Abstract
In +1 programmed ribosomal frameshifting (PRF), ribosomes skip one nucleotide toward the 3′-end during translation. Most of the genes known to demonstrate +1 PRF have been discovered by chance or by searching homologous genes. Here, a bioinformatic framework called FSscan is developed to perform a systematic search for potential +1 frameshift sites in the Escherichia coli genome. Based on a current state of the art understanding of the mechanism of +1 PRF, FSscan calculates scores for a 16-nt window along a gene sequence according to different effects of the stimulatory signals, and ribosome E-, P- and A-site interactions. FSscan successfully identified the +1 PRF site in prfB and predicted yehP, pepP, nuoE and cheA as +1 frameshift candidates in the E. coli genome. Empirical results demonstrated that potential +1 frameshift sequences identified promoted significant levels of +1 frameshifting in vivo. Mass spectrometry analysis confirmed the presence of the frameshifted proteins expressed from a yehP-egfp fusion construct. FSscan allows a genome-wide and systematic search for +1 frameshift sites in E. coli. The results have implications for bioinformatic identification of novel frameshift proteins, ribosomal frameshifting, coding sequence detection and the application of mass spectrometry on studying frameshift proteins.
INTRODUCTION
Translation is a highly accurate process. The frequency of decoding error is estimated to be on the order of 10−5 per codon (1). Programmed ribosomal frameshifting (PRF) is a coded shift in the reading frame during translation. Consequently, mRNAs with PRF features may yield two different protein products, an inframe product and a frameshifted product. In +1 PRF, the ribosome skips over one nucleotide toward the 3′ direction. Today, 88 cases of +1 PRF have been found in different organisms in the RECODE database (2). +1 PRF has been observed to occur during the translation of prfB to produce release factor 2 (RF2) in Escherichia coli (3). In Saccharomyces cerevisiae four retrotransposable elements, Ty1, Ty2, Ty3 and Ty4 (4–6), and three genes, ABP140 (7), EST3 (8) and OAZ1 (9) use +1 PRF. The expression of mammalian antizyme has also been shown to involve +1 PRF (10).
A genome-wide prediction of +1 frameshift sites is currently a difficult task because the sequence elements for +1 frameshifting are diverse among the organisms. To date, most of the known genes involving +1 PRF have been discovered by chance, and in some cases, by searching homologous genes. Several computer programs have been developed to identify +1 frameshift sites (11,12). Shah et al. (11) hypothesized that selective pressure would have rendered potential frameshift sites under-abundant in protein coding sequences. In that study, a computer program was developed to identify oligos that are over- or underrepresented for reasons other than codon bias. Their result suggested that the heptanucleotides CUU AGG C and CUU AGU U, +1 PRF sites for the production of ABP140 and EST3, respectively, rank among the least represented of the heptanucleotides in the coding sequence of S. cerevisiae. While the approach is able to identify novel sequences, the method did not account for stimulatory signals. The program ‘FSFinder’ by Moon et al. (12) used known components of a frameshift cassette for predicting both −1 and +1 PRF sites. This method achieves a high sensitivity and a high specificity (0.88 and 0.97, respectively) for predicting +1 PRF. However, FSFinder does not predict novel +1 frameshift sites in E. coli. A novel antizyme gene, whose expression requires +1 frameshifting, was found in the zebra fish Danio rerio by a protein BLAST search against the translated nucleotide database of the known antizyme family sequence (13). While the method successfully identified novel genes requiring +1 frameshifting, the approach is limited to the antizyme family in eukaryotic cells.
Recently, a mathematical model revealed that destabilization of the deacylated tRNA in the ribosomal E-site, rearrangement of the peptidyl-tRNA in the ribosomal P-site, and availability of the cognate aminoacylated tRNA (aa-tRNA) corresponding to the ribosomal A-site act synergistically to promote efficient +1 PRF in E. coli (14). Motivated by this result, one might identify potential +1 frameshift sites in the E. coli genome by searching sequences with a combination of stimulatory, E-, P- and A-site features. In this study, FSscan is developed to perform a systematic and genome-wide search for potential +1 frameshift sites in E. coli. Based on a current state-of-art understanding of the mechanism of +1 PRF, FSscan looks for a 16-nt sequence with possible synergistic effects in the E. coli genome. Potential +1 frameshift sequences so identified are shown to promote significant levels of +1 frameshifting in vivo. The mass spectrometry data obtained from a multiple reaction monitoring assay (MRM), a specific and sensitive mass scan method (15), experimentally confirms the expression of the predicted frameshift protein. Importantly, current methods of coding sequence detection generally do not take into account the shift of the reading frames and only a few algorithms assign a frameshift as a possible regulatory process (16). FSscan presented in the study provides an algorithm to predict potential +1 frameshift products in E. coli.
FSscan algorithm
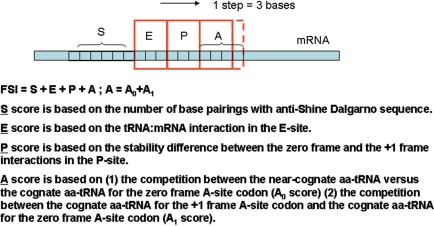
FSscan is developed in Python (v2.4.3, Python Software Foundation, Hampton, NH) to search for potential +1 frameshift sites in the E. coli genome. The program assigns scores for a 16-nt window along a gene sequence according to different effects of the stimulatory signals (S score) and interactions of the E-, P- and A-site in the ribosome (E, P and A scores, respectively) (Figure 1). A stimulatory signal in E. coli for +1 PRF can be a Shine–Dalgarno (SD)—like sequence upstream of the frameshift site (17). FSscan assigns zero to the S score if <4 base pairings can be formed between the 6 nt upstream of the E-site position and the anti-SD sequence (3′UCCUCC5′); otherwise, FSscan assigns the number of base pairings divided by three to the S score [Equation (1)].
 |
1 |
Figure 1.
