Abstract
To cleave DNA, the single polypeptide restriction–modification enzyme LlaGI must communicate between a pair of indirectly repeated recognition sites. We demonstrate that this communication occurs by a 1-dimensional route, namely unidirectional dsDNA loop translocation rightward of the specific recognition sequence 5′-CTnGAyG-3′ as written (where n is either A, G, C or T and y is either C or T). Motion across thousands of base pairs is catalysed by the helicase domain and requires the hydrolysis of 1.5-2 ATP per base pair. DNA loop extrusion is accompanied by changes in DNA twist consistent with the motor following the helical pitch of the polynucleotide track. LlaGI is therefore an example of a polypeptide that is a completely self-contained, multi-functional molecular machine.
INTRODUCTION
A central requirement of the biological role of restriction–modification (RM) enzymes is the ability of an endonuclease to introduce a double-strand (ds) break into DNA molecules that contain an unmethylated binding sequence. For many of the endonucleases classified as Type II RM enzymes, recognition of a solitary sequence is sufficient to elicit this activity. For example, the extensively studied endonuclease EcoRV is a homodimer that binds to one copy of the palindromic sequence 5′-GAT↓ATC-3′ and introduces a dsDNA break as indicated by the arrow (1). However, for many other Type II RM enzymes (2), and for all of the RM enzymes classified as Types I or III (3), DNA cleavage activity requires the presence of at least two copies of the recognition site. These sites can be separated in solution by distances much larger than the dimensions of the interacting protein(s), and therefore ‘long-range communication’ events must be used to signal the presence of both sites. For the ATP-independent Type II enzymes, this communication is brought about by an enzyme binding simultaneously to two (or more) DNA sequences (4). Where the sites are on the same DNA molecule (in cis), this will result in the formation of a DNA loop. Where the sites are on separate DNA molecules (in trans), this will result in the formation of a so-called sandwich complex. The efficiency of communication is therefore dependent on the local concentration of the DNA sites rather than their connectivity per se (5). In contrast, the ATP-dependent Type I and Type III enzymes will only communicate with sites in cis (6,7). Proteins from both families contain domains with amino acid motifs and structures characteristic of Superfamily 2 DNA helicases (8,9). These domains are not involved in DNA unwinding, but are used to couple ATP hydrolysis to long-range 1-D communication (10–12). A number of models, which are described below, have been proposed for how this can occur.
DNA loop translocation
In this model, the helicase domain binds non-specific DNA adjacent to the recognition site, hydrolyses adenosine triphosphate (ATP) and translocates along the intact duplex strand away from the site. At the same time, the helicase domain interacts with other protein components of the RM complex that are tightly bound to the recognition site. The result is that a growing loop of DNA is formed (13). When the translocating helicase collides head-on with another translocating RM motor, dsDNA cleavage is activated. It is likely that during translocation the nuclease domain is inactivated, and cleavage can only occur upon interaction with the second enzyme. Loop translocation has been observed for the Type I RM systems using a variety of assays both in vitro and in vivo (e.g. 14–16). These enzymes have two helicase domains per RM complex, each motor translocating along the DNA from either side of the site, resulting in the simultaneous translocation of two DNA loops (14–18). There is therefore no preference for relative site orientations between a pair of sites (19). The motor steps one base pair for each ATP hydrolysed, with the DNA being twisted by 360° for each 10 bp moved (20). Significant amounts of ATP must be consumed during translocation, particularly as these enzymes can move over thousands of base pairs.
A similar loop translocation model has also been proposed for the Type III enzymes, except that the motor moves in just one direction on DNA governed by the initial binding of the enzyme to its asymmetric recognition site (21). This model explains the preference during cleavage for pairs of sites in specific inverted orientations, as only motors that collide head-on will cleave the DNA. Whilst evidence from several independent AFM studies is consistent with loop formation (22–24), other studies have failed to find DNA looping (7), changes in DNA topology (25) or stepwise motion on DNA (26) consistent with a 1-D tracking model. Additionally, the Type III enzymes have ATPase activities more than two orders of magnitude slower than the Type I enzymes yet similar DNA cleavage rates (12,21,26), inconsistent with the same 1-bp-one-ATP-tracking mechanism.
DNA tracking without loop formation
It has also been proposed that the Type III RM complexes could release their recognition sequences during translocation, moving on DNA without loop formation (27). The direction of translocation would still be dictated by the orientation of the DNA sequence, so giving a site orientation preference. Whilst consistent with the evidence against the formation of DNA loops, this model is still inconsistent with the current models for ATP coupling by helicase motors given the small amounts of ATP consumed before cleavage occurs.
DNA sliding
A third model proposed recently for the Type III enzymes is that the role of ATP hydrolysis is to induce a conformational change in the enzyme from a DNA-recognition mode to a 1-D DNA sliding mode, the latter being a passive mechanism driven by thermal motion (26,28). Because the initial site recognition event is asymmetric and DNA sliding proceeds without the enzyme releasing the DNA, ‘memory’ of site orientations can be maintained during communication. As the motion is thermally driven and thus random, the enzyme will search both up- and down-stream of the site with equal probability. This model can account for the low ATP usage of Type III enzymes—as little as one ATP per sliding event could be required. It also explains the inhibitory effect of DNA ends upstream from a site on linear DNA as the enzyme will be more likely to fall off the DNA. DNA ‘hopping/jumping’, where an enzyme dissociates from and re-binds to the same DNA molecule, cannot account for the 1-D movement of Type III enzymes as DNA release would lose the memory of the site orientation and all site pairings would be cut equally well.
Passive DNA looping
Whilst the formation of DNA loops between pairs of Type I or Type III enzymes by 3-D capture has been observed (22,23,29,30), this mechanism alone cannot account for the activity of these enzymes. Instead, the formation of these loops may be an early step that stabilises a multi-site nucleoprotein complex so facilitating the subsequent 1-D communication events.
In the accompanying paper (31), we demonstrate that dsDNA cleavage by the single polypeptide RM enzyme LlaGI requires ATP hydrolysis-dependent communication between two DNA sites in a specific head-to-head orientation. Therefore, which of the mechanisms described above could LlaGI use to communicate between the sites? On the basis of primary amino acid sequence analysis, the helicase and methyltransferase (MTase) domains of LlaGI are most similar to the classical Type I enzymes (32). However, the requirement for a pair of DNA sites with a specific relative orientation and the methylation of only one strand of those sites (31), are more reminiscent of the Type III enzymes [and some recently-identified Type II enzymes (33)]. Here we demonstrate that LlaGI communicates between distant DNA sites using a unidirectional loop translocation mechanism that is most consistent with a Type I-like behaviour. This activity can be readily incorporated into a model for how LlaGI cleaves DNA. Multi-domain proteins such as LlaGI are the most abundant proteins in existence—over 50% of prokaryotic proteins and 70% of eukaryotic proteins have more than one domain (34)—and the fusion of nuclease and helicase domains play a number of important genetic roles (35). Despite this, very little mechanistic analysis has been carried out on such systems and it is not clear how these modular enzyme activities are coordinated and controlled on DNA. The data presented here and in the accompanying papers show that LlaGI is an ideal model system to address these questions.
MATERIALS AND METHODS
DNA
To prepare DNA for biochemical assays, Escherichia coli Top10 (Invitrogen, CA, USA) or XL10-Gold (Stratagene) were transformed with the required plasmid, grown in M9 minimal medium supplemented with 37 MBq/l [3H-methyl] thymidine and the DNA extracted by density gradient centrifugation in CsCl-ethidium bromide (31). Linear DNA substrates were generated by digestion with commercial restriction enzymes and purified by phenol/chloroform and chloroform extraction followed by ethanol precipitation. DNA concentrations were determined from UV absorbance at 260 nm, assuming that an optical density of 1 corresponds to 50 µg/ml DNA and a molecular weight of 6.6 × 105 Da/kb.
