Abstract
Kink turns (k-turns) are important structural motifs that create a sharp axial bend in RNA. Most conform to a consensus in which a three-nucleotide bulge is followed by consecutive G•A and A•G base pairs, and when these G•A pairs are modified in vitro this generally leads to a failure to adopt the k-turn conformation. Kt-23 in the 30S ribosomal subunit of Thermus thermophilus is a rare exception in which the bulge-distal A•G pair is replaced by a non-Watson–Crick A•U pair. In the context of the ribosome, Kt-23 adopts a completely conventional k-turn geometry. We show here that this sequence is induced to fold into a k-turn structure in an isolated RNA duplex by Mg2+ or Na+ ions. Therefore, the Kt-23 is intrinsically stable despite lacking the key A•G pair; its formation requires neither tertiary interactions nor protein binding. Moreover, the Kt-23 k-turn is stabilized by the same critical hydrogen-bonding interactions within the core of the structure that are found in more conventional sequences such as the near-consensus Kt-7. T. thermophilus Kt-23 has two further non-Watson–Crick base pairs within the non-canonical helix, three and four nucleotides from the bulge, and we find that the nature of these pairs influences the ability of the RNA to adopt k-turn conformation, although the base pair adjacent to the A•U pair is more important than the other.
INTRODUCTION
Kink turns (k-turns) are commonly found structural elements in the architecture of RNA–protein complexes and large RNA species. The k-turn motif introduces a sharp kink into the axis of the helix, thereby exerting an important influence on the long-range structure of RNA. k-turns were first identified in both subunits of the ribosome (1). They have also been found in the nucleolar RNA species that guide RNA modification (2–5), U4 snRNA (6,7) and in untranslated regions of mRNA (8,9), including within riboswitches (10–12). Therefore, k-turns seem to be involved in almost every aspect of RNA function, including the translation and modification of RNA, spliceosome assembly and the control of gene expression.
The great majority of k-turn motifs conform to a consensus secondary structure comprising a three-nucleotide bulge flanked by regular base pairing on its 5′ side (the C helix) and two consecutive A•G pairs on its 3′ side (the NC helix) (Figure 1). The k-turn Kt-7 in the 23S rRNA of the Haloarcula marismortui ribosome is close to this consensus (1); its structure is very similar to those of a number of other k-turns (2,6,10). The kinking juxtaposes the two minor grooves, with an included angle between the axes of ∼60°, and the conformation appears to be stabilized by interactions between the stacked adenosines of the A•G pairs and the C stem, and by stacking of the 5′ and central bases of the bulge on the ends of the C and NC helices, respectively.
Figure 1.
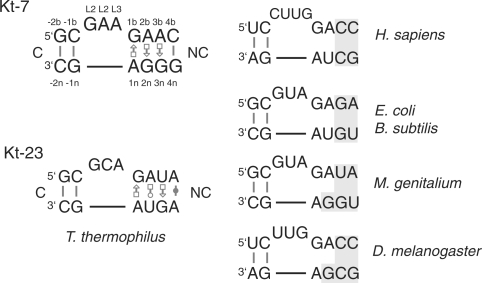
Comparison of ribosomal k-turn sequences. The sequence of Kt-7 is shown since it closely approximates the consensus k-turn sequence, with the nucleotides numbered according to the scheme used throughout (19), together with Kt-23 of T. thermophilus 16S rRNA. The designation of base pairs 3b•3n and 4b•4n is ambiguous since these are only connected by a single hydrogen bond (refer to Figure 2C) (22). Kt-23 sequences are compared for a variety of organisms, with important differences shaded. The sequence information was taken from the Gutell comparative RNA data base (31). Base pairing of non-Watson–Crick pairs is designated using the convention of Leontis and Westhof (32).
k-turn motifs do not form a stably kinked structure in the absence of metal ions (13,14), or the binding of proteins of the L7Ae family (15). The kinked conformation can be stabilized by the addition of either divalent or monovalent metal ions (13) and folding can be treated as a two-state process. Thus, it is likely that electrostatic interactions must be screened to allow the folded structure to achieve stability. This suggests a dynamic character for k-turn structures, where they sample both the kinked and a more extended geometry. Computer modelling studies have suggested that k-turns undergo hinge-like motions on a fast timescale (16–18). We have made a detailed study of the hydrogen-bonding interactions that are important in stabilizing the kinked conformation of Kt-7 in the presence of Mg2+ ions (19,20). These include critical hydrogen bonds between the 2′O of the L1 ribose and N1 of the adenine nucleobase in the 1n position, and between the 2′O of the L3 ribose and the proS non-bridging O of the phosphate group linking L1 and L2 (19).
We have also studied the importance of the two A•G pairs adjacent to the bulge of the archetypal k-turn (20). Both are trans sugar edge (G)/Hoogsteen edge (A) pairs, linked by potential hydrogen bonds G:N2 to A:N7 and A:N6 to G:N3. The 1b•1n pair is strongly buckled, while the bases of the 2n•2b pair are much closer to coplanarity. These are conserved in the great majority of k-turns (21), and sequence substitution of any of the four nucleotides within Kt-7 (13), or disruption of hydrogen bonding (20), completely prevents ion-induced folding. On this basis, it would appear that the two A•G pairs are an indispensible feature of the k-turn motif. It is therefore surprising that within the 16S RNA of the T. thermophilus ribosome k-turn Kt-23 lacks the A•G pair at the 2b•2n position (1) (Figure 1), being replaced by a reverse Hoogsteen A•U pair (22). This is also true in Kt-23 of E. coli (23); the two Kt-23 structures are closely similar. We have previously shown that the introduction of the equivalent G2nU substitution in Kt-7 completely prevented detectable ion-induced folding by that sequence (13). Yet Kt-23 adopts a normal k-turn geometry in the ribosome (Figure 2A), with an inter-axial angle of 60° between the C and NC helices. Despite the absence of the A•G pair at the 2b•2n position, the 1n and 2b adenine nucleobases of Kt-23 become located in virtually identical positions if H. marismortui Kt-7 and T. thermophilus Kt-23 are superimposed by their backbones (Figure 2B), and are thus able to participate fully in the A-minor interaction (24) with the C helix. The L1 and L2 nucleobases cap the C and NC helices, respectively, and the key hydrogen bonds involving 2′-hydroxyl groups (19) are present in the Kt-23 structures (22,23). The corresponding Kt-23 sequences of human, mouse and Bacillus subtilis preserve the A•U pair at the 2b•2n position, while it is replaced by conventional A•G pairs in Drosophila melanogaster and Mycoplasma genitalium (Figure 1).
