Abstract
LlaGI is a single polypeptide restriction–modification enzyme encoded on the naturally-occurring plasmid pEW104 isolated from Lactococcus lactis ssp. cremoris W10. Bioinformatics analysis suggests that the enzyme contains domains characteristic of an mrr endonuclease, a superfamily 2 DNA helicase and a γ-family adenine methyltransferase. LlaGI was expressed and purified from a recombinant clone and its properties characterised. An asymmetric recognition sequence was identified, 5′-CTnGAyG-3′ (where n is A, G, C or T and y is C or T). Methylation of the recognition site occurred on only one strand (the non-degenerate dA residue of 5′-CrTCnAG-3′ being methylated at the N6 position). Double strand DNA breaks at distant, random sites were only observed when two head-to-head oriented, unmethylated copies of the site were present; single sites or pairs in tail-to-tail or head-to-tail repeat only supported a DNA nicking activity. dsDNA nuclease activity was dependent upon the presence of ATP or dATP. Our results are consistent with a directional long-range communication mechanism that is necessitated by the partial site methylation. In the accompanying manuscript [Smith et al. (2009) The single polypeptide restriction–modification enzyme LlaGI is a self-contained molecular motor that translocates DNA loops], we demonstrate that this communication is via 1-dimensional DNA loop translocation. On the basis of this data and that in the third accompanying manuscript [Smith et al. (2009) An Mrr-family nuclease motif in the single polypeptide restriction–modification enzyme LlaGI], we propose that LlaGI is the prototype of a new sub-classification of Restriction-Modification enzymes, named Type I SP (for Single Polypeptide).
INTRODUCTION
LlaGI is a restriction–modification (RM) enzyme isolated from Lactococcus lactis ssp. cremoris W10 (1,2). It is encoded on the epigenetic element pEW104 and comprises the fusion of four putative protein domains (Figure 1A and B): an mrr-family nuclease (3,4); a Superfamily 2 (SF2) helicase (1; a γ-type adenine methyltransferase (MTase) (1); and a target recognition domain (TRD) (1). Comparing the LlaGI primary amino acid sequences to the bacterial and archaeal sequences currently listed in the REBASE database (2) identifies 69 additional single polypeptide, helicase-containing RM enzymes (Table 1). Of these, two distinct groupings were noted on the basis of characteristic Walker A box sequences in their helicase domains: those with a GTGKT sequence which includes LlaGI (56 members) and those with an RFGKT sequence (14 members). Studies of Lactococcus bacteriophage infection show that LlaGI has efficient RM activity, producing at least a thousand-fold decrease in infectivity (1). However, details of the reaction mechanism of LlaGI, or any of the related enzymes, are not known. Whilst helicase-nuclease domain fusions are also found in the ATP-dependent Type I and Type III RM enzymes (5,6), and nuclease-MTase domain fusions are typical to many sub-classes of Type II RM enzymes (7–9), the fusion of all three elements is unique to LlaGI and related enzymes. Thus LlaGI can be considered the prototype of a new sub-type of RM enzymes. On the basis of similarities in amino acid motifs and domain organisation, Madsen and Josephsen (1) suggested that LlaGI is a variant of the Type I RM enzymes. Here, and in the accompanying papers (10,11), we provide an in-depth biochemical analysis of the in vitro enzymatic activities of LlaGI. On the basis of this data we discuss below the relationship of LlaGI to other RM enzymes of Types I, II and III, and suggest a suitable familial classification that is consistent with both domain organisation and function.
Figure 1.
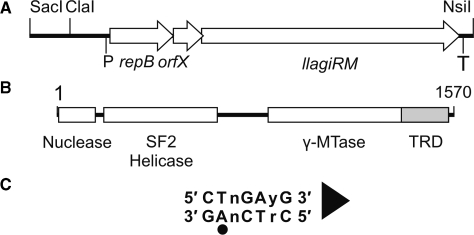
The LlaGI RM enzyme. (A) The SacI-NsiI region of pEW104 identified previously as sufficient for in vivo RM activity (1). The approximate locations of the putative promoter (P) and terminator (T) of the operon are indicated. (B) The llagiRM gene product is a single polypeptide, multi-domain protein (1). Protein domain boundaries were estimated using secondary structure prediction and alignment with LlaBIII (not shown). Target Recognition Domain (TRD). (C) Recognition sequence of LlaGI. The adenine residue that is methylated is indicated by a circle. The arrowhead defines the directionality of the site according to cleavage (Figure 4) and translocation assays (10). The ‘top strand’ sequence (i.e. that starting with CT) is quoted throughout.
Table 1.
LlaGI-like sequences identified from REBASE (2)
| GTGKT subgroup (56 sequences) | ||
| AauTCORF264P | CdpORF453P | LlaSKORF6P |
| AavORF578P | CglORF3009P | LmoDORFAP |
| AspFBORF4323P | CsyMAORFGP | MboAORF2049P |
| AviDJORF22890P | DvuORF2175P | MboBCGORF2043P |
| BfrYORF1980P | EsaRM24P | MboBCGTORF2038P |
| BheHORF15450P | GthNGORF2804P | MpePMORF102P |
| BovEORF2337P | HacSORF1335P | MspELB17ORFDP |
| BtrCIPORF1006P | Hpy99ORF612P | MtuCTORF2082P |
| BtrCIPORF1021P | HpyAORF668P | MtuFORF12055P |
| BtrCIPORF1035P | HpyGORF629P | Mxa6833447ORFDP |
| BtrCIPORF1053P | HpyHORF652P | Mxa6833447ORFFP |
| BtrCIPORF1064P | HpyPORF680P | MxaDKORF1808P |
| BtrCIPORF1080P | HpySORF3515P | NeuCORF2561P |
| BtrCIPORF1105P | Hso2336ORF1665P | NfaORF2220P |
| BtrCIPORF14P | LbiPAORF5032P | Pdi8503ORF1520P |
| BtrCIPORF164P | LbiPORF33P | PsyTORF37P |
| BtrCIPORF2491P | LinCORF21P | RspKDORF4062P |
| BtrCIPORF455P | LlaBIII | Tha33315P |
| BtrCIPORF541P | LlaGI | |
| RFGKT subgroup (14 sequences) | ||
| BthAHORF843P | LdeBBORF1654P | PpeRS5ORF3P |
| BthVORF4013P | LlaBIIP | Sth1066ORF1376P |
| DolHORF905P | NspJCORF1588P | Sth18311ORF1376P |
| LacORF475P | PgiCORF415P | SthLMDORF13303P |
| Lbr367ORF22P | PgiTORF1696P | |
Two distinct subgroups were observed with distinct sequences at helicase Motif I (the Walker A box). Note that some of these sequences have N-terminal deletions and/or mutations that may result in an inactive nuclease.
MATERIALS AND METHODS
DNA
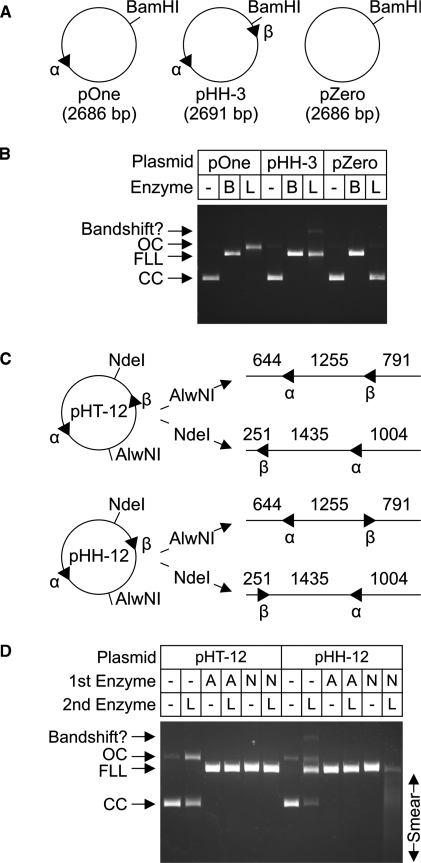
All oligonucleotides were supplied by MWG biotech (Germany). Plasmid pOne is pUC19 (12), and is re-named here for clarity. pZero was derived from pOne by QuikChange mutagenesis (Stratagene, La Jolla, CA) using primers 5′-CCCGGCAACAATTAATAGATTGGATGGAGGCGGATAAAGTTGC-3′ and 5′-GCAACTTTATCCGCCTCCATCCAATCTATTAATTGTTGCCGGG-3′ (where the LlaGI sequence is highlighted in bold and the mutated base pair underlined). The library of two-site plasmids (Figure 3) was generated by directional cloning of duplex oligonucleotides into the XbaI/HindIII fragment of plasmid pOne. One strand of each duplex oligonucleotide is shown in Figure 3B (white background). Where the sites are in direct repeat (where the ‘top strand’ sequences in Figure 3 are on the same DNA strand), the plasmids are named pHT for head-to-tail. Where the sites are in indirect repeat, the plasmids are named pHH for head-to-head (note that these DNA have sites in both head-to-head and tail-to-tail orientation). The sequence of Site β is denoted by a numeric suffix (1–14), Site α being constant (Figure 3B). Linear DNA substrates for the site identification experiments were produced by PCR using Pfu polymerase as directed by the manufacturer’s recommendations. Sequences of the primers used are available from M.D.S. on request.