Triplex substrates
Quikchange mutagenesis (Stratagene) was used to introduce an NcoI site into pZero (31) to create plasmid pZeroNcoI using the oligonucleotides: 5′-CGATCTGTCTATTTCGTTCATCCATGGTTGCCTGACTCCCCGTCG-3′ and 5′-CGACGGGGAGTCAGGCAACCATGGATGAACGAAATAGACAGATCG-3′. QuikChange mutagenesis was also used to mutate an EcoRI site in pLKS5 [a plasmid containing four triplex binding sites (TBSs) (36)] to create plasmid pLKS5dE using the oligonuceotides: 5′-GCGAGTCGACGGCGGCCGCGAACTCCTCGAGCCCGGGTGACTGC-3′ and 5′-GCAGTCACCCGGGCTCGAGGAGTTCGCGGCCGCCGTCGACTCGC-3′. pLKS5dE was digested with AfeI and EcoRV. The 2148-bp fragment containing the TBSs was purified and ligated to pZeroNcoI that had been linearized with SmaI, to generate plasmid pRMA03. Oligonucleotides 5′AGCTCTAGCTAATAGACTGGATGGAGGCG-3′ and 5′-CTAGCGCCTCCATCCAGTCTATTAGCTAG-3′ were annealed and ligated into pRMA03 that had been digested with XbaI and HindIII to generate pRMA03F. pRMA03R was generated in the same way using oligonucleotides 5′-CTAGCTAGCTAATAGACTGGATGGAGGCG-3′ and 5′-AGCTCGCCTCCATCCAGTCTATTAGCTAG-3′. RMA03, RMA03F and RMA03R are EcoRI-linearized versions of the plasmids.
Catenane substrates
pSH1 (37) was digested with BamHI and the 3480-bp fragment re-ligated to form pSH1-F. One round of QuikChange mutagenesis was used to remove one of the three LlaGI sites in pSH1-F to generate pSH1-G. The 742-bp BamHI fragment from pSH1 (containing a LlaGI site) was re-inserted into pSH1-G in the original orientation at the BamHI site to generate plasmid pSH1-H. Further rounds of QuikChange mutagenesis were used to remove either one or two of the LlaGI sites to produce pC2 and pC1, respectively. C2cat and C1cat are [-2] singly interlinked catenane versions of the plasmids generated using Tn21 resolvase as described previously (5–7).
Proteins
Wild-type LlaGI and LlaGIDA078 were produced and purified as described in the accompanying papers (31,38). Wheat germ topoisomerase I, E. coli topoisomerase I and purine nucleoside phosphorylase (PNP) were purchased from Promega (WI, USA), New England Biolabs (MA, USA) and Sigma-Aldrich (Dorset, UK), respectively. Coumarin-labelled phosphate binding protein (PBP) and Tn21 resolvase were produced as described previously (5,20).
DNA cleavage assay
Cleavage assays contained 4 nM DNA, 4 mM ATP (or other nucleotide where indicated) and 200 nM LlaGI in TMD buffer [50 mM Tris–HCl pH 8.0, 10 mM MgCl2, 1 mM DTT]. Reactions were started by adding LlaGI to a mix of ATP and DNA in TMD buffer and incubated at 20°C for the times indicated. Reactions were stopped with 0.5 vol of 3 × STEB [0.1 M Tris (pH 7.5), 0.2 M EDTA, 40% (w/v) sucrose, 0.4 mg/ml bromophenol blue]. Samples were analysed by agarose gel electrophoresis and the percentage of 3H-labelled DNA in each band per lane ascertained by scintillation counting (31).
DNA triplex displacement assay
Triplex displacement measurements were carried out in an SF61-DX2 stopped-flow fluorimeter as described previously (20). The sample temperature was maintained at 20°C by a waterbath connected to the chamber housing the syringes and flow cell. Reactions were started by adding ATP to a DNA/enzyme mix with the final reaction conditions as 1 nM linear DNA (0.5 nM tetramethylrhodamine triplex), 10 nM LlaGI unless indicated otherwise, and 0–4000 µM ATP in TMD Buffer. Reactions containing <4 mM ATP were supplemented with 25 mM phosphocreatine and 5 U/ml creatine phosphokinase (both Sigma-Aldrich). The exponential phases of the triplex displacement profiles were fitted to a single or double exponential increase with time offset (double shown):
| 1 |
Where A1 and A2 are the amplitudes of the phases and, k1 and k2 are the corresponding rates of those phases, t is the time after mixing with ATP and Tapp is the lag time corresponding to translocation of the fastest enzyme population. The variation of Tapp (s) with distance (d, bp) was fitted to:
| 2 |
Where kstep is the rate of translocation and Ti is a macroscopic reaction time that is governed by either the initiation phase of the reaction or by steps at triplex displacement (20). Although the distance between the LlaGI site and the triplexes defines an upper bound for the translocation distance (see above), the data in Figure 2E are independent of this uncertainty. The dependence of kstep on the ATP concentration was fitted to a hyperbolic relationship:
| 3 |
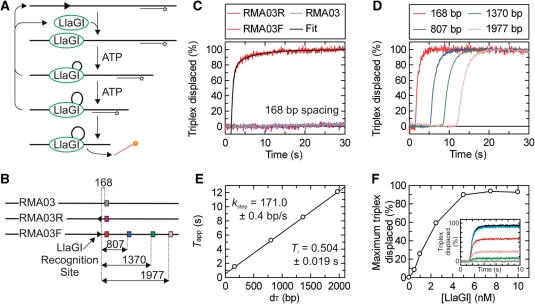
Figure 2.
LlaGI translocase activity measured using the DNA triplex displacement assay. (A) Schematic of the translocation assay showing substrate binding, translocation and triplex displacement. DNA is shown as a thick line, the LlaGI binding site is shown as an arrowhead as in Figure 1, the TFO is shown as a line with a star indicating the position of the TAMRA label and LlaGI is shown as a green oval. It is assumed in this sketch that LlaGI is a DNA loop translocase—this is demonstrated in Figure 4. (B) DNA substrates for the triplex assays: a substrate without a LlaGI site (RMA03), and two 1-site substrates in which the LlaGI site faces either away (RMA03R) or towards (RMA03F) the TBSs. The distances (bp) between the LlaGI site (arrowhead) and the TBSs (coloured squares) are indicated. For clarity, only one of the four TBSs is shown on RMA03 and RMA03R. (C) Triplex displacement is dependent upon the LlaGI sequence. The three DNA substrates (at 1 nM) with the 168-bp spacing triplex bound (0.5 nM) were pre-incubated with LlaGI (100 nM) and translocation initiated by mixing with 4 mM ATP. Triplex displacement is only observed on RMA03F, where the LlaGI site faces towards the TBSs (as defined by the arrowhead). The black line through the RMA03F data represents the fitted line from Equation (1). (D) The rate of triplex displacement is distance dependent. The translocation of LlaGI on RMA03F was investigated individually for each of the four triplex spacings as indicated. Reaction conditions as in (C). (E) The reaction profiles in (D) were fitted to Equation (1) to obtained an estimate of Tapp (not shown). A linear fit of the relationship between Tapp and distances [Equation (2)] gives kstep and Ti. (F) The dependence of triplex displacement on LlaGI concentration. 1 nM RMA03F bound by 0.5 nM 168 bp triplex was incubated with varying concentrations of LlaGI and the reaction initiated by adding 4 mM ATP. (inset) Triplex displacement time course profiles at (from bottom to top) 0, 0.5, 1.0, 2.5, 5.0 and 7.5 nM LlaGI. (Main graph) Relationship of the maximum triplex displaced as a function of LlaGI concentration. Maximum values were calculated from the total amplitudes of the exponential fits.