The scoring system for FSscan program. FSscan calculates scores for a 16-nt window along the gene sequence. Each step is 3 nt. FS index (FSI) = S + E + P + A.
Sanders et al., (18) suggested that zero frame condon:anticodon interactions in the E-site can affect frameshifting. The E score is calculated as exp (−ΔGc), where ΔGc is the codon:anticodon interaction (19) in the ribosome E-site. For the P-site, both zero frame and +1 frame interactions can influence +1 frameshifting (20). The P score in the program represents the stability difference between the zero frame and the +1 frame interactions for the P-site tRNA, normalized with the maximum stability difference obtained among 256 possible P-site sequences (Supplementary Data). The A score is the combination of the A0 score and the A1 score. The A0 score is the ratio of the arrival frequency, on the basis of transport by diffusion, of the near-cognate aa-tRNA versus the cognate aa-tRNA corresponding to the zero frame A-site codon (21), normalized with the maximum ratio of the arrival frequency obtained among 64 possible zero frame A-site codons. The A1 score is the ratio between the concentration of the cognate aa-tRNA for the +1 frame A-site codon to that of the cognate aa-tRNA for the zero frame A-site codon (21), normalized with the maximum concentration ratio obtained among 256 possible A-site sequences. For a stop codon in the zero frame A-site, the A0 and A1 scores were set to be 0.9 for TAG and TGA, and 0.6 for TAA. If the summation of the E, P and A scores is <3, the S score is then reset to zero [Equation (2)].
 |
2 |
Equation (2) has a higher priority than Equation (1), which means, as long as the summation of the E, P and A score is <3, the program assigns zero to the S score no matter how many base pairings can be formed between the mRNA sequence and the anti-SD sequence.
The frameshift index (FSI) for a 16-nt window is calculated as Equation (3).
| 3 |
A higher FSI suggests the sequence contains more features for +1 frameshifting. It is important to note that FSI is not set for quantitatively predicting the level of the +1 frameshifting, but rather how likely a sequence is a frameshift site.
MATERIALS AND METHODS
Plasmids and bacterial strains
Escherichia coli XL1 blue MRF’ (Stratagene, La Jolla, CA, USA) was used in all experimental studies. All constructs were verified by DNA sequencing. The construction of the dual fluorescence reporter was performed as described previously (14). The control strain has both DsRed and enhanced green fluorescence protein (EGFP) coding sequences in frame. For the test strain, the linker sequences inserted between the two reporters contained predicted frameshift sequences followed by an in-frame stop codon and the downstream egfp in the +1 frame. The control strain expressed the DsRed-EGFP fusion protein from the reporter. The test strains expressed DsRed proteins as non-frameshift proteins (due to the stop codon in the linker sequence) and DsRed-EGFP fusion protein as frameshift proteins (because the stop codon is bypassed by +1 frameshifting). Table 1 lists the nucleotide sequences incorporated into the dual fluorescence reporter for testing +1 frameshift efficiency in vivo in this study. A negative control strain, ran1, was transformed with a plasmid containing a randomly designed linker (rand) inserted between the two fluorescence reporters with egfp in the +1 frame.
Table 1.
Nucleotide sequences incorporated into the dual fluorescence reporter system for testing +1 frameshift efficiency in vivo in this study
| Original gene | 16-nt window with max FSI in the gene (the P-site position is underlined) | Strain (transformed with corresponding reporter plasmids) |
|---|---|---|
| yehP | GTG GAG TAT GGT CGG C | yehP6 |
| nuoE | GAG CGG TAT AAA TGA A | nuoE6 |
| pepP | AGT GAG ATA TCC CGG C | pepP6 |
| cheA | AGT CGC TAT CCC CGG C | cheA6 |
| ygcH | CCA CTC TAT TTT CGG C | ygcH6 |
| yeaI | AAT ATT TAT AAT CGG C | yeaI6 |
| pspD | CAG CGT TAT AAA AGG T | pspD6 |
| glnD | GGT GGG ATA AAA GCC C | glnD6 |
| yjgN | GAG AGA TAT TTT CTT A | yjgN6 |
| cysD | CAG GGG TAT TTT TAA G | cysD6 |
| rand | TCT GGC TCT GGC TGA G | ran1 |
| yehP | GTG GAG TTAGGT CGG C (mutated sequence shown in bold) | yehP7 |
yehP, nuoE, pepP, cheA, ygcH and yeaI are the top ranking candidates identified by FSscan.
glnD, yjgN and cysD are selected genes with one or two frameshifting features. rand is a randomly designed sequence to serve as a negative control.
The first 915 nt in yehP were PCR-amplified with the forward primer, yehPf, 5′-AAACTGCAGAATGTCTGAACTGAACGATCTTCTG-3′ (PstI site underlined) and two reverse primers, yehPr0 5′-ATTGGTACCACGAGGATAATGACGCTTTTCGCTGG-3′ and yehPr1 5′-ATTGGTACCCACGAGGATAATGACGCTTTTCGCTGG-3′ (KpnI site underlined) using E. coli genomic DNA as a template. The PstI/KpnI restricted PCR products were ligated with a PstI/KpnI-restricted pEGFP (Clontech, Mountain View, CA, USA) vector to yield pYehP0 (using yehPr0 as the reverse primer for PCR) and pYehP1 (using yehPr1 as the reverse primer for PCR). The predicted frameshift sequence in pYehP1 was mutated by using QuikChange II site-directed mutagenesis kit (Stratagene) to create pYehPC. BsrGI/EcoRI restricted pYehP0, pYehP1 and pYehPC were ligated with a nucleotide sequence, 5′-GTACAAGCATCATCATCATCATCATTAAG-3′, to create pYehP20, pYehP21 and pYehP2C to add a 6X-histidine tag downstream of egfp. KpnI/NcoI restricted pYehP20, pYehP21 and pYehP2C were ligated with a nucleotide sequence, 5′-CGTCTAGCTCTGGCTCTGGCTCTGGCAC-3′, to create pYehP40, pYehP41 and pYehP4C to incorporate an in-frame stop codon and a flexible linker between yehP and egfp. Escherichia coli strains transformed with pYehP40, pYehP41 and pYehP4C are named yehP40, yehP41 and yehP4C, respectively.