Figure 3.
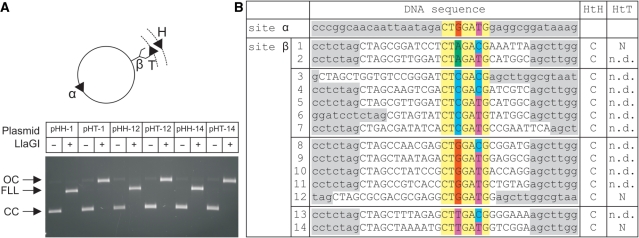
Cleavage of LlaGI recognition sequences. (A) A library of two-site DNA substrates were generated from pOne (Figure 4) by cloning different LlaGI sequences (Site β – see B). Site α was constant in each case. For three of the sequences both directly-repeated and indirectly-repeated copies of Site β were tested. Example agarose gel shown where CC is Covalently closed Circular substrate DNA, OC is Open Circle nicked intermediate and FLL is the Full Length Linear product cleaved in both strands. DNA and LlaGI were incubated for 10 min. (B) Sequences of sites tested with outcome for indirectly-repeated (HtH) and directly-repeated (HtT) arrangements of sites: C, dsDNA cleavage; N, nicking only; n.d., not determined. pOne sequence is highlighted in grey, cloned sequence in white, constant LlaGI sequence in yellow and degenerate positions in red (dG), green (dA), blue (dC), purple (dT). Sequence shown is the ‘top strand’ as defined in Figure 1C.
pRSFLlaGI was generated as follows: The llagiRM gene was amplified from pEW104 (1) by PCR using oligonucleotides 5′-GCGTAAGTCCCATGGTGGCATTTTGGAAGG-3′ and 5′-GCGTAAGTCGGATCCTTATTCTTGAATTTCAAATTCTGGTAA-3′; The PCR product was digested with NcoI and BamHI and inserted into pETDuet-1 (Novagen, Germany) digested with NcoI and BglII; The NcoI-XhoI fragment containing the llagiRM gene was then inserted into pRSFDuet-1 (Novagen, Germany); The llagiRM gene was fully sequenced. pET28OrfX was generated as follows: The orfx gene was amplified from pEW104 by PCR using oligonucleotides 5′-CGAGCCCATATGATGAGTGAAAAGTTAAAG-3′ and 5′-CTAGCTCTCGAGTCATTTTACATTCCTTCC-3′; The PCR product was digested with NdeI and XhoI and inserted into pET28a (Novagen, Germany); The his6-orfX gene was fully sequenced.
To prepare DNA for biochemical assays, E. coli Top10 (Invitrogen, CA, USA) or XL10-Gold (Stratagene) were transformed with the required plasmid, grown in LB medium and the DNA extracted using either commercial protocols (Qiagen, Hilden, Germany) or by density gradient centrifugation in CsCl-ethidium bromide (13). For experiments requiring 3H-labelled DNA, transformants were grown in M9 minimal medium supplemented with 37 MBq/l [3H-methyl] thymidine. Linear DNA substrates were generated by incubating 4 nM plasmid DNA with 1 U/μl of AlwNI or 2 U/μl of NdeI in NEBuffer 2 (New England Biolabs, MA, USA). The DNA was purified by phenol/chloroform and chloroform extraction followed by ethanol precipitation. DNA concentrations were determined from UV absorbance at 260 nm, assuming that an optical density of 1 corresponds to 50 µg/ml DNA and a molecular weight of 6.6 × 105 Da/kb.
Protein expression and purification
Wild type LlaGI was purified from E. coli BL21 (DE3) cells (Novagen, Germany) and transformed with pRSFLlaGI. Cultures were grown in 4 L of LB medium containing kanamycin (50 μg/ml) at 37°C and 250 r.p.m. until the OD600 reached ∼0.4. IPTG was added to 1 mM and the cultures were incubated further at 27°C for an hour (final OD600 of ∼1.0). Cells were harvested, re-suspended in 45 ml of Buffer A [50 mM Tris–Cl (pH 8.0), 150 mM NaCl, 5 mM MgCl2, 2.5 mM EDTA, 10% (v/v) Glycerol, 3 mM DTT, 51.7 µM phenylmethylsulfonyl fluoride, and EDTA free protease inhibitor tablets according to the manufacturer’s instructions (Roche, UK)] and lysed using a cell disruptor (Constant Systems Ltd, Northants, UK). The cell extract was clarified by centrifugation at 7358g for 10 min followed by 106 255g for 1 h 50 min. The supernatant was dialysed against Buffer B [50 mM Tris–Cl (pH 8.0), 1 mM EDTA, 50 mM NaCl, 1 mM DTT], 0.45 µm filtered and loaded onto two 20 ml HiPrep Heparin 16/10 FF columns (GE Healthcare UK Ltd) connected in series and equilibrated in Buffer B. Bound proteins were eluted with a linear gradient of NaCl in Buffer B (0.05–1.0 M, 400 ml). LlaGI was eluted at ∼0.40 M NaCl. Fractions containing LlaGI were pooled and, concentrated and equilibrated into Buffer B using 50 kDa cut-off centrifugal filter units (Millipore, MA, USA). The resulting sample was loaded onto an 8 ml Mono Q 10/10 column (GE Healthcare) pre-equilibrated with Buffer B. Bound proteins were eluted with a linear gradient of NaCl in Buffer B (0.05–1.0 M, 160 ml). LlaGI eluted at ∼0.38 M NaCl. Fractions containing LlaGI were pooled and concentrated as above. The resulting sample was further purified over a HiLoad 16/60 Superdex 200 pg column (GE Healthcare) pre-equilibrated in Buffer B containing 400 mM NaCl. Fractions containing LlaGI were pooled and concentrated as above, supplemented with glycerol to 50% (v/v) and stored at −20°C. The protein concentration was determined by the Bradford dye-binding procedure (Bio-Rad Protein Assay, Bio-Rad, Hemel Hempstead, UK) using bovine serum albumin (BSA) as standard. Similar concentrations (within experimental error) were determined by UV absorption at 280 nm using an extinction coefficient derived from the aromatic amino acid composition of the predicted amino acid sequences. The production and purification of the LlaGI nuclease mutant DA078 is described in the accompanying paper (11).
His6-tagged OrfX was purified from E. coli Tuner (DE3) cells (Novagen) transformed with pET28OrfX. Cultures were grown in 1 L LB medium containing 50 μg/ml kanamycin at 37°C and 250 r.p.m. until the OD600 reached ∼0.4. IPTG was added to 25 μM and the cultures were incubated for a further 4 h at 37°C (final OD600 of ∼4.5). Cells were harvested, re-suspended in 45 ml of Buffer C [50 mM Tris–Cl (pH 8.0), 200 mM NaCl, 10 mM Imidazole, 11.1 mg/ml lysozyme], incubated on ice for 10 min, and then lysed by cell disruption (as above). The cell extract was clarified by centrifugation at 7358g for 10 min followed by 106 255g for 37 min. The supernatant was 0.45 µm filtered, loaded onto a 5 ml HiTrap Chelating column that had been charged with NiSO4 (GE Healthcare), and washed in Buffer C. Bound proteins were eluted with a linear gradient of Imidazole in Buffer C (0.01–0.5 M, 50 ml). Fractions containing OrfX were pooled, and diluted 10-fold with Buffer B. The resulting sample was then loaded onto a 1 ml Mono Q 5/50 GL column (GE Healthcare) pre-equilibrated with Buffer B. Bound proteins were eluted with a linear gradient of NaCl in Buffer B (0.05–1.0 M, 20 ml). OrfX was eluted at ∼0.25 M NaCl. Fractions containing OrfX were pooled and dialysed against Buffer D [50 mM Tris–Cl (pH 8.0), 500 mM NaCl, 10 mM Imidazole]. The sample was then added to 20 mg Protino Ni-IDA Resin (Macherey-Nagel) and incubated at 4°C for 1 h. The beads were washed three times in Buffer D to remove unbound protein. OrfX was eluted in Buffer E [50 mM Tris–Cl (pH 8.0), 500 mM NaCl, 500 mM Imidazole] and then dialysed into Buffer B containing 400 mM NaCl. OrfX was supplemented with glycerol to 50% (v/v) and stored at −20°C. The protein concentration was determined by the Bradford dye-binding procedure using BSA as a standard.
