Abstract
The conserved mRNA export receptor NXF1 (Mex67 in yeast) assembles with messenger ribonucleoproteins (mRNP) in the nucleus and guides them through the nuclear pore complex into the cytoplasm. The DEAD family RNA helicase Dbp5 is essential for nuclear export of mRNA and is thought to dissociate Mex67 from mRNP upon translocation, thereby generating directional passage. However, the molecular mechanism by which Dbp5 recognizes Mex67-containing mRNP is not clear. Here we report that the human NXF1-binding protein RBM15 binds specifically to human DBP5 and facilitates its direct contact with mRNA in vivo. We found that RBM15 is targeted to the nuclear envelope, where it colocalizes extensively with DBP5 and NXF1. Gene silencing of RBM15 leads to cytoplasmic depletion and nuclear accumulation of general mRNA as well as individual endogenous transcripts, indicating that RBM15 is required for efficient mRNA export. We propose a model in which RBM15 acts locally at the nuclear pore complex, by facilitating the recognition of NXF1–mRNP complexes by DBP5 during translocation, thereby contributing to efficient mRNA export.
INTRODUCTION
In the course of mRNA metabolism, the protein composition of messenger ribonucleoprotein complexes (mRNP) changes in a specific order and in a strictly regulated manner. Such changes are assisted by RNP remodeling enzymes that facilitate the reorganization of RNA:RNA and RNA:protein contacts [for a recent review, see ref. (1)]. Of these, the helicase superfamily 2 (SF2) proteins, DEAD/DExH RNA helicases, act to unwind RNA helices and/or displace the RNA-bound proteins, in an ATP-dependent fashion. SF2 factors have been implicated at all mRNA metabolic steps from transcription to decay (2–5). In the nucleus, the DExH/D protein eIF4AIII (6,7) and the splicing/nuclear export factor Sub2/UAP56 (8–12) participate in the assembly of export-ready mRNP, and DEAD-box protein Dbp5 (DBP5/DDX19 in humans) is thought to assist the translocation of mRNP through the nuclear pore complex (NPC). Dbp5 is essential for mRNA export in yeast (13–15), and studies in metazoa revealed a remarkably conserved network of interactions between its orthologs and their binding partners, suggesting a conserved mechanism by which Dbp5 acts on translocating mRNP to generate directional passage (16–18). In a current model, Dbp5 is targeted to the cytoplasmic fibrils of NPC via interactions with the nucleoporin Nup159/NUP214, while its binding partner Gle1 interacts with the nucleoporin-like protein Nup42/Rip1/hCG1 at an adjacent site on the NPC, leading to the facilitated formation of Dbp5–Gle1 complex. The RNA binding and ATPase activities of Dbp5 are enhanced by Gle1 and inositol hexakisphosphate, and hence the complex formation results in Dbp5’s activation at the precisely defined locale, the cytoplasmic side of the NPC (19–21). Dbp5 is then thought to dissociate the export receptor Mex67/NXF1 from mRNP complexes as they emerge at the cytoplasmic side of NPC, and thus confer direction to the NPC passage (19,20,22,23). In this scenario, the spatial restriction of Dbp5 activity prevents premature mRNP remodeling. Recent work showed that Dbp5 can promote the dissociation of Nab2 mRNA export factor as well as polyadenylate-binding protein Pab1 from the bound RNA in vitro (24), confirming its predicted RNP remodeling activity and suggesting that it can act on a variety of RNP substrates. A recently discovered role of Dbp5 in translation termination (25) further supports its promiscuity. The mechanism that directs Dbp5 specifically to the Mex67/NXF1-containing mRNP during export and prevents its non-productive effects on other exported RNP remains to be elucidated. There is no evidence of direct recognition between the yeast Mex67 and Dbp5, and we previously reported that the human orthologs NXF1 and DBP5 do not bind directly in vitro (26), supporting the lack of constitutive interaction. We reasoned that the DBP5–NXF1 recognition could be enabled transiently, by a cofactor that only acts at the NPC, thereby allowing DBP5 to target the NXF1-containing mRNP selectively and at a proper location.
Here, we report that the human RNA binding motif protein 15 (RBM15) has properties expected from a factor that facilitates the contacts of DBP5 with mRNA at NPC. RBM15 belongs to the Spen (split end) family of proteins, which share domain architecture including three N-terminal RNA recognition motifs (RRM) and a C-terminal SPOC (Spen paralogue and orthologue C-terminal) domain, and are conserved across metazoa. In humans, the Spen proteins are represented by SHARP, RBM15 (also referred to as OTT1) and RBM15B/OTT3 (herein referred to as OTT3). The mammalian RBM15 orthologs have been implicated in hematopoiesis (27), transcription regulation (28), mRNA export and splicing (29,30). Our previous work showed that RBM15 binds to NXF1 and serves as receptor for the RNA export element RTE (30). RTE utilizes the NXF1 export pathway in microinjected X. laevis oocytes (30), synergizes in cis with NXF1’s high affinity RNA ligand CTE (31) and is essential for propagation of murine LTR-IAP retrotransposons (32), suggesting that RBM15 is a bona fide general mRNA export factor that is hijacked by mobile elements to achieve the efficient export of their otherwise defective, nuclear-retained transcripts (32). While studies of retroelements revealed the mRNA export activity of RBM15, its role in the general mRNA metabolism remains to be elucidated. In this work, we show that RBM15 is required for the efficient mRNA export in human cultured cells, and propose the underlying mechanism.
MATERIALS AND METHODS
Cell culture, antibodies, immunoprecipitation and in vivo cross-linking
The mammalian expression plasmids for RBM15, DBP5 and NXF1, and GST fusion plasmids for NXF1 and DBP5 were described (15,30,33). Transfections in human 293 or HeLa-derived HLtat cells were performed as described (33). Cell fixation and permeabilization were performed as in ref. (34). For indirect immunofluorescence, RBM15 pAb (10587-1-AP, Proteintech), NXF1 mAb (53H8, Santa Cruz), DBP5 pAb (A300-547A, Bethyl Labs), SC-35 mAb (SC-35, Sigma), HA epitope mAb (HA.11, Covance) and FLAG-epitope mAb (M2, Sigma) were used as primary antibodies, followed by detection with Alexa-conjugated secondary antibodies (Molecular Probes), according to the manufacturers’ instructions. The endogenous SR proteins were detected on western blots using 1H4 mAb (Zymed) that recognizes a phospho-epitope common for all members of the SR protein family. For coimmunoprecipitation assays, cells were extracted under mild conditions (200 mM NaCl, 0.5% Triton X100), treated with RNase A prior to immunoprecipitation with anti-FLAG agarose (Sigma), and the complexes were eluted with 3XFLAG peptide (Sigma) to ensure that only soluble, RNA-independent complexes were analyzed. UV-crosslinking in vivo was performed as in ref. (35) with 400 mJ energy dose, and poly(A)+ RNA was isolated using Dynabeads mRNA DIRECT procedure.
Image analysis
The wide-field epifluorescence images were acquired using Axio Observer Z1 microscope equipped with AxioCam MRM CCD camera, PlanApo 100X objective, appropriate filter sets and AxioVision software (Carl Zeiss Microimaging, Thornwood, NY). The multi-color experiments were performed using appropriate controls to exclude leakage between the channels. Some images were acquired as Z-stacks and subjected to 3D digital deconvolution using AutoDeblur software (Bitplane, Saint Paul, MN). Quantitative colocalization analyses (QCA) were performed using the orthogonal regression as well as expectation maximization algorithms, implemented in MIPAV software (CIT, NIH) with default parameters.
Posttranscriptional gene silencing
Shortly after seeding in 60-mm plates, 293 cells were transfected with ON-TARGETplus SMARTpool siRNAs (Dharmacon) for RBM15 (sense strands: ACGAGAAUUUGAUCGAUUUUU, GGUGAUAGUUGGGCAUAUAUU, UAGCAGGGCCCAAUGGUUAUU, GCAGUAGCCGGGAUCGUUAUU), OTT3 (sense strands: GGGAGCAGUCGGCGAAGUAUU, CCAUAUGAGGAACGGAGUAUU, CUACAGAGACGGCCGAAAUUU, UGAGAAGGGAAUCCGGUUAUU) or non-targeting control, at 100 nM, by using HiPerFect reagent (Qiagen). At day 2 post-transfection, cells were trypsinized, split at 1:3 and transfected again under the same conditions.