Fluorescence assay
Cells with the appropriate plasmids were cultured in 1 ml Luria-Bertani (LB) medium containing 100 µg/ml ampicillin in a 24-well plate for 24 h at 37°C. The fluorescence was then measured by a plate reader (SpectraMax M5, Molecular Devices, Sunnyvale, CA, USA). The fluorescence measurement was performed as described previously (14). Frameshift efficiency (FS%) was obtained as the ratio of the green fluorescence to the red fluorescence for the test strains, normalized against the fluorescence ratio of the control strain. Statistical analysis was applied to all data sets according to Jacobs and Dinman (22). Eleven to twelve replicates for test strains and control strains were performed to satisfy the minimum sample requirement for statistical significance.
Western analysis
Cells with the appropriate plasmids were cultured in 3 ml LB medium containing 100 µg/ml ampicillin in 17 ml round-bottom tubes at 37°C. Aliquots of cells were harvested after 24-h cultivation and pelleted by centrifugation for 20 min at 4°C and 4000 g. The cell pellet was resuspended in 50 µl phosphate-buffered saline per OD600 and resolved by SDS–PAGE (10% w/v Tris–HCl). Immunoblot was performed as described by Gupta and Lee (23), except rabbit anti-GFP (1:5000, Clontech) and alkaline phosphatase conjugated mouse anti-rabbit IgG antibody (1:10 000; Sigma, St. Louis, MO, USA) were used as the primary and secondary antibodies, respectively.
Protein digestion
yeh41 cell lysate was purified by Ni–NTA under denaturing conditions according to the manufacturer's protocol (Qiagen, Valencia, CA, USA). The purified protein sample was exchanged into 0.2 M ammonium bicarbonate using Amicon Ultra 10-kDa molecular cutoff filter (Millipore, Billerica, MA, USA). The buffer-exchanged sample was denatured and reduced by 6 M urea and 200 mM dithiothreitol (DTT) at room temperature for an hour. Then, the sample was alkylated by 200 mM iodoacetamide at room temperature for an hour in the dark. The remaining iodoacetamide in the sample was quenched by 200 mM DTT at room temperature for an hour and the sample was digested by trypsin (Promega, Madison, WI, USA) at 37°C for 14 h. The digestion was stopped by decreasing the pH of the solution with 88% formic acid (FA) and vacuum dried, and the digested sample was reconstituted with 25 µl of 0.1% FA.
Liquid chromatography tandem mass spectrometry
Of the digested sample, 1.2 µl was separated by Dionex 3000 nLC system (Sunnyvale, CA, USA) with an Acclaim PepMap 100 C18 trap column (300 µm × 5 mm, 5 µm, for the online desalting at a flow rate of 30 µl/min for 3 min) and an Acclaim PepMap 100 C18 analytical column (75 µm × 15 cm, 3 µm) at a flow rate of 250 nl/min. Peptides were eluted with gradients of 2–90% acetonitrile with 0.1% FA and the eluent was directly introduced into 4000 QTRAP MS through Nanospray II source (Applied Biosystems, Foster City, CA, USA) for MRM study. To determine the appropriate MRM transitions that would be specific to the peptide of interest, the frameshift protein sequence was imported into the MIDAS Workflow software system (Applied Biosystems). The software generates a list of possible MRM transitions (Table S2), including mass to charge ratios of precursor ions, fragment ions and collision energy values for fragmentation. MS and MS/MS data obtained through MRM were searched within a custom sequence database that included the addition of the frameshift protein sequence. The spectral assignment of MS/MS were performed using ProteinPilot (v1.2 Applied Biosystems).
RESULTS
FSscan identifies a +1 frameshift hot spot in prfB gene
FSscan successfully identifies the +1 frameshift site in prfB. Figure 2 shows the FSI along the prfB gene sequence. The FSI is at maximum when the ribosome P-site is positioned at the 25th codon in the coding sequence, the frameshift site for prfB in the literature (3).
Figure 2.
FSscan identifies the +1 frameshift site in prfB. A peak FSI is observed as the ribosome P-site is positioned at the 25th codon.
Analysis of 4132 protein coding sequences in the E. coli genome reveals additional potential +1 frameshift candidates
To identify potential +1 framshifting sites, FSscan analyzed 4132 protein coding sequences in E. coli K12 MG1655 genome (Genbank: U00096). As the FSI calculation requires an additional nucleotide downstream of the A-site codon, the 4132 coding sequences were adjusted to include one more nucleotide downstream of the stop codon. The maximum FSI obtained in each protein coding sequence is plotted in Figure 3. prfB, whose expression has been shown to involve +1 PRF (3), has the highest FSI among all tested coding sequences (maximum FSI in prfB = 5.05). The next four highest ranking genes are yehP, nuoE, pepP and cheA, with a maximum FSI 4.47, 4.39, 4.39 and 3.54 in their coding sequences, respectively. The potential +1 frameshift sequences in these genes are listed in Table 1. None of these candidates has been reported by previous approaches to identify +1 PRF genes (11,12). The other 4127 protein-coding sequences all have a maximum FSI <3.50.
Figure 3.
Maximum FSI in each of the 4132 E. coli protein-coding sequences. Five genes with a maximum FSI above 3.5 are indicated in red. prfB has the maximum FSI 5.05. yehP has the maximum FSI 4.47. nuoE has the maximum FSI 4.39. pepP has the maximum FSI 4.39. cheA has the maximum FSI 3.55.