EcoAI and EcoR124I methyltransferases (denoted by the prefix M) and HsdRs (denoted by the prefix R) were purified and characterised as described previously (data not shown) (14).
Gel filtration and N-terminal sequencing
N-terminal sequencing of LlaGI was carried out in-house at the Proteomics Facility. Analytical gel filtration was carried out using a 24 ml Superose 6 column (GE Healthcare). The column was pre-equilibrated at 0.4 ml/min with TMD buffer [50 mM Tris–HCl (pH 8.0), 10 mM MgCl2, 1 mM DTT] supplemented with 15 mM NaCl. A 100 µl loop was filled with protein sample in storage buffer [30 mM Tris–Cl (pH 8.0), 0.6 mM EDTA, 240 mM NaCl, 0.6 mM DTT, 50% (v/v) glycerol]. LlaGI was eluted with TMD plus 15 mM NaCl at 0.4 ml/min and monitored by measuring the absorbance at 280 nm. The Superose column was calibrated using thyroglobulin (669 kDa), apoferritin (443 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), BSA (66 kDa) and carbonic anhydrase (29 kDa). Blue dextran was used to calculate the void volume.
DNA cleavage assays
Cleavage assays contained 4 nM DNA (supercoiled or linear), 4 mM ATP (or other nucleotide where indicated) and LlaGI (at the concentrations indicated) in TMD buffer. Where included, S-adenosyl methionine (AdoMet) was at 100 µM. Reactions were started by adding LlaGI to a mix of ATP, DNA and AdoMet (where present) in TMD buffer and incubated at 20°C for the times indicated. Other Type II RM endonucleases were added at concentrations recommended by the suppliers. Reactions were stopped with 0.5 volumes of 3× STEB [0.1 M Tris (pH 7.5), 0.2 M EDTA, 40% (w/v) sucrose, 0.4 mg/ml bromophenol blue]. Samples were analysed by agarose gel electrophoresis and, the percentage of 3H-labelled DNA in each band per lane was ascertained by scintillation counting (13), where required.
DNA methylation assays
Competition assays using EcoAI and EcoR124I
As indicated, 4 nM pHH-7 was methylated with 200 nM LlaGIDA078 (11), 40 nM M.EcoAI (14) or 100 nM M.EcoR124I (14) in TMD buffer supplemented with 4 mM ATP and 100 µM AdoMet. For methylation by M.EcoAI, the TMD buffer was supplemented with 50 mM NaCl. The reactions were incubated for 4 h on ice, at 20°C, at 25°C or at 37°C for untreated, LlaGIDA078, M.EcoAI or M.EcoR124I, respectively. Additional AdoMet was added after 1, 2 and 3 h and additional enzyme, where required, was added after 2 h. To each sample 0.1 volumes of 0.5 M EDTA was added and the DNA purified in three steps: (i) phenol/chloroform extraction and ethanol precipitation; (ii) by Qiagen PCR kit; and (iii) phenol/chloroform extraction and ethanol precipitation. The untreated and methylated DNA were then mixed with 4 mM ATP and 100 µM AdoMet in TMD buffer (supplemented with 50 mM NaCl for the EcoAI reactions), in the presence or absence of: 200 nM LlaGI; 400 nM M.EcoAI and 600 nM R.EcoAI; or, 40 nM M.EcoR124I and 300 nM R.EcoR124I. The reactions were incubated for 10 min on ice, at 20°C, at 25°C or at 37°C for no enzyme, LlaGI, EcoAI and EcoR124I, respectively. The reactions were stopped by the addition of 0.5 volumes of 3 x STEB containing 1 mg/ml Proteinase K and heated to 67°C for 20 min. The DNA substrates and products were separated by agarose gel electrophoresis.
Relative methylation of the eight LlaGI recognition sequences
As indicated, 4 nM DNA (pZero, pOne, pHH-1, -2, -3, -6, -8, -9, -13, -14) and 200 nM LlaGIDA078 were incubated for 1 h at 37°C in NEBuffer 4 supplemented with 4 mM ATP and 100 µM [3H-methyl] AdoMet. AlwNI (0.5 U/μl) and NdeI (1 U/μl) were added to each tube and incubated further at 37°C for 1 h. The reactions were stopped by the addition of 0.1 volume 6 x STEB [0.2 M Tris (pH 7.5), 0.4 M EDTA, 80% (w/v) sucrose, 0.8 mg/ml bromophenol blue] and the DNA fragments separated by agarose gel electrophoresis. Fragment 1 containing Site α and Fragment 2 containing Site β were quantified by scintillation counting. Gel background counts were subtracted from all samples. The counts (dpm) from pZero fragment 1 (i.e. background) were subtracted from the counts of all other fragment 1 bands, and the counts from pZero Fragment 2 (i.e. background) were subtracted from the counts of all other Fragment 2 bands. The counts from each Fragment were then divided by the counts from pOne Fragment 1. The log2 of the resulting values were then calculated to give a linear scale (see legend to Figure 7).
Figure 7.
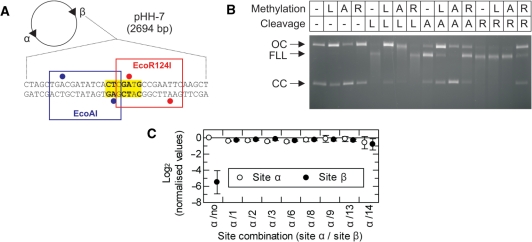
Methylation specificity of LlaGI. (A) Plasmid pHH-7 (Figure 3) showing the overlapping EcoAI (blue), LlaGI (bold and yellow) and EcoR124I (red) sequences at Site β. Adenine residues methylated by EcoAI and EcoR124I are indicated by blue and red circles, respectively (2). (B) pHH-7 was treated with LlaGIDA078 (L), M.EcoAI (A) or M.EcoR124I (R), as indicated, in the presence of AdoMet. These DNA substrates were then treated with LlaGI (L), EcoAI (A) or EcoR124I (R), as indicated, in the presence of ATP and AdoMet. The DNA was then separated by agarose gel electrophoresis. See ‘Materials and Methods’ section and main text for full details. Note, cleavage by all three enzymes produces linear DNA that is further processed to generate a DNA smear. (C) pZero, pOne and pHH−1, −2, −3, −6, −8, −9, −13 and −14 (Figure 3) were incubated with LlaGIDA078 and [3H-methyl] AdoMet for 1 h. The extent of 3H-labelling of each site was then assessed by scintillation counting (‘Materials and Methods’ section). For each plasmid, the labelling at each site was corrected for the gel background and the background from the corresponding fragments of pZero. Values were then normalised relative to Site α of pOne. These values are presented as a linear scale where positive values indicate increased labelling relative to Site α and negative values indicate reduced labelling relative to Site α. In pOne the region of Site β is random DNA and therefore acts as a non-specific control. The value of this should tend towards an infinitely small negative value. The observed value of ∼−6 therefore suggests a small background labelling of non-specific sites when a specific site is present on the DNA (i.e. the background labelling is higher than on pZero). In each plasmid the sequence at Site α is the same. Therefore, the values should all be zero, as in plasmid pOne. Given that the values for Sites α and β were very similar on each DNA, small variations between plasmids most likely represent different efficiencies of methylation that arise from variations in DNA preparation quality. Error bars represent the standard error from three repeat experiments.
RESULTS
Site specificity of DNA cleavage by LlaGI
The earlier study of LlaGI identified a region of pEW104 encompassed by the SacI and NsiI restriction sites as sufficient for in vivo RM activity (1). This region contains three genes: repB, orfX and llagiRM that appear to be co-transcribed from a single promoter (Figure 1A). Whilst a similar gene arrangement is seen in some enzymes that are closely related to LlaGI (e.g. LlaBIII), it is not observed in other cases (e.g. RM.GthNGORF2804P) (2). Genes within a single operon may participate in a common metabolic pathway, but this is not always the case: co-transcribed proteins often simply perform broadly similar general functions, and may even have completely unrelated functions (15,16). repB produces a protein associated with plasmid replication (17). orfX is often found associated with repB and a similar ORF has been identified in many lactococcal theta-replicating plasmids (18–21). The precise function of the orfX product remains mostly unknown: in some plasmids it may participate in copy number, stability, or both (19,21,22); however, it has been shown to be dispensable for plasmid replication in other cases and is sometimes completely absent (23). To determine whether the product of the llagiRM gene alone was sufficient for site-specific DNA recognition, methylation and cleavage, and whether orfX participated in these processes, we produced recombinant clones of native llagiRM and His-tagged orfX (‘Materials and Methods’ section).