Cell fractionation and quantitative RT–PCR
Human 293 cells were extracted sequentially with 10 mM HEPES pH 7.9, 1 mM MgCl2, 2 mM NaCl, 0.5 mM DTT and 0.004% digitonin (C fraction, soluble cytoplasmic); and 20 mM HEPES pH 7.9, 1 mM MgCl2, 100 mM NaCl, 0.5 mM DTT, 0.5% Triton X100 and 10% glycerol (N1 fraction, soluble nuclear). The remaining pellet was resuspended in the N1 buffer and solubilized by sonication as in ref. (36) yielding the N2 fraction (insoluble nuclear). All extractions were performed at +4°C. Alternatively, cells were extracted with 15 mM HEPES, pH 7.9, 50 mM KCl, 200 mM NaCl, 0.1 mM EDTA and 0.2% Triton X100 (‘soluble cell fraction’ corresponding to C+N1). Poly(A)+ RNA was purified using Dynabeads mRNA DIRECT procedure. After reverse transcription with SuperScript III (Invitrogen) and random hexadeoxynucleotide primers, real time quantitative PCR (qPCR) was performed using ABI PRISM 7700 Sequence Detection System (Applied Biosystems). The TaqMan gene expression assays (primer sets) for β-actin (ACTB, cat #4333762), glyceraldehyde 3-phosphate dehydrogenase (GAPDH, cat #4333764T) and inositol hexakisphosphate kinase 1 (IP6K1, assay ID: Hs00384812_m1) were purchased from Applied Biosystems. Reactions were performed in triplicate using the default cycling conditions as recommended by the manufacturer, and data were analyzed using the relative quantification method, in which the experimental Ct values (the number of cycles required for the amplification signal to surpass a set threshold) were used to calculate the relative cDNA input concentrations (I) : I = 2−(Ct − Co) where Co is a calibration constant The background levels were obtained from control reactions with omitted template, and typically were at least two orders of magnitude lower than the mRNA signal. Datasets were compared using one-way analysis of variance (ANOVA) (α = 0.05) with Bonferroni or Newman–Keuls post-tests.
Detection of general mRNA, tRNA and U snRNP
For general mRNA detection, poly(A)+ RNA was 3′-labeled with [32P] pCp (NEN) using T4 RNA ligase (Fermentas), followed by complete digestion with RNase T1 (Worthington). After additional round of oligo(dT) capture using Dynabeads mRNA DIRECT procedure, the isolated poly(A) tails were separated on 15% TBE–urea gels and detected by phosphoimager. For tRNA and U snRNP detection, total RNA was purified from subcellular extracts by proteinase K digestion and RNAzol extraction, and separated on 15% TBE–urea gels. tRNA was visualized by ethidium bromide staining, and U snRNAs were detected on northern blots as described (37).
RESULTS
RBM15 binds specifically to DBP5 in vivo and in vitro
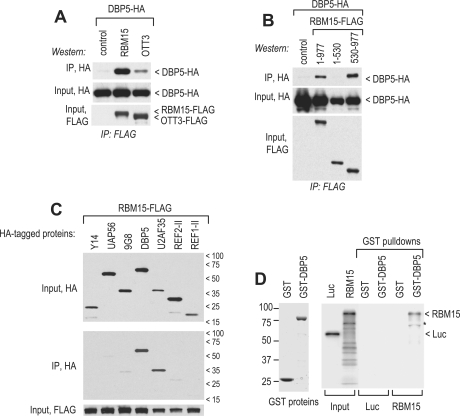
To probe the association between RBM15 and DBP5 in vivo, we immunoprecipitated the epitope-tagged proteins from the extracts of transfected cells, which were pretreated with RNase A to exclude RNA-mediated interactions. We found that DBP5 associated efficiently with RBM15, while OTT3 exhibited a weaker association (Figure 1A). This interaction was mediated by RBM15’s C-terminal region (aa 530–977) that also binds NXF1 (30), while the N-terminal region containing the RRM motifs (aa 1–530) did not associate (Figure 1B). We further evaluated the selectivity of the RBM15–DBP5 binding by studying RBM15’s interactions with other mammalian RNA-binding proteins implicated in the assembly and/or remodeling of mRNA export complexes: Y14 (38–40), 9G8 (41–43), U2AF35 (37,44,45), REF1-II and REF2-II (46–50). As negative control, we used the DExH/D RNA helicase UAP56 (8,11,12,51), which is essential for mRNA export in human cells (52) and was shown not to bind RBM15 (30). These factors were HA-tagged and expressed in the presence of FLAG-tagged RBM15. Immunoprecipitations with FLAG antibodies were performed from RNase-digested extracts in the presence of 2 M urea and 400 mM NaCl to ensure stringent conditions. We found that RBM15 discriminated efficiently between DBP5 and a structurally similar (32% identity) UAP56 (Figure 1C), indicating that DBP5’s ability to bind RBM15 was selective and not a generic property of SF2 proteins. Under these conditions, RBM15 did not bind appreciably to Aly/REF proteins REF1-II and REF2-II, or Y14 and 9G8. We noted a strong association between RBM15 and U2AF35, an SR family RRM motif-containing splicing factor that binds NXF1 directly (37), but it was not shared by other SR proteins (9G8), NXF1 binders (9G8, Aly/REF) or the RRM motif-containing proteins (9G8, Y14, Aly/REF) (Figure 1C). The interaction with U2AF35 was consistent with the previously reported association of RBM15 with the spliceosome (53) and was not further addressed in this study. We further examined the RBM15-DBP5 binding in vitro, using pulldowns of reticulocyte-produced RBM15 with bacterially expressed, purified GST–DBP5. RNaseA was added to the reactions to exclude the RNA-mediated interactions, and firefly luciferase protein was used as a control for non-specific binding. These experiments showed that RBM15 bound specifically to DBP5 in vitro (Figure 1D). In summary, our data revealed that the interactions between RBM15 and DBP5 are RNA-independent, persist under stringent conditions and are highly selective. These data suggested that RBM15 could serve to bridge NXF1 to DBP5. However, exogenously expressed RBM15 did not stimulate significantly the co-precipitation of NXF1 and DBP5 proteins from crude extracts (data not shown), leading us to speculate that RBM15’s bridging activity could be spatially regulated and restricted to a subpopulation of NXF1 complexes such as mRNP in transit through NPC.
Figure 1.
RBM15 associates with DBP5 in vivo and in vitro. The expression plasmids encoding epitope-tagged proteins (1–5 µg each) were transiently transfected in human 293 cells as indicated, and coprecipitation assays were performed from RNase A-treated extracts (30). (A–C) The extracts were adjusted to 400 mM NaCl before adding the antibodies, and the precipitates were washed in the presence of 400 mM NaCl and 2 M urea prior to elution. The epitopes detected on western blots and the positions of detected proteins are indicated. Input, 1:100 of the extract before IP; Control, omission of bait protein and n/t, non-transfected cells. (D) Binding of E.coli-produced, immobilized GST-tagged DBP5 or isolated GST moiety (left panel, GST proteins) to metabolically radiolabeled RBM15 and luciferase (Luc) proteins which were synthesized in reticulocyte lysates (input) (26). Prior to binding, RNase A was added to reticulocyte samples to exclude the RNA-mediated interactions. Right panel (GST pulldowns), SDS–PAGE/radiofluorography analysis of the bound fractions. Asterisk shows a truncated RBM15 protein.