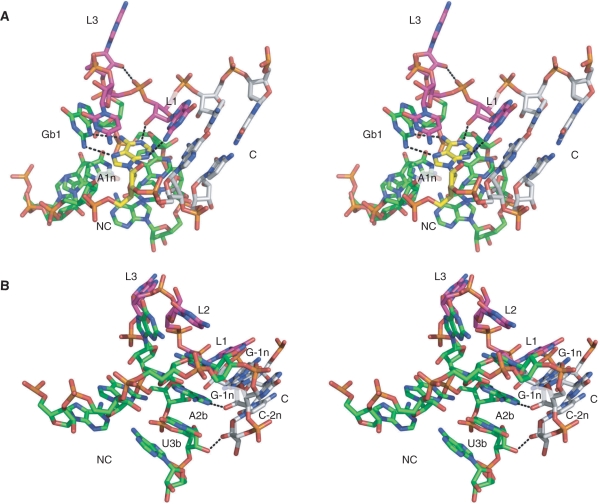
Figure 2.
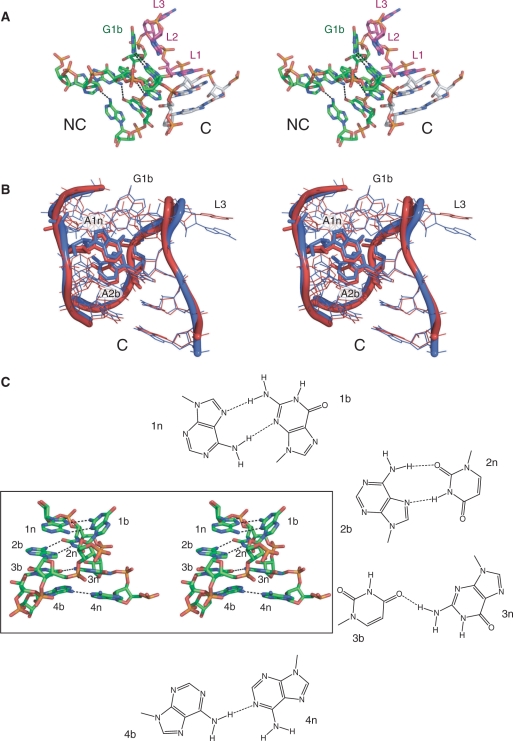
The molecular structure of Kt-23. (A) Parallel-eye stereoscopic view of the structure of Kt-23 taken from the crystal structure of the T. thermophilus 30S ribosomal subunit. The view is from the side of the strand not containing the three-nucleotide loop. The NC helix is coloured green, the C helix grey and the nucleotides of the loop purple. Hydrogen bonds forming the base pairs of the NC helix are shown as broken lines. All images of the Kt-23 structure were made using PDB file 1J5E (22). (B) Superposition of the crystallographic structures of Kt-7 and T. thermophilus Kt-23. The two structures were superimposed by their backbone atoms (O5′, C5′, C4′, C3′, O3′, P and the non-bridging phosphate O). The structures are in parallel-eye stereoscopic view, with ribbons to indicate the position of the backbones. Kt-23 is drawn in red and Kt-7 in blue. The structure of Kt-7 was taken from the H. marismortui 50S ribosomal subunit, PDB file 1FFK (30). (C) Base pairing in the NC helix of T. thermophilus Kt-23. Parallel-eye stereoscopic view of the NC helix of Kt-23, with hydrogen bonds forming the base pairs of the NC helix are shown as broken lines. The structures of the four non-Watson–Crick base pairs are drawn out.
Thus, Kt-23 breaks what appeared to be a cardinal rule for k-turn stability, yet it adopts a normal k-turn conformation within the ribosome. This provides a challenge to our understanding of the k-turn structure, and raises two questions. First, can metal ions induce Kt-23 to adopt the conventional k-turn structure in an isolated RNA duplex, unaided by any potential tertiary interactions or RNA–protein interactions that might stabilize the structure within the context of the 30S ribosomal subunit? In this work, we find that T. thermophilus Kt-23 readily adopts the kinked geometry of the k-turn in isolation, with preservation of the key hydrogen-bonding interaction in the core of the structure. The second question is, therefore, what features of the complete Kt-23 sequence allow it to fold normally, despite the presence of U at position 2n, when G2nU substitution completely prevented Kt-7 from adopting k-turn geometry in the presence of Mg2+ ions in vitro (13)?