Phosphate release assay
The preparation and calibration of the Coumarin-labelled PBP, and its utilisation in a stopped-flow fluorimeter assay was as described previously (20). Reactions were started by adding ATP to a DNA/enzyme mix with the final conditions as 0.5 nM linear DNA, 10 nM LlaGI, 0–1050 µM ATP, 200 µM 7-methylguanosine, 0.01 U/µl PNP and 12 µM PBP in TMD buffer. The concentration of PBP used allowed reliable linear measurement of up to 2 µM Pi and all steady-state fits were made within this range (data not shown). The photo multiplier tube response was calibrated using titration of a Pi standard according to Webb (39). As we measured a steady-state accumulation of Pi (i.e. the translocating LlaGI concentration being ∼24 000-fold less than that of the PBP), the observed rates were not limited by the KD of PBP for Pi. At the maximal ATPase rate measured under our experimental conditions, PBP was not rate-limited by the Pi-binding rate (39). LlaGI showed a non-specific ATPase rate that was 6.5–13% of the specific signal. We corrected the profiles on the specific DNA by subtracting the non-specific DNA data collected at the same ATP concentration.
The ATPase rate profiles were fitted to an equation describing a single exponential approach to a steady state (40):
| 4 |
Where V is the steady-state velocity of Pi production and kini is the initial exponential rate. The use of this fit assumes that the steady state is close to 100% occupied and that initiation comprises a unimolecular rate-limiting step (see main text). The linear steady-state phases were also fitted to:
| 5 |
The dependence of the V on ATP concentration was fitted to a hyperbolic relationship [equivalent to Equation (3)].
Error propagation in the triplex displacement and ATPase assays
For the purposes of estimating the ATP coupling ratio, we need to be able to estimate the influence of both statistical/random errors and systematic errors. This is considered in detail in (20). In brief, for the determination of KM and Vmax using both assays, the statistical errors are smaller than the scatter in the data points. Since the scatter makes a greater contribution to the statistics of the fit, the fitting to Equation (3) was not weighted to the statistical errors. For the ATPase data, additional systematic errors that arise due to uncertainties in the DNA concentration (which defines the maximum possible concentration of translocating species) and in the absolute occupancy of the steady state will affect all data points to the same extent and with the same sign and therefore are not useful in weighting the fit. Where the systematic errors are more critical are in the calculation of the coupling ratio. Estimates determined previously using the Type I enzyme EcoR124I for the total systematic error from the ATPase values are +11% and −6% (20). We currently do not have enough information on the LlaGI enzyme to accurately estimate the steady-state occupancy during translocation. However, the triplex data is consistent with a re-initiation of translocation that is not rate limiting. In addition, the initiation times of LlaGI are faster than that of EcoR124I, indicating that the steady state will be occupied to a greater extent. We are therefore confident that the errors here are within this range. For the coupling ratio of EcoR124I (20), the total precision of the sample mean that incorporates both statistical and systematic errors was +15% and –10%. We suggest that because similar or smaller errors will arise here, we will apply the same error range to our coupling data when considering the data precision.
DNA topoisomerase assay
Relaxed DNA was generated by treating 42 nM DNA (pRMA03 and pRMA03F) with 0.25 U/µl wheat germ topoisomerase I in NEBuffer 2 (New England Biolabs). Reactions were incubated at 37°C for 30 min. The DNA was purified by phenol/chloroform and chloroform extraction followed by ethanol precipitation. Topology experiments contained 10 nM DNA, 4 mM ATP, 50 nM LlaGIDA078, 1 U/µl wheat germ topoisomerase I or 0.5 U/µl E. coli topoisomerase I in NEBuffer 4 containing 0.1 mg/ml bovine serum albumin. Reactions were incubated at 37°C for 30 min and stopped by the addition of 0.5 volumes 3 × STEB [0.1 M Tris, pH 7.5, 0.2 M EDTA, 40% (w/v) sucrose, 0.4 mg/ml bromophenol blue]. Samples were heated to 67°C and then run on a 1% (w/v) agarose TAE gel in the absence of ethidium bromide (EtBr) at 6.25 V/cm for ∼3 h. Gels were post-stained with EtBr to visualise the DNA.
RESULTS
Based on the alternative models for long-range communication described above, we can make predictions about the activities of LlaGI that we can test using assays that have been successfully applied to Type I, II and III RM enzymes.
Endonuclease activity on DNA catenanes
As noted above, whilst enzymes that utilize 3-D DNA looping do not necessarily require covalent connectivity between a pair of sites to activate DNA cleavage, local site concentration can limit the reaction. For example, the Type II enzyme SfiI cleaves a two-site DNA faster that a one-site DNA as the intramolecular collisions between two sites on the same DNA are more frequent than the intermolecular interactions between two sites on separate DNA molecules (5). However, if separate one-site DNA molecules are topologically linked to form a catenane, the local intermolecular site concentration is very similar to the intramolecular site concentration. Accordingly, SfiI cuts two sites on separate rings of a catenane as rapidly as two sites on the same DNA (5). In contrast, the Type III RM enzymes cannot cut a catenane when the recognition sites are on separate rings as the 1-D connectivity is lost (7). Therefore, by comparing the DNA cleavage activity of LlaGI on both plasmid and catenane substrates we can distinguish between 1-D and 3-D mechanisms of communication (41).
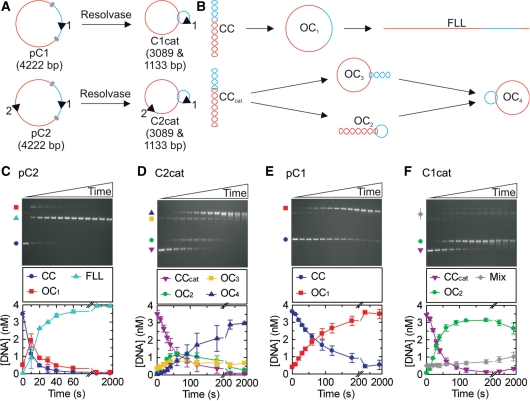
We first compared a plasmid with two LlaGI sites in indirect repeat (pC2, Figure 1A) with a [-2] singly-interlinked catenane in which the sites are on separate DNA rings (C2cat, Figure 1A). Intermediates and products of DNA cleavage by LlaGI that can be separated by agarose gel electrophoresis are shown in Figure 1B. The rate of cleavage of pC2 was measured at a saturating concentration of LlaGI (Figure 1C) (rate here refers to the initial rate of substrate loss). The profile obtained matches closely that observed with pHH-3 in Figure 6D of Smith et al. (31). Under the same reaction conditions, C2cat was cleaved more slowly (Figure 1D), first generating intermediates in which one or other DNA ring is nicked (OC2 and OC3), followed by the final product in which both rings are nicked (OC4). The linear products resulting from dsDNA cleavage of either ring were never observed. We also observed that upon extended incubation, the nicked product became smeared on the agarose gel. We interpret this as further strand-specific cleavage at the nick sites to produce ssDNA gaps that will have faster electrophoretic mobilities (Stanley,L. and Szczelkun,M.D, unpublished data).
Figure 1.
LlaGI endonuclease activity measured on DNA catenanes. (A) The two plasmid substrates used to form catenanes in the presence of Tn21 resolvase. LlaGI recognition sequences are represented as arrowheads where the orientation is 5′-CTnGAyG-3′. Res sites for Tn21 resolvase are shown as grey blocks whilst the different DNA domains that are separated by recombination are shown in red and blue. (B) Possible DNA species formed when LlaGI is incubated with the plasmid or catenane substrates. Each arrow represents the cleavage of one DNA strand. CC, covalently closed circular DNA (negatively supercoiled); OC1, open circle DNA (‘nicked’); FLL, full-length linear DNA; CCcat, covalently closed circular catenane DNA. For the catenanes, OC2 is nicked in the small ring only, OC3 is nicked in the large ring only and OC4 is nicked in both rings. Example gels and average cleavage profiles are shown for: (C) two-site plasmid DNA pC2; (D) two-site catenane DNA C2cat; (E) one-site plasmid DNA pC1; and (F) one-site catenane DNA C1cat. Reactions contained 200 nM LlaGI, 4 nM DNA and 4 mM ATP (‘Materials and Methods’ section). ‘Mix’ in (F) represents the combination of OC3, OC4 and dimeric DNA products. Time points were collected at 0, 10, 20, 30, 40, 60, 90, 120, 180, 300, 600, 1200 and 1800 s.