RBM15 promotes the access of DBP5 to mRNP complexes
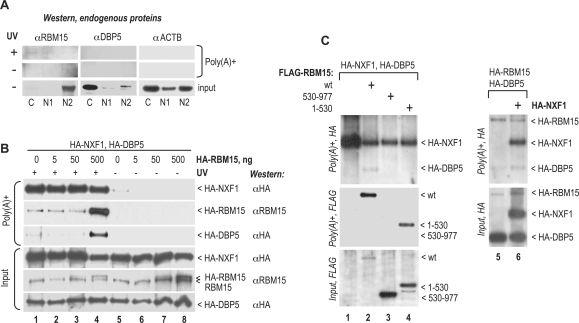
To examine the association of endogenous RBM15 and DBP5 with general mRNP complexes, we UV-irradiated human 293 cells and analyzed the proteins that crosslinked to poly(A)+ RNA, in the cytoplasmic (C), soluble nuclear (N1) and insoluble nuclear (N2) cell extracts (Figure 2A). This sequential fractionation protocol allows dissecting the mRNP maturation stages: N1 contains the mature, export-ready spliced mRNP, whereas pre-mRNP are confined to the N2 fraction, which also contains the bulk of spliceosomal components (36,37,42,54–57). To analyze the UV-crosslinked proteins, the extracts were adjusted to high salt and detergent concentrations so that all non-covalently bound factors were dissociated, and the covalent poly(A)+ RNA–protein adducts were purified by oligo(dT) capturing. The bound proteins were then released from RNA complexes by RNase digestion and analyzed on western blots. Figure 2A, input, shows that, in crude extracts, the endogenous RBM15 was strongly enriched in the N2 fraction, consistent with its spliceosomal association (53), whereas endogenous DBP5 was mostly cytoplasmic, as expected (15). We found that RBM15 crosslinked detectably to poly(A)+ RNA, whereas DBP5 as well as β-actin (ACTB) did not [Figure 2A, poly(A)+]. However, the crosslinked RBM15 was not enriched in the N2 fraction, suggesting that RBM15 was added to the mature mRNP after splicing. The above sequential extraction protocol allowed dissecting the cytoplasmic and soluble nuclear complexes, but led to poor yields of crosslinked product (Figure 2A). In subsequent crosslinking experiments, we used a simultaneously extracted C + N1 fraction (‘soluble cell fraction’), that provided higher yields (see below, Figure 2B). In summary, these data showed that endogenous RBM15 has access to export-ready mRNP and prompted us to address RBM15’s interactions with its cofactors within such complexes.
Figure 2.
RBM15 stimulates crosslinking of DBP5 to mRNA. (A) Exponentially growing human 293 cells were UV-irradiated with a dose of 400 mJ when indicated (UV+) and fractionated into the C, N1 and N2 extracts. Poly(A)+RNA was isolated under denaturing conditions, and the endogenous RBM15, DBP5 and, as negative control, ACTB proteins were analyzed in the poly(A)+ fractions on Western blots, after RNase digestion (polyA+); and in 1:1000 aliquots of crude extracts (input). (B and C) Human 293 cells were transiently transfected with 1 µg of HA-NXF1, 1 µg of HA-DBP5, and the indicated amounts of HA-RBM15 plasmids (B) or with 1 µg of each of the indicated plasmid (C). At day 1 posttransfection, cells were UV-irradiated (B, lanes 1–4; C, lanes 1–6) as in (A). Poly(A)+ RNA was isolated from soluble cell fractions (C + N1), and the crosslinked proteins were analyzed as in (A). Input, a 1:200 aliquot of crude extract. The positions of proteins and the antibodies used are indicated. In panel B, staining with the RBM15 antibody (pAb 10587-1-AP, Proteintech) detected both the exogenous and endogenous proteins with the expected mobility difference [endogenous, isoform 2-L, 100 kDa; exogenous, HA-tagged isoform 1 − S + AE, 108 kDa (30)]. The identity of the crosslinked DBP5 bands was verified by staining with protein-specific antibodies (data not shown). Similar data were obtained in three independent transfection experiments.
We coexpressed the epitope-tagged NXF1, DBP5 and RBM15 proteins, and analyzed their interactions with poly(A)+ RNA in the soluble cell fraction (C + N1) using UV crosslinking in vivo (Figure 2B and C). We found that, in the absence of exogenous RBM15, HA-DBP5 did not crosslink to mRNA efficiently (Figure 2B, lane 1), whereas NXF1 crosslinked strongly, as expected (35). Remarkably, a modest (∼2-fold; Figure 2B, lane 4; input) increase in RBM15 levels upon exogenous expression caused a strong increase in both RBM15 and DBP5 crosslinking to mRNA (Figure 2B, lane 4). Using RBM15 and DBP5 that were both HA-tagged, we found that they yielded comparable signals in poly(A)+ fraction, suggesting that they crosslinked at comparable molar amounts (Figure 2C, lane 6), which is consistent with DBP5’s recruitment via stoichiometric RBM15 binding. The crosslinking of RBM15 and DBP5 was detectable in the absence of exogenous NXF1 and did not change significantly in the presence of cotransfected NXF1 (Figure 2C, lanes 5 and 6), suggesting that the endogenous levels were saturating in our assays. Because RBM15 binds to NXF1 directly (30), it could access mRNP via direct binding to NXF1, while interactions with other mRNP proteins and/or naked RNA could also play a role. Our recent work (58) showed that RBM15 binds selectively to NXF1’s NTF2-like domain (59) that is not engaged in mRNP association (37), and thus, RBM15 may be able to bind mRNP-associated NXF1. We concluded that RBM15 acts as a limiting factor enabling the access of DBP5 to general mRNA. It cannot be excluded that the stimulation by RBM15 was indirect, e.g. reflected slower mRNA export rates in the presence of exogenous RBM15, leading to extend the dwelling time of DBP5 in the vicinity of translocating mRNA. However, the exogenous expression of RBM15 or its deletion mutants did not cause mRNA export inhibition (30) or excessive accumulation of poly(A)+ RNA or DBP5 at the nuclear envelope (data not shown), and hence there is no evidence to support an indirect stimulation. Taken together with the direct binding and highly selective, stable interactions between DBP5 and RBM15 (Figure 1), our results strongly favor the direct recruitment.
To understand the recruitment mechanism, we examined the responsible determinants within RBM15. We found that the N-terminal region of RBM15 comprising the RRM motifs (aa 1–530; see Figure 1A) crosslinked to mRNA efficiently (Figure 2C, lane 4), possibly reflecting its ability to bind RNA directly (30), but failed to recruit DBP5. Thus, the presence of RBM15’s C-terminal region (aa 530–977) that contains the NXF1- and DBP5-binding determinants was critical, further suggesting that DBP5 was recruited via direct binding. Our data are compatible with two mechanisms: (i) DBP5 binds directly to the mRNP-bound RBM15; (ii) RBM15 acts allosterically to induce the direct binding between DBP5 and mRNP components other than RBM15. Importantly, the isolated C-terminal region did not crosslink or recruit DBP5 (Figure 2C, lane 3), demonstrating a critical role of the N-terminal region (aa 1–530). While this role could be limited to the maintenance of direct contacts with mRNA (Figure 2C, lane 4), it is also possible that RBM15’s N-terminal region could contribute to DBP5 recruitment by targeting RBM15 to the vicinity of mRNP remodeling.
RBM15 is targeted to the nuclear envelope via its N-terminal region
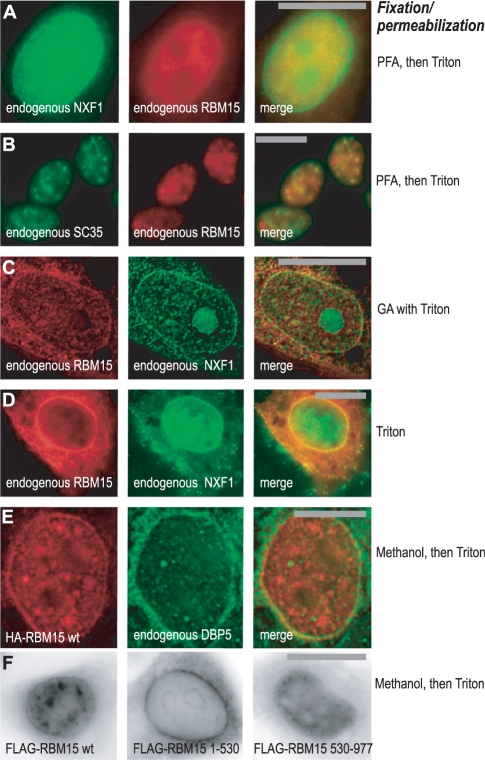
To further address the mechanism of DBP5 recruitment, we examined the subcellular localization of RBM15 in cultured human cells, and compared it to that of NXF1 and DBP5. Using indirect fluorescence to detect the endogenous proteins, we found that in paraformaldehyde (PFA)-fixed HeLa cells (Figure 3A) RBM15 localized to the nucleoplasm and was undetectable at the nuclear envelope (NE), whereas NXF1 displayed both nucleoplasmic and NE localization as expected (33,35). Similar localization was observed after methanol fixation (not shown). Under the same conditions, in 293 cells (Figure 3B), endogenous RBM15 also localized to the nucleoplasm and partially colocalized with SC35 antigen in nuclear speckles, indicating its enrichment in splicing factor compartment (SFC), as expected from the previous characterization of RBM15 as a spliceosome-associated protein (53). Previously, we observed that in HeLa cells, the endogenous RBM15 was not enriched in SFC at steady state, but the exogenously expressed protein accumulated readily in SFC, confirming that RBM15 has access to this compartment (58).