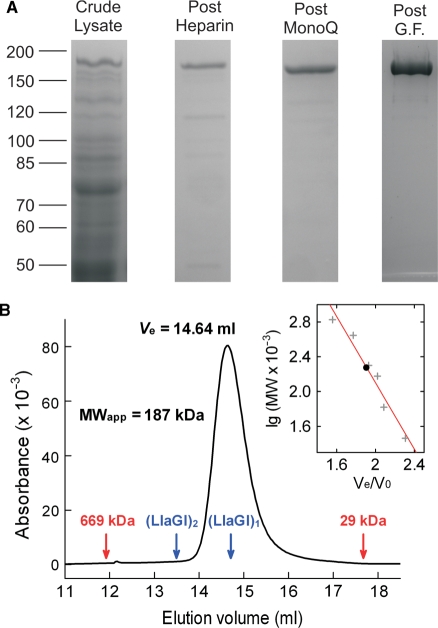
Native LlaGI was over-expressed and purified to >94% homogeneity using a three column method (Figure 2A; ‘Materials and Methods’ section). (Induction of LlaGI expression was carried out at 27°C to optimise enzyme yield—data not shown). Analytical gel filtration under reaction buffer conditions (see below, ‘Materials and Methods’ section) resulted in a single peak, suggesting a single species. Using molecular weight standards to calibrate the column, we calculated an apparent MW for LlaGI of 187 kDa, similar to the predicted MW of 179 kDa and consistent with a monomeric species. N-terminal sequencing of the purified LlaGI indicated two protein populations; full-length protein and a nine amino acid N-terminal truncation (data not shown). These two species could not be resolved by gel filtration.
Figure 2.
Purification and solution stoichiometry of LlaGI. (A) Lanes from separate SDS polyacrylamide gels showing stages in the protein purification protocol. G.F., gel filtration. The Post G.F. lane represents 5 µg of purified RM.LlaGI. See ‘Materials and Methods’ section for full details. (B) Elution profile of 263 µg (14.7 µM) LlaGI from a 24 ml Superose 6 column. 100 µl of protein sample was eluted in TMD Buffer plus 15 mM NaCl at 0.4 ml/min. Protein was monitored using absorbance at 280 nm. The elution volumes of the thyroglobulin (669 kDa) and carbonic anhydrase (29 kDa) standards are indicated in red. Indicated in blue are the putative elution volumes of a LlaGI monomer and dimer assuming a theoretical MW of 179 kDa (calculated from the full length amino acid sequence). (Inset) The apparent molecular weight of LlaGI was evaluated from the elution volume using a series of standards (‘Materials and Methods’ section). The red line indicates a least squares linear fit.
His-tagged OrfX was purified to >80% homogeneity (data not shown; ‘Materials and Methods’ section). We tested if the DNA cleavage pattern of LlaGI was altered by the inclusion of the orfx gene product. No change in DNA cleavage specificity was observed (data not shown) and site-specific DNA cleavage, methylation and loop-translocation were obtained using LlaGI in isolation [see below and refs (10,11)]. We therefore find no evidence that the close association of llagiRM-related genes to repB- and/or orxX-like genes reflects the participation of repB and the orfX gene products in the LlaGI RM system. However, the close linkage may still be important during plasmid replication so that maintenance methylation and replication are temporally linked.
To ascertain the recognition sequence of LlaGI we used an in vitro approach similar to that used previously to analyse the Type III enzyme PstII (24). A library of arbitrary DNA plasmids (data available from M.D.S. on request) were screened for susceptibility to cleavage by a >20-fold molar excess of LlaGI in the presence of ATP (predicted to be a necessary co-factor because of the presence of the helicase domain) (‘Materials and Methods’ section). Cleavage activity was largely independent of the buffer components and AdoMet was not obligatory, although some stimulation of the cleavage reaction was observed (see below and Figure 6). We did not note any differences in the cleavage patterns when we tested DNA purified from a dam-/dcm- strain (GM2929—ref. 25) (data not shown). Secondary cleavage with suitably located Type II restriction enzymes revealed that cleavage by LlaGI resulted in a smear of DNA fragments of variable lengths reminiscent of the cleavage patterns of Type I RM enzymes (see below) (26–28). We therefore, assumed that LlaGI used a similar mechanism; i.e. DNA cleavage occurs as the result of communication and collision between two enzymes originating from two recognition sites located on either side of the cleavage loci. Using primer walking, a library of PCR substrates was then generated in which one end of the DNA was iteratively shortened. These DNA substrates were tested for dsDNA cleavage. When DNA cleavage activity was lost, it was assumed that the recognition site was no longer copied by the PCR and that the LlaGI site resided in the region of DNA up to the previous primer in the series. (As part of this process, we found that LlaGI would not cut a DNA if its site was within 30 bp of a free DNA end; data not shown). Several DNA regions that were indentified as containing a possible site were analysed to identify common sequences of 6 ± 1 bp in length. To make sense of our results we also had to make the assumption that dsDNA cleavage only occurred when the sites were found in an indirectly-repeated orientation (the directionality of the site is defined in Figure 1C). This process identified the sequence 5′-CTnGAyG-3′, giving a total of eight possible LlaGI sequences.
Figure 6.
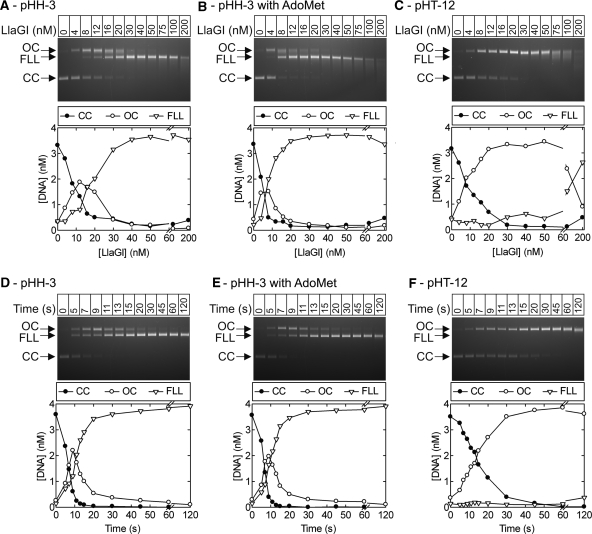
Stoichiometry and rate of DNA cleavage by LlaGI. Points are the mean of at least two repeat experiments. Note that where the FLL product becomes smeared due to end-processing, this can cause an overestimation of the CC band which overlaps with the smear. Similarly, smearing of the OC intermediate in part C causes the CC and FLL bands to be overestimated. This smearing most likely results from processing of the nicked DNA to form ssDNA gaps of different lengths (e.g. by exonucleolytic digestion from the nick). (A) pHH-3 (Figure 3) was incubated with LlaGI at the concentrations indicated for 1 hr. (B) As (A) except in the presence of 100 µM AdoMet. (C) pHT-12 (Figure 3) was incubated with LlaGI at the concentrations indicated for 1 h. (D) Time course of cleavage of pHH-3 with a saturating concentration of LlaGI (200 nM). (E) as (D) except in the presence of 100 µM AdoMet. (F) Time course of cleavage of pHT-12 with a saturating concentration of LlaGI (200 nM). Gels labelled as in Figure 3.
To confirm the recognition sequence, we created a library of two-site DNA substrates using as the parental plasmid pOne, which contained only one putative sequence, named site α (Figure 3A, ‘Materials and Methods’ section). The 14 DNA sequences tested were designed to represent the eight variations of the putative LlaGI site (i.e. all combinations of the degenerate positions) and to also test variations in adjacent non-specific sequence (Figure 3B). dsDNA cleavage (as judged by the appearance of linear DNA) was only observed when the pair of LlaGI sites were in an indirectly-repeated orientation (Figures 3A and B). All eight versions of the LlaGI site resulted in dsDNA cleavage, with no measurable preference for any other bases in the flanking DNA sequences. Plasmids are named hereafter with a numeric suffix (1–14) to indicate the ‘top strand’ sequence of Site β (and adjacent DNA), Site α being constant.