The key difference is unlikely to reside in the unpaired loop. These nucleotides are poorly conserved and generally only make stacking interactions. We therefore turned our attention to the NC helix beyond the 2b•2n position distal to the bulge. The four base–base interactions of the NC helix of T. thermophilus Kt-23 are shown in Figure 2C. Many, though not all, k-turn sequences include a non-Watson–Crick pair at the 3b•3n position; in T. thermophilus Kt-23, this is a U•G pair with a single hydrogen bond from G3n:N2 to U3b:O4. Unusually, the 4b•4n position is also non-Watson–Crick, a cis A•A pair hydrogen bonded from A4b:N6 to A4n:N1. The C1′–C1′ distance of the A•U pair in Kt-23 at 2b•2n is longer than that of an A•G pair (Table 1), and the width of the NC helix could be a factor in stabilizing the k-turn geometry. We have therefore examined the effect of functional group substitutions designed to probe the effect of the stability of the 3b•3n and 4b•4n positions on the ability of T. thermophilus Kt-23 to undergo ion-induced folding.
Table 1.
Comparison of C1′–C1′ distances for the base pairs of the NC helix in Kt-23 of T. thermophilus (T. th) and E. coli (E. co), Kt-7 and the U4 snRNA k-turn
| Kt-23 (T. th) | Kt-23 (E. co) | Kt-7 | U4 | |||||
|---|---|---|---|---|---|---|---|---|
| 1b•1n | G•A | 9.3 Å | G•A | 9.3 Å | G•A | 9.2 Å | G•A | 9.6 Å |
| 2b•2n | A•U | 9.9 Å | A•U | 9.6 Å | A•G | 9.4 Å | A•G | 9.0 Å |
| 3b•3n | U•G | 11.0 Å | G•G | 11.4 Å | A•G | 10.0 Å | G–C | 10.8 Å |
| 4b•4n | A•A | 12.3 Å | A–U | 10.8 Å | G-C | 10.4 Å | G–C | 10.9 Å |
MATERIALS AND METHODS
RNA synthesis and construction of k-turn species
The majority of oligonucleotides were synthesized using t-BDMS phosphoramidite chemistry (27), as described in Wilson et al. (28), with nucleobase and 2′-deoxyribo substitution as required. Fluorescein and Cy3-conjugated oligonucleotides were attached at 5′-termini as phosphoramidites during synthesis as required. Oligoribonucleotides were deprotected in 25% ethanol/ammonia solution at 55°C for 2 h, and evaporated to dryness. Oligoribonucleotides were redissolved in 300 µl 1 M tetrabutylammonium fluoride (Aldrich) in tetrahydrofuran to remove t-BDMS protecting groups, and agitated at 20°C for 16 h prior to desalting by G25 Sephadex (NAP columns, Pharmacia) and ethanol precipitation. All oligonucleotides were purified by gel electrophoresis in polyacrylamide, and recovered from gel fragments by electroelution or diffusion in buffer followed by ethanol precipitation. Fluorescently labelled RNA was subjected to further purification by reversed phase HPLC on a C18 column (ACE10, Advanced Chromatography Technologies) using an acetonitrile gradient with an aqueous phase of 100 mM triethylammonium acetate pH 7.0. Oligonucleotides required for electrophoretic analysis were radioactively 5′-32P-labelled using T4 polynucleotide kinase and [γ-32P] ATP (Amersham).
Duplex species were prepared by mixing equimolar quantities of the appropriate oligoribonucleotides and annealing them in 50 mM Tris–HCl (pH 7.5), 25 mM NaCl by slow cooling from 90 to 4°C. They were purified by electrophoresis in 12% polyacrylamide under non-denaturing conditions, and recovered by either electroelution or diffusion into buffer, followed by ethanol precipitation.
Analysis of k-turn folding by comparative gel electrophoresis
Radioactively [5′-32P]-labelled RNA species were electrophoresed in 13 % polyacrylamide (29:1, acrylamide:bis) gels in 90 mM Tris–borate (pH 8.3), 2 mM Mg2+. Electrophoresis was performed at 120 V at room temperature for up to 68 h, with recirculation of the buffer at >1 l h−1. Gels were dried onto Whatman 3 MM paper, exposed to storage phosphor plates and imaged using a Fuji BAS-1500 phosphorimager.
The sequences used for the electrophoretic experiments were (written 5′ to 3′) as follows.
Kt-7 upper strand: CGCAAGCGACAGGAACCTCGCC AGUCAGUGGCGAAGAACCAUGUC AGGGGACTGTCAAGTTGAACAGG
Kt-7 lower strand: CCTGTTCAACTTGACAGTCCCCUG ACAUGGGGAGCCACUGAC UGGCGAGGTTCCTGTCGCTTGCG
Kt-23 upper strand: CGCAAGCGACAGGAACCTCGCC AGUCAGAUGCGCAGAUACCGGGA GAGGGACTGTCAAGTTGAACAGG
Kt-23 lower strand: CCTGTTCAACTTGACAGTCCCUC UCCCGGAGUAGCAUCUGAC UGGCGAGGTTCCTGTCGCTTGCG
The DNA sections of these oligonucleotides are shown underlined. Modified nucleotides were introduced into the RNA sections as required.
Study of Mg2+-induced folding by fluorescence resonance energy transfer
Absorption spectra were measured in 90 mM Tris–borate (pH 8.3) in 120 µl volumes using a Cary 1E spectrophotometer. Spectra were deconvoluted using a corresponding RNA species labelled only with Cy3, and fluorophore absorption ratios calculated using a MATLAB program. Fluorescence spectra were recorded in 90 mM Tris–borate (pH 8.3) at 4°C using an SLM-Aminco 8100 fluorimeter. Spectra were corrected for lamp fluctuations and instrumental variations, and polarization artefacts were avoided by setting excitation and emission polarizers crossed at 54.7°. Values of fluorescence resonance energy transfer (FRET) efficiency (EFRET) were measured using the acceptor normalization method (29) implemented in MATLAB. EFRET as a function of metal ion concentration was analyzed on the basis of a model in which the fraction of folded molecules corresponds to a simple two-state model for ion-induced folding, i.e.
| 1 |
where E0 is the FRET efficiency of the RNA in the absence of added metal ions, ΔEFRET is the increase in FRET efficiency at saturating metal ion concentration, [M] is the prevailing Mg2+ or Na+ ion concentration, KA is the apparent association constant for metal ion binding and n is a Hill coefficient. Data were fitted to this equation by non-linear regression. The metal ion concentration at which the transition is half complete is given by [M]1/2 = (1/KA)1/n.