Figure 3.
RBM15 is targeted to SFC and nuclear envelope. (A–E) Detection by indirect immunofluorescence: (A) Endogenous RBM15 and NXF1 in HLtat cells after PFA fixation followed by Triton X100 permeabilization. (B) Endogenous RBM15 and SC35 in human 293 cells after PFA fixation followed by Triton X100 permeabilization. (C) Endogenous RBM15 and NXF1 in HLtat cells after fixation with 4% glutaraldehyde in the presence of 0.5 % Triton X100. (D) Endogenous RBM15 and NXF1 in permeabilized HLtat cells. Preformed complexes of the primary and secondary antibodies in PBS were added to live cells, followed by addition in situ of Triton X100 to 0.5%, and images were captured after 15 min incubation at room temperature. (E) HA-tagged RBM15 and endogenous DBP5 in HLtat cells after methanol fixation and Triton X100 permeabilization. (F) FLAG-tagged RBM15 (wild type and mutants spanning aa 1–530 and aa 530–977) in HLtat cells after methanol fixation and Triton X100 permeabilization. Inverted images are shown. Signals were visualized by widefield epifluorescence microscopy, and sections through the centre of the nucleus are presented. Raw (A, B, D, F) and digitally deconvoluted (C, E) images are shown. Typical fields are presented, and similar results were obtained in three independent experiments. Bars, 20 µm (A, B, C, D and F), 10 µm (E).
The nuclear envelope localization of NPC-associated proteins is best viewed upon cell extraction, which removes the loosely bound fraction while preserving the strong association. Permeabilization of cells during or prior to fixation is commonly used to study the strong NPC binders, e.g. NXF1 (60). We therefore performed indirect immunofluorescence in HeLa cells fixed with glutaraldehyde (GA) in the presence of Triton X100 (Figure 3C). We found that, in all cells, a fraction of the endogenous RBM15 associated with NE, where it colocalized with NXF1 (Figure 3C). Staining of unfixed Triton X100-permeabilized cells, a technique that has previously been used to visualize the NPC-bound DBP5 (15) and NXF1 (60), provides an even higher degree of extraction. Remarkably, using this technique, an extensive and persistent association of RBM15 as well as of NXF1 with the NE was revealed in all cells (Figure 3D). No signal was detected at the NE after staining for nuclear non-NPC antigens including SC35 or upon omission of primary antibodies, and the integrity of nuclear membranes was confirmed by staining with fluorescent lipophilic dye DiOC6 (61) (data not shown). We concluded that the endogenous RBM15 resides mostly in the nucleoplasm at steady state, while a fraction of the protein is present at the nuclear envelope. It is possible that poor epitope accessibility contributed to the lack of RBM15 signal at the NE of cells that were fixed prior to permeabilization (Figure 3A and B), prompting us to examine the epitope-tagged proteins. We found that the exogenously expressed HA-tagged as well as FLAG-tagged RBM15 accumulated strongly at the NE (Figure 3E and F). The nuclear envelope-targeting determinant resided entirely within its N-terminal region, whereas the C-terminal region accounted for the nucleoplasmic localization (Figure 3F). Under these conditions, the endogenous DBP5 was found in the cytoplasm and at the NE as expected (15) and colocalized with HA-RBM15 at the NE (Figure 3E). Post-embedment immunoelectron microscopy of the same cells confirmed the colocalization of HA-RBM15 and DBP5 at NPC (data not shown). RBM15’s isolated N-terminal region also associated prominently with the perinuclear compartment (PN) flanking the cytoplasmic side of the nuclear envelope (Figure 3F), but it did not colocalize significantly with the known PN components such as the ER and the perinuclear actin filaments (62), and these conclusions were confirmed by using quantitative colocalization analyses (data not shown).
In summary, RBM15 colocalized at the nuclear envelope with the NPC-associated DBP5 and NXF1, strongly suggesting the presence of a dedicated NPC targeting signal. The NPC association does not require the presence of its C-terminal region, which is responsible for binding to DBP5–NXF1. However, only a fraction of the endogenous protein resides at the NPC at steady state, which could be due to short dwelling time and/or low affinity. The exogenously expressed HA-RBM15 accumulated extensively at the NPC, suggesting that this compartment could be relevant for the observed RBM15-driven recruitment of DBP5 to mRNP (Figure 2A and B). These findings further indicate that the DBP5–mRNA recognition could be facilitated by high local concentrations of RBM15 at the NPC. We conclude that RBM15 has properties of a factor that acts at the NPC to facilitate the access of DBP5 to mRNA, and further address its relevance for general mRNA export.
Gene silencing of RBM15 and OTT3 lead to general mRNA export inhibition
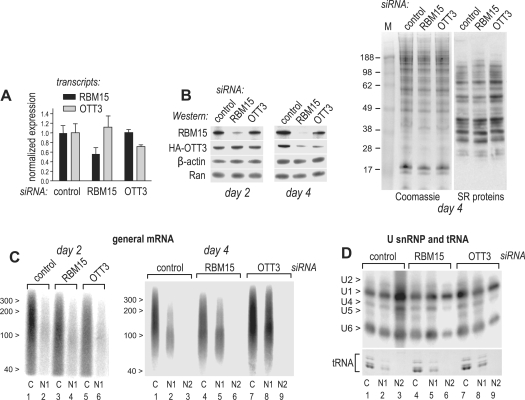
To examine the participation of RBM15 and OTT3 in general mRNA export, we performed posttranscriptional gene silencing in human 293 cells with siRNAs directed either to RBM15 or to its cognate protein OTT3 or using a non-targeting siRNA as control. To evaluate the mRNA target specificity, we used quantitative RT–PCR (RT–qPCR) to measure the levels of the endogenous RBM15 and OTT3 transcripts. We found that RBM15 and OTT3 siRNAs caused an appreciable reduction of their target transcript levels, whereas those of non-targeted transcripts were not reduced (Figure 4A), suggesting the absence of off-target effects on mRNA level. To analyze the protein depletion, OTT3 was tagged with an HA epitope to facilitate detection and was analyzed on Western blots along with the endogenous RBM15. We found that both RBM15 and OTT3 siRNAs caused strong, specific depletions of their targeted proteins, by day 2 and 4, respectively (Figure 4B), and similar data were obtained with different siRNA doses (data not shown). Interestingly, RBM15 siRNA also led to codepletion of OTT3 protein by day 4, while the levels of β-actin and Ran control proteins were not affected (Figure 4B). We further analyzed the proteins in raw extracts by SDS–PAGE/Coomassie staining, which is a standard approach to assess siRNA specificity, allowing to visualize multiple proteins simultaneously. We found that in the same samples as in Figure 4B day 4, there were no appreciable off-target effects of RBM15 or OTT3 siRNAs (Figure 4B, Coomassie). To ask whether the depletions affected generically the factors that are, like RBM15 and OTT3, involved in mRNA metabolism, we analyzed the SR proteins, a family of RNA-binding factors that have been implicated at all mRNA metabolic steps [for recent review, see ref. (63)]. Using a monoclonal antibody to a family-specific shared epitope, we detected multiple SR factors, as expected, and found that neither of those were depleted by day 4 after RBM15 or OTT3 siRNA transfection (Figure 4B, SR proteins). Together, these studies established that the targeted RBM15 and OTT3 depletions as well as the OTT3 codepletion by RBM15 siRNA occurred under the conditions that did not affect numerous other proteins. Codepletions on the protein but not mRNA level have been observed for binding partners of siRNA-targeted proteins, and are attributable to protein destabilization upon loss of binding (64–68). In support of such possibility, we recently found that RBM15 coprecipitates with OTT3 in vivo, pointing to their presence in the same complexes (58). However, it is also possible that codepletion reflected indirect effects of generalized mRNA export inhibition (Figures 4C and 5).
Figure 4.