The recognition site requirements for DNA cleavage by LlaGI are summarised in Figure 4. Cleavage of circular plasmid DNA was not observed in the absence of a recognition site (pZero, Figure 4A and B). In the presence of a single site (pOne), efficient DNA nicking was observed without the formation of linear DNA. In the presence of two indirectly-repeated sites (pHH-3), dsDNA cleavage resulted in the formation of linear DNA (also for pHH-12 in Figure 4C and D). If the two sites were directly repeated (pHT-12), only DNA nicking was observed (Figure 4D), similar to the result on pOne. On the pHH-3 and pHH-12 substrates, cleavage could result from interactions between the sites via the head-to-head and/or tail-to-tail DNA arcs. We therefore also generated a series of linear DNA substrates in which the pair of LlaGI sites were oriented either head-to-tail (HT), head-to-head (HH) or tail-to-tail (TT), as defined by the site orientation in Figure 1C (Figure 4C). The plasmid DNA was cut first by a Type II RM enzymes (‘1st enzyme’, as indicated) and then treated with LlaGI (‘2nd enzyme’, as indicated) (Figure 4D). dsDNA cleavage by LlaGI was only observed when the sites were in a head-to-head orientation. Note that we cannot measure DNA nicking of the linear DNA in this assay. The smear of DNA products observed with the HH substrate had a size distribution consistent with cleavage within the region between the sites. A similar distribution of cleavage loci exclusively in the HH DNA arc was also observed following cleavage of plasmid DNA (data not shown).
Figure 4.
DNA site requirements for cleavage by LlaGI. (A and B) Plasmid substrates with no sites, one-site or two indirectly-repeated sites were incubated with either saturating BamHI (B) or LlaGI (L) for 1 h. Substrates and products were separated by agarose gel electrophoresis as indicated. (C and D) Plasmid substrates with two directly-repeated sites (pHT-12) or two indirectly-repeated sites (pHH-12) were cleaved with either AlwNI (A) or NdeI (N) to produce the linear DNA indicated. Sequences of the LlaGI sites are in Figure 3. The parental plasmids and linear DNA were then incubated with saturating LlaGI for 1 h. Substrates and products were separated by agarose gel electrophoresis as indicated. See main text for full details. Under these assay conditions, an additional slowly-migrating band was observed which we assign to a LlaGI-DNA bandshift. Gels labelled as in Figure 3.
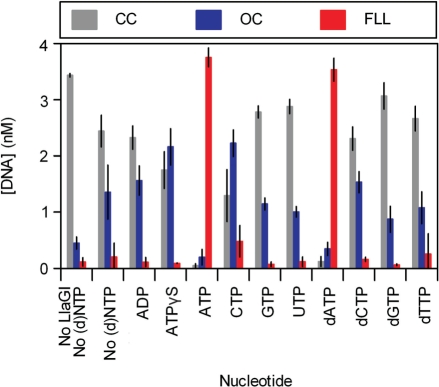
Nucleotide-dependence of the DNA cleavage activity of LlaGI
Using the two-site plasmid pHH-3, we tested the nucleotide-dependence of the cleavage reaction (Figure 5). LlaGI was incubated with 3H-labelled pHH-3 for 10 min under the conditions indicated, and the relative levels of substrate (CC), intermediate (OC) and full length linear product (FLL) quantified (‘Materials and Methods’ section). Efficient dsDNA cleavage was only observed in the presence of either ATP or dATP. In the complete absence of nucleotide, incomplete DNA nicking was observed. This nicking activity was stimulated moderately by ATPγS and CTP, but still at a reduced level compared to the activity with ATP or dATP. All subsequent assays here and in the accompanying manuscripts (10,11) were carried out with ATP.
Figure 5.
Nucleotide dependence of DNA cleavage by LlaGI. Error bars represent standard errors from at least two repeat experiments. pHH-3 was incubated with excess LlaGI for 10 min in the absence or presence of 4 mM nucleotide, as indicated. Reaction products were separated by agarose gel electrophoresis and the extent of DNA cleavage quantified (‘Materials and Methods’ section).
Rate and stoichiometry of DNA cleavage by LlaGI
The reactions above used an excess of LlaGI over DNA and were incubated until no further DNA cleavage occurred (reaction time of ≥10 min). To assess further the DNA cleavage mechanism of LlaGI, we measured the extent of DNA cleavage at the reaction ‘end point’ as a function of the LlaGI:DNA molecular ratio and measured the rate of one- and two-strand DNA cleavage at a saturating concentration of LlaGI. The effect of the MTase cofactor AdoMet was assessed in each case.
Cleavage stoichiometry
Titration of LlaGI with 3H-labelled pHH-3 in the absence of AdoMet is shown in Figure 6A. Complete DNA cleavage (as judged by the appearance of >90% FLL DNA) was only obtained with >10 LlaGI molecules per DNA molecule (five monomers per site). Below this concentration, nicked DNA accumulated. At the highest enzyme concentrations examined the linear DNA was further processed to produce a smear. The distribution of the DNA products indicates an exonuclease activity. A similar reaction profile was observed in the presence of AdoMet except that complete DNA cleavage was obtained with >5 LlaGI monomers per DNA molecule (two to three monomers per site) (Figure 6B). In addition to its role as a methyl donor, AdoMet has been widely demonstrated to have a second role for methyltransferases as an allosteric activator of DNA binding (29,30). The data in Figure 6B is consistent with such a role in LlaGI. The exonucleolytic processing of the linear product DNA also occurred at lower protein concentrations with AdoMet (note, this produces an artificial increase in the CC substrate—see legend to Figure 6).
A requirement for multiple LlaGI molecules per site can be explained by a number of different models which are not mutually exclusive: (i) dsDNA cleavage requires the interaction of multiple LlaGI molecules. For example, a requirement for a dimer of LlaGI at each recognition site would be consistent with the data in Figure 6B; (ii) The DNA-binding affinity is relatively low under our reaction conditions; or (iii) the proportion of inactive LlaGI in our preparations is relatively high (i.e. a low specific activity).
Titration of LlaGI with 3H-labelled pHT-12 in the absence of AdoMet is shown in Figure 6C. Full DNA nicking activity (>80% OC produced) was observed with >2 LlaGI molecules per site. Full nicking activity was also seen on pHH-3 at the same molar ratio (Figure 6A). This suggests that the nicking of both plasmids requires the same amount of LlaGI molecules per site but that cleavage of the second strand of pHH-3 to generate a dsDNA break requires additional enzyme molecules. At the higher enzyme concentrations, the nicked DNA band becomes smeared, producing an apparent increase in the CC concentration because of overlapping DNA mobilities.
Cleavage rate
A time course of pHH-3 cleavage in the presence of excess LlaGI (25 per site) and in the absence of AdoMet is shown in Figure 6D. The reaction profile shows that nicked DNA is a significant intermediate in the reaction, reaching a maximum of >50%. This is higher than the ∼33% expected for a reaction in which each strand cleavage rate is identical. This could arise because, either the rate of the first strand cleavage is faster than the second or, the long-range communication between the LlaGI sites collapses before both strand cleavages can occur. We cannot distinguish between these alternatives given our current data. In the presence of AdoMet, the rate and profile of DNA cleavage were very similar (Figure 6E). Whilst AdoMet can enhance protein–DNA interactions, once DNA binding is saturated, AdoMet does not alter the reaction pathway.
The rate of DNA nicking of pHT-12 in the presence of excess LlaGI and in the absence of AdoMet is shown in Figure 6F. The nicking rate was ∼3-fold slower than that observed on pHH-3 under the same conditions. The difference in these values is consistent with a different cleavage pathway for sites in direct or indirect repeat.