The sequences used in the FRET analyses were (written 5′ to 3′) as follows.
Kt-23 upper strand: Fluorescein-CCAGUCAGAUGCGCAGAUACCGGGAGAGG
Kt-23 lower strand: Cy3-CCUCUCCCGGAGUAGCAUCUGACUGG
Single nucleotide substitutions were introduced as required.
RESULTS
Analysis of the ion-induced folding of Kt-23 into k-turn conformation
We can readily detect the formation of the k-turn in RNA by virtue of the markedly increased kinking of the helical axis with folding. In this study, we have explored the ion-induced folding of Kt-23 and variants using two approaches, as in our previous studies of k-turns (13,19,20). In comparative gel electrophoresis, we place the Kt-23-containing RNA into the centre of a 65 bp duplex, of which the outer 20 bp on each side are DNA for synthetic convenience. When the RNA adopts the k-turn geometry, the axial kinking results is a marked retardation of electrophoretic mobility in a polyacrylamide gel. The mobilities can be compared with duplexes of the same length containing oligoadenine bulges of different length, that exhibit progressively greater kinking and hence retardation (25,26). The second method is FRET. This employs a 26 bp RNA duplex that includes a central Kt-23 sequence, with fluorescein and Cy3 fluorophores attached to the 5′-termini. Formation of the k-turn geometry significantly reduces the separation between the fluorophores, and thus leads to an increase in the efficiency of FRET. All the experimental work described here has been directed at T. thermophilus Kt-23 and its derivatives.
Kt-23 is induced to fold into the kinked geometry by the presence of metal ions
Using comparative gel electrophoresis, we have previously shown that Kt-7, whose sequence is close to the k-turn consensus, exhibits a mobility similar to a corresponding duplex with a 7-nt bulge. Electrophoresis of Kt-7 and Kt-23 side by side in the presence of 2 mM Mg2+ ions shows them to have closely similar mobility, suggesting that they fold to similar extents under these conditions (Figure 3). Ion-induced kinking of Kt-23 can also be observed by FRET (Figure 4), with a change in FRET efficiency (ΔEFRET) of 0.25 in both Mg2+ and Na+ ions. The data are well fitted by a two-state folding model. Mg2+-induced folding is consistent with non-cooperative ion binding (Hill coefficient n = 1.2), while that for Na+-induced folding indicates more cooperative binding of the monovalent ions (n = 1.7) although this is not well defined by the data. The half magnesium concentration ([Mg2+]1/2) obtained by fitting to the two-state folding model (Table 2) is 35 µM, approximately half the value for Kt-7 (19), indicating that the k-turn geometry of Kt-23 is a little more stable than that of Kt-7 (i.e. it requires a lower concentration of Mg2+ ions to allow folding into the kinked geometry). However, the [Na+]1/2 values for Kt-23 and Kt-7 are closely similar. Both the electrophoretic and FRET results show that the Kt-23 sequence folds into the k-turn geometry with similar stability as Kt-7, despite not having the A•G pair at the 2b•2n position. Evidently, Kt-23 can adopt the k-turn geometry without requiring assistance from either protein binding or tertiary interactions.
Figure 3.
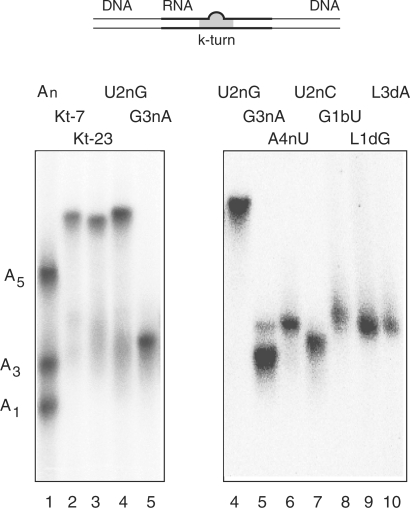
Electrophoretic analysis of Mg2+-induced kinking of Kt-23 and variants. The scheme (top) shows the 65 bp DNA/RNA/DNA duplex species with a central k-turn sequence used in this analysis. Radioactively [5′-32P]-labelled k-turn species (with or without modifications in the k-turn) were electrophoresed in 13% polyacrylamide gels in 90 mM Tris–borate (pH 8.3) containing 2 mM Mg2+. An-bulge-containing duplexes were made using the same lower strand, hybridized to complement the non-Watson–Crick pairings, and with the three-nucleotide bulge replaced by an A1, A3 or A5 bulge. A mixture of the three species was electrophoresed in track 1, alongside Kt-7 in track 2. The remaining tracks contain: track 3, unmodified Kt-23; track 4, Kt-23 U2nG; track 5, Kt-23 G3nA; track 6, Kt-23 A4nU; track 7, Kt-23 U2nC; track 8, Kt-23 G1bU; track 9, Kt-23 GL12′H; track 10, Kt-23 AL32′H. The samples were electrophoresed in two equivalent gels, with the samples Kt-23 U2nG and G3nA common to both.
Figure 4.