RBM15 and OTT3 are required for general mRNA export. Human 293 cells were transfected with siRNAs targeting RBM15, OTT3 or non-targeting siRNA (control) and analyzed at day 2 or 4 posttransfection as indicated. (A) RT–qPCR detection of RBM15 and OTT3 transcripts at day 2. Expression levels were calculated from real-time PCR values (Ct) using relative quantitation method and are plotted on the y-axis after normalization to those obtained in the cells transfected with the non-targeting siRNA control (normalized expression). Mean (n=3) values are presented, and bars show one SEM. (B) Cells were transfected with the indicated siRNAs and, next day, with a plasmid expressing HA-OTT3 (1 μg). At day 2 or day 4 after siRNA transfection, cell pellets were boiled in Laemmli sample buffer, proteins separated on 10% SDS–PAGE and analyzed on western blots with antibodies to RBM15, HA, Ran, β-actin or SR proteins as indicated; or by Coomassie staining. (C) Cells at day 2 (left panel) or day 4 (right panel) posttransfection were separated into the C, N1 and N2 fractions, mRNA poly(A) tails were 3′-radiolabeled, cut off by RNase T1 digestion, separated by urea–PAGE and detected by phosphoimager. Positions of size markers (nt) are shown. (D) U snRNAs from the same fractions as in (D) were separated by urea–PAGE and detected on northern blots, and positions of the individual U snRNAs are indicated to the left. tRNA was detected on the same gels, by ethidium bromide staining prior to blotting (tRNA).
Figure 5.
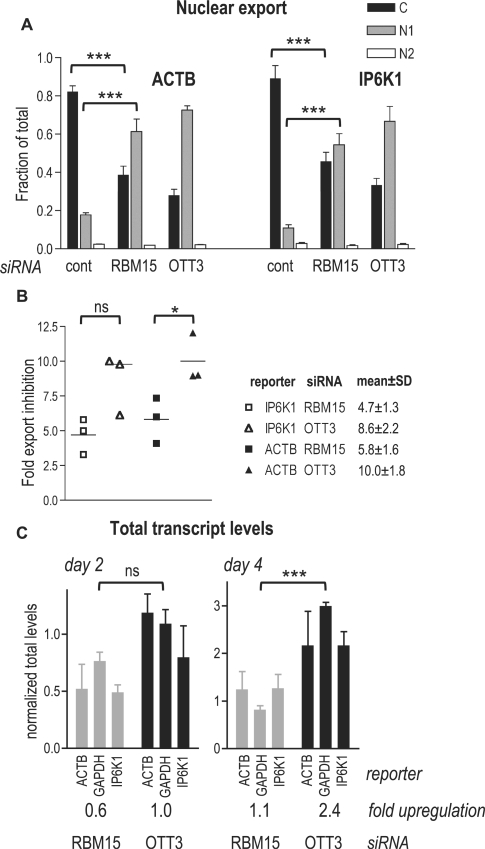
Quantification of individual transcripts upon RBM15 and OTT3 silencing. (A) Gene silencing and cell fractionation into the C, N1 and N2 subcellular extracts were performed as in Figure 4, and the endogenous IP6K1 and ACTB transcripts were detected by RT–qPCR. Expression levels were calculated as in Figure 4A, and, for each subcellular extract, are plotted on the y-axis as a fraction of mean total cell levels (C + N1 + N2) (fraction of total). Means (n = 3) are presented, and bars show one SEM. ***P < 0.001, one-way ANOVA (α = 0.05), Bonferroni post-test. (B) A summary of three independent siRNA experiments. For each transcript, the fold export inhibition values are nucleocytoplasmic ratios ((N1 + N2)/C) upon siRNA treatment (mean of triplicate reactions), normalized to those obtained with the non-targeting siRNA. For individual experiments, these values are plotted on the y-axis as shown in the legend, and their means and standard deviations are presented. *P < 0.05; ns, P > 0.05, one-way ANOVA (α = 0.05), Newman–Keuls post-test. (C) After gene silencing as in Figure 4, total RNA was isolated by RNAzol procedure and analyzed by RT–qPCR as in panel A. For each transcript, mean total levels (n = 3) are plotted on the y-axis after normalization to those obtained with the non-targeting siRNA (normalized total levels) and bars show one SEM. For each siRNA, a mean of normalized total levels for the three transcripts is presented (fold up-regulation). ***P < 0.001; ns, P > 0.05, one-way ANOVA (α = 0.05), Newman–Keuls post-test.
To evaluate the effects of RBM15 and OTT3 depletions on mRNA export, we compared the levels of general mRNA in the cytoplasmic (C), soluble nuclear (N1) and insoluble nuclear (N2) cell extracts (Figure 2A). The poly(A)+ RNA from these fractions was radiolabeled at the 3′ ends of poly(A) tails and digested with RNase T1, leading to the complete hydrolysis of mRNA body while leaving the poly(A) tail intact. These isolated, radiolabeled poly(A) tails were further purified by oligo(dT) capturing and separated by urea–PAGE (Figure 4C). This procedure ensured that only the genuine 3′-polyadenylated transcripts were analyzed, and resulted in efficient detection of poly(A) tails with sizes typical of mature mRNA (∼60–200 nt) in the C and N1 fractions (Figure 4C), whereas considerably longer (∼200–250 nt) tails likely belonging to pre-mRNA were detected in the N2 fractions upon overexposure (data not shown). Because the analyzed RNA samples consisted of pure polyadenine and did not contain any internal controls with which to normalize the purification yields and gel loading, this approach is semiquantitative. We note that Figure 4D (see below) shows other RNA classes in the same cell extracts as used in Figure 4C, right panel, providing external controls.
At day 2 posttransfection, we found that in cells transfected with non-targeting siRNA, mRNA was enriched in the C fraction (Figure 4C, left panel), confirming its efficient export from the nucleus, and similar distributions were observed in the absence of siRNA or when using siRNAs targeted to irrelevant genes (data not shown). In contrast, silencing of RBM15 and OTT3 led to cytoplasmic reduction, while the nuclear mRNA levels were not affected appreciably (Figure 4C, left panel). At day 4, silencing of RBM15 and OTT3 further led to mRNA accumulation in the N1 fractions that represent the post-splicing, export-ready mRNP (Figure 4C, right panel, compare lanes 2, 5 and 8), but not in the N2 fractions, indicating that the accumulation was not due to spliceosomal sequestration. Because the poly(A) length distributions in the accumulated species were not altered detectably (Figure 4C, right panel, compare lanes 2, 5 and 8), these RNAs likely represented intact transcripts and not decay intermediates. OTT3 silencing also led to an increase of total mRNA levels at day 4, but not day 2 (Figure 4C, right panel). These data indicated that OTT3 silencing primarily affected mRNA export, while the net mRNA increase reflected the long-term effects, and they were further studied using individual reporter mRNAs (Figure 5).
To understand how RBM15 and OTT3 silencing affected the export of other RNA classes, we analyzed the transfer RNA (tRNA) and U snRNP, which are exported independently of mRNA and of each other (69–72). In the absence of silencing, tRNA was found in the C and N1 fractions and exhibited a C/N1 distribution comparable to that of mRNA, whereas U snRNPs U1, U2, U4 and U5 were enriched in the N2, yet they were also prominent in the C and N1 fractions, and U6 was distributed evenly between the C, N1 and N2 fractions (Figure 4D, lanes 1–3). This pattern, particularly the high levels of U6 in the non-spliceosomal compartments, is characteristic of 293 cells and was expected from our previous studies (37), further validating the quality of cell fractionation. We found that RBM15 and OTT3 silencing did not considerably affect the nucleocytoplasmic distribution of tRNA (Figure 4D, tRNA), but led to a marked reduction of all U snRNPs in the N2 fraction (Figure 4D, compare lanes 3, 6 and 9). Indirect immunofluorescence analysis of the endogenous proteins did not reveal any abnormal localization of NXF1, DBP5 and SC35 proteins, confirming the integrity of subcellular compartments and indicating that the reduction of U snRNPs in the N2 fraction was not accompanied by morphological changes in SFC (data not shown). While RBM15 silencing caused some decrease in C/N1 ratios of U1, U2, U4, U6 but not U5 (lanes 4 and 5), the silencing of OTT3 did not lead to changes (lanes 7 and 8). Since RBM15 is part of the spliceosome (53), associates with U2AF35 (Figure 1D), and is present in the SFC (Figure 3B), there could be a direct effect on U snRNP metabolism. However, the observed redistributions were not accompanied by a reduction in the nuclear levels of general mRNA (Figure 4C) or individual spliced transcripts (Figure 5), indicating that general splicing was not inhibited. In summary, we concluded that RBM15 and OTT3 silencing led to general mRNA export inhibition, whereas the nucleocytoplasmic ratios of other RNA classes were not significantly affected (tRNA) or were reduced (U snRNPs).