In vivo examination of +1 frameshift sequences agrees with the program predictions
Several +1 frameshift candidates were examined in vivo by using a dual fluorescence reporter system. A randomly designed sequence with FSI = 1.70 (rand, Table 1) was constructed to serve as a negative control strain (see ‘Materials and Methods’ section). Potential frameshift sequences from yehP, nuoE, pepP and cheA resulted in FS% significantly higher than rand (Figure 4). A lower FS% was observed for sequences with FSI <3.5, suggesting that FSI 3.5 may serve as a threshold for identifying potential frameshift cassettes.
Figure 4.
Frameshift efficiency (FS%) for potential frameshift sequences identified by FSscan. The histogram indicates the experimentally observed FS% for different test strains listed in Table 1. Error bars show the standard deviation. Diamonds demonstrate the program calculated FSI for the potential frameshift cassettes (sequences are shown in Table 1).
FSscan identifies yehP as a +1 frameshift candidate
yehP contains a potential +1 frameshift sequence with the second highest FSI, only after prfB. The predicted frameshifting sequence is GTG GAG TATGGT CGG C (where each zero frame codon is separated by a space and the P-site position for obtaining the maximum FSI is underlined). In this sequence, an ATG in the +1 frame (shown in bold in the sequence above) together with an upstream GGAG may result in internal translation, causing non-frameshifting based EGFP expression in the dual reporter system. To further confirm yehP as a candidate +1 PRF gene, the sequence was mutated to GTG GAG TTAGGT CGG C (mutation shown in bold) to remove ATG in the +1 frame while keeping a weaker E-site interaction (yehP7 in Table 1). A small decrease in FS% was observed (Figure 5), but the mutation still resulted in a significantly higher FS% as compared to the negative control strain, ran1 (Figure 4). This observation suggests that the higher FS% for yehP6 is not likely due to the internal translation of EGFP starting from the linker sequence.
Figure 5.
Frameshift efficiency (FS%) for yehP6 and yehP7. In yehP6, the linker inserted between the two fluorescence reporters contains the predicted yehP frameshift sequence: GTG GAG TAT GGT CGG C. In yehP7, the frameshift sequence is mutated to GTG GAG TTA GGT CGG C (where zero frame codons are separated by spaces).
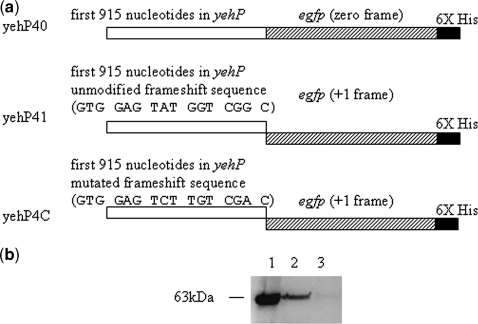
To study the frameshift site in yehP, the fusion constructs yehP40, yehP41 and yehP4C were made with egfp 3′ to yehP (Figure 6a). Proteins from cell lysate were subjected to western analysis. Protein bands with molecular weight 63 kDa, the expected mass for the fusion protein, were observed for yehP40 and yehP41. Interestingly, no or very few proteins with this mass were observed when the potential frameshift sequence was mutated to GTG GAG TCT TGT CGA C to remove frameshifting features (yehP4C, mutated nucleotides shown in bold) (Figure 6a and b). The result suggests that the +1 frameshift event is specific to the predicted sequence.
Figure 6.
(a) The nucleotide sequence design for yehP40, yehP41 and yehP4C. (b) Western blot for the cell lysate to detect the frameshift protein. Lane 1: total lysate from yehP40; lane 2: total lysate from yehP41; lane 3: total lysate from yehP4C. The amount of the protein loaded for yehP40 is one-third of the amount of the protein for yehP41 and yehP4C.
Proteins from yehP41 cell lysate were purified, buffer-exchanged and digested by trypsin. The digest was analyzed by liquid chromatography tandem mass spectrometry (LC-MS/MS) using MRM. MRM is a highly sensitive scanning technique for peptide identification. The greater specificity is achieved by fragmenting the analyte and monitoring both parent and one or more product ions simultaneously [see review by Kitteringham et al. (24)]. Figure 7 presents the amino acid sequence derived from the frameshift site and the tryptic peptides observed by MRM. The presence of the peptide VQLGGGTNIASAVEYGGNLLNNQR (Figure S3 in the Supplementary Data), whose coding sequence spans the potential frameshift site, is a result of the +1 frameshifting at the 291st codon, GTT CGG C (where the P-site position is underlined), in yehP. This result further confirms the frameshift site in yehP, as suggested by FSscan.
Figure 7.
Nucleotide and amino acid sequence for the YehP-EGFP frameshift protein in yehP41. (a) The nucleotide and amino acid sequence for the predicted frameshift region in YehP-EGFP. The predicted frameshift sequence is shown in bold, with the P-site codon underlined. The zero frame and the +1 frame amino acid sequences are shown under the nucleotide sequence. The peptide spanning the frameshift site, with the zero frame translation before the site and the +1 frame translation after the site, is shown in red. (b) Amino acid sequence for the frameshift protein in yehP41 strain. The YehP-EGFP was expressed as a result of +1 frameshifting. Tryptic peptides observed by MRM are marked in red (>95% confidence level). The sequence coverage is 21.7%.
For +1 frameshifting at the 291st codon in yehP, the ribosome encounters a stop codon 15 codons downstream of the frameshift site. As a result, the frameshift product is 303 amino acids in length, which is 75 amino acids shorter than the non-frameshift yehP product. Importantly, yehP is highly conserved in different E. coli strains and is also observed in several other eubacteria (Table 2). The consensus of the yehP frameshift cassette for the 31 sequences in Table 2 is shown by a sequence logo (Figure 8) (25,26). Only a minor diversity is observed at position 1, 6, 12 and 14 in the 16-nt frameshifting window.