Site specificity of DNA methylation by LlaGI
The amino acid sequence of the MTase domain of LlaGI has an arrangement of catalytic motifs consistent with the adenine-specific γ sub-family (1,31). The eight LlaGI recognition sequences (Figures 1C and 3B) contain two invariant adenine residues, one on each strand, which are potential targets for methylation. In some sites adenine residues also occur at the degenerate positions and it is possible that these are also targets for modification. We first tested the invariant adenine residues. One of the two-site indirectly repeated plasmids described earlier (pHH-7) had been designed such that site β overlaps with the recognition sequences for the Type I RM enzymes EcoR124I and EcoAI (Figure 7A). The methylation specificity of one half site of EcoR124I or EcoAI corresponds to each putative target adenine residue in the LlaGI site. pHH-7 was first methylated using full-length LlaGIDA078 [an endonuclease mutant that cannot cleave DNA but which has wild type translocation properties (11)], M.EcoAI or M.EcoR124I (‘Materials and Methods’ section). Each methylated DNA was then incubated separately with each endonuclease and the extent of DNA cleavage ascertained (Figure 7B). In each case endonuclease activity was almost completely blocked by the cognate MTase, as expected. (The small amount of cleavage observed with EcoR124I represents the inefficient de novo methylation of the Type I MTases.) However, whilst methylation by EcoR124I had no effect on LlaGI (and vice versa), the methylation activities of LlaGI and EcoAI blocked both endonucleases. These results are consistent with methylation by LlaGI of the adenine residue on the bottom strand at the second base pair (Figure 1C) but not at the adenine residue on the top strand at the fifth base pair. Attempts to confirm this result using oligoduplexes containing a synthetic N6-methyl deoxyadenosine residue were unsuccessful (data not shown). This may be the result of the large amount of adjacent DNA required for LlaGI activity/binding (noted above).
The data in Figure 7B deals exclusively with the constant adenine residues found in all the possible LlaGI recognitions sequences (Figure 3B). For two of the eight sites (CTCGACG and CTGGACG), there are no other adenine residues and the sites can only ever be hemi-methylated. However, for the other six versions of the site, adenine residues can also be found at one or both of the degenerate positions. It is possible that these residues may also be targets for methylation by LlaGI. Using the library of two-site DNA generated to test the DNA cleavage activity (Figure 3, ‘Materials and Methods’ section), we monitored the extent of DNA methylation of each of the eight sites relative to a constant site (Site α, Figure 3) using [3H-methyl] AdoMet (Figure 7C, ‘Materials and Methods’ section). All of the eight sites were methylated to the same extent. The sequence at site α contains one degenerate adenine (Figure 3B). The other sites contain either 0, 1 or 2 degenerate adenines. Therefore, if any of the degenerate residues were targets we would have expected a two-fold increase or decrease to be observed on some substrates. As this was not observed, these data show that all eight LlaGI sites were methylated at the same single adenine, i.e. LlaGI only hemi-methylates DNA regardless of the recognition sequence. Note, the data in Figure 7B also shows that hemi-methylation of only one of the two sites (by pre-treatment with M.EcoAI) is sufficient to prevent dsDNA cleavage (but not the Site α-dependent nicking activity).
DISCUSSION
Our characterisation of the DNA cleavage and methylation activities of the single polypeptide RM enzyme LlaGI shows that whilst this enzyme shares some features with the classical Type I RM enzymes, it also has activities and features in common with Type II and III RM enzymes.
The 7-nt DNA recognition sequence of LlaGI (Figure 1C) is distinct from the bipartite sequences of the classical Type I enzymes [e.g. GAANNNNNNRTCG for EcoR124I, where R is any purine, (2)]. This is perhaps unsurprising, given that bioinformatics analysis does not indicate the presence of an HsdS (or half HsdS) domain in LlaGI. Where some Type II enzymes contain HsdS-like domains they also recognise bipartite sequences (2). The LlaGI recognition sequence is also distinct from the short asymmetric sequences of the Type III enzymes which are 6 bp or less in length and do not contain degenerate base pairs [e.g. CTGATG for PstII (2,24)]. However, similar seven base pair semi-redundant sequences have been identified for some ATP-independent, single polypeptide Type IIS RM enzymes [e.g. CGRGGAC for RpaB5I and CAAGNAC for DraRI, (2)]. Outside of highly conserved MTase amino acid motifs, there does not appear to be any sequence relationship between LlaGI and these enzymes (data not shown).
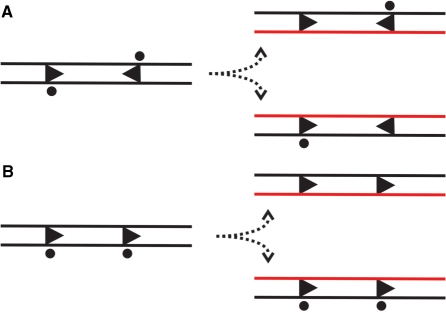
LlaGI only hemi-methylates its recognition sequences (Figure 7) which could cause a problem following semi-conservative replication as unmethylated sites will occur on the daughter DNA [Figure 8 (32,33)]. Hemi-methylation is also a feature of all Type III enzymes studied to date whereas in contrast, the Type I enzymes methylate both DNA strands (2). Where ATP-independent Type II enzymes recognise an asymmetric sequence, methylation of both DNA strands is achieved by using either two separate, independent MTases (e.g. HphI), or a single polypeptide that is the fusion of two independent MTases (e.g. FokI) (2). More recently however, the single polypeptide Type IIS RM enzyme MmeI and related enzymes have been identified that can only hemi-methylate their recognition sequences (e.g. TCCRAC for MmeI, where the adenine underlined is modified whilst the adenine complementary to the first T residue is not) (2,9,34). The consequences of hemi-methylation are considered in more detail below.
-
Whilst a DNA nicking activity is observed in the presence of a single LlaGI site, full dsDNA cleavage only occurs when two LlaGI sites are present in a head-to-head orientation (Figures 3 and 4). This is different to the classical Type I enzymes where dsDNA cleavage is independent of the relative site orientation (27), and only requires a single site on circular DNA (35). Instead the activity is more similar to the Type III enzymes (5,24,32,33). It has been suggested that the requirement for site orientation specificity by Type III enzymes is the direct consequence of the incomplete methylation of the recognition site (32). By requiring the presence of two indirectly repeated sites, at least one of the pair remains hemi-methylated following replication (Figure 8). We directly demonstrate here that methylation of one LlaGI site of a pair is sufficient for protection from dsDNA cleavage (Figure 7B). We suggest that the requirement for a pair of directional sites for DNA cleavage by LlaGI evolved to compensate for the incomplete methylation of the site that would otherwise result in lethal dsDNA breaks. However, the single unmethlyated site still results in DNA nicking. Whilst not as lethal as a dsDNA break, nicks could result in fork arrest if they are not repaired before the next round of replication (36).
Although LlaGI and the Type III enzymes share the same requirement for ATP hydrolysis and a pair of indirectly-repeated recognition sites, LlaGI cleaves at random loci in the DNA between a pair of head-to-head sites whilst the Type III enzymes cleave ∼25–27 bp downstream of one or other site (2). The random cleavage profile of LlaGI is very similar to the distributive cleavage pattern of the classical Type I enzymes (26,27), which results from the convergent collision of enzymes that are translocating from the sites (28). In the accompanying manuscript we demonstrate that LlaGI has ATPase and loop translocation activities similar to that of the classical Type I RM enzymes and quite distinct to the Type III RM enzymes (10,11). However, whilst the classical Type I enzymes translocate bidirectionally from their sites (37), LlaGI catalyses unidirectional translocation. A model for how translocation leads to DNA cleavage is illustrated in the accompanying paper (10).
The DNA cleavage profiles shows that nicked DNA is a major intermediate during dsDNA cleavage (Figure 6). A similar accumulation of nicked DNA is seen for some classical Type I enzymes, except that the maximum percentage observed is usually <33% (35, Frank Peske unpublished observations). Type III RM enzymes accumulate virtually no nicked DNA intermediate (30), which is most likely related to the rate-limiting ATPase activity that precedes DNA cleavage (33). Since we show in the accompanying manuscript that LlaGI communicates between its sites using a mechanism similar to that of the classical Type I enzymes (10), the differences in the reaction profiles most likely reflects differences in the DNA cleavage steps rather than communication steps. In the third accompanying manuscript (11) we show that LlaGI uses an Mrr nuclease domain distinct to the RecB-family nuclease domain of the classical Type I RM enzymes. Although both are part of the PD(D/E)xK superfamily of nucleases (3), it is possible that dsDNA cleavage proceeds by different mechanisms.
Figure 8.
The consequences of recognition site hemi-methylation following replication, adapted from the model of Meisel et al.(32,33). The LlaGI site is indicated as an arrowhead (Figure 1C), the methylated site by a circle and the newly synthesised DNA in red. (A) Where two sites are in head-to-head repeat, one site of the pair is methylated in each of the daughter DNAs and both are protected from dsDNA cleavage (Figure 7). (B) With two sites in head-to-tail repeat, both sites in one daughter DNA are completely unmethylated. However, this arrangement of sites does not result in a dsDNA break (Figures 3 ands 4). DNA nicking that result from the unmethylated sites is likely to be repaired before the replication fork next passes.