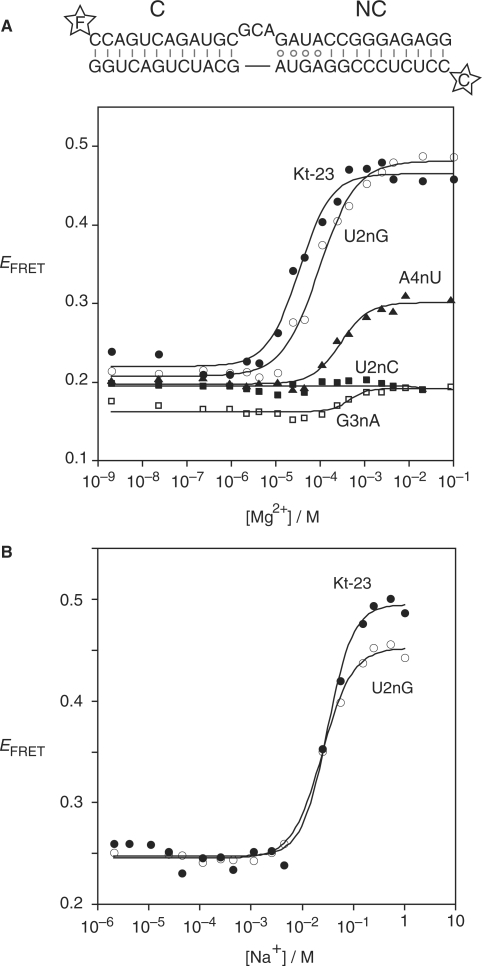
Analysis of ion-induced folding of Kt-23 and variants studied by FRET. The scheme (top) shows the 26 bp RNA duplex with a central Kt-23 sequence, with fluorescein (F) and Cy3 (C) fluorophores attached to the 5′-termini. Base substitutions were introduced into this sequence as required. Plots of FRET efficiency (EFRET) as a function of ionic concentration are presented, fitted to two-state transitions (lines). Symbols: closed circles, unmodified Kt-23; open circles, Kt-23 U2nG; closed squares, Kt-23 U2nC; open squares, Kt-23 G3nA; closed triangles, Kt-23 A4nU. (A) Plot of EFRET as a function of Mg2+ ion concentration. (B) Plot of EFRET as a function of Na+ ion concentration.
Table 2.
Summary of FRET data for folding of Kt-23 and variant k-turns induced by Mg2+ and Na+ ions
| [Mg2+]1/2 (µM) | ΔEFRET | [Na+]1/2 (mM) | ΔEFRET | ||
|---|---|---|---|---|---|
| Kt-23 | 35 | 0.25 | 32 | 0.25 | |
| Kt-7a | 78 | 0.33 | 30 | 0.33 | |
| G1b U | NF | ||||
| U2n G | 97 | 0.27 | 26 | 0.22 | |
| U2n C | NF | ||||
| G3n A | 770 | 0.034 | |||
| G3n inosine | 560 | 0.095 | |||
| A4n U | 290 | 0.10 | 240 | 0.12 | |
| A4n G | 140 | 0.23 | 68 | 0.16 | |
| GL1 2′H | NF | ||||
| AL3 2′H | NF | ||||
| GL1 inosine | NF | ||||
| C-2n 2′H | 240 | 0.29 | |||
| U3b 2′H | 130 | 0.30 | |||
| G-1n 2′H | 200 | 0.12 |
ΔEFRET was measured as a function of ionic concentration and the data fitted to a two-state model in which folding occurs as a result of ion binding (Equation 1).
NF, no folding detected.
aData for Kt-7 are taken from Liu & Lilley (19). The absolute value of ΔEFRET is not comparable between Kt-7 and Kt-23 since differences in the exact structure adopted in the helical arms could alter the relative placement of the fluorophores.
Conversion of the A•U pair at the 2b•2n position of Kt-23 to A•G allows normal k-turn folding to occur
The context of the A•U pair at the 2b•2n position in Kt-23 allows stable formation of the k-turn despite the absence of the normal A•G pair. This raises the question of whether an A•G pair would in fact be tolerated at that position in the Kt-23 k-turn. We therefore made a U2nG change in order to create the conventional A•G pair at this position. Comparative gel electrophoresis shows that the U2nG species has closely similar mobility to that of Kt-7 in the presence of 2 mM Mg2+ ions (Figure 3). The change in FRET efficiency for the U2nG species was similar to Kt-23, with [Mg2+]1/2 = 97 µM and [Na+]1/2 = 26 mM (Figure 4). Thus, Kt-23 U2nG folds into the k-turn geometry in a closely similar manner to Kt-7, indicating that the sequence context can tolerate an A•G pair at the 2b•2n position. Indeed, an A•G pair is present at the 2b•2n position of Kt-23 from M. genitalium and D. melanogaster.
Other changes at the 2b•2n position of Kt-23 prevent ion-induced folding
In marked contrast to Kt-23 U2nG, the substitution U2nC completely prevented folding into the k-turn geometry. No retardation was observed in comparative gel electrophoresis (Figure 3), and no change of FRET efficiency was induced by either Mg2+ or Na+ ions (Figure 4, Table 2). This substitution would prevent formation of one of the two hydrogen bonds stabilizing the trans Hoogsteen/Watson–Crick A•U pair, removing the U:N3H imino proton donated to A:N7.
The nature of the base pair at the 3b•3n position of Kt-23 is very important
The ability of the T. thermophilus Kt-23 sequence to adopt the k-turn geometry in the presence of Mg2+ ions, unaided by protein binding, despite the presence of the A•U pair at the 2b•2n position raises an interesting question. Since the introduction of an A•U pair at the corresponding position of Kt-7 completely prevents ion-induced folding (13), what is different about the sequence of Kt-23 that tolerates this change? This is likely to be the sequence context. In both the cases, the 1b•1b pair is G•A, and this cannot be changed in either Kt-7 (13,20) or Kt-23 (Figure 3; Table 2). On the kink-distal side of the 2b•2n pair in H. marismortui Kt-7, there is a further A•G pair at the 3b•3n position, followed by Watson–Crick pairing. By contrast, in T. thermophilus Kt-23 there is a U•G pair at 3b•3n and an A•A pair at 4b•4n. However, in the E. coli K-23 these base pairs are Hoogsteen/sugar edge G•G and Watson–Crick A–U, respectively.