Table 2.
BLAST result for yehP. blastn was used as the algorithm to search the nucleotide collection database in National Center for Biotechnology Information's website
| Accession | Description | Max score | Total score | Query coverage (%) | E-value | Max ident (%) |
|---|---|---|---|---|---|---|
| CP000948.1 | Escherichia coli str. K12 substr. DH10B, complete genome | 2254 | 2290 | 100 | 0.0 | 100 |
| AP009048.1 | Escherichia coli str. K12 substr. W3110 DNA, complete genome | 2254 | 2290 | 100 | 0.0 | 100 |
| U00096.2 | Escherichia coli str. K-12 substr. MG1655, complete genome | 2254 | 2290 | 100 | 0.0 | 100 |
| U00007.1 | 47 to 48 centisome region of E. coli K12 BHB2600 | 2254 | 2254 | 100 | 0.0 | 100 |
| CU928160.2 | Escherichia coli str. IAI1 chromosome, complete genome | 2119 | 2155 | 100 | 0.0 | 100 |
| AP009240.1 | Escherichia coli SE11 DNA, complete genome | 2095 | 2132 | 100 | 0.0 | 100 |
| CP000800.1 | Escherichia coli E24377A, complete genome | 2095 | 2132 | 100 | 0.0 | 100 |
| CP000036.1 | Shigella boydii Sb227, complete genome | 2095 | 2168 | 100 | 0.0 | 100 |
| AB426057.1 | Escherichia coli O111:H- DNA, genomic island GEI2.21 | 2087 | 2087 | 100 | 0.0 | 98 |
| CP000034.1 | Shigella dysenteriae Sd197, complete genome | 2087 | 2160 | 100 | 0.0 | 100 |
| CP000946.1 | Escherichia coli ATCC 8739, complete genome | 2056 | 2092 | 100 | 0.0 | 100 |
| CP000802.1 | Escherichia coli HS, complete genome | 2032 | 2068 | 100 | 0.0 | 100 |
| AE005674.1 | Shigella flexneri 2a str. 301, complete genome | 1992 | 2065 | 100 | 0.0 | 100 |
| AE014073.1 | Shigella flexneri 2a str. 2457T, complete genome | 1992 | 2065 | 100 | 0.0 | 100 |
| AE014075.1 | Escherichia coli CFT073, complete genome | 1976 | 2085 | 100 | 0.0 | 100 |
| CU928164.2 | Escherichia coli str. IAI39 chromosome, complete genome | 1961 | 2033 | 100 | 0.0 | 100 |
| BA000007.2 | Escherichia coli O157:H7 str. Sakai DNA, complete genome | 1961 | 2033 | 100 | 0.0 | 100 |
| AE005174.2 | Escherichia coli O157:H7 EDL933, complete genome | 1961 | 2033 | 100 | 0.0 | 100 |
| CP001164.1 | Escherichia coli O157:H7 str. EC4115, complete genome | 1953 | 2025 | 100 | 0.0 | 100 |
| CP000970.1 | Escherichia coli SMS-3-5, complete genome | 1937 | 2009 | 100 | 0.0 | 100 |
| CU928162.2 | Escherichia coli str. ED1a chromosome, complete genome | 1913 | 2021 | 100 | 0.0 | 100 |
| FM180568.1 | Escherichia coli 0127:H6 E2348/69 complete genome, strain E2348/69 | 1905 | 1977 | 100 | 0.0 | 100 |
| CU928161.2 | Escherichia coli str. S88 chromosome, complete genome | 1897 | 2006 | 100 | 0.0 | 100 |
| CP000468.1 | Escherichia coli APEC O1, complete genome | 1897 | 2006 | 100 | 0.0 | 100 |
| CP000243.1 | Escherichia coli UTI89, complete genome | 1897 | 2006 | 100 | 0.0 | 100 |
| CU928158.2 | Escherichia fergusonii str. ATCC 35469T chromosome, complete genome | 1850 | 1924 | 100 | 0.0 | 95 |
| CP000247.1 | Escherichia coli 536, complete genome | 1850 | 1958 | 100 | 0.0 | 100 |
| CU928163.2 | Escherichia coli str. UMN026 chromosome, complete genome | 1842 | 1914 | 100 | 0.0 | 100 |
| CU651637.1 | Escherichia coli LF82 chromosome, complete sequence | 1818 | 1926 | 100 | 0.0 | 100 |
| AP000400.1 | Enterobacteria phage VT1-Sakai genomic DNA, prophage inserted region in Escherichia coli O157:H7 | 1542 | 1542 | 81 | 0.0 | 96 |
| CP000038.1 | Shigella sonnei Ss046, complete genome | 603 | 675 | 29 | 8e-169 | 100 |
The search was optimized for highly similar sequences
Max ident, Maximum identities.
Figure 8.
Sequence conservation of the predicted frameshift cassette in yehP. The sequence logo was generated by aligning 31 sequences in Table 2.
DISCUSSION
The scoring system
In FSscan, the S score represents the stimulatory effect on +1 frameshifting. FSscan assigns zero to the S score for <4 base pairings between the six nucleotides upstream of the E-site and the anti-SD sequence [Equation (1)]. Equation (1) implies that at least four base pairing between mRNA and the anti-SD sequence are required to reveal the stimulatory effect. FSscan identifies yehP as the second best candidate for +1 frameshifting by using four as a threshold value in Equation (1), while the program identifies cheA as the second best candidate by using five as a threshold value. The in vivo observation that yehP6 results in higher frameshift efficiency than cheA6 (Figure 4) suggests that four base pairings could be sufficient to induce a stimulatory effect. In addition, FSscan assigns zero to the S score if the summation of the E, P and A scores is <3 [Equation (2)]. Equation (2) implies that for a less prominent synergic effect of the E-, P- and A-site for +1 frameshifting, the stimulatory effect by SD:anti-SD interaction is negligible.