From the data in this paper and the accompanying manuscripts (10,11), we can compare the domain structure and enzyme activities of LlaGI to that of Type I, II and III RM enzymes (Table 2). The Type II RM enzymes represent a diverse class with varied domain/subunit architectures and a broad repertoire of enzyme activities. Here we have chosen to only compare a sub-grouping that are most similar to LlaGI, the Type IIL enzymes, as exemplified by MmeI (34). Whilst some features of LlaGI activity are clearly Type I-like (the presence of similar helicase and MTase domains, an extensive ATPase activity, dsDNA loop translocation, distributive non-specific DNA cleavage loci), others are Type III-like (a requirement for two sites with a particular orientation, single-strand methylation) or Type IIL-like (short degenerate recognition sequence, single-strand methylation, single polypeptide multi-domain arrangement). The classification of Type I and III enzymes was historically on the basis of gene arrangement (Type I enzymes comprising HsdS, HsdM and HsdR subunits, Type III enzymes comprising Mod and Res subunits). Subsequent biochemical analysis has shown that both Types have enzyme behaviour that is largely family-specific and distinct. We suggest that given the presence of a helicase domain that couples extensive ATP hydrolysis to loop translocation (10), LlaGI is most closely related to the Type I enzymes and should be classified as such. However, to distinguish the single polypeptide enzymes from the classical multi-subunit enzymes, we suggest the nomenclature Type I SP (for single polypeptide). This avoids confusion with the single letter suffices that have already been assigned to sub-families of Type I enzymes from enterobacteria (on the basis of gene complementation and antibody cross-reactivity) and to the Type II enzymes (on the basis of gene and enzyme properties) (38). Our classification redefines the Type I enzymes in more general terms as dsDNA loop translocases that cleave DNA at random sites distant from their recognition sequences. The classification of the Type III enzymes remains open to discussion, as DNA looping, DNA loop translocation, DNA translocation and bi-directional DNA sliding models have all been proposed and a consistent mechanism has yet to emerge.
Table 2.
Genetic and biochemical classification of LlaGI
| Type I | Type III | Type IIL | Type ISP | |
|---|---|---|---|---|
| e.g. EcoR124I | e.g. EcoPI | e.g. MmeI | e.g. LlaGI | |
| Genes/protein | ||||
| Genes | HsdR, hsdM, hsdS | Res, mod | RM | RM |
| Nuclease Complex | R2M2S1 | Res2Mod2 | n.d. | RM1 (RM2 for cleavage?) |
| Recognition sequence | Asymmetric, bipartite GAAnnnnnnRTCG | Short, asymmetric AGACC | Short, asymmetric, degenerate TCCrAC | Short, asymmetric, degenerate CTnGAyG |
| DNA methylation | ||||
| Methylation | Both strands | One strand | One strand | One strand |
| DNA cleavage | ||||
| Cofactors | ATP, Mg2+, AdoMet | ATP, Mg2+ | Mg2+ | ATP, Mg2+ |
| Multiple sites required? | Yes, Linear DNANo, circular DNA | Yes (+AdoMet) No (-AdoMet)a | Yes | Yes (dsDNA break) No (nicking) |
| Sites required in cis? | Yes | Yes | No | Yes |
| Communication mode | 1D | 1D | 3D | 1D |
| Preference for site directionality? | None | Indirect repeatb | None | Indirect Repeat (head-to-head only) |
| Cleavage loci? | Non-specific random | Non-specific | Non-specific | Non-specific random |
| Distant and proximal | Fixed proximal | Fixed proximal | Distant and proximal | |
| Helicase motor activity | No helicase domain | |||
| DNA Loop translocation required | Yes | No | Yes | |
| DNA Supercoiling | Yes | No | Yes | |
| Directionality | Bidirectional (two unidirectional motors per complex) | Bidirectionalb | Unidirectional | |
| Translocation rate | 463 bp/s (20°C) | n.d. | 173 ATP/s (20°C) | |
| ATPase rate | 503 ATP/s/motor(20°C) | ∼0.2 ATP/s/per complex (20°C) | 250 ATP/s (20°C) | |
| ATP per bp | 1–1.5 | «1 | 1.5–2 | |
| Step size | 1–2 | n.d. | n.d. | |
| Average distance moved per event | ∼5000 bp | n.d. | n.d. | |
| Motor polarity | 3′–5′ | n.d. | n.d. | |
| Triplex displacement | Yes | No | Yes |
Data for EcoR124I was collated from refs (14,27,35,39–41). Data for EcoPI was collated from refs (30,42–45). Data for MmeI was collated from refs (2,9,34). Data for LlaGI was from this paper and refs. 1,10,11. n.d. is where data is not determined.
aRelaxed cleavage conditions with high enzyme:DNA ratios.
bDogma states that Type III enzymes only cut sites in head-to-head repeat (33) but more recent analysis suggests that tail-to-tail repeats are cut equally well (Kara van Aelst, M.D.S. and Ralf Seidel, unpublished data).
FUNDING
The BBSRC (BB/D009715/1) and The Wellcome Trust (067439) to M.D.S. The BBSRC (BB/F007361/1) to N.J.S. Funding for open access charge: Wellcome Trust Value in People Award.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Will Mawby of the Bristol University Proteomics Facility and the Dundee Sequencing Service for protein and DNA sequencing, respectively.
Footnotes
This article is linked to 10.1093/nar/gkp794 by Smith et al. and 10.1093/nar/gkp795 by Smith et al.
REFERRENCES
- 1.Madsen A, Josephsen J. The LlaGI restriction and modification system of Lactococcus lactis W10 consists of only one single polypeptide. FEMS Microbiol. Lett. 2001;200:91–96. doi: 10.1111/j.1574-6968.2001.tb10698.x. [DOI] [PubMed] [Google Scholar]
- 2.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE – enzymes and genes for DNA restriction and modification. Nucleic Acids Res. 2007;35:D269–D270. doi: 10.1093/nar/gkl891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aravind L, Makarova KS, Koonin EV. SURVEY AND SUMMARY: Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 2000;28:3417–3432. doi: 10.1093/nar/28.18.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bujnicki JM, Rychlewski L. Identification of a PD-(D/E)XK-like domain with a novel configuration of the endonuclease active site in the methyl-directed restriction enzyme Mrr and its homologs. Gene. 2001;267:183–191. doi: 10.1016/s0378-1119(01)00405-x. [DOI] [PubMed] [Google Scholar]
- 5.Bourniquel AA, Bickle TA. Complex restriction enzymes: NTP-driven molecular motors. Biochimie. 2002;84:1047–1059. doi: 10.1016/s0300-9084(02)00020-2. [DOI] [PubMed] [Google Scholar]
- 6.McClelland SE, Szczelkun MD. Molecular motors that process DNA in Restriction Enzymes. In: Pingound A, editor. Nucleic Acids and Molecular Biology. Vol. 14. Germany: Springer; 2004. pp. 111–135. [Google Scholar]
- 7.Cesnaviciene E, Petrusyte M, Kazlauskiene R, Maneliene Z, Timinskas A, Lubys A, Janulaitis A. Characterization of AloI, a restriction-modification system of a new type. J. Mol. Biol. 2001;314:205–216. doi: 10.1006/jmbi.2001.5049. [DOI] [PubMed] [Google Scholar]
- 8.Marshall JJ, Gowers DM, Halford SE. Restriction endonucleases that bridge and excise two recognition sites from DNA. J. Mol. Biol. 2007;367:419–431. doi: 10.1016/j.jmb.2006.12.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morgan RD, Bhatia TK, Lovasco L, Davis TB. MmeI: a minimal Type II restriction-modification system that only modifies one DNA strand for host protection. Nucleic Acids Res. 2008;36:6558–6570. doi: 10.1093/nar/gkn711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith RM, Josephsen J, Szczelkun MD. The single polypeptide Restriction Modification enzyme LlaGI is a self-contained molecular motor that translocates DNA loops. 2009 doi: 10.1093/nar/gkp794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith RM, Josephsen J, Szczelkun MD. An Mrr-family nuclease motif in the single polypeptide Restriction-Modification enzyme LlaGI. 2009 doi: 10.1093/nar/gkp795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 13.Vipond IB, Baldwin GS, Oram M, Erskine SG, Wentzell LM, Szczelkun MD, Nobbs TJ, Halford SE. A general assay for restriction endonucleases and other DNA-modifying enzymes with plasmid substrates. Mol. Biotechnol. 1995;4:259–268. doi: 10.1007/BF02779019. [DOI] [PubMed] [Google Scholar]