Gene silencing of RBM15 and OTT3 inhibits the export of endogenous spliced transcripts
Since our general mRNA analysis (Figure 4C) was semi-quantitative, we quantified the observed effects using real-time RT–PCR (RT–qPCR) detection of the endogenous transcripts representing the abundant (β-actin, ACTB) and lower abundance (inositol hexakisphosphate kinase 1, IP6K1) species (Figure 5). At day 4, we found that in the cells transfected with non-targeting siRNA, both reporters were enriched in the cytoplasmic fraction, as expected, while silencing of RBM15 as well as of OTT3 led to a significant cytoplasmic depletion/nuclear accumulation of IP6K1 and ACTB transcripts (Figure 5A). Under the same conditions, the export of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA was affected to lesser extent, not allowing to evaluate the significance (data not shown). By comparing the nucleocytoplasmic ratios, we found that OTT3 silencing caused ∼10-fold, and RBM15 silencing ∼5-fold inhibition of both IP6K1 and ACTB mRNA export (Figure 5B, fold export inhibition).
To study the effects on the total transcript levels, we purified RNA from unfractionated cells and quantified the IP6K1, ACTB and GAPDH mRNAs with RT–qPCR (Figure 5C). These analyses showed an increase of all reporters at day 4 upon OTT3 depletion, while RBM15 depletion had a lesser effect (Figure 5C, day 4), whereas at day 2 neither of the reporters showed an appreciable increase (Figure 5C, day 2). Since all transcripts analyzed exhibited similar trends, we averaged their up-regulation values for each siRNA treatment (Figure 5C, fold up-regulation) and found them in good agreement with the general mRNA data (Figure 4C), verifying an increase of general mRNA levels at later stages of OTT3 depletion. In summary, our analyses of individual transcripts confirmed that RBM15 and OTT3 silencing led to a specific inhibition of mRNA export. The observed mRNA export phenotypes strongly suggest that RBM15 and OTT3 are necessary to support the physiological mRNA export rates in human cultured cells.
DISCUSSION
This work shows that a previously described mRNA export factor RBM15 (30) and its cognate OTT3 protein (29) are necessary for the efficient nuclear export of general mRNA, and that RBM15 can facilitate the access of DBP5 to mRNP. The mRNA export phenotype of OTT3 is in agreement with our work showing that both RBM15 (30) and OTT3 (58) possess mRNA export activity. While RBM15 and OTT3 might have related but independent roles in mRNA metabolism, it is also possible that the two proteins are integral parts of one functional entity, so that a depletion of either component affects the function. Our recent work supports the latter view and suggests that RBM15 and OTT3 can form mixed complexes in vivo (58). Further studies are required to understand the molecular details and functional relevance of RBM15–OTT3 association. We noted that RBM15 and OTT3 silencing did not lead to a complete block of mRNA export and cell death. Although this could be attributed to incomplete depletions, we favor the view that RBM15 and OTT3 act to increase the efficiency of mRNA export rather than being indispensable for the basic translocation mechanism. In support of this idea, stable shRNA-mediated depletions of the RBM15 ortholog in mouse cultured cells were non-lethal (28); whereas mouse genetic knockouts caused drastic changes of mRNA expression profiles in homozygous embryos, but these embryos survived until a relatively late developmental stage (27,58,73), pointing to a defect less severe than that of e.g. NXF1 depletion (74,75). Regarding the contribution of Dbp5/Gle1 to metazoan mRNA export, RNAi interference with the orthologues of Dbp5 and Gle1 as well as Dbp5-binding nucleoporin Nup159 did not lead to general mRNA export block in Drosophila cultured cells (11,76). In mammals, the Dbp5/Gle1 mechanism remains to be studied systematically, yet it is interesting to note that siRNA silencing of Nup42/Rip1/hCG1 as well as Nup159/NUP214 in human cells had subtle mRNA phenotypes and did not block the general export (23,77), whereas homozygosity for mutant human GLE1 alleles led to a neurological condition that was late embryonic lethal, and was accompanied by changes in mRNA expression profiles (78,79). Thus, some components of the Dbp5/Gle1 machinery in metazoa could be dispensable for the basic, constitutive mRNA export, suggesting their more specialized or/and regulatory roles. Similarly, the strong yet non-lethal mRNA export phenotypes of RBM15 and OTT3 knockdowns are compatible with regulatory roles. Plausibly, the ability of RBM15 to bind DBP5 and NXF1 and to recruit DBP5 to mRNA could be key to such roles. We propose a model in which RBM15 directs DBP5 to NXF1-mRNP complexes at the NPC and precludes DBP5’s non-productive engagement with other RNP, thereby acting to promote the NXF1-mRNP remodeling and serving as a specificity control of export substrate. In this scenario, RBM15’s loss of function is expected to reduce the access of DBP5 to mRNA at NPC and consequently lower the export rates, in agreement with our gene silencing data. Because DBP5 did not crosslink to mRNA significantly at physiological RBM15 concentrations, our assays could not address directly the effects of RBM15 depletion on DBP5–mRNA interactions, and further studies are needed to delineate the affected steps. We noted that the transcripts that responded to silencing were spliced, had poly(A) tails indicative of intact, mature mRNA and accumulated in the soluble nuclear fraction, suggesting they were part of post-splicing, export-ready mRNP complexes (36,37,42,54–56) and pointing to a likely defect in NPC translocation. Our model further implies that a direct, high affinity binding of pre-mRNA to RBM15 may allow targeting to DBP5 in a manner that is independent of splicing-mediated NXF1 deposition, resulting in export prior to splicing. Indeed, our previous work showed that the otherwise nuclear-retained, unspliced CAT transcripts were efficiently exported from the nucleus upon tethering of RBM15’s NXF1 and DBP5-binding region (30), and similar activity was demonstrated for OTT3 (58). Importantly, our model was drawn from studies of mammalian proteins. Because there are no known lethal alleles or RNAi for the putative RBM15 and OTT3 orthologs in C. elegans and D. melanogaster, they could be non-essential, and there are no obvious structural homologs in yeast. We therefore speculate that RBM15 could be a recent acquisition by the ancient Mex67/Dbp5 machinery, and future studies may reveal older mechanism(s) ensuring Dbp5’s selective action on Mex67-containing mRNA. Our study has identified a crucial function for human RBM15 protein in general mRNA export and provides a new mechanism by which RBM15 facilitates the access of RNP remodeling factor DBP5 to its substrate, general mRNA.
FUNDING
Funding for open access charge: Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank K. Nagashima and S. Lockett for microscopy analyses, E. Izaurralde for reagents, J. Hanover and G. R. Pilkington for comments, and T. Jones for editorial assistance.