We next compared a plasmid with one LlaGI site (pC1, Figure 1A) with a [-2] singly-interlinked catenane in which the LlaGI site is on the smaller of the two rings (C1cat, Figure 1A). The cleavage rate profile for pC1 shows the DNA nicking expected of a one-site DNA (Figure 1E). The rate is slower than that on either C2cat (Figure 1D) or pHT-12 (Figure 6F in (31)). The rate of cleavage of C1cat is faster than the corresponding pC1 (Figure 1E) but is similar to that of C2cat (Figure 1D).
In the accompanying paper we showed that on a one-site DNA, LlaGI could only produce a nicked product (31). The cleavage profiles on the DNA catenanes show that even when linked, each DNA ring acts like an isolated one-site substrate and only supports DNA nicking. Therefore, dsDNA cleavage by LlaGI must require two sites on the ‘same’ DNA molecule consistent with a 1-D communication mechanism. We note that some site-specific recombinases will only recombine DNA catenanes that have multiple interlinks (42–46). In these cases, the multiple catenane nodes substitute for supercoil nodes that are usually required for successful long-range communication. However, we can rule this out for LlaGI as cleavage activity is independent of DNA topology (7,26) and the subsequent assays presented here confirm a 1-D tracking mechanism (see below).
The cleavage rate profile for C1cat shows that the DNA nicking activity is confined to the small ring that contains the site, with the large ring remaining intact (Figure 1F). This indicates that the non-specific nicking activity is also linked by a 1-D communication mechanism. A model for the nicking activity that explains the different DNA cleavage rates is considered in the Discussion. Although we can dismiss passive 3-D DNA looping as playing a direct role in the cleavage pathways, we cannot rule out that looping between pairs of LlaGI molecules may play an indirect role in stabilising a higher-order nucleoprotein complex, as suggested for the classical Type I enzymes (29,30) and the Type III enzymes (23).
Translocation activity measured by the triplex displacement assay
The catenane data demonstrates that LlaGI most likely communicates between sites in 1-D, which could occur by either tracking or sliding modes. Previously, we developed an assay to validate and measure tracking by means of triplex displacement [Figure 2A (14)]. Whilst Type I RM enzymes and other bona fide DNA translocases can displace a specifically bound triplex upon collision (14,20), the Type III RM enzymes cannot (26). To test the activity of LlaGI, we utilized three linear DNA substrates each of which has four independent TBSs regularly-spaced along the DNA (Figure 2B). The first DNA (RMA03) does not carry an LlaGI recognition site and we do not expect any displacement from this DNA. The second and third substrates have a single LlaGI in alternative orientations (RMA03R and RMA03F). Given the preference for DNA with two sites in head-to-head repeat (31), we predicted that the communication process has a directional preference. We therefore expected triplex displacement from RMA03F but not from RMA03R, as on the latter DNA the enzyme will move away from the triplexes and towards the free DNA end.
LlaGI was pre-incubated with linear DNA in which a single DNA triplex had been pre-bound (see ‘Materials and Methods’ section). The reaction was then started by rapid mixing with ATP (to 4 mM) in a stopped-flow fluorimeter and the triplex binding monitored by changes in fluorescence. Reaction profiles for RMA03, RMA03R and RMA03F are shown for the 168-bp spacing in Figure 2C. Triplex displacement was only observed on RMA03F, in which the triplex was located on the 3′ side of the LlaGI recognition site, as written (5′-CTnGAyG-3′). The profile gives a good fit to Equation (1) as expected for a translocation process (see below). When the triplex was located on the 5′ side of the site (RMA03R) or in the absence of an LlaGI site (RMA03), triplex displacement was not observed. This result is consistent with unidirectional translocation by LlaGI from its site. The direction of motion coincides with that predicted from the cleavage assays in the accompanying paper (31). It differs from observations with the classical Type I enzymes in which bi-directional translocation produces triplex displacement from both sides of the site simultaneously (14).
The triplex displacement profile on RMA03F (168 bp) in Figure 2C shows an exponential displacement rate preceded by a distinct lag phase with a good fit to Equation (1). This is characteristic of motion of an enzyme population in a stepwise manner along the DNA starting at the LlaGI site (14,47). To explore this further, we measured the LlaGI activity on RMA03F using each of the four triplexes in turn (Figure 2D). Similar profiles were seen with all spacings: a lag phase followed by an exponential displacement phase. To obtain an accurate estimate of the lag time (Tapp), each profile was fitted to a single exponential increase with time offset (see ‘Materials and Methods’ section, modified version of Equation (1); data not shown). The good fit to a single exponential (data not shown) suggests that dissociation during translocation or other off-pathway events are rare. The Tapp values as a function of distance could be fitted by Equation (2) (Figure 2E). This linear relationship is entirely consistent with a unidirectional translocating motor (14). We can estimate the rate of translocation as ∼171 bp/s. The y-axis intercept gives Ti = ∼0.5 s. This value can relate to either the translocation initiation rate or to the triplex dissociation rate (20)—we cannot distinguish between these from the triplex assay alone.
In the experiments above, LlaGI was present at a concentration sufficient to fully displace the triplex at all spacings (100 nM—see ‘Materials and Methods’ section). To explore the effect of protein concentration in more detail, we measured triplex displacement on RMA03F (168 bp) at different LlaGI concentrations (Figure 2F, inset). Below 5 nM LlaGI (with the DNA at 1 nM), we observed a decrease in the maximum amount of triplex displaced. The profiles show double exponential displacement phases with no further displacement observed, even with extended incubation. A simple explanation for this data is that below 5 nM LlaGI the DNA is no longer saturated with active protein and on some DNA translocation cannot occur. Where translocation does occur, the LlaGI motors become irreversibly associated with that DNA so that steady-state turnover of LlaGI does not occur (at least on a minute timescale). An apparent lack of turnover has also been attributed to the classical Type I and Type III RM enzymes (3). Figure 2F shows the maximum percentage triplex displaced as a function of LlaGI concentration. The data cannot be described by one-site binding relationships (data not shown). Two alternative models are consistent with the data: (i) Cooperative binding of n LlaGI monomers to make an active translocating machine; or (ii) interaction of a LlaGI monomer under ‘tight-binding conditions’ but where the active concentration of LlaGI is lower than the input concentration (i.e. a low specific activity). We cannot currently distinguish between these alternatives.
ATP coupling during DNA translocation
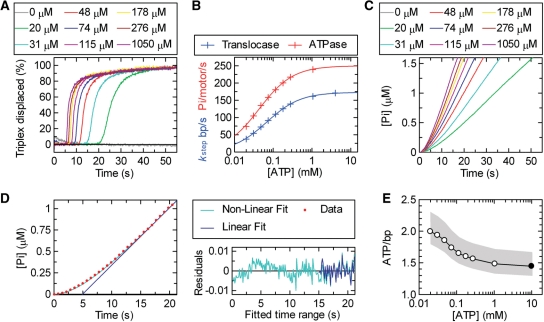
The above-mentioned triplex data are consistent with stepwise DNA translocation. In this case, the hydrolysis of ATP should be coupled to the stepping process. We first tested this by measuring the effect of ATP concentration on the triplex profiles. At a saturating concentration of ATP (4 mM; Figure 2D), the bimolecular rate of the nucleotide binding is significantly faster than the translocation rate. If the concentration of ATP were lowered however, the bimolecular binding rate would start to become rate limiting and the observed stepping rate would decrease (20). This is exactly what we observed (shown for the 807 bp spacing in Figure 3A). As the ATP concentration decreases, the lag phase increases (indicative of slower translocation) whilst the exponential displacement phase gets slower (indicative of a broadening of the enzyme population distribution on the DNA). By measuring the lag phases on RMA03F with each triplex at different ATP concentrations, we obtained a series of linear lag time–distance relationships (data not shown) from which we could estimate the stepping rate as a function of ATP (Figure 3B, blue line). This relationship could be fitted by a hyperbolic relationship to give a KM of 67.8 ± 1.2 µM and a Vmax of 173 ± 1 bp/s. This relationship is similar to that seen for the Type I RM enzyme EcoR124I and indicates that ATP binding is not cooperative during the motor cycle. We cannot however rule out the action of multiple independent motors.