The U•G pair at the 3b•3n position of T. thermophilus Kt-23 is a Hoogsteen/sugar edge pair that forms a single hydrogen bond donated from G3n:N2 to U3b:O4. It significantly widens the helix, with a C1′–C1′ distance of 11.1 Å, compared to 10.0 Å in Kt-7 (Table 1). G3nA substitution removes the 2-NH2 thus preventing formation of the hydrogen bond. This substitution almost completely prevented folding into the k-turn geometry as judged by both comparative gel electrophoresis (Figure 3) and FRET (Figure 4; Table 2). This could result from formation of a Watson–Crick U–A base pair, but selective removal of the 2-NH2 from the guanine (inosine substitution, i.e. G3nI) also led to extremely impaired in k-turn folding (Table 2).
The nature of the base pair at the 4b•4n position of Kt-23 is less important
The 4b•4n base pair is usually Watson–Crick in the majority of k-turns. The cis Watson–Crick/Watson–Crick A•A pair in T. thermophilus Kt-23 again widens the helix, with a C1′–C1′ distance of 12.3 Å compared to 10.4 Å in Kt-7 (Table 1). The adenine bases are connected by a single hydrogen bond donated from A4b:N6 to A4n:N1. Restoration of a potential Watson–Crick pair by an A4nU substitution significantly impaired folding, with a major loss of electrophoretic retardation (Figure 3), and ΔEFRET = 0.10 with a [Mg2+]1/2 = 290 µM (Table 2). Disruption of the A•A pair by A4nG substitution (creating a potential A•G pair) was less disruptive, with ΔEFRET = 0.23 with a [Mg2+]1/2 = 140 µM. In general, it appears that changing the 4b•4n base pair impairs the folding of Kt-23 less than substitutions at the 3b•3n pair. In the E. coli Kt-23, this position has a regular A–U base pair.
Critical contacts arising from the bulge loop are very important in Kt-23
The two most conserved hydrogen bonds in crystallographic structures of k-turn RNA involve 2′-hydroxyl groups of the bulge (19). These are formed between the L1 ribose 2′OH and the N1 of the adenine at the invariant 1n position, and between the L3 ribose O2′ and the proS O of the L1/L2 phosphate. Both are present in the Kt-23 crystal structures (22,23) (Figure 5A), with distances of 2.7 (2.6) Å and 3.0 (2.7) Å, respectively (E. coli values in parenthesis), and with near-tetrahedral geometries in both the cases. Removal of the O2′ (by deoxyribose substitution) at either L1 or L3 led to almost complete loss of electrophoretic retardation (Figure 3), and no k-turn folding detectable by FRET (Table 2). We conclude that both interactions are critical to the stability of the k-turn geometry of Kt-23 in the presence of metal ions.
Figure 5.
Hydrogen bonding in the Kt-23 k-turn. Parallel-eye stereoscopic views of the structure of Kt-23 taken from the crystal structure of the T. thermophilus 30S ribosomal subunit, with key hydrogen bonds indicated by broken lines. (A) Hydrogen bonding in the core of the k-turn. The A1n nucleotide is highlighted in yellow. This is held by four hydrogen bonds; in addition to the two bonds with G1b that form the trans sugar edge/Hoogsteen edge base pair, there is the critical, conserved A1n:N1–GL1:O2′H interaction, and the A1n:N3–GL1:N2H hydrogen bond. The hydrogen bond between AL3:O2′H and the L1/L2 phosphate proS O is also visible in this image. (B) Hydrogen bonding between the C and NC helices. Two hydrogen bonds are shown. One is from G-1n:O2′H to A2b:N1, and between U3b:O2′ and C-2n:O2′H.
A possible L1–1n base–base interaction
In T. thermophilus Kt-23, the nucleobase of A1n makes another hydrogen bond. A1n:N3 accepts a proton from GL1:N2H, with a length of 3.3 Å and a near-tetrahedral angle of 114° (Figure 5A) (22). This plainly cannot be a universal interaction, because the L1 nucleotide is not constrained to be guanine in other Kt-23 sequences, and even when L1 is guanine the distance is often long. For example, this interaction is also present in the E. coli Kt-23, but the putative bond length is 3.5 Å (23). We examined the importance of this in Kt-23 by replacing the guanine by inosine (GL1I), selectively removing the exocyclic amine that is the putative donor. The modified RNA exhibited no ion-induced folding detectable by FRET (Table 2), consistent with an important role for this interaction. This hydrogen bond plus the L1:2′O to A1n:N1 bond together hold the nucleobase of adenine 1n by both ring nitrogen atoms. Perhaps its tight positioning becomes important in Kt-23 in order to hold A2b (on which it is stacked) in place in the absence of the A•G pair at 2b•2n.
A base–backbone interaction in Kt-23 involving A2b
Inspection of the structures of both T. thermophilus and E. coli Kt-23 in the crystal suggested a further hydrogen bonding interaction from the 2′-hydroxyl of G-1n and A2b:N1 (Figure 5B). This has a length of 2.6 Å and almost perfect tetrahedral geometry (110°) in both structures. Removal of the 2′-hydroxyl group of G-1n led to impaired folding, with ΔEFRET = 0.12 (Table 2). Formation of this hydrogen bond would help to fix the position of the adenine nucleobase at 2b; this again could assume greater important in the absence of the usual A•G pair at 2b•2n.