REFERENCES
- 1.Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, Jantsch MF, Konrat R, Blasi U, Schroeder R. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4:118–130. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- 2.Linder P. Dead-box proteins: a family affair—active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanner NK, Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 4.Jankowsky E, Bowers H. Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases. Nucl. Acids Res. 2006;34:4181–4188. doi: 10.1093/nar/gkl410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 6.Chan CC, Dostie J, Diem MD, Feng W, Mann M, Rappsilber J, Dreyfuss G. eIF4A3 is a novel component of the exon junction complex. RNA. 2004;10:200–209. doi: 10.1261/rna.5230104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ballut L, Marchadier B, Baguet A, Tomasetto C, Seraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat. Struct. Mol. Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- 8.Jensen TH, Boulay J, Rosbash M, Libri D. The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr. Biol. 2001;11:1711–1715. doi: 10.1016/s0960-9822(01)00529-2. [DOI] [PubMed] [Google Scholar]
- 9.Herold A, Teixeira L, Izaurralde E. Genome-wide analysis of nuclear mRNA export pathways in Drosophila. EMBO J. 2003;22:2472–2483. doi: 10.1093/emboj/cdg233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linder P, Stutz F. mRNA export: travelling with DEAD box proteins. Curr. Biol. 2001;11:R961–R963. doi: 10.1016/s0960-9822(01)00574-7. [DOI] [PubMed] [Google Scholar]
- 11.Gatfield D, Le Hir H, Schmitt C, Braun IC, Kocher T, Wilm M, Izaurralde E. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr. Biol. 2001;11:1716–1721. doi: 10.1016/s0960-9822(01)00532-2. [DOI] [PubMed] [Google Scholar]
- 12.Luo ML, Zhou Z, Magni K, Christoforides C, Rappsilber J, Mann M, Reed R. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature. 2001;413:644–647. doi: 10.1038/35098106. [DOI] [PubMed] [Google Scholar]
- 13.Tseng SS, Weaver PL, Liu Y, Hitomi M, Tartakoff AM, Chang TH. Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J. 1998;17:2651–2662. doi: 10.1093/emboj/17.9.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snay-Hodge CA, Colot HV, Goldstein AL, Cole CN. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 1998;17:2663–2676. doi: 10.1093/emboj/17.9.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitt C, von Kobbe C, Bachi A, Pante N, Rodrigues JP, Boscheron C, Rigaut G, Wilm M, Seraphin B, Carmo-Fonseca M, et al. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J. 1999;18:4332–4347. doi: 10.1093/emboj/18.15.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cole CN, Scarcelli JJ. Transport of messenger RNA from the nucleus to the cytoplasm. Curr. Opin. Cell Biol. 2006;18:299–306. doi: 10.1016/j.ceb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 17.Stewart M. Ratcheting mRNA out of the nucleus. Mol. Cell. 2007;25:327–330. doi: 10.1016/j.molcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 18.Linder P. mRNA export: RNP remodeling by DEAD-box proteins. Curr. Biol. 2008;18:R297–R299. doi: 10.1016/j.cub.2008.02.027. [DOI] [PubMed] [Google Scholar]
- 19.Weirich CS, Erzberger JP, Flick JS, Berger JM, Thorner J, Weis K. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat. Cell Biol. 2006;8:668–676. doi: 10.1038/ncb1424. [DOI] [PubMed] [Google Scholar]
- 20.Alcazar-Roman AR, Tran EJ, Guo S, Wente SR. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat. Cell Biol. 2006;8:711–716. doi: 10.1038/ncb1427. [DOI] [PubMed] [Google Scholar]
- 21.Alcazar-Roman AR, Wente SR. Inositol polyphosphates: a new frontier for regulating gene expression. Chromosoma. 2008;117:1–13. doi: 10.1007/s00412-007-0126-4. [DOI] [PubMed] [Google Scholar]
- 22.Lund MK, Guthrie C. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol. Cell. 2005;20:645–651. doi: 10.1016/j.molcel.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 23.Kendirgi F, Rexer DJ, Alcazar-Roman AR, Onishko HM, Wente SR. Interaction between the shuttling mRNA export factor Gle1 and the nucleoporin hCG1: a conserved mechanism in the export of Hsp70 mRNA. Mol. Biol. Cell. 2005;16:4304–4315. doi: 10.1091/mbc.E04-11-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tran EJ, Zhou Y, Corbett AH, Wente SR. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol. Cell. 2007;28:850–859. doi: 10.1016/j.molcel.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 25.Gross T, Siepmann A, Sturm D, Windgassen M, Scarcelli JJ, Seedorf M, Cole CN, Krebber H. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science. 2007;315:646–649. doi: 10.1126/science.1134641. [DOI] [PubMed] [Google Scholar]
- 26.Tan W, Zolotukhin AS, Tretyakova I, Bear J, Lindtner S, Smulevitch SV, Felber BK. Identification and characterization of the mouse nuclear export factor (Nxf) family members. Nucleic Acids Res. 2005;33:3855–3865. doi: 10.1093/nar/gki706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raffel GD, Mercher T, Shigematsu H, Williams IR, Cullen DE, Akashi K, Bernard OA, Gilliland DG. Ott1(Rbm15) has pleiotropic roles in hematopoietic development. Proc. Natl Acad. Sci. USA. 2007;104:6001–6006. doi: 10.1073/pnas.0609041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma X, Renda MJ, Wang L, Cheng EC, Niu C, Morris SW, Chi AS, Krause DS. Rbm15 modulates Notch-induced transcriptional activation and affects myeloid differentiation. Mol. Cell Biol. 2007;27:3056–3064. doi: 10.1128/MCB.01339-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hiriart E, Gruffat H, Buisson M, Mikaelian I, Keppler S, Meresse P, Mercher T, Bernard OA, Sergeant A, Manet E. Interaction of the Epstein-Barr virus mRNA export factor EB2 with human Spen proteins SHARP, OTT1, and a novel member of the family, OTT3, links Spen proteins with splicing regulation and mRNA export. J. Biol. Chem. 2005;280:36935–36945. doi: 10.1074/jbc.M501725200. [DOI] [PubMed] [Google Scholar]
- 30.Lindtner S, Zolotukhin AS, Uranishi H, Bear J, Kulkarni V, Smulevitch S, Samiotaki M, Panayotou G, Felber BK, Pavlakis GN. RNA-binding motif protein 15 binds to the RNA transport element RTE and provides a direct link to the NXF1 export pathway. J. Biol. Chem. 2006;281:36915–36928. doi: 10.1074/jbc.M608745200. [DOI] [PubMed] [Google Scholar]
- 31.Smulevitch S, Bear J, Alicea C, Rosati M, Jalah R, Zolotukhin AS, von Gegerfelt A, Michalowski D, Moroni C, Pavlakis GN, et al. RTE and CTE mRNA export elements synergistically increase expression of unstable, Rev-dependent HIV and SIV mRNAs. Retrovirology. 2006;3:6–15. doi: 10.1186/1742-4690-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zolotukhin AS, Schneider R, Uranishi H, Bear J, Tretyakova I, Michalowski D, Smulevitch S, O'K;eeffe S, Pavlakis GN, Felber BK. The RNA transport element RTE is essential for IAP LTR-retrotransposon mobility. Virology. 2008;377:88–99. doi: 10.1016/j.virol.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 33.Bear J, Tan W, Zolotukhin AS, Tabernero C, Hudson EA, Felber BK. Identification of novel import and export signals of human TAP, the protein that binds to the constitutive transport element of the type D retrovirus mRNAs. Mol. Cell Biol. 1999;19:6306–6317. doi: 10.1128/mcb.19.9.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tretyakova I, Zolotukhin AS, Tan W, Bear J, Propst F, Ruthel G, Felber BK. NXF family protein participates in cytoplasmic mRNA trafficking. J. Biol. Chem. 2005;280:31981–31990. doi: 10.1074/jbc.M502736200. [DOI] [PubMed] [Google Scholar]
- 35.Katahira J, Strasser K, Podtelejnikov A, Mann M, Jung JU, Hurt E. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinol-Roma S, Choi YD, Matunis MJ, Dreyfuss G. Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev. 1988;2:215–227. doi: 10.1101/gad.2.2.215. [DOI] [PubMed] [Google Scholar]
- 37.Zolotukhin AS, Tan W, Bear J, Smulevitch S, Felber BK. U2AF participates in the binding of TAP (NXF1) to mRNA. J. Biol. Chem. 2002;277:3935–3942. doi: 10.1074/jbc.M107598200. [DOI] [PubMed] [Google Scholar]