Figure 3.
The coupling of ATP hydrolysis to DNA translocation. (A) Translocation profiles measured using triplex displacement at different ATP concentrations as indicated using 1 nM RMA03F (0.5 nM 807 bp triplex) and 10 nM LlaGI. By using data collected at each ATP concentration for each of the triplex spacings, kstep was calculated as in Figure 2E (data not shown). (B) The relationship between kstep (blue) and kATP (red) as a function of ATP concentration. The solid lines represent least-squares fits to Equation (3). (C) Steady-state phosphate release measured using the PBP assay (‘Materials and Methods’ section); 0.5 nM RMA03F was pre-incubated with 10 nM LlaGI and the reaction initiated with ATP at the concentrations indicated. Profiles have been corrected for a non-specific background measured on RMA03 (data not shown). (D) Example of the fits used to estimate the ATPase rate (V) from the phosphate release data. For clarity, a reduced number of data points (red circles) are shown for the 48 µM profile from (C). Fitting was carried out using the complete data set. The data was fitted to both Equation (4) (green line) and Equation (5) (blue line). The residuals for each fit are shown. There are systematic non-random variations from Equation (4) during the initiation phase, whilst both equations return the same linear fit beyond 15 s (see text for more details). kATP was calculated from the linear steady-state rate (V) assuming one active motor per DNA molecule. (E) The coupling of ATP to translocation. The measured and fitted values for kATP were divided by the corresponding kstep values to give the apparent number of ATP molecules hydrolysed per base pair translocated as a function of ATP concentration. The filled circle indicates the value calculated using the Vmax values from (B). The grey shading indicates the extent of the estimated error range (‘Materials and Methods’ section).
We measured the ATPase rate of LlaGI using the PBP assays as used previously with the Type I enzyme EcoR124I (20). In brief, LlaGI was pre-incubated with linear DNA in the presence of coumarin-labelled PBP (see ‘Materials and Methods’ section). The reaction was then started by rapid mixing with ATP in a stopped-flow fluorimeter and the fluorescence monitored (Figure 3C). We measured the rate profiles using both specific (RMA03F) and non-specific (RMA03) substrates. The data for the specific DNA were then corrected for the non-specific background.
The resulting profiles show a slow acceleration phase leading to a linear steady state (Figure 3C). (Over the timescales and sensitivity used here, we did not observe any changes in the ATPase rate that could be related to the LlaGI motor reaching the free DNA end.) Both the acceleration and linear rates increased with ATP concentration. These data can be described by the model for loop initiation and translocation in Figure 2A. The first step (kini) is the initial formation of a DNA loop catalysed by ATP binding. Since the ATPase activity is measured under ‘steady-state’ conditions ([DNA] « [PBP]), we cannot determine if ATP is hydrolysed at this step. Thereafter, loop translocation is dependent upon ATP hydrolysis (at a rate kATP). If we assume that dissociation from the DNA during translocation (koff) is negligible, the accumulation of Pi in this model can be described by a single exponential approach to a linear steady state [Equation (4) in ‘Materials and Methods’ section; (40)], where the steady-state rate is kATP. However, fitting the LlaGI data with Equation (4) reveals small but systematic variations in the exponential phase that are indicative of a higher-order relationship during initiation (Figure 3D). Similar deviations were observed with EcoR124I (20), and can be attributed to either: (i) multiple rate-limiting steps during loop formation whose rates are similar in magnitude; (ii) off-pathway inhibited states during initiation; and/or (iii) an acceleration of the translocation rate as the loop grows beyond a topologically constrained size. We cannot distinguish between these models from our data. The LlaGI data were also fitted to a simple linear relationship [Equation (5)] (Figure 3D). The convergence of Equations (4) and (5) for points above 15 s shows that the linear phase has been reached and therefore both equations can yield an accurate estimate of the steady-state rate.
The rate of phosphate released was converted to phosphates released per motor per second by dividing by the DNA concentration. This assumes that all DNA molecules have a single translocating LlaGI motor and that all motors are active in the steady state. The former is likely to be correct as the binding is saturated (Figure 2F). However, if termination of translocation is significant, the steady-state occupancy would be <100% as some enzymes would be in the processes of re-initiation. In the previous study of EcoR124I, the steady-state occupancy could be calculated with a degree of accuracy from the known rates for motor dissociation and re-initiation (20). Although the triplex data in Figure 2D are consistent with a negligible off rate, the ATPase rates obtained here are a lower boundary because the other kinetic values are not currently known.
The relationship between kATP and the ATP concentration is show in Figure 3B. The data can be fitted to a hyperbolic relationship to give a KM of 43.6 ± 1.3 µM and a Vmax of 250 ± 2 bp/s. The difference between the KM values from the triplex and ATPase assays most likely reflect experimental and systematic errors in the assays (see ‘Materials and Methods’ section). The experimental and fitted translocation and ATPase rates at each ATP concentration (Figure 3B) were then compared to estimate the coupling ratio (Figure 3E). The coupling ratio has a range of 1.5–2 ATP/bp, with a systematic trend in the data, where an increase in ATP concentration produced a decrease in the ratio. The meaning and accuracy of this is discussed below.
LlaGI is a DNA loop translocase
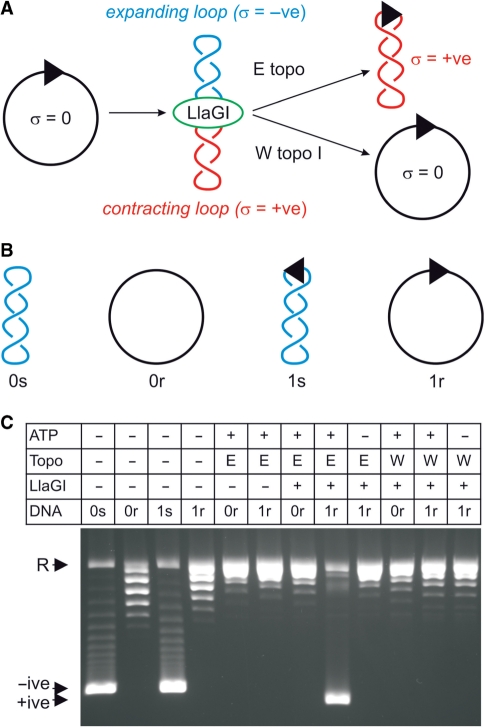
The data presented above are consistent with the ATP-dependent stepwise motion of LlaGI along DNA in a directional manner. This could occur in one of two ways: (i) LlaGI could leave its recognition site during translocation (the ‘tracking model’, above); or (ii) LlaGI could remain bound to its recognition site during translocation, so extruding a growing loop of DNA (the ‘loop translocation’ model, above). We tested for the formation of a DNA loop during translocation using a topoisomerase assay that was successfully used to investigate the classical Type I enzymes [Figure 4A (48)]. In brief, if a motor translocates a loop of DNA by tracking along the DNA backbone, changes in DNA twist will be trapped in the DNA loop(s). Starting with a relaxed circular DNA, this produces an expanding loop with reduced twist (and therefore negative supercoiling) and a contracting loop with increased twist (and therefore positive supercoiling). Note that the changes in supercoiling are transient—if the enzyme dissociates, the DNA returns to a relaxed state. However, by including a DNA topoisomerase in the reaction, the changes in supercoiling can be permanently fixed (Figure 4A). Using wheat germ topoisomerase I, both supercoiled domains are relaxed. However, by using E. coli topoisomerase I, only the negatively supercoiled domain is relaxed such that the DNA becomes positively supercoiled overall. These changes would not be observed for a translocase that leaves its site and tracks on DNA without loop formation.