- 14.McClelland SE, Dryden DT, Szczelkun MD. Continuous assays for DNA translocation using fluorescent triplex dissociation: application to type I restriction endonucleases. J. Mol. Biol. 2005;348:895–915. doi: 10.1016/j.jmb.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 15.de Daruvar A, Collado-Vides J, Valencia A. Analysis of the cellular functions of Escherichia coli operons and their conservation in Bacillus subtilis. J. Mol. Evol. 2002;55:211–221. doi: 10.1007/s00239-002-2317-1. [DOI] [PubMed] [Google Scholar]
- 16.Price MN, Arkin AP, Alm EJ. The life-cycle of operons. PLoS Genet. 2006;2:e96. doi: 10.1371/journal.pgen.0020096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan SA. Rolling-circle replication of bacterial plasmids. Microbiol. Mol. Biol. Rev. 1997;61:442–455. doi: 10.1128/mmbr.61.4.442-455.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gravesen A, Josephsen J, von Wright A, Vogensen FK. Characterization of the replicon from the lactococcal theta-replicating plasmid pJW563. Plasmid. 1995;34:105–118. doi: 10.1006/plas.1995.9996. [DOI] [PubMed] [Google Scholar]
- 19.Sánchez C, Hernández de Rojas A, Martínez B, Argüelles ME, Suárez JE, Rodríguez A, Mayo B. Nucleotide sequence and analysis of pBL1, a bacteriocin-producing plasmid from Lactococcus lactis IPLA 972. Plasmid. 2000;44:239–249. doi: 10.1006/plas.2000.1482. [DOI] [PubMed] [Google Scholar]
- 20.Alvarez-Martín P, O'C;onnell-Motherway M, van Sinderen D, Mayo B. Functional analysis of the pBC1 replicon from Bifidobacterium catenulatum L48. Appl. Microbiol. Biotechnol. 2007;76:1395–1402. doi: 10.1007/s00253-007-1115-5. [DOI] [PubMed] [Google Scholar]
- 21.Frère J, Novel M, Novel G. Molecular analysis of the Lactococcus lactis subsp. lactis CNRZ270 bidirectional theta replicating lactose plasmid pUCL22. Mol. Microbiol. 1993;10:1113–1124. doi: 10.1111/j.1365-2958.1993.tb00981.x. [DOI] [PubMed] [Google Scholar]
- 22.Kiewiet R, Bron S, de Jonge K, Venema G, Seegers JF. Theta replication of the lactococcal plasmid pWVO2. Mol. Microbiol. 1993;10:319–327. [PubMed] [Google Scholar]
- 23.van Kranenburg R, de Vos WM. Characterization of multiple regions involved in replication and mobilization of plasmid pNZ4000 coding for exopolysaccharide production in Lactococcus lactis. J. Bacteriol. 1998;180:5285–5290. doi: 10.1128/jb.180.20.5285-5290.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sears A, Peakman LJ, Wilson GG, Szczelkun MD. Characterization of the Type III restriction endonuclease PstII from Providencia stuartii. Nucleic Acids Res. 2005;33:4775–4787. doi: 10.1093/nar/gki787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmer BR, Marinus MG. The dam and dcm strains of Escherichia coli – a review. Gene. 1994;143:1–12. doi: 10.1016/0378-1119(94)90597-5. [DOI] [PubMed] [Google Scholar]
- 26.Studier FW, Bandyopadhyay PK. Model for how type I restriction enzymes select cleavage sites in DNA. Proc. Natl Acad. Sci. USA. 1988;85:4677–4681. doi: 10.1073/pnas.85.13.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szczelkun MD, Janscak P, Firman K, Halford SE. Selection of non-specific DNA cleavage sites by the type IC restriction endonuclease EcoR124I. J. Mol. Biol. 1997;271:112–123. doi: 10.1006/jmbi.1997.1172. [DOI] [PubMed] [Google Scholar]
- 28.Szczelkun MD. Kinetic models of translocation, head-on collision, and DNA cleavage by type I restriction endonucleases. Biochemistry. 2002;41:2067–2074. doi: 10.1021/bi011824b. [DOI] [PubMed] [Google Scholar]
- 29.Sistla S, Rao DN. S-Adenosyl-L-methionine-dependent restriction enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:1–19. doi: 10.1080/10409230490440532. [DOI] [PubMed] [Google Scholar]
- 30.Peakman LJ, Antognozzi M, Bickle TA, Janscak P, Szczelkun MD. S-adenosyl methionine prevents promiscuous DNA cleavage by the EcoP1I type III restriction enzyme. J. Mol. Biol. 2003;333:321–335. doi: 10.1016/j.jmb.2003.08.042. [DOI] [PubMed] [Google Scholar]
- 31.Malone T, Blumenthal RM, Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol. 1995;253:618–632. doi: 10.1006/jmbi.1995.0577. [DOI] [PubMed] [Google Scholar]
- 32.Meisel A, Bickle TA, Krüger DH, Schroeder C. Type III restriction enzymes need two inversely oriented recognition sites for DNA cleavage. Nature. 1992;355:467–469. doi: 10.1038/355467a0. [DOI] [PubMed] [Google Scholar]
- 33.Meisel A, Mackeldanz P, Bickle TA, Kruger DH, Schroeder C. Type III restriction endonucleases translocate DNA in a reaction driven by recognition site-specific ATP hydrolysis. EMBO J. 1995;14:2958–2966. doi: 10.1002/j.1460-2075.1995.tb07296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morgan RD, Dwinell EA, Bhatia TK, Lang EM, Luyten YA. The MmeI family: type II restriction-modification enzymes that employ single-strand modification for host protection. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp534. doi:10.1093/nar/gkp534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szczelkun MD, Dillingham MS, Janscak P, Firman K, Halford SE. Repercussions of DNA tracking by the type IC restriction endonuclease EcoR124I on linear, circular and catenated substrates. EMBO J. 1996;15:6335–6347. [PMC free article] [PubMed] [Google Scholar]
- 36.McGlynn P, Lloyd RG. Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell Biol. 2002;3:859–870. doi: 10.1038/nrm951. [DOI] [PubMed] [Google Scholar]
- 37.Firman K, Szczelkun MD. Measuring motion on DNA by the type I restriction endonuclease EcoR124I using triplex displacement. EMBO J. 2000;19:2094–2102. doi: 10.1093/emboj/19.9.2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev SKh, Dryden DT, Dybvig K, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janscak P, Abadjieva A, Firman K. The type I restriction endonuclease R. EcoR124I: over-production and biochemical properties. J. Mol. Biol. 1996;257:977–991. doi: 10.1006/jmbi.1996.0217. [DOI] [PubMed] [Google Scholar]
- 40.Seidel R, van Noort J, van der Scheer C, Bloom JG, Dekker NH, Dutta CF, Blundell A, Robinson T, Firman K, Dekker C. Real-time observation of DNA translocation by the type I restriction modification enzyme EcoR124I. Nat. Struct. Mol. Biol. 2004;11:838–843. doi: 10.1038/nsmb816. [DOI] [PubMed] [Google Scholar]
- 41.Seidel R, Bloom JG, Dekker C, Szczelkun MD. Motor step size and ATP coupling efficiency of the dsDNA translocase EcoR124I. EMBO J. 2008;27:1388–1398. doi: 10.1038/emboj.2008.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bachi B, Reiser J, Pirrotta V. Methylation and cleavage sequences of the EcoP1 restriction-modification enzyme. J. Mol. Biol. 1979;128:143–163. doi: 10.1016/0022-2836(79)90123-2. [DOI] [PubMed] [Google Scholar]
- 43.Humbelin M, Suri B, Rao DN, Hornby DP, Eberle H, Pripfl T, Kenel S, Bickle TA. Type III DNA restriction and modification systems EcoP1 and EcoP15. J. Mol. Biol. 1988;200:23–29. doi: 10.1016/0022-2836(88)90330-0. [DOI] [PubMed] [Google Scholar]
- 44.Saha S, Rao DN. ATP hydrolysis is required for DNA cleavage by EcoPI restriction enzyme. J. Mol. Biol. 1995;247:559–567. doi: 10.1016/s0022-2836(05)80137-8. [DOI] [PubMed] [Google Scholar]
- 45.Janscak P, Sandmeier U, Szczelkun MD, Bickle TA. Subunit assembly and mode of DNA cleavage of the type III restriction endonucleases EcoP1I and EcoP15I. J. Mol. Biol. 2001;306:417–431. doi: 10.1006/jmbi.2000.4411. [DOI] [PubMed] [Google Scholar]