- 38.Le Hir H, Gatfield D, Braun IC, Forler D, Izaurralde E. The protein Mago provides a link between splicing and mRNA localization. EMBO Rep. 2001;2:1119–1124. doi: 10.1093/embo-reports/kve245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim VN, Yong J, Kataoka N, Abel L, Diem MD, Dreyfuss G. The Y14 protein communicates to the cytoplasm the position of exon-exon junctions. EMBO J. 2001;20:2062–2068. doi: 10.1093/emboj/20.8.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kataoka N, Yong J, Kim VN, Velazquez F, Perkinson RA, Wang F, Dreyfuss G. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell. 2000;6:673–682. doi: 10.1016/s1097-2765(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 41.Huang Y, Steitz JA. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell. 2001;7:899–905. doi: 10.1016/s1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- 42.Huang Y, Yario TA, Steitz JA. A molecular link between SR protein dephosphorylation and mRNA export. Proc. Natl Acad. Sci. USA. 2004;101:9666–9670. doi: 10.1073/pnas.0403533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hargous Y, Hautbergue GM, Tintaru AM, Skrisovska L, Golovanov AP, Stevenin J, Lian LY, Wilson SA, Allain FHT. Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8. EMBO J. 2006;25:5126–5137. doi: 10.1038/sj.emboj.7601385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu S, Romfo CM, Nilsen TW, Green MR. Functional recognition of the 3' splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- 45.Zhang M, Zamore PD, Carmo-Fonseca M, Lamond AI, Green MR. Cloning and intracellular localization of the U2 small nuclear ribonucleoprotein auxiliary factor small subunit. Proc. Natl Acad. Sci. USA. 1992;89:8769–8773. doi: 10.1073/pnas.89.18.8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Z, Luo MJ, Straesser K, Katahira J, Hurt E, Reed R. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 2000;407:401–405. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]
- 47.Zenklusen D, Vinciguerra P, Strahm Y, Stutz F. The yeast hnRNP-like proteins Yra1p and Yra2p participate in mRNA export through interaction with Mex67p. Mol. Cell Biol. 2001;21:4219–4232. doi: 10.1128/MCB.21.13.4219-4232.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stutz F, Bachi A, Doerks T, Braun IC, Seraphin B, Wilm M, Bork P, Izaurralde E. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA. 2000;6:638–650. doi: 10.1017/s1355838200000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hautbergue GM, Hung ML, Golovanov AP, Lian LY, Wilson SA. Mutually exclusive interactions drive handover of mRNA from export adaptors to TAP. Proc. Natl Acad. Sci. USA. 2008;105:5154–5159. doi: 10.1073/pnas.0709167105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Golovanov AP, Hautbergue GM, Tintaru AM, Lian LY, Wilson SA. The solution structure of REF2-I reveals interdomain interactions and regions involved in binding mRNA export factors and RNA. RNA. 2006;12:1933–1948. doi: 10.1261/rna.212106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kiesler E, Miralles F, Visa N. HEL/UAP56 binds cotranscriptionally to the Balbiani ring pre-mRNA in an intron-independent manner and accompanies the BR mRNP to the nuclear pore. Curr. Biol. 2002;12:859–862. doi: 10.1016/s0960-9822(02)00840-0. [DOI] [PubMed] [Google Scholar]
- 52.Kapadia F, Pryor A, Chang TH, Johnson LF. Nuclear localization of poly(A)+ mRNA following siRNA reduction of expression of the mammalian RNA helicases UAP56 and URH49. Gene. 2006;384:37–44. doi: 10.1016/j.gene.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 53.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 54.Lai MC, Tarn WY. Hypophosphorylated ASF/SF2 binds TAP and is present in mRNPs. J. Biol. Chem. 2004;279:31745–31749. doi: 10.1074/jbc.C400173200. [DOI] [PubMed] [Google Scholar]
- 55.Mili S, Shu HJ, Zhao Y, Pinol-Roma S. Distinct RNP complexes of shuttling hnRNP proteins with pre-mRNA and mRNA: candidate intermediates in formation and export of mRNA. Mol. Cell Biol. 2001;21:7307–7319. doi: 10.1128/MCB.21.21.7307-7319.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin S, Xiao R, Sun P, Xu X, Fu XD. Dephosphorylation-dependent sorting of SR splicing factors during mRNP maturation. Mol. Cell. 2005;20:413–425. doi: 10.1016/j.molcel.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 57.Zolotukhin AS, Michalowski D, Bear J, Smulevitch SV, Traish AM, Peng R, Patton J, Shatsky IN, Felber BK. PSF acts through the human immunodeficiency virus type 1 mRNA instability elements to regulate virus expression. Mol. Cell Biol. 2003;23:6618–6630. doi: 10.1128/MCB.23.18.6618-6630.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uranishi H, Zolotukhin AS, Lindtner S, Warming S, Zhang GM, Bear J, Copeland NG, Jenkins NA, Pavlakis GN, Felber BK. The RNA binding motif protein 15B (RBM15B/OTT3) acts as co-factor of the nuclear export receptor NXF1. J. Biol. Chem. 2009;284:26106–26116. doi: 10.1074/jbc.M109.040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Herold A, Suyama M, Rodrigues JP, Braun IC, Kutay U, Carmo-Fonseca M, Bork P, Izaurralde E. TAP (NXF1) belongs to a multigene family of putative RNA export factors with a conserved modular architecture. Mol. Cell Biol. 2000;20:8996–9008. doi: 10.1128/mcb.20.23.8996-9008.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bachi A, Braun IC, Rodrigues JP, Pante N, Ribbeck K, von Kobbe C, Kutay U, Wilm M, Gorlich D, Carmo-Fonseca M, et al. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA. 2000;6:136–158. doi: 10.1017/s1355838200991994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Terasaki M, Reese TS. Characterization of endoplasmic reticulum by co-localization of BiP and dicarbocyanine dyes. J. Cell Sci. 1992;101:315–322. doi: 10.1242/jcs.101.2.315. [DOI] [PubMed] [Google Scholar]
- 62.Munter S, Enninga J, Vazquez-Martinez R, Delbarre E, David-Watine B, Nehrbass U, Shorte S. Actin polymerisation at the cytoplasmic face of eukaryotic nuclei. BMC Cell Biol. 2006;7:23. doi: 10.1186/1471-2121-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 64.van Dijk EL, Schilders G, Pruijn GJ. Human cell growth requires a functional cytoplasmic exosome, which is involved in various mRNA decay pathways. RNA. 2007;13:1027–1035. doi: 10.1261/rna.575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Machida YJ, Dutta A. The APC/C inhibitor, Emi1, is essential for prevention of rereplication. Genes Dev. 2007;21:184–194. doi: 10.1101/gad.1495007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chattopadhyay S, Bielinsky AK. Human Mcm10 regulates the catalytic subunit of DNA polymerase-alpha and prevents DNA damage during replication. Mol. Biol. Cell. 2007;18:4085–4095. doi: 10.1091/mbc.E06-12-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boehmer T, Enninga J, Dales S, Blobel G, Zhong H. Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc. Natl Acad. Sci. USA. 2003;100:981–985. doi: 10.1073/pnas.252749899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Osmundson EC, Ray D, Moore FE, Gao Q, Thomsen GH, Kiyokawa H. The HECT E3 ligase Smurf2 is required for Mad2-dependent spindle assembly checkpoint. J. Cell Biol. 2008;183:267–277. doi: 10.1083/jcb.200801049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jarmolowski A, Boelens WC, Izaurralde E, Mattaj IW. Nuclear export of different classes of RNA is mediated by specific factors. J. Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rodriguez MS, Dargemont C, Stutz F. Nuclear export of RNA. Biol. Cell. 2004;96:639–655. doi: 10.1016/j.biolcel.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 71.Shaw DJ, Eggleton P, Young PJ. Joining the dots: Production, processing and targeting of U snRNP to nuclear bodies. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2008;1783:2137–2144. doi: 10.1016/j.bbamcr.2008.07.025. [DOI] [PubMed] [Google Scholar]
- 72.Kutay U, Lipowsky G, Izaurralde E, Bischoff FR, Schwarzmaier P, Hartmann E, Gorlich D. Identification of a tRNA-specific nuclear export receptor. Mol. Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- 73.Raffel GD, Chu GC, Jesneck JL, Cullen DE, Bronson RT, Bernard OA, Gilliland DG. Ott1(Rbm15) is essential for placental vascular branching morphogenesis and embryonic development of the heart and spleen. Mol. Cell Biol. 2009;29:333–341. doi: 10.1128/MCB.00370-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Herold A, Klymenko T, Izaurralde E. NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA. 2001;7:1768–1780. [PMC free article] [PubMed] [Google Scholar]
- 75.Tan W, Zolotukhin AS, Bear J, Patenaude DJ, Felber BK. The mRNA export in Caenorhabditis elegans is mediated by Ce-NXF-1, an ortholog of human TAP/NXF and Saccharomyces cerevisiae Mex67p. RNA. 2000;6:1762–1772. doi: 10.1017/s1355838200000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Farny NG, Hurt JA, Silver PA. Definition of global and transcript-specific mRNA export pathways in metazoans. Genes Dev. 2008;22:66–78. doi: 10.1101/gad.1616008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hutten S, Kehlenbach RH. Nup214 is required for CRM1-dependent nuclear protein export in vivo. Mol. Cell Biol. 2006;26:6772–6785. doi: 10.1128/MCB.00342-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nousiainen HO, Kestila M, Pakkasjarvi N, Honkala H, Kuure S, Tallila J, Vuopala K, Ignatius J, Herva R, Peltonen L. Mutations in mRNA export mediator GLE1 result in a fetal motoneuron disease. Nat. Genet. 2008;40:155–157. doi: 10.1038/ng.2007.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pakkasjarvi N, Gentile M, Saharinen J, Honkanen J, Herva R, Peltonen L, Kestila M. Indicative oligodendrocyte dysfunction in spinal cords of human fetuses suffering from a lethal motoneuron disease. J. Neurobiol. 2005;65:269–281. doi: 10.1002/neu.20185. [DOI] [PubMed] [Google Scholar]