Figure 4.
LlaGI produces transient changes in DNA supercoiling consistent with loop translocase activity. (A) Schematic of the topoisomerase assay (48). DNA and enzyme shown as in previous figures. See main text for further details. (B) The four different starting substrates for the topology assay derived from pRMA03 and pRMA03F (‘Materials and Methods’ section). 0s is zero-site supercoiled, 0r is zero-site relaxed, 1s is one-site supercoiled and 1r is one-site relaxed. (C) LlaGIDA078, a nuclease mutant of LlaGI (38), was used to prevent DNA cleavage. Reactions contained 10 nM DNA and, as indicated, 4 mM ATP, 5 U E. coli topoisomerase I (E), 10 U wheat germ topoisomerase I (W) and/or 50 nM LlaGIDA078. Reactions were incubated for 30 min at 37°C and separated by agarose gel electrophoresis.
Zero- and one-site plasmids (pRMA03 and pRMA03F; see ‘Materials and Methods’ section) were pre-treated with wheat germ topoisomerase I to produce relaxed circular DNA (re-named for clarity in Figure 4B). Some nicked DNA was produced during this reaction. These DNA were then treated with LlaGIDA078 and ATP as indicated, in the presence of either wheat germ topoisomerase I or E. coli topoisomerase I (Figure 4C). LlaGIDA078 is a nuclease mutant with normal translocation properties that was used to prevent DNA nicking during the reaction [see accompanying paper for more details (38)]. In the absence of an LlaGI site or in the absence of ATP, permanent changes in the DNA topology were not observed. Using the one-site DNA and 4 mM ATP, the combined action of LlaGI and E. coli topoisomerase I produced a supercoiled product. This was confirmed as positively supercoiled DNA by subsequent topoisomerase treatment (data not shown). This product was not observed if wheat germ topoisomerase I was used instead. These data ares entirely consistent with DNA loop translocation by LlaGI as shown in Figure 4A and as observed previously for the classical Type I enzyme EcoAI (48).
DISCUSSION
The data presented in this paper show that the single polypeptide RM enzyme LlaGI is a DNA loop translocase with features in common with the classical, multi-subunit Type I RM enzymes (3). The steps in the reaction process can be described as follows: (i) LlaGI binds to its recognition site via the target recognition domain; (ii) the helicase domain binds the non-specific DNA rightward of the recognition site, trapping one DNA loop; (iii) the ATPase activity then causes DNA translocation past the helicase domain; (iv) a growing negatively supercoiled DNA loop is formed; and (v) loop translocation continues for thousands of base pairs until collision with another LlaGI enzyme triggers DNA cleavage (see below). On the basis of this mechanism, we suggest that LlaGI can be defined as a sub-class of the Type I RM enzymes. It highlights that almost exactly the same translocation mechanism can be evolved by both single and multi-subunit systems. In fact, it could be argued that loop translocation is simply a consequence of a motor protein moving on DNA whilst an attached domain remains bound to the DNA; the attachment could be either via protein–protein contacts or a direct covalent linkage. The trapping of DNA domains as expanding and contracting loops is therefore very likely to be a common feature of molecular motors in a cell, where they will most often be part of higher-order nucleoprotein complexes. How helicase modules are coordinated and controlled within such machines is therefore a key question. LlaGI offers a very simple and tractable test system in this regard.
For the classical Type I RM enzymes, a combination of bulk solution and single molecule experiments have shown that the hydrolysis of one ATP is coupled to the movement of a monomeric motor by one base pair along the 3′-5′ strand of intact dsDNA (20,49). Whilst the translocation activity of LlaGI is consistent with a Type I scheme, we do not currently have sufficient data to directly compare all these features. The range of ATP coupling ratios observed here (1.5–2.0; Figure 3E) are higher than those measured for EcoR124I (1–1.5) (20). Because of uncertainties in the concentration of the steady-state translocating species, both these ranges may be underestimates. Whereas the coupling of EcoR124I appeared to be independent of ATP concentration (at least within experimental error) (20), the coupling of LlaGI shows a systematic increase in the ratio as the ATP concentration drops below the KM. One explanation for a coupling ratio >1:1 is that LlaGI occasionally enters an uncoupled state in which ATP hydrolysis no longer leads to motion on DNA. If this state were entered when the helicase motor is in a nucleotide-unbound state, then lowering the ATP concentration would increase its occupancy and thus increase the ratio, as observed (Figure 3E). Alternatively, the variation we observe may simply reflect systematic error differences between the two assays. Another explanation for a coupling ratio >1:1 is that multiple, independent LlaGI motors might be required to hydrolyse ATP and to move 1 bp. This could explain why multiple LlaGI monomers are required per site for cleavage. However, we cannot rule out an inactive population of enzyme in our preparations. Further analysis of the LlaGI translocation activity will be required, for example, using single molecule techniques, to fully compare LlaGI to the other dsDNA motors.
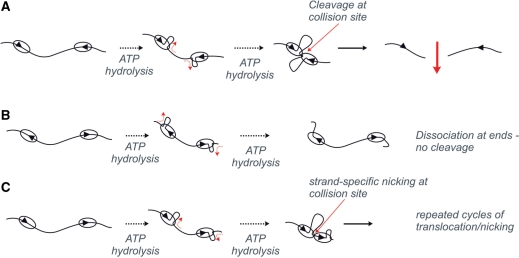
In the accompanying paper we show that dsDNA cleavage by LlaGI requires the presence of two sites on the same DNA in a specific head-to-head orientation (31). DNAs with other arrangements of sites are only nicked. The DNA translocation activity measured here can readily explain these observations (Figure 5). Convergent translocation by two LlaGI motors on a head-to-head substrate results in collision between the proteins (Figure 5A). This interaction forms the active endonuclease which then cuts the non-specific DNA on both strands at the interaction site. Consequently, dsDNA cleavage is randomized to locations between the head-to-head sites (50), as observed. It is possible that the collision complex also recruits further LlaGI molecules from solution to complete the cleavage reaction. As translocation is unidirectional, dsDNA cleavage cannot occur if the sites are orientated tail-to-tail because the enzymes travel divergently towards the DNA ends (Figure 5B).
Figure 5.
How DNA translocation by LlaGI leads to DNA cleavage. The consequences of unidirectional loop translocation by LlaGI are shown for linear DNA with two sites in: (A) head-to-head repeat; (B) tail-to-tail repeat; and (C) head-to-tail repeat. See main text for full details.