Possible inter-helical hydrogen bonding in Kt-23
In studies of other k-turns, we have noted that there is hydrogen bonding between the C and NC stems observed in crystal structures, mainly involving 2′-hydroxyl groups (19). However these are variable from one structure to another. In one class (exemplified by Kt-7), there is bonding between O2′ of −2n and 3b, and in the other class (a bigger group, that includes the box C/D k-turn) there is a hydrogen bond formed between O2′ of −1n and 2b. However, removal of selected 2′-hydroxyl groups has suggested that none of these interactions is important to the stability of the k-turn (19). In the Kt-23 crystal structures, there is a hydrogen bond between U3b:O2′ and C-2n:O2′ with a distance of 2.7 Å (T. thermophilus) or 3.1 Å (E. coli) and excellent tetrahedral geometry (Figure 5B). We investigated the importance of this interaction to the stability of the k-turn by removing the 2′-hydroxyl from U3b. The resulting RNA folded well, with a ΔEFRET = 0.3, and a slightly elevated requirement for Mg2+ ions ([Mg2+]1/2 = 130 µM) (Table 2). Removal of the 2′-hydroxyl at C-2n led to relatively unperturbed folding, with ΔEFRET = 0.29 and [Mg2+]1/2 = 240 µM. As with other k-turns (19), it appears that the interhelical hydrogen bonding outside the core region is not very important for the stability of the kinked geometry of Kt-23. In general, these hydrogen bonds form adventitiously, but contribute little stabilization to k-turn conformation.
DISCUSSION
Since Kt-23 breaks the seemingly critical requirement for an A•G pair at the 2b•2n position in both T. thermophilus and E. coli, it might have been thought that it would be unable to adopt the k-turn conformation unaided. While both Kt-23 sequences clearly adopt normal k-turn structure in the context of the 30S ribosomal subunits, we suspected that either long-range tertiary interactions, protein-RNA interactions or both might provide sufficient stabilization to overcome an inherent instability. Our results show that that is not the case; T. thermophilus Kt-23 can adopt the k-turn structure in a simple RNA duplex in the absence of any proteins and with no possibility for tertiary structure. Stable k-turn formation only requires charge neutralization by either divalent or monovalent metal ions. Indeed, folding of Kt-23 occurs at a lower ion concentration compared to Kt-7 with its near canonical k-turn sequence, indicating than the Kt-23 is if anything more stable as a k-turn than is Kt-7. This is supported by a consistently greater electrophoretic retardation of Kt-23 compared to Kt-7, although the effect is small.
This surprising result shows that the sequence requirements for k-turn stability are more complex than we previously supposed. It leaves a question outstanding. Why can Kt-23 adopt the k-turn conformation despite possessing an A•U pair at the 2b•2n position, when conversion of the 2b•2n position in Kt-7 to A•U prevents any detectable ion-induced folding? Clearly, some aspect of the sequence environment of Kt-23 permits the RNA to form a stable k-turn despite the A•U pair at the 2b•2n position, although this does not prevent folding from occurring when an A•G pair is restored at this site. Kt-23 makes the previously identified key hydrogen-bonding interactions within the core of the k-turn involving the 2′-hydroxyl groups of the loop nucleotides L1 and L2 (19). A2b also makes an important interaction with the ribose of G-1n, with a hydrogen bond from G-1n:2′OH to A2b:N1. The A•U pair does not make any additional direct contacts with its neighbours in the NC helix, so perhaps the overall geometry of the NC helix is better able to accommodate this pair. We have shown that the nature of both the 3b•3n and 4b•4n pairs can influence the stability of the k-turn conformation of T. thermophilus Kt-23. The trans Hoogsteen/Watson–Crick edge A•U pair is significantly wider than the usual A•G pair (Table 1), and it is possible that the additional width of the Hoogsteen/sugar edge U•G pair at the 3b•3n position is an important factor. The Hoogsteen/sugar edge G•G pair at the corresponding position in E. coli Kt-23 is also significantly wider, at 11.4 Å. Our results suggest that the nature of the base pair at the 3b•3n position is more important than that at the 4b•4n position, and that changes that should destabilize this base pair markedly impair folding into the k-turn geometry. Normal k-turn sequences with an A•G pair at the 2b•2n position appear less sensitive to the sequence on the bulge-distal side; in the U4 snRNA k-turn there are conventional Watson–Crick G–C base pairs at both 3b•3n and 4b•4n positions (6).
Kt-23 reveals a surprising tolerance to departure from the consensus sequence. It folds into a near perfect k-turn geometry that places the critical 1n and 2b adenine bases into the required positions. The key hydrogen-bonding interactions in the core of the k-turn are preserved. The nucleobase at position 2 is exposed on the splayed-out major groove face of the k-turn, and most generally it is this face that interacts with L7Ae (2) and related (6) proteins. L7Ae can induce k-turn folding upon binding to Kt-7 and other k-turns (15), and so it is possible that the folding of k-turns (such as Kt-23) that differ in this region might be induced by different proteins. We are in the process of investigating this possibility.
FUNDING
Wellcome Trust and Cancer Research UK. Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Ben Turner and Jonathan Ouellet for discussion, Scott McPhee for RNA synthesis, and the Wellcome Trust and Cancer Research UK for financial support.
REFERENCES
- 1.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore T, Zhang Y, Fenley MO, Li H. Molecular basis of box C/D RNA-protein Interactions; Cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure. 2004;12:807–818. doi: 10.1016/j.str.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 3.Hamma T, Ferré-D'A;maré AR. Structure of protein L7Ae bound to a K-turn derived from an archaeal box H/ACA sRNA at 1.8 Å resolution. Structure. 2004;12:893–903. doi: 10.1016/j.str.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 4.Szewczak LB, Gabrielsen JS, Degregorio SJ, Strobel SA, Steitz JA. Molecular basis for RNA kink-turn recognition by the h15.5K small RNP protein. RNA. 2005;11:1407–1419. doi: 10.1261/rna.2830905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Youssef OA, Terns RM, Terns MP. Dynamic interactions within sub-complexes of the H/ACA pseudouridylation guide RNP. Nucleic Acids Res. 2007;35:6196–6206. doi: 10.1093/nar/gkm673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vidovic I, Nottrott S, Hartmuth K, Luhrmann R, Ficner R. Crystal structure of the spliceosomal 15.5 kD protein bound to a U4 snRNA fragment. Mol. Cell. 2000;6:1331–1342. doi: 10.1016/s1097-2765(00)00131-3. [DOI] [PubMed] [Google Scholar]
- 7.Wozniak AK, Nottrott S, Kuhn-Holsken E, Schroder GF, Grubmuller H, Luhrmann R, Seidel CA, Oesterhelt F. Detecting protein-induced folding of the U4 snRNA kink-turn by single-molecule multiparameter FRET measurements. RNA. 2005;11:1545–1554. doi: 10.1261/rna.2950605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mao H, White SA, Williamson JR. A novel loop-loop recognition motif in the yeast ribosomal protein L30 autoregulatory RNA complex. Nature Struct. Biol. 1999;6:1139–1147. doi: 10.1038/70081. [DOI] [PubMed] [Google Scholar]
- 9.White SA, Hoeger M, Schweppe JJ, Shillingford A, Shipilov V, Zarutskie J. Internal loop mutations in the ribosomal protein L30 binding site of the yeast L30 RNA transcript. RNA. 2004;10:369–377. doi: 10.1261/rna.2159504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- 11.Blouin S, Lafontaine DA. A loop loop interaction and a K-turn motif located in the lysine aptamer domain are important for the riboswitch gene regulation control. RNA. 2007;13:1256–12567. doi: 10.1261/rna.560307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heppell B, Lafontaine DA. Folding of the SAM aptamer is determined by the formation of a K-turn-dependent pseudoknot. Biochemistry. 2008;47:1490–1499. doi: 10.1021/bi701164y. [DOI] [PubMed] [Google Scholar]
- 13.Goody TA, Melcher SE, Norman DG, Lilley DMJ. The kink-turn motif in RNA is dimorphic, and metal ion dependent. RNA. 2003;10:254–264. doi: 10.1261/rna.5176604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsumura S, Ikawa Y, Inoue T. Biochemical characterization of the kink-turn RNA motif. Nucleic Acids Res. 2003;31:5544–5551. doi: 10.1093/nar/gkg760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turner B, Melcher SE, Wilson TJ, Norman DG, Lilley DMJ. Induced fit of RNA on binding the L7Ae protein to the kink-turn motif. RNA. 2005;11:1192–1200. doi: 10.1261/rna.2680605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Razga F, Koca J, Sponer J, Leontis NB. Hinge-like motions in RNA Kink-turns: The role of the second A-minor motif and nominally unpaired bases. Biophys. J. 2005;5:3466–3485. doi: 10.1529/biophysj.104.054916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cojocaru V, Klement R, Jovin TM. Loss of G-A base pairs is insufficient for achieving a large opening of U4 snRNA K-turn motif. Nucleic Acids Res. 2005;33:3435–3446. doi: 10.1093/nar/gki664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Razga F, Zacharias M, Reblova K, Koca J, Sponer J. RNA kink-turns as molecular elbows: hydration, cation binding, and large-scale dynamics. Structure. 2006;14:825–835. doi: 10.1016/j.str.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 19.Liu J, Lilley DMJ. The role of specific 2′-hydroxyl groups in the stabilization of the folded conformation of kink-turn RNA. RNA. 2007;13:200–210. doi: 10.1261/rna.285707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turner B, Lilley DM. The importance of G.A hydrogen bonding in the metal ion- and protein-induced folding of a kink turn RNA. J. Mol. Biol. 2008;381:431–442. doi: 10.1016/j.jmb.2008.05.052. [DOI] [PubMed] [Google Scholar]
- 21.Lescoute A, Leontis NB, Massire C, Westhof E. Recurrent structural RNA motifs, isostericity matrices and sequence alignments. Nucleic Acids Res. 2005;33:2395–2409. doi: 10.1093/nar/gki535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 23.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 24.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc. Natl Acad. Sci. USA. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhattacharyya A, Murchie AIH, Lilley DMJ. RNA bulges and the helical periodicity of double-stranded RNA. Nature. 1990;343:484–487. doi: 10.1038/343484a0. [DOI] [PubMed] [Google Scholar]
- 26.Gohlke C, Murchie AIH, Lilley DMJ, Clegg RM. The kinking of DNA and RNA helices by bulged nucleotides observed by fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA. 1994;91:11660–11664. doi: 10.1073/pnas.91.24.11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beaucage SL, Caruthers MH. Deoxynucleoside phosphoramidites – a new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett. 1981;22:1859–1862. [Google Scholar]
- 28.Wilson TJ, Zhao Z-Y, Maxwell K, Kontogiannis L, Lilley DMJ. Importance of specific nucleotides in the folding of the natural form of the hairpin ribozyme. Biochemistry. 2001;40:2291–2302. doi: 10.1021/bi002644p. [DOI] [PubMed] [Google Scholar]
- 29.Clegg RM. Fluorescence resonance energy transfer and nucleic acids. Meth. Enzymol. 1992;211:353–388. doi: 10.1016/0076-6879(92)11020-j. [DOI] [PubMed] [Google Scholar]
- 30.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 31.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'S;ouza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]