The E score in the program represents the effect of E-site interaction on +1 frameshifting. FSscan calculates the E score as exp (−ΔGc), where ΔGc is the codon:anticodon interaction (19) in the ribosome E-site. The interaction in ribosome E-site has been shown to affect the reading frame maintenance (14,18,27–30). Weaker codon:anticodon interactions in the ribosome E-site have also been observed to result in a higher +1 frameshift efficiency (14,18). Notably, FSscan does not account for different tRNA:ribosome interactions in the E-site. While the tRNA:ribosome interactions are important for the E-site interaction, there has not been a well-established method to estimate these interactions. Previously, it has been suggested that a major fraction of the E-site tRNA binding is contributed by the binding of the 3′-terminal adenine to the ribosome (31). As the 3′-terminal adenine is conserved in all E. coli tRNAs, FSscan assumes a similar level of tRNA:ribosome interactions for different tRNAs and considers only codon:anticodon interactions in the E-site.
The P score represents the stability difference between the +1 frame and the zero frame interaction for the P-site tRNA. FSscan assumes the stability difference between the +1 frame and the zero frame interaction (Δstability*) as M1S1 − M2S0, where S1 is the stability of the +1 frame interaction, S0 is the stability of the zero frame interaction, and M1 and M2 are weighting factors. A separate data fitting program suggests M1 and M2 as 0.63 and 0.26, respectively, for the best linear correlation between the Δstability* and the logarithm of +1 frameshift efficiency observed by Curran (20) (Supplementary Data). The weighting factor for the +1 frame stability is 2.4-fold larger than that for the zero frame stability. Interestingly, zero frame duplexes are in general cognate but the realigned complexes contain a much wilder array of pairing and stabilities. Taken together, a favorable +1 frame interaction in the P-site may contribute more than an unstable zero frame interaction to a higher +1 frameshift efficiency.
FSscan accounts for two A-site features that enhance +1 frameshifting: (i) the competition between the cognate and the near cognate aa-tRNA for the zero frame A site codon (A0 score); (ii) the competition between the cognate aa-tRNA for the zero frame A-site codon and the cognate aa-tRNA for the +1 frame A-site codon (A1 score). A ribosome pause because of a stop codon or a rare codon in the A-site is a key factor for +1 frameshifting (32,33). It has been shown that the competition between the near-cognate aa-tRNA and the cognate aa-tRNA to the ribosome A-site plays an important role on the translation rate (21). The imbalance of the zero frame A-site tRNA and the +1 frame A-site tRNA was also shown to enhance +1 frameshifting (34). Three +1 frameshift candidates, yehP, pepP and cheA, all have CGG C in the A-site (where the zero frame codon is separated by the space). While the average A score is 0.44, the A score for CGG C is 1.58. CGG has one cognate tRNA,  , with 639 molecules per cell, and four near-cognate tRNAs,
, with 639 molecules per cell, and four near-cognate tRNAs,  ,
,  ,
,  and
and  , with 4752, 881, 4470 and 900 molecules per cell, respectively (21). The fact that near-cognate tRNAs outnumber cognate tRNAs for CGG results in a competition between these tRNAs for the ribosome A-site. In addition, the concentration of the cognate tRNA for the +1 frame A-site codon (GGC) is about 7-fold higher than that for the zero frame A-site codon (CGG). These two features may result in a longer pause during translation, making CGG C a likely A-site codon for +1 frameshifting. The other +1 frameshift candidate, nuoE, has TGA A in the A-site. The A score for TGA A is 1.8, which is also much higher than the average A score.
, with 4752, 881, 4470 and 900 molecules per cell, respectively (21). The fact that near-cognate tRNAs outnumber cognate tRNAs for CGG results in a competition between these tRNAs for the ribosome A-site. In addition, the concentration of the cognate tRNA for the +1 frame A-site codon (GGC) is about 7-fold higher than that for the zero frame A-site codon (CGG). These two features may result in a longer pause during translation, making CGG C a likely A-site codon for +1 frameshifting. The other +1 frameshift candidate, nuoE, has TGA A in the A-site. The A score for TGA A is 1.8, which is also much higher than the average A score.
FSI for a 16-nt window sums up S, E, P and A scores. The S score ranges from 0 to 2. The E score ranges from 0 to 1. The P score ranges from −1 to 1. The A score ranges from 0 to 2 because it combines A0 and A1, each ranging from 0 to 1. As a result, FSscan weighs the stimulatory, P-site, and A-site effects more than the E-site effect. This algorithm is supported by the kinetic model of +1 PRF, which suggested that +1 frameshift efficiency is more sensitive to the change in the stimulatory signal, P-site, and A-site effects (14).
Analysis of six reading frames and pseudogenes
Analysis of the six reading frames of the E. coli genome by FSscan reveals that 192 sequences have FSI higher than 3.5. Eighty-three of these sequences are located in the annotated coding regions, but only five sequences are in-frame with the start codon. The five cassettes are in prfB, yehP, nuoE, pepP and cheA. This result is consistent with the analysis of the 4132 protein-coding sequences (Figure 3). The function of intergenic sequence with FSI higher than 3.5 is not clear and requires further investigation. In addition, none of the 163 pseudogenes in the E. coli genome had a maximum FSI higher than 3.5 (data not shown).
yehP
yehP contains a potential +1 frameshift site with the second highest FSI, only after prfB. The predicted frameshift site in yehP is highly conserved in different E. coli strains (Table 2 and Figure 8). The potential cassette, GTG GAG TAT GGT CGG C (the zero frame is separated by a space and the P-site position is underlined), forms four base pairings with the anti-SD sequence and allows a weaker interaction in the E-site. In the P-site,  may form two canonical base pairings with the +1 frame although a central position mismatch can also occur. Notably, it has been proposed that <2 base parings in the shifted codon : anticodon complex may be sufficient for the efficient frameshifting (35). In a more extreme case, mRNA sites with little or no potential for canonical base pairing with the peptidyl-tRNA in the ribosome can also be used as landing positions for ribosomal bypassing (36). In the A-site, CGG is one of the four codons with the highest near-cognate tRNA competition (21). All of these features make yehP a potential +1 frameshifting candidate.
may form two canonical base pairings with the +1 frame although a central position mismatch can also occur. Notably, it has been proposed that <2 base parings in the shifted codon : anticodon complex may be sufficient for the efficient frameshifting (35). In a more extreme case, mRNA sites with little or no potential for canonical base pairing with the peptidyl-tRNA in the ribosome can also be used as landing positions for ribosomal bypassing (36). In the A-site, CGG is one of the four codons with the highest near-cognate tRNA competition (21). All of these features make yehP a potential +1 frameshifting candidate.
To date, the function of the yehP product is not well described in the literature. A known +1 PRF case in E. coli is the expression of RF2 from prfB gene (3). RF2 frameshifting is auto-regulated, meaning higher frameshift efficiency is driven by a lower level of the frameshifted products (3). It is suggested that this auto-regulation property may be evolved to evade a newly discovered fidelity control system: the ribosome would trigger a premature termination of protein synthesis when a mismatch P-site interaction is presented (37). RF2 frameshifting occurs more frequently when RF2 level is low, making it more difficult for ribosomes to trigger early termination in the presence of mismatch P-site. Whether yehP has involved in any regulation feedback loop or other mechanisms to escape from this fidelity control mechanism is uncertain. A yehP knockout E. coli strain was previously shown to result in a different swarming phenotype (38). yehP was suggested to have been introduced to the E. coli genome by the horizontal gene transfer (39). The predicted frameshifted product is 75 amino acids shorter than the standard decoding product. The function of the yehP frameshift protein remains unclear and needs to be investigated further.
Other frameshift-prone sequences
FSscan did not identify several shift-prone sequences observed experimentally in previous studies (40,41). argI was found to have a high level of +1 frameshifting at the very beginning of the coding sequence, UUU UAU (40). However, the maximum FSI in the gene is relatively low (2.0 for the P-site at the 110th codon). For the P-site positioned at the fourth codon UUU, FSI equals 0.38. Because argI frameshifting does not involve ribosomal pausing at a stop codon or a hungry codon in the A-site, the recoding may be achieved through mechanisms not considered by FSscan. In addition, CCC TGA containing genes, pheL, yjeF, ykgD and yrhB, were also shown to result in a higher level of +1 frameshifting (41). Notably, these sequences do not form >3 base pairings with the anti-SD sequence and their E-site interactions are relatively strong, which result in lower FSI. It is possible that a slippery sequence in the P-site (i.e. P-site tRNA can form complementary interactions with the +1 frame) along with a stop codon in the A-site can efficiently induce +1 frameshifting, which FSscan does not consider. On the other hand, not all of the CCC TGA containing genes promotes efficient +1 frameshifing, suggesting different mechanisms may be involved for pheL, yjeF, ykgD and yrhB framshifting. As growing numbers of the +1 frameshifting features are discovered, these features can be incorporated into FSscan to better predict frameshift sites.
FSscan as a bioinformatic program to search for novel +1 frameshift sequences
FSscan locates a 16-nt sequence with features for stimulatory signals, E-, P- and A-site effects in the E. coli genome. As compared to previous +1 frameshift site searching programs (11,12), FSscan differs in several major ways. (i) FSscan is not limited to a specific P- or A-site codon. Instead, FSscan looks for any P-site codon with a higher opportunity for tRNA rearrangement and any A-site codon with a higher possibility for a ribosome pausing during translation. (ii) The algorithm does not search for overlapping genes. Thus, it is not necessary that predicted frameshifting cassettes yield C-terminally extended fusion products. (iii) FSscan is intended for searching the E. coli genome, because the tRNA data for the score calculation and the experimental system are specific to E. coli. FSscan may be directly applied to screen the genome of E. coli bacteriophage, whose proteins can be translated by using E. coli ribosomes and tRNA pool. The strategy can be extended to other organisms with minor adjustments for the scoring system. (iv) FSscan predicts how likely a sequence is a frameshift site, but not the +1 frameshift efficiency. (v) FSscan needs no prior knowledge of the mRNA secondary structure involved in recoding. This method can be modified by varying the size of the recoding window to include mRNA structures serving as stimulatory signals.
CONCLUSION
FSscan performs a mechanistic-based genetic algorithm search for potential +1 frameshift sites in E. coli. The program successfully identifies prfB as a +1 frameshift candidate and predicts the frameshift site in this gene. Other predicted frameshift cassettes are shown to result in frameshift efficiency higher than a randomly designed sequence in vivo. These results suggest that the synergistic effects of ribosome E-, P- and A-sites are functionally important for +1 frameshifting. Importantly, FSscan provides the ability to perform a genome-wide systematic search for +1 frameshift sites. Further investigation of the predicted +1 frameshift sequences are in progress. The knowledge of different frameshift sites will enable researchers to better understand translational control.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The University of Delaware. Funding for open access charge: University of Delaware internal funds.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge Robert S. Kuczenski for advice in developing the Python and Matlab program. We are thankful to Dr. Jonathan D. Dinman for his insightful comments of this work.
REFERENCES
- 1.Kurland CG. Translational accuracy and the fitness of bacteria. Annu. Rev. Genet. 1992;26:29–50. doi: 10.1146/annurev.ge.26.120192.000333. [DOI] [PubMed] [Google Scholar]
- 2.Baranov PV, Gurvich OL, Hammer AW, Gesteland RF, Atkins JF. Recode 2003. Nucleic Acids Res. 2003;31:87–89. doi: 10.1093/nar/gkg024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Craigen WJ, Caskey CT. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986;322:273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- 4.Belcourt MF, Farabaugh PJ. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell. 1990;62:339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farabaugh PJ, Zhao H, Vimaladithan A. A novel programed frameshift expresses the POL3 gene of retrotransposon Ty3 of yeast: frameshifting without tRNA slippage. Cell. 1993;74:93–103. doi: 10.1016/0092-8674(93)90297-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janetzky B, Lehle L. Ty4, a new retrotransposon from Saccharomyces cerevisiae, flanked by tau-elements. J. Biol. Chem. 1992;267:19798–19805. [PubMed] [Google Scholar]
- 7.Asakura T, Sasaki T, Nagano F, Satoh A, Obaishi H, Nishioka H, Imamura H, Hotta K, Tanaka K, Nakanishi H, et al. Isolation and characterization of a novel actin filament-binding protein from Saccharomyces cerevisiae. Oncogene. 1998;16:121–130. doi: 10.1038/sj.onc.1201487. [DOI] [PubMed] [Google Scholar]
- 8.Morris DK, Lundblad V. Programmed translational frameshifting in a gene required for yeast telomere replication. Curr. Biol. 1997;7:969–976. doi: 10.1016/s0960-9822(06)00416-7. [DOI] [PubMed] [Google Scholar]
- 9.Palanimurugan R, Scheel H, Hofmann K, Dohmen RJ. Polyamines regulate their synthesis by inducing expression and blocking degradation of ODC antizyme. EMBO J. 2004;23:4857–4867. doi: 10.1038/sj.emboj.7600473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsufuji S, Matsufuji T, Miyazaki Y, Murakami Y, Atkins JF, Gesteland RF, Hayashi S. Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell. 1995;80:51–60. doi: 10.1016/0092-8674(95)90450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah AA, Giddings MC, Parvaz JB, Gesteland RF, Atkins JF, Ivanov IP. Computational identification of putative programmed translational frameshift sites. Bioinformatics. 2002;18:1046–1053. doi: 10.1093/bioinformatics/18.8.1046. [DOI] [PubMed] [Google Scholar]
- 12.Moon S, Byun Y, Kim HJ, Jeong S, Han K. Predicting genes expressed via -1 and +1 frameshifts. Nucleic Acids Res. 2004;32:4884–4892. doi: 10.1093/nar/gkh829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivanov IP, Pittman AJ, Chien CB, Gesteland RF, Atkins JF. Novel antizyme gene in Danio rerio expressed in brain and retina. Gene. 2007;387:87–92. doi: 10.1016/j.gene.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 14.Liao PY, Gupta P, Petrov AN, Dinman JD, Lee KH. A new kinetic model reveals the synergistic effect of E-, P- and A-sites on +1 ribosomal frameshifting. Nucleic Acids Res. 2008;36:2619–2629. doi: 10.1093/nar/gkn100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Harrison P, Kumar A, Lan N, Echols N, Snyder M, Gerstein M. A small reservoir of disabled ORFs in the yeast genome and its implications for the dynamics of proteome evolution. J. Mol. Biol. 2002;316:409–419. doi: 10.1006/jmbi.2001.5343. [DOI] [PubMed] [Google Scholar]
- 17.Weiss RB, Dunn DM, Dahlberg AE, Atkins JF, Gesteland RF. Reading frame switch caused by base-pair formation between the 3′ end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J. 1988;7:1503–1507. doi: 10.1002/j.1460-2075.1988.tb02969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanders CL, Curran JF. Genetic analysis of the E site during RF2 programmed frameshifting. RNA. 2007;13:1483–1491. doi: 10.1261/rna.638707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klump HH. Exploring the energy landscape of the genetic code. Arch. Biochem. Biophys. 2006;453:87–92. doi: 10.1016/j.abb.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 20.Curran JF. Analysis of effects of tRNA:Message stability on frameshift frequency at the Escherichia coli RF2 programmed frameshift site. Nucleic Acids Res. 1993;21:1837–1843. doi: 10.1093/nar/21.8.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fluitt A, Pienaar E, Viljoen H. Ribosome kinetics and aa-tRNA competition determine rate and fidelity of peptide synthesis. Comput. Biol. Chem. 2007;31:335–346. doi: 10.1016/j.compbiolchem.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobs JL, Dinman JD. Systematic analysis of bicistronic reporter assay data. Nucleic Acids Res. 2004;32:e160. doi: 10.1093/nar/gnh157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta P, Lee KH. Silent mutations result in HlyA hypersecretion by reducing intracellular HlyA protein aggregates. Biotechnol. Bioeng. 2008;101:967–974. doi: 10.1002/bit.21979. [DOI] [PubMed] [Google Scholar]
- 24.Kitteringham NR, Jenkins RE, Lane CS, Elliott VL, Park BK. Multiple reaction monitoring for quantitative biomarker analysis in proteomics and metabolomics. J. Chromatogr. B. Analyt Technol. Biomed. Life Sci. 2009;877:1229–1239. doi: 10.1016/j.jchromb.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 25.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marquez V, Wilson DN, Tate WP, Triana-Alonso F, Nierhaus KH. Maintaining the ribosomal reading frame: the influence of the E site during translational regulation of release factor 2. Cell. 2004;118:45–55. doi: 10.1016/j.cell.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 28.Sergiev PV, Lesnyak DV, Kiparisov SV, Burakovsky DE, Leonov AA, Bogdanov AA, Brimacombe R, Dontsova OA. Function of