The DNA nicking activity of LlaGI observed in Figure 1 and in the accompanying paper (31) can also be explained by a translocation/collision model in which the nuclease domains will cut DNA upon collision with an immovable roadblock (Figure 5C). On a one-site circular DNA highly processive unidirectional translocation will eventually cause the motor to travel the full length of the DNA and collide with the ‘back’ of its own MTase-TRD domains bound at the site. Triplex displacement profiles on circular DNA suggest that LlaGI can complete translocation of >90% of a circular DNA despite the associated topological constraints (Smith, R.M., unpublished data). Similarly, where there are two sites in direct repeat, one LlaGI motor will collide with the back of the other (e.g. Figure 5C). The stalling of the motor in both instances may cause the ‘half’ endonuclease to engage with one of the two strands. The other ‘half’ is not available to bind the opposite strand and consequently only one of the two strands can be cut. This suggests that the tracking motor is targeted to one of the two strands. This model is consistent with our data because the observed rate of nicking is dependent upon the distance to collision. On HT-12 [Figure 6F in (31)], C2cat and C1cat (Figure 1D and F), LlaGI has to travel 1.1–1.3 kb before the first collision and complete nicking is observed within ∼100 s. On pC1 however, LlaGI must translocate 4.2 kb and complete nicking is only observed after 200 s. Repeated rounds of translocation, collision and nicking of the same DNA strand might lead to the dissociation of short oligonucleotides and the formation of DNA gaps. This would also account for the smearing of nicked DNA seen here (e.g. Figure 1) and in the accompanying paper [e.g. Figure 6 in (31)].
It is notable that the model in Figure 5B is essentially the same as that suggested previously for the Type III enzymes to account for directional communication between their sites (21). Moreover, it is highly likely that LlaGI requires this directional communication mechanism to overcome problems associated with site hemimethylation [see accompanying paper (31)], exactly as predicted for the Type III enzymes (51). In agreement with the predictions of the model (Figure 5B), LlaGI translocates on DNA, forms supercoiled DNA loops and has an ATPase activity that is consistent with a stepwise helicase-based translocase activity. In contrast, experiments with the Type III enzymes provide evidence both for and against the model (7,12,21–28). It remains to be seen if the Type III enzymes and LlaGI share a similar mechanism, or if two completely different mechanisms have evolved to deal with the same biological problem of incomplete site methylation.
FUNDING
The BBSRC (BB/D009715/1); and The Wellcome Trust (067439). Funding for open access charge: Wellcome Trust Value in People Award.
Conflict of interest statement. None declared.
Footnotes
This article is linked to 10.1093/nar/gkp790 by Smith et al. and 10.1093/nar/gkp795 by Smith et al.
REFERENCES
- 1.Winkler FK, Prote AE. Structure and function of EcoRV endonuclease in restriction enzymes. In: Pingound A, editor. Nucleic Acids and Molecular Biology. Vol. 14. Germany: Springer; 2004. pp. 179–210. [Google Scholar]
- 2.Gowers DM, Bellamy SR, Halford SE. One recognition sequence, seven restriction enzymes, five reaction mechanisms. Nucleic Acids Res. 2004;32:3469–3479. doi: 10.1093/nar/gkh685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourniquel AA, Bickle TA. Complex restriction enzymes: NTP-driven molecular motors. Biochimie. 2002;84:1047–1059. doi: 10.1016/s0300-9084(02)00020-2. [DOI] [PubMed] [Google Scholar]
- 4.Halford SE, Welsh AJ, Szczelkun MD. Enzyme-mediated DNA looping. Annu. Rev. Biophys. Biomol. Struct. 2004;33:1–24. doi: 10.1146/annurev.biophys.33.110502.132711. [DOI] [PubMed] [Google Scholar]
- 5.Szczelkun MD, Halford SE. Recombination by resolvase to analyse DNA communications by the SfiI restriction endonuclease. EMBO J. 1996;15:1460–1469. [PMC free article] [PubMed] [Google Scholar]
- 6.Szczelkun MD, Dillingham MS, Janscak P, Firman K, Halford SE. Repercussions of DNA tracking by the type IC restriction endonuclease EcoR124I on linear, circular and catenated substrates. EMBO J. 1996;15:6335–6347. [PMC free article] [PubMed] [Google Scholar]
- 7.Peakman LJ, Szczelkun MD. DNA communications by Type III restriction endonucleases-confirmation of 1D translocation over 3D looping. Nucleic Acids Res. 2004;32:4166–4174. doi: 10.1093/nar/gkh762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorbalenya AE, Koonin EV. Endonuclease (R) subunits of type-I and type-III restriction-modification enzymes contain a helicase-like domain. FEBS Lett. 1991;291:277–281. doi: 10.1016/0014-5793(91)81301-n. [DOI] [PubMed] [Google Scholar]
- 9.McClelland SE, Szczelkun MD. Molecular motors that process DNA in restriction enzymes. In: Pingound A, editor. Restriction Endonucleases, Nucleic Acids and Molecular Biology. Vol. 14. Germany: Springer; 2004. pp. 111–135. [Google Scholar]
- 10.Webb JL, King G, Ternent D, Titheradge AJ, Murray NE. Restriction by EcoKI is enhanced by co-operative interactions between target sequences and is dependent on DEAD box motifs. EMBO J. 1996;15:2003–2009. [PMC free article] [PubMed] [Google Scholar]
- 11.Davies GP, Powell LM, Webb JL, Cooper LP, Murray NE. EcoKI with an amino acid substitution in any one of seven DEAD-box motifs has impaired ATPase and endonuclease activities. Nucleic Acids Res. 1998;26:4828–4836. doi: 10.1093/nar/26.21.4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saha S, Rao DN. ATP hydrolysis is required for DNA cleavage by EcoPI restriction enzyme. J. Mol. Biol. 1995;247:559–567. doi: 10.1016/s0022-2836(05)80137-8. [DOI] [PubMed] [Google Scholar]
- 13.Studier FW, Bandyopadhyay PK. Model for how type I restriction enzymes select cleavage sites in DNA. Proc. Natl Acad. Sci. USA. 1988;85:4677–4681. doi: 10.1073/pnas.85.13.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Firman K, Szczelkun MD. Measuring motion on DNA by the type I restriction endonuclease EcoR124I using triplex displacement. EMBO J. 2000;19:2094–2102. doi: 10.1093/emboj/19.9.2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seidel R, van Noort J, van der Scheer C, Bloom JG, Dekker NH, Dutta CF, Blundell A, Robinson T, Firman K, Dekker C. Real-time observation of DNA translocation by the type I restriction modification enzyme EcoR124I. Nat. Struct. Mol. Biol. 2004;11:838–843. doi: 10.1038/nsmb816. [DOI] [PubMed] [Google Scholar]
- 16.García LR, Molineux IJ. Translocation and specific cleavage of bacteriophage T7 DNA in vivo by EcoKI. Proc. Natl Acad. Sci. USA. 1999;96:12430–12435. doi: 10.1073/pnas.96.22.12430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosamond J, Endlich B, Linn S. Electron microscopic studies of the mechanism of action of the restriction endonuclease of Escherichia coli B. J. Mol. Biol. 1979;129:619–635. doi: 10.1016/0022-2836(79)90472-8. [DOI] [PubMed] [Google Scholar]
- 18.Endlich B, Linn S. The DNA restriction endonuclease of Escherichia coli B. I. Studies of the DNA translocation and the ATPase activities. J. Biol. Chem. 1985;260:5720–5728. [PubMed] [Google Scholar]
- 19.Szczelkun MD, Janscak P, Firman K, Halford SE. Selection of non-specific DNA cleavage sites by the type IC restriction endonuclease EcoR124I. J. Mol. Biol. 1997;271:112–123. doi: 10.1006/jmbi.1997.1172. [DOI] [PubMed] [Google Scholar]
