators, and specific chromatin remodelers, as well as chromatin remodeling of endogenous genes have been extensively conducted, mostly in model organisms such as fly and yeast (27–29). Trans-acting regulatory factors for mammalian genes have also been examined (30,31). More recently, gene activation or inhibition has been shown to involve epigenetic alterations (32,33). With respect to chromatin remodeling of mammalian hormone target genes, studies have examined hormonal activation and repression (24,34), but mechanism by which chromatin remodeling occurred on the same gene that could be activated and repressed by the same hormone is not clearly established. This current study provides evidence for potential opposing effects of certain hormones in both activating and repressing the same target genes in different physiological contexts, and establishes the physiological role for RIP140 in T3 repression of Crabp1 gene during adipocyte differentiation.
MATERIALS AND METHODS
Cell culture, silencing of RIP140 and luciferase reporter assay are described in Supplementary Data.
Reverse transcriptase polymerase chain reaction, immunoprecipitation and western blot analyses
Reverse transcriptase polymerase chain reaction (RT–PCR), immunoprecipitation (IP) and western blot (WB) assays were performed as described (24). Gene-specific primer sequences are in Supplementary Table S1. Two hundred micrograms of whole cell extracts were subjected to IP with the indicated antibodies, and the precipitated protein complex was analyzed by WB.
ChIP and repeated ChIP assays
Antibody sources are described in Supplementary data. Chromatin immunoprecipitation (ChIP) assays were performed as described (24). For repeated ChIP assays (ReChIP), immunoprecipitated complex was eluted with 10 mM dithiothreitol, diluted in 20 volumes of ReChIP dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl and 20 mM Tris–HCl, pH 8.1), and subjected to ChIP procedures. For PCR amplification, DNA precipitated by glucocorticoid receptor binding protein 1 (GRIP1) and p300/CBP-associated factor (PCAF) antibodies was amplified for 32 cycles, and others for 30 cycles. The captured DNA fragments were amplified by using primer sets for the TRE and GC box regions (Supplementary Table S2).
MNase nucleosome mapping and restriction enzyme accessibility assays
Micrococcal nuclease (MNase) digestion and ligation-mediated PCR (LM-PCR) were performed as described (24). Nuclei isolated from differentiating 3T3-L1 cells were digested with MNase (Worthington) for 5 min at 37°C followed by proteinase K treatment at 37°C overnight. The purified DNA was subjected to Southern blot analysis.
Restriction enzyme accessibility assay was carried out as described (24). Isolated nuclei from differentiating 3T3-L1 cells were digested with 100 U of PstI, XhoI, SmaI, SpeI and ApaLI (New England Biolabs) for 30 min. The purified genomic DNA was re-digested with 100 U of ApaI completely (for first digestion with XhoI, PstI and SmaI) or 100 U of PstI (for first digestion with SpeI and ApaLI). The digested fragments were analyzed by Southern blot using 32P-labeled probe 1 (for SmaI digestion) or 2 (for XhoI, PstI, SpeI and ApaLI digestion).
LM-PCR
Nuclei were digested with 45 U of MNase (Worthington) for 30 min at 37°C followed by proteinase K treatment overnight (24). Mononucleosomal DNAs (∼150 bp) were recovered from agarose gels and the purified fragments (1 µg) were phosphorylated at 5′-termini and ligated to the universal linker adaptor with T4 polynucleotide kinase and T4 DNA ligase (24). Purified DNA was amplified by PCR using the universal linker oligonucleotide (25 bp) and 32P-labeled Crabp1-specific primers: for N5, 5′-CCG AGG AAA GTA ATC TGC TTA GGA CCT AAA C-3′; N3, 5′-AAT TAG AGT GGC GGG AAA GGC CCA GCC C-3′; N-1, 5′-AAT TCT CGC TGC TGC GCA TCT TCC AGG TAC-3′.
RESULTS
RIP140 is required for T3-repression of Crabp1 in differentiating/differentiated adipocyte cultures
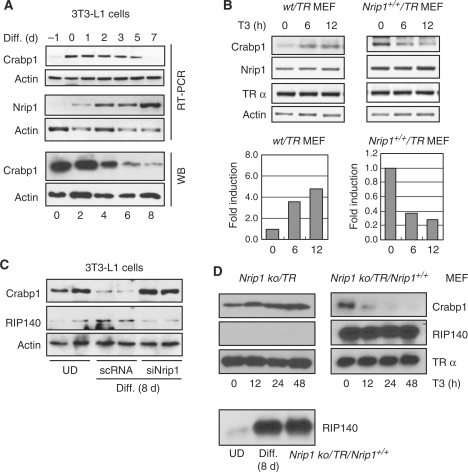
The behavior of the endogenous Crabp1 gene and the expression of endogenous RIP140 during the course of 3T3-L1-adipocyte differentiation (with insulin, dexamethasone, isobutylmethylxanthine and T3) assessed by oil red O staining (Supplementary Figure S1) were monitored (Figure 1). Crabp1 gene expression, initially very weak in subconfluent cultures (panel B) (24), was upregulated in pre-differentiation cultures but was gradually repressed at both mRNA and protein levels during differentiation (panel A). As speculated, the expression of the endogenous Nrip1 gene (encoding corepressor RIP140) was gradually elevated and peaked on the final day (7 days) of examination (panel A, middle) (35).
Figure 1.
Crabp1 is down regulated during adipocyte differentiation, which requires RIP140. (A) Expression patterns of Crabp1 and Nrip1 (RIP140) during differentiation of 3T3-L1 cells were monitored by RT-PCR and Crabp1 by WB analyses. (B) Expression patterns of Crabp1, Nrip1 and TRα in stable MEF cell lines transfected with hTRα alone (wt/TR) or RIP140 and hTRα (Nrip1+/+/TR) in the presence of T3, were monitored by RT-PCR. Fold inductions of Crabp1 were plotted after normalizing its levels with the levels of actin (lower panels). Nrip1 expression was quantified by qPCR (Supplementary Figure S2) (C) Protein expression of Crabp1 and RIP140 in 3T3-L1 cells transfected with scrambled RNA or siRNA of RIP140 (Nrip1) and then differentiated for 8 days. UD,undifferentiated cells. Duplicated sets of data are shown. (D) Protein expression of Crabp1, RIP140 and TRα in the RIP140-deficient MEF line (Nrip1 ko/TR) and the line that were reconstituted with constitutively expressed hTRα and RIP140 (Nrip1 ko/TR/Nrip1+/+), without differentiation, was monitored on WB for up to 48 h in the presence of T3. Relative protein expression level of RIP140 before and during 3T3-L1 differentiation and Nrip1 ko/TR/Nrip1+/+ MEFs is shown at the bottom.
RIP140 was known to be associated with TR in a ligand-dependent manner (23). Gain- and loss-of-function experiments were conducted in MEF and 3T3-L1 cells to determine the functional role of RIP140. As shown in Figure 1B, forced expression [9–10-folds higher, according to qPCR result (Supplementary Figure S2)] of RIP140 in pre-differentiating MEF cells rendered endogenous Crabp1 rapidly repressed by T3 without differentiation (Figure 1B, right). But in wt/TR pre-differentiating cells, Crabp1 was activated by T3 (Figure 1B, left). According to siRNA-mediated knockdown of RIP140 in differentiated cells (Figure 1C), it was clear that Crabp1 was no longer repressed by T3 when cells were depleted of RIP140. In fact, Crabp1 expression level was even higher than that detected in undifferentiated cells (UD). In RIP140-knockout MEF (Nrip1 ko/TR), Crabp1 was effectively induced by T3; in knockout cells rescued with a constitutive RIP140-expression vector (Nrip1 ko/TR/Nrip1+/+) (Figure 1D, left), Crabp1 was very effectively repressed by T3 even in the pre-differentiating cells (Figure 1D, right). The expression level of RIP140 in reconstituted MEFs was similar to that in differentiated 3T3-L1 cells (Figure 1D, lower).
These results unambiguously show that RIP140 plays a functional role in mediating T3 repression of Crabp1 gene in differentiating cells, consistent with its gradual elevation during differentiation.
Maintenance of juxtaposed chromatin on Crabp1 promoter
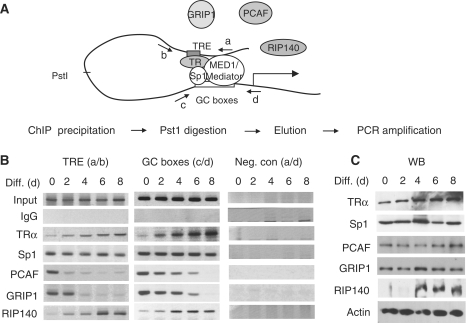
In proliferating cells (before the differentiation cocktail is added), the chromatin segment spanning the TRE to TIS region of Crabp1 promoter adopts a juxtaposed, or folded, conformation in the presence of T3 (24). The question was whether this chromatin segment remained folded in differentiating cells when Crabp1 expression was gradually repressed, by employing reciprocal chromatin immunoprecipitation (ChIP) analyses of disconnected chromatin fragments as described (24). The procedure would monitor the simultaneous occupancy of TRE and GC box regions by both TRα and Sp1 (Figure 2A), which would indicate chromatin folding/juxtaposition. PstI digestion would disconnect TRE region from the GC box region and remove contiguous fragments. As shown in Figure 2B, TRα, weakly detected on the TRE (amplified by primers a and b) and GC box (amplified by primers c and d) regions in UD, began to be highly detected on both regions around day 2, and even more so in later time points, whereas Sp1 constantly occupied both regions. Negative controls showed no amplified products (amplified by primers a and d), ruling out DNA contamination (Figure 2B, right panels). Figure 2C showed the expression levels of relevant endogenous components. The expression of both TRα and RIP140 was gradually, but quite obviously, increased as cells were differentiating. All together, the results show that TRE still makes contact with the GC box region during differentiation, i.e. chromatin remains juxtaposed or folded. For this promoter, TRα and Sp1, the primary transacting factors binding to their cognitive elements on Crabp1 gene regulatory region (24), likely continue to provide the platform to recruit RIP140 and other factors (see following) in differentiating cells, thereby contributing to the maintenance of the juxtaposed configuration of this chromatin segment during differentiation.
Figure 2.
Chromatin juxtaposition between TRE and GC box regions of Crabp1 promoter and associated protein complexes. (A) Schematic depiction of Crabp1 promoter/regulatory region harboring the TRE and GC boxes, to which TRα and Sp1 bind, respectively (23,24). Primers for PCR amplification were marked with arrows and the Pst1 site to disconnect TRE from GC boxes was shown. The previously developed procedure for ChIP analysis of chromatin juxtaposition is shown under the figure. (B) ChIP data. Protein/chromatin fragments were precipitated with the indicated antibodies and DNAs were digested with PstI (20 U, 30 min) and washed before the elution of captured chromatin fragments. DNA fragments were amplified with primer set a/b for TRE, c/d for GC boxes, and a/d for the negative control. The occupancy of TRα, PCAF and GRIP1 on both regions was quantitatively monitored by qPCR (Supplementary Figure S3) (C) The expression levels of the relevant endogenous proteins during adipocyte differentiation were analyzed on WBs.
Consistent with the Crabp1 expression pattern, coactivators GRIP1 and PCAF were detected at both regions on Day 0, but began to leave this chromatin segment around Day 2 and almost completely disappeared from both regions on Day 4. qPCR result (Supplementary Figure S3) confirmed the occupancy of TRα on both regions, presumably as a platform to recruit PCAF and GRIP1 on Day 0, and RIP140 later in differentiation. Thus, RIP140 recruitment to this chromatin was almost entirely parallel to that of TRα. These results agree with the notion of coactivators being replaced by corepressor RIP140 during the process of gene repression.
Recruitment of RIP140 to form a repressive module on Crabp1 promoter
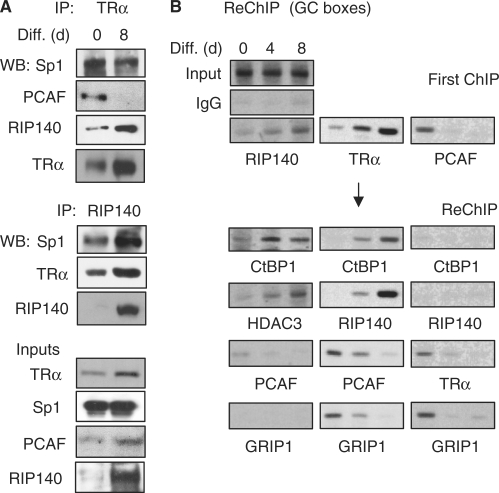
We examined the endogenous protein complexes in co-immunoprecipitation tests to determine the effects of T3 on the endogenous TRα/coregulator complex formation (Figure 3A). On Day 0, TRα was primarily associated with coactivator PCAF; but on Day 8, it was preferentially associated with RIP140. Throughout differentiation, TRα remained associated with Sp1. This was further supported in the reciprocal co-immunoprecipitation test with anti-RIP140, which co-precipitated more abundant Sp1 and TRα on day 8 (with a high RIP140 level).
Figure 3.
Replacement of the coactivator complex with the RIP140 repressive complex on Crabp1 promoter during adipocyte differentiation. (A) Endogenous complexes detected by co-immunoprecipitation. Nuclear extracts were immunoprecipitated with antibodies against TRα or RIP140 and the precipitated proteins were detected with antibodies indicated on the side. Input shows the endogenous levels of proteins in undifferentiated (0 day) and differentiated cells (8 days). (B) ReChIP to monitor protein complexes on the basal promoter region. First ChIP was performed with RIP140, TRα or PCAF antibodies (First ChIP), followed by ReChIP with antibodies indicated under the panels. 3T3-L1 cells differentiated at different time points (0, 4 and 8 days) were examined.
We then carried out ReChIP assays to monitor molecular complexes recruited to the GC box region (which would contribute to the promoter activity), with PstI digestion to disconnect the TRE from the GC box region (Figure 3B). Three sets of ReChIP were conducted to examine complexes containing RIP140 (corepressor), PCAF (coactivator) and TRα (for T3 response). Apparently, RIP140 was increasingly recruited to this region in more differentiated cells (first ChIP). ReChIP showed other repressive components such as CtBP1 and HDAC3, but not the activating components such as GRIP1 and PCAF, increasingly associated with RIP140 on this promoter (left panels). Consistently, first ChIP with anti-PCAF showed its strong association with this promoter and its forming complex with TRα and GRIP1 only on Day 0 (right panels). The first ChIP with TRα showed its increased association with this promoter (because of its elevated expression later in differentiation, Figure 2C), and ReChIP confirmed increased association of repressive components such as RIP140 and CtBP1 with, and decreased association of active components such as PCAF and GRIP1 from, TRα on this promoter (middle panels). These results support that, on the GC box region, coactivating components are replaced by corepressive components as cells undergo differentiation. One important player contributing to the switch of the coregulatory complexes on this promoter appears to be RIP140 that is highly elevated in more differentiated cells.
Bi-directional nucleosome sliding on Crabp1 promoter
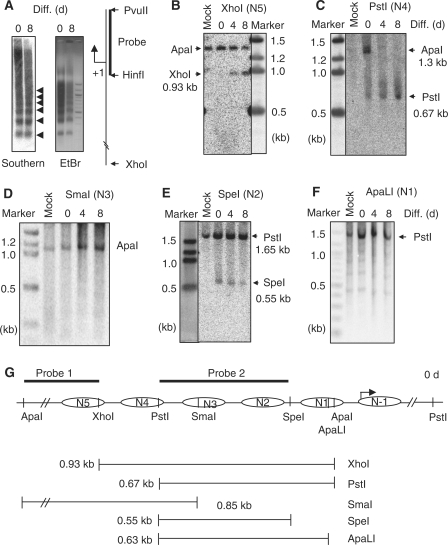
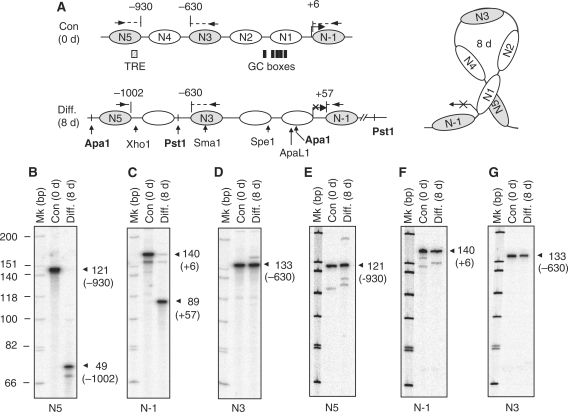
The issue now was if and how this segment of chromatin might be remodeled in differentiating cells where the gene would be repressed. MNase digestion-coupled Southern blot was conducted using the probe covering TIS (Figure 4A). The experiment detected a regular nucleosome array on the promoter/regulatory region of Crabp1 gene (left, marked by arrowheads), both before and after differentiation, suggesting that nucleosomes were likely to be continuously present on this region during differentiation. The juxtaposed chromatin segment covers five nucleosomes and one additional nucleosome adjacent to the TIS (Figures 4G and 5A). We then observed if these nucleosomes changed their positions during the course of differentiation, first by using restriction enzyme accessibility assay to examine nucleosomes that could be accessed by specific restriction enzymes. The 5′-terminal nucleosome (N5) in this segment was monitored by XhoI following ApaI digestion, with 0.93 kb (ApaI/XhoI) fragment indicating accessible XhoI site. Figure 4B showed the detection of this fragment only after differentiation (Days 4 and 8), indicating gradual opening of this region during differentiation. Since the nucleosome array is retained before and after differentiation, this result would suggest either upward or downward sliding of N5, which was examined later (see later Figure 5B and Supplementary Figures S4 and S5).
Figure 4.
Alterations of restriction enzyme accessibility in the juxtaposed region of Crabp1 promoter along the course of adipocyte differentiation. (A) Detection of nucleosome array. Chromatin of MNase partially digested nuclei was separated on agarose gels followed by Southern blot analysis (left panel) and EtBr staining (right panel). A probe used covering the TIS (HinfI-PvuII) was used for Southern blot analysis. (B–F) Nuclei (differentiation Days 0, 4, 8) were digested with the indicated restriction enzymes and the recovered chromatin DNAs were completely re-digested with ApaI (B–D) or PstI (E and F) (the relationship of specific restriction site and each individual nucleosome was depicted in panel G), followed by Southern blot analyses using probe 1 (D) or probe 2 (B–C and E–F). (G) Schematic description of restriction enzyme digestion, nucleosomes, and predicted fragments detected with probes on the Southern blots. Complete digestion with ApaI produced 1.3 kb, and complete digestion with PstI produced 1.65 kb fragments. The generated fragments by digestion with XhoI, PstI, SmaI, SpeI and ApaLI for the corresponding nucleosome are 0.93, 0.67, 0.86, 0.55 and 0.65 kb, respectively. Nucleosomes on this array were named N5 to N-1, from the 5′- to the 3′-ends of this chromatin segment. Experiments were performed for least three times.
Figure 5.
Bi-directional nucleosome sliding during differentiation assessed by LM-PCR. (A) The comparison of nucleosome locations before (0 day) and after (8 days) differentiation (left panel). Primers used in LM-PCR for nucleosomes N5, N3 and N-1 are shown above these nucleosomes. Primer and nucleosome border positions are determined relative to TIS. The positions of TRE and GC boxes are marked as shaded and closed boxes, respectively, below the nucleosome map. The juxtaposed chromatin and nucleosome arrangement on this segment are depicted on the right. LM-PCR data in 3T3-L1 cells (B–D) and Nrip ko/TR MEFs (E–G). Mononucleosomal DNA fragments were ligated with the universal linker followed by amplification with the linker and the labeled specific primers for N5 (B and E), N-1 (C and F) and N3 (D and G). Reproducible results were obtained from at least two independent experiments.
With the same principle, N4 opening or sliding away would be predicted by the appearance of 0.67 kb ApaI/PstI fragment. Figure 4C showed increasing accessibility of this site in more differentiated cells, suggesting sliding of N4. N3 covers a SmaI site, and the lack of 0.85 kb ApaI/SmaI fragment throughout differentiation (Figure 4D) would rule out movement, or opening, of N3 during differentiation. No restriction site is located in N2; therefore, we monitored an initially accessible SpeI site in the immediate 3′-flanking region of N2 (indicated by the appearance of 0.55 kb PstI/SpeI fragment, Figure 4E), which became less accessible in differentiating cells. This suggested that this site was protected by either downward sliding of N2 or upward sliding of N1 during differentiation. As the site is located immediately outside the 3′-end of N2, we suspected its protection by downward sliding of N2, which was supported by data of a nucleosome scanning experiment (Supplementary Figure S5). N1 spans one ApaLI site which remained closed (lacking 0.63 kb ApaLI/PstI fragment, Figure 4F). This series of restriction enzyme accessibility assays support the MNase data (Figure 4A) and suggest nucleosome moving during the course of differentiation. To provide more support, we evaluated the accessibility of enzyme sites by monitoring fragment spanning these sites using PCR amplification of digested chromatin DNA collected at different time points of differentiation (Supplementary Figure S4), which are in agreement with the notion of nucleosome sliding.
To determine the precise positioning of nucleosomes and the direction of their moving, we then employed LM-PCR to map several key nucleosomes. The nucleosome-specific primers are shown in Figure 5A, marked by arrows depicted on the nucleosomes. We first determined the positions of two terminal nucleosomes, N5 and N-1. Figure 5B showed that the 3′-border of N5 moved from −930 to −1002 positions (relative to TIS), indicating its upward sliding for 72 nucleotides. Interestingly, N-1 moved from the positions of +6 to +57, indicating its downward sliding for 51 bp (Figure 5C). More interestingly, the center nucleosome (N3) did not move, because its 5′-border remained at −630 both before and after differentiation (Figure 5D). Importantly, LM-PCR data of RIP140-deficient MEF showed no shift of nucleosomes N5 (Figure 5E), N-1 (Figure 5F) and N3 (Figure 5G) during differentiation. Taken together, these results confirm that, before and after differentiation, the nucleosomes are maintained on this gene promoter and its regulatory region, and that two terminal nucleosomes move away from the central nucleosome that stays at the same position before and after differentiation. This requires RIP140. Further evidence for the bi-directional nucleosome sliding during the course of differentiation was obtained from a less laborious PCR-based nucleosome scanning (Supplementary Figure S5).
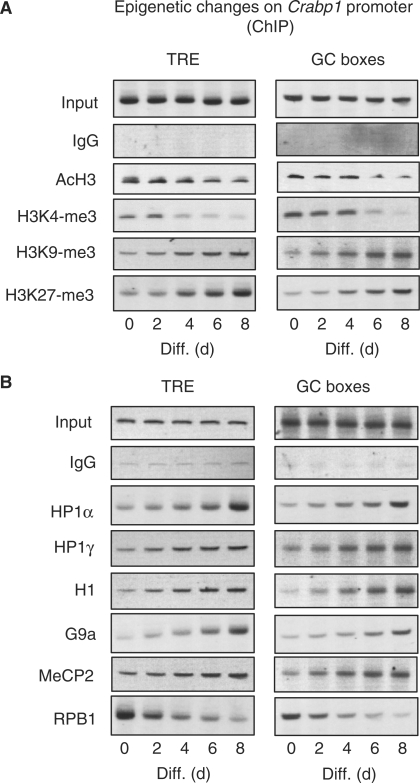
Changes of several potential epigenetic marks on Crabp1 promoter
RIP140 is known to recruit repressive cofactors such as HDACs (36) and CtBP (37). According to our data, we predicted progressive formation of repressive chromatin on the Crabp1 gene promoter in differentiating/differentiated cells. We conducted ChIP analyses of TRE and GC box regions along the course of 8-days differentiation (Figure 6). Figure 6A showed decrease in acetylation of histone H3 (AcH3) and lysine methylation on histone H3K4 on these regions, with corresponding increase in several repressive chromatin marks such as methylation at Lys9 and Lys27 of histone H3 (38). H3K9-me3 was known to recruit heterochromatin proteins and H3K27-me3 was found enriched in facultative heterochromatin regions. We then examined possible recruitment of several known heterochromatin markers on these regions (Figure 6B). Indeed, heterochromatin proteins 1α (HP1α) and 1γ (HP1γ) and histone H1 (H1) were increasingly recruited to this promoter, and so was G9a, the H3K9 methyltransferase. The promoter of this gene is rich in clustered CpG islands and its repression is known to be related to the level of cytosine-methylation (39). Consistently, MeCP2 was increasingly detected on both TRE and GC box regions and RNA polymerase II (RPB1) gradually disappeared from this promoter in more differentiated cultures.
Figure 6.
Epigenetic changes on TRE and GC box regions of Crabp1 promoter. ChIP analysis was performed with antibodies against several epigenetic markers (A), and heterochromatin proteins and proteins involved in DNA methylation (B).
DISCUSSION
The initial studies of hormonal regulation of gene expression have focused on target gene activation by hormones. In the past decade, gene repression by hormones has been increasingly reported (10,11,19,40). But it was less clear if the same gene could be subjected to the opposing effects of the same hormone, and in what context this might occur. We have observed that RA could activate and then repress TR2 gene through coregulators’ competition in the 3T3-L1 model (41). The current study reports mechanistic details of biphasic regulation of Crabp1 gene, primarily at the level of transcription through hormone-induced chromatin remodeling, and reveals a physiological role for RIP140 in the repressive phase of T3 regulation of this gene. However, we do not rule out potential effects of differentiation on other regulatory events such as controlling the stability of Crabp1 mRNA or its protein. Since RIP140, unlike N-CoR and SMRT (1,2), is a known ligand-dependent coregulator that renders gene repression in the presence of hormones (6,23,41,42), we propose RIP140 as a counteracting molecule, with respect to hormone-dependent coactivators, to modulate hormonal signals for proper control of certain hormone-sensitive genes. To this end, it is important that RIP140 is highly elevated only in differentiating/differentiated cells. In these cells, it could stoichiometrically compete with coactivators such as PCAF and GRIP1 for interaction with TRα (Figures 1–3).
This study also provides some mechanistic insights into chromatin remodeling during the hormone-repressed phase, and possible players for, and specific changes occurred on, the increasingly repressed chromatin of this promoter during cell differentiation. However, it remains to be tested if this can be generalized for many other hormonal target genes in different experimental systems. As to the specific physiological needs for biphasic regulation of Crabp1, more studies are needed to determine whether and why shutting down Crabp1 would be important for differentiating or differentiated adipocytes. To this end, Crabp1 is known to titrate intracellular RA concentrations, which might be crucial to the differentiation process of adipocyte. It is also interesting that Crabp1 gene has been implicated as a tumor suppressor in esophageal squamous-cell carcinoma (43).
Chromatin juxtaposition/folding on the Crabp1 promoter and regulatory region and the nucleosome array on this chromatin are maintained throughout the differentiation process, despite the dramatically changed expression of Crabp1 gene. MED1/Mediator complex mediates chromatin juxtaposition by serving as a nexus for TRα and Sp1 during the phase of T3-activation of Crabp1 gene in undifferentiated MEFs (24). As cells undergo differentiation, the increasingly recruited RIP140 would maintain this chromatin feature. In addition, nucleosomes slide bi-directionally in this repressive phase, anchored by a central nucleosome within the juxtaposed segment. It is tempting to speculate that this bidirectional sliding might occur as a result of, or contribute to, the formation of a more compacted chromatin configuration on this region in differentiated cells. Probably, longer linker regions, as a result of nucleosome sliding away from the center, would, at least partially, release tension introduced into the much tighter chromatin and to maintain the folded/juxtaposed configuration. LM-PCR data (Figure 5) of wild type and RIP140-deficient cells confirm RIP140-requiring bidirectional nucleosome sliding during the process of differentiation. Increase in putative heterochromatin marks and recruitment of certain predicted heterochromatin enzyme machineries on this region would suggest that this region becomes heterochromatinized after differentiation. It would be important to determine the driving force for sliding of nucleosomes and compacting of chromatin.
It has been shown that euchromatin and compacted heterochromatin are distinguished by several histone signatures and the recruitment of HP1. Transcriptionally active genes are generally associated with hyperacetylation at Lys residues of N-terminal tails of histones H3 and H4. In addition, genome-wide ChIP-on-Chip experiments in yeast have shown that H3K4-me of euchromatin and H3K9-me of heterochromatin are mutually exclusive (44). H3K9-me could provide a recognition site for HP1 binding and propagate heterochromatin locally or genome-wide. In this process, histone methyltranferases such as Suv39h (45,46) and G9a (47) are key regulators for H3K9-me. Furthermore, methylation of cytosine residues of CpG islands, which is facilitated by DNA methyltranserases following H3K27-me, is a common feature of most heterochromatin (48). Changes in histone modification on Crabp1 promoter would suggest that RIP140 could probably contribute to the possible heterochromatinization on the Crabp1 promoter, as indicated by changes in the signatures of AcH3, H3K9-me3, H3K27-me3, HP1α, HP1γ and histone H1 on this promoter during differentiation (Figure 6). RIP140 is known to be present in certain complexes containing histone-modifying enzymes such as arginine and lysine methyltransferases (49,50), which could methylate histone H4 on Arg3 (51). RIP140 is also extensively modified post-translationally (52), and acetylation at lysine residues by Erk2-mediated phosphorylation has been shown to increase its repressive activity (53). Further studies are required to decipher how might other post-translational modifications of RIP140 affect its gene-repressive activity, and whether and how RIP140 indeed plays a specific role in the formation of heterochromatin.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health grants DA11190, DA11806, DK54733, DK60521, K02-DA13926 to L.-N.W. Funding for open access charge: XXX.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank M. Parker for RIP140 null MEF. They also thank Dr R. G. Roeder for discussion of this study.
REFERENCES
- 1.Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 2.Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 3.Dressel U, Thormeyer D, Altincicek B, Paululat A, Eggert M, Schneider S, Tenbaum SP, Renkawitz R, Baniahmad A. Alien, a highly conserved protein with characteristics of a corepressor for members of the nuclear hormone receptor superfamily. Mol. Cell Biol. 1999;19:3383–3394. doi: 10.1128/mcb.19.5.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 5.Panning B, Taatjes DJ. Transcriptional regulation: it takes a village. Mol. Cell. 2008;31:622–629. doi: 10.1016/j.molcel.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavailles V, Dauvois S, L’Horset F, Lopez G, Hoare S, Kushner PJ, Parker MG. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandes I, Bastien Y, Wai T, Nygard K, Lin R, Cormier O, Lee HS, Eng F, Bertos NR, Pelletier N, et al. Ligand-dependent nuclear receptor corepressor LCoR functions by histone deacetylase-dependent and -independent mechanisms. Mol. Cell. 2003;11:139–150. doi: 10.1016/s1097-2765(03)00014-5. [DOI] [PubMed] [Google Scholar]
- 8.Epping MT, Wang L, Edel MJ, Carlee L, Hernandez M, Bernards R. The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell. 2005;122:835–847. doi: 10.1016/j.cell.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 9.Delage-Mourroux R, Martini PG, Choi I, Kraichely DM, Hoeksema J, Katzenellenbogen BS. Analysis of estrogen receptor interaction with a repressor of estrogen receptor activity (REA) and the regulation of estrogen receptor transcriptional activity by REA. J. Biol. Chem. 2000;275:35848–35856. doi: 10.1074/jbc.M001327200. [DOI] [PubMed] [Google Scholar]
- 10.Sasaki S, Lesoon-Wood LA, Dey A, Kuwata T, Weintraub BD, Humphrey G, Yang WM, Seto E, Yen PM, Howard BH, et al. Ligand-induced recruitment of a histone deacetylase in the negative-feedback regulation of the thyrotropin beta gene. EMBO J. 1999;18:5389–5398. doi: 10.1093/emboj/18.19.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weitzel JM. To bind or not to bind—how to down-regulate target genes by liganded thyroid hormone receptor? Thyroid Res. 2008;1:4. doi: 10.1186/1756-6614-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schoorlemmer J, van Puijenbroek A, van Den Eijnden M, Jonk L, Pals C, Kruijer W. Characterization of a negative retinoic acid response element in the murine Oct4 promoter. Mol. Cell Biol. 1994;14:1122–1136. doi: 10.1128/mcb.14.2.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sylvester I, Schöler HR. Regulation of the Oct-4 gene by nuclear receptors. Nucleic Acids Res. 1994;22:901–911. doi: 10.1093/nar/22.6.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Z, Zheng H, Dong KW. Identification of negative and positive estrogen response elements in human GnRH upstream promoter in the placental JEG-3 cells. Mol. Cell Endocrinol. 2001;184:125–134. doi: 10.1016/s0303-7207(01)00612-8. [DOI] [PubMed] [Google Scholar]
- 15.Ng Y, Wolfe A, Novaira HJ, Radovick S. Estrogen regulation of gene expression in GnRH neurons. Mol. Cell Endocrinol. 2009;303:25–33. doi: 10.1016/j.mce.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei LN, Chang L, Hu X. Studies of the type I cellular retinoic acid-binding protein mutants and their biological activities. Mol. Cell Biochem. 1999;200:69–76. doi: 10.1023/a:1006906415388. [DOI] [PubMed] [Google Scholar]
- 17.Wei LN, Blaner WS, Goodman DS, Nguyen-Huu MC. Regulation of the cellular retinoid-binding proteins and their messenger ribonucleic acids during P19 embryonal carcinoma cell differentiation induced by retinoic acid. Mol. Endocrinol. 1989;3:454–463. doi: 10.1210/mend-3-3-454. [DOI] [PubMed] [Google Scholar]
- 18.Wei LN, Lee CH, Chang L. Retinoic acid induction of mouse cellular retinoic acid-binding protein-I gene expression is enhanced by sphinganine. Mol. Cell Endocrinol. 1995;111:207–211. doi: 10.1016/0303-7207(95)03570-w. [DOI] [PubMed] [Google Scholar]
- 19.Wei LN, Lee CH, Filipcik P, Chang L. Regulation of the mouse cellular retinoic acid-binding protein-I gene by thyroid hormone and retinoids in transgenic mouse embryos and P19 cells. J. Endocrinol. 1997;155:35–46. doi: 10.1677/joe.0.1550035. [DOI] [PubMed] [Google Scholar]
- 20.Wei LN, Chang L. Promoter and upstream regulatory activities of the mouse cellular retinoic acid-binding protein-I gene. J. Biol. Chem. 1996;271:5073–5078. doi: 10.1074/jbc.271.9.5073. [DOI] [PubMed] [Google Scholar]
- 21.Chang L, Wei LN. Characterization of a negative response DNA element in the upstream region of the cellular retinoic acid-binding protein-I gene of the mouse. J. Biol. Chem. 1997;272:10144–10150. doi: 10.1074/jbc.272.15.10144. [DOI] [PubMed] [Google Scholar]
- 22.Bi J, Hu X, Zhou FC, Wei LN. Upregulation of cellular retinoic acid-binding protein I expression by ethanol. Dev. Growth Differ. 2001;43:553–561. doi: 10.1046/j.1440-169x.2001.00591.x. [DOI] [PubMed] [Google Scholar]
- 23.Wei LN, Hu X. Receptor interacting protein 140 as a thyroid hormone-dependent, negative co-regulator for the induction of cellular retinoic acid binding protein I gene. Mol. Cell Endocrinol. 2004;218:39–48. doi: 10.1016/j.mce.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 24.Park SW, Li G, Lin YP, Barrero MJ, Ge K, Roeder RG, Wei LN. Thyroid hormone-induced juxtaposition of regulatory elements/factors and chromatin remodeling of Crabp1 dependent on MED1/TRAP220. Mol. Cell. 2005;19:643–653. doi: 10.1016/j.molcel.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 25.Collins CA, Watt FM. Dynamic regulation of retinoic acid-binding proteins in developing, adult and neoplastic skin reveals roles for beta-catenin and Notch signalling. Dev. Biol. 2008;324:55–67. doi: 10.1016/j.ydbio.2008.08.034. [DOI] [PubMed] [Google Scholar]
- 26.Lane MA, Xu J, Wilen EW, Sylvester R, Derguini F, Gudas LJ. LIF removal increases CRABPI and CRABPII transcripts in embryonic stem cells cultured in retinol or 4-oxoretinol. Mol. Cell Endocrinol. 2008;280:63–74. doi: 10.1016/j.mce.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King-Jones K, Thummel CS. Nuclear receptors—a perspective from Drosophila. Nat. Rev. Genet. 2005;6:311–323. doi: 10.1038/nrg1581. [DOI] [PubMed] [Google Scholar]
- 28.Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol. Cell. 2006;24:735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 29.Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- 30.Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 31.Lonard DM, Lanz RB, O’Malley BW. Nuclear receptor coregulators and human disease. Endocr. Rev. 2007;28:575–587. doi: 10.1210/er.2007-0012. [DOI] [PubMed] [Google Scholar]
- 32.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 33.Mohn F, Schübeler D. Genetics and epigenetics: stability and plasticity during cellular differentiation. Trends Genet. 2009;25:129–136. doi: 10.1016/j.tig.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 34.Kobrossy L, Rastegar M, Featherstone M. Interplay between chromatin and trans-acting factors regulating the Hoxd4 promoter during neural differentiation. J. Biol. Chem. 2006;281:25926–25939. doi: 10.1074/jbc.M602555200. [DOI] [PubMed] [Google Scholar]
- 35.Leonardsson G, Steel JH, Christian M, Pocock V, Milligan S, Bell J, So PW, Medina-Gomez G, Vidal-Puig A, White R, et al. Nuclear receptor corepressor RIP140 regulates fat accumulation. Proc. Natl Acad. Sci. USA. 2004;101:8437–8442. doi: 10.1073/pnas.0401013101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei LN, Hu X, Chandra D, Seto E, Farooqui M. Receptor-interacting protein 140 directly recruits histone deacetylases for gene silencing. J. Biol. Chem. 2000;275:40782–40787. doi: 10.1074/jbc.M004821200. [DOI] [PubMed] [Google Scholar]
- 37.Castet A, Boulahtouf A, Versini G, Bonnet S, Augereau P, Vignon F, Khochbin S, Jalaguier S, Cavaillès V. Multiple domains of the Receptor-Interacting Protein 140 contribute to transcription inhibition. Nucleic Acids Res. 2004;32:1957–1966. doi: 10.1093/nar/gkh524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sims RJ, Reinberg D. Is there a code embedded in proteins that is based on post-translational modifications? Nat. Rev. Mol. Cell Biol. 2008;9:815–820. doi: 10.1038/nrm2502. [DOI] [PubMed] [Google Scholar]
- 39.Wei LN, Lee CH. Demethylation in the 5′-flanking region of mouse cellular retinoic acid binding protein-I gene is associated with its high level of expression in mouse embryos and facilitates its induction by retinoic acid in P19 embryonal carcinoma cells. Dev. Dyn. 1994;201:1–10. doi: 10.1002/aja.1002010102. [DOI] [PubMed] [Google Scholar]
- 40.Bretschneider N, Brand H, Miller N, Lowery AJ, Kerin MJ, Gannon F, Denger S. Estrogen induces repression of the breast cancer and salivary gland expression gene in an estrogen receptor alpha-dependent manner. Cancer Res. 2008;68:106–114. doi: 10.1158/0008-5472.CAN-07-5647. [DOI] [PubMed] [Google Scholar]
- 41.Gupta P, Park SW, Farooqui M, Wei LN. Orphan nuclear receptor TR2, a mediator of preadipocyte proliferation, is differentially regulated by RA through exchange of coactivator PCAF with corepressor RIP140 on a platform molecule GRIP1. Nucleic Acids Res. 2007;35:2269–2282. doi: 10.1093/nar/gkl1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Augereau P, Badia E, Balaguer P, Carascossa S, Castet A, Jalaguier S, Cavaillès V. Negative regulation of hormone signaling by RIP140. J. Steroid Biochem. Mol. Biol. 2006;102:51–59. doi: 10.1016/j.jsbmb.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka K, Imoto I, Inoue J, Kozaki K, Tsuda H, Shimada Y, Aiko S, Yoshizumi Y, Iwai T, Kawano T, et al. Frequent methylation-associated silencing of a candidate tumor-suppressor, CRABP1, in esophageal squamous-cell carcinoma. Oncogene. 2007;26:6456–6468. doi: 10.1038/sj.onc.1210459. [DOI] [PubMed] [Google Scholar]
- 44.Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- 45.Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB, et al. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999;18:1923–1938. doi: 10.1093/emboj/18.7.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rea S, Eisenhaber F, O'C;arroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 47.Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- 48.Attema JL, Papathanasiou P, Forsberg EC, Xu J, Smale ST, Weissman IL. Epigenetic characterization of hematopoietic stem cell differentiation using miniChIP and bisulfite sequencing analysis. Proc. Natl Acad. Sci. USA. 2007;104:12371–12376. doi: 10.1073/pnas.0704468104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mostaqul Huq MD, Gupta P, Tsai NP, White R, Parker MG, Wei LN. Suppression of receptor interacting protein 140 repressive activity by protein arginine methylation. EMBO J. 2006;25:5094–5104. doi: 10.1038/sj.emboj.7601389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huq MD, Ha SG, Barcelona H, Wei LN. Lysine methylation of nuclear co-repressor receptor interacting protein 140. J. Proteome Res. 2009;8:1156–1167. doi: 10.1021/pr800569c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barrero MJ, Malik S. Two functional modes of a nuclear receptor-recruited arginine methyltransferase in transcriptional activation. Mol. Cell. 2006;24:233–243. doi: 10.1016/j.molcel.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huq MDM, Gupta P, Wei LN. Post-translation Modification of Nuclear Co-repressor RIP140: potential targets of therapeutics. Curr. Med. Chem. 2008;15:386–392. doi: 10.2174/092986708783497382. [DOI] [PubMed] [Google Scholar]
- 53.Ho PC, Gupta P, Tsui YC, Ha SG, Huq M, Wei L-N. Modulation of lysine acetylation-stimulated repressive activity by Erk2-mediated phosphorylation of RIP140 in adipocyte differentiation. Cell Signal. 2008;20:1911–1919. doi: 10.1016/j.cellsig.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.