- 20.Seidel R, Bloom JG, Dekker C, Szczelkun MD. Motor step size and ATP coupling efficiency of the dsDNA translocase EcoR124I. EMBO J. 2008;27:1388–1398. doi: 10.1038/emboj.2008.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meisel A, Mackeldanz P, Bickle TA, Kruger DH, Schroeder C. Type III restriction endonucleases translocate DNA in a reaction driven by recognition site-specific ATP hydrolysis. EMBO J. 1995;14:2958–2966. doi: 10.1002/j.1460-2075.1995.tb07296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reich S, Gössl I, Reuter M, Rabe JP, Krüger DH. Scanning force microscopy of DNA translocation by the Type III restriction enzyme EcoP15I. J. Mol. Biol. 2004;341:337–343. doi: 10.1016/j.jmb.2004.06.031. [DOI] [PubMed] [Google Scholar]
- 23.Crampton N, Roes S, Dryden DT, Rao DN, Edwardson JM, Henderson RM. DNA looping and translocation provide an optimal cleavage mechanism for the type III restriction enzymes. EMBO J. 2007;26:3815–3825. doi: 10.1038/sj.emboj.7601807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crampton N, Yokokawa M, Dryden DT, Edwardson JM, Rao DN, Takeyasu K, Yoshimura SH, Henderson RM. Fast-scan atomic force microscopy reveals that the type III restriction enzyme EcoP15I is capable of DNA translocation and looping. Proc. Natl Acad. Sci. USA. 2007;104:12755–12760. doi: 10.1073/pnas.0700483104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janscak P, Sandmeier U, Szczelkun MD, Bickle TA. Subunit assembly and mode of DNA cleavage of the type III restriction endonucleases EcoP1I and EcoP15I. J. Mol. Biol. 2001;306:417–431. doi: 10.1006/jmbi.2000.4411. [DOI] [PubMed] [Google Scholar]
- 26.Ramanathan SP, van Aelst K, Sears A, Peakman LJ, Diffin FM, Szczelkun MD, Seidel R. Type III restriction enzymes communicate in 1D without looping between their target sites. Proc. Natl Acad. Sci. USA. 2009;106:1748–1753. doi: 10.1073/pnas.0807193106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raghavendra NK, Rao DN. Unidirectional translocation from recognition site and a necessary interaction with DNA end for cleavage by Type III restriction enzyme. Nucleic Acids Res. 2004;32:5703–5711. doi: 10.1093/nar/gkh899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peakman LJ, Szczelkun MD. S-adenosyl homocysteine and DNA ends stimulate promiscuous nuclease activities in the Type III restriction endonuclease EcoPI. Nucleic Acids Res. 2009;37:3934–3945. doi: 10.1093/nar/gkp267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berge T, Ellis DJ, Dryden DT, Edwardson JM, Henderson RM. Translocation-independent dimerization of the EcoKI endonuclease visualized by atomic force microscopy. Biophys. J. 2000;79:479–484. doi: 10.1016/S0006-3495(00)76309-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neaves KJ, Cooper LP, White JH, Carnally SM, Dryden DT, Edwardson JM, Henderson RM. Atomic force microscopy of the EcoKI Type I DNA restriction enzyme bound to DNA shows enzyme dimerization and DNA looping. Nucleic Acids Res. 2009;37:2053–2063. doi: 10.1093/nar/gkp042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith RM, Diffin FM, Savery NJ, Josephsen J, Szczelkun MD. DNA cleavage and methylation specificity of the single polypeptide restriction modification enzyme LlaGI. 2009 doi: 10.1093/nar/gkp790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Madsen A, Josephsen J. The LlaGI restriction and modification system of Lactococcus lactis W10 consists of only one single polypeptide. FEMS Microbiol. Lett. 2001;200:91–96. doi: 10.1111/j.1574-6968.2001.tb10698.x. [DOI] [PubMed] [Google Scholar]
- 33.Morgan RD, Dwinell EA, Bhatia TK, Lang EM, Luyten YA. The MmeI family: type II restriction-modification enzymes that employ single-strand modification for host protection. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp534. doi:10.1093/nar/gkp534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han JH, Batey S, Nickson AA, Teichmann SA, Clarke J. The folding and evolution of multidomain proteins. Nat. Rev. Mol. Cell Biol. 2007;8:319–330. doi: 10.1038/nrm2144. [DOI] [PubMed] [Google Scholar]
- 35.Aravind L, Walker DR, Koonin EV. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stanley LK, Szczelkun MD. Direct and random routing of a molecular motor protein at a DNA junction. Nucleic Acids Res. 2006;34:4387–4394. doi: 10.1093/nar/gkl569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hall SC, Halford SE. Specificity of DNA recognition in the nucleoprotein complex for site-specific recombination by Tn21 resolvase. Nucleic Acids Res. 1993;21:5712–5719. doi: 10.1093/nar/21.24.5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith RM, Josephsen J, Szczelkun MD. An Mrr-family nuclease motif in the single polypeptide restriction-modification enzyme LlaGI. 2009 doi: 10.1093/nar/gkp795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Webb MR. Development of fluorescent biosensors for probing the function of motor proteins. Mol. Biosyst. 2007;3:249–256. doi: 10.1039/b614154d. [DOI] [PubMed] [Google Scholar]
- 40.Frieden C. Slow transitions and hysteretic behavior in enzymes. Annu. Rev. Biochem. 1979;48:471–489. doi: 10.1146/annurev.bi.48.070179.002351. [DOI] [PubMed] [Google Scholar]
- 41.Adzuma K, Mizuuchi K. Interaction of proteins located at a distance along DNA: mechanism of target immunity in the Mu DNA strand-transfer reaction. Cell. 1989;57:41–47. doi: 10.1016/0092-8674(89)90170-0. [DOI] [PubMed] [Google Scholar]
- 42.Krasnow MA, Cozzarelli NR. Site-specific relaxation and recombination by the Tn3 resolvase: recognition of the DNA path between oriented res sites. Cell. 1983;32:1313–1324. doi: 10.1016/0092-8674(83)90312-4. [DOI] [PubMed] [Google Scholar]
- 43.Kanaar R, van de Putte P, Cozzarelli NR. Gin-mediated recombination of catenated and knotted DNA substrates: implications for the mechanism of interaction between cis-acting sites. Cell. 1989;58:147–159. doi: 10.1016/0092-8674(89)90411-x. [DOI] [PubMed] [Google Scholar]
- 44.Stark WM, Sherratt DJ, Boocock MR. Site-specific recombination by Tn3 resolvase: topological changes in the forward and reverse reactions. Cell. 1989;58:779–790. doi: 10.1016/0092-8674(89)90111-6. [DOI] [PubMed] [Google Scholar]
- 45.Benjamin HW, Cozzarelli NR. Geometric arrangements of Tn3 resolvase sites. J. Biol. Chem. 1990;265:6441–6447. [PubMed] [Google Scholar]
- 46.Benjamin KR, Abola AP, Kanaar R, Cozzarelli NR. Contributions of supercoiling to Tn3 resolvase and phage Mu Gin site-specific recombination. J. Mol. Biol. 1996;256:50–65. doi: 10.1006/jmbi.1996.0067. [DOI] [PubMed] [Google Scholar]
- 47.McClelland SE, Dryden DT, Szczelkun MD. Continuous assays for DNA translocation using fluorescent triplex dissociation: application to type I restriction endonucleases. J. Mol. Biol. 2005;348:895–915. doi: 10.1016/j.jmb.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 48.Janscak P, Bickle TA. DNA supercoiling during ATP-dependent DNA translocation by the type I restriction enzyme EcoAI. J. Mol. Biol. 2000;295:1089–1099. doi: 10.1006/jmbi.1999.3414. [DOI] [PubMed] [Google Scholar]
- 49.Stanley LK, Seidel R, van der Scheer C, Dekker NH, Szczelkun MD, Dekker C. When a helicase is not a helicase: dsDNA tracking by the motor protein EcoR124I. EMBO J. 2006;25:2230–2239. doi: 10.1038/sj.emboj.7601104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szczelkun MD. Kinetic models of translocation, head-on collision, and DNA cleavage by type I restriction endonucleases. Biochemistry. 2002;41:2067–2074. doi: 10.1021/bi011824b. [DOI] [PubMed] [Google Scholar]
- 51.Meisel A, Bickle TA, Krüger DH, Schroeder C. Type III restriction enzymes need two inversely oriented recognition sites for DNA cleavage. Nature. 1992;355:467–469. doi: 10.1038/355467a0. [DOI] [PubMed] [Google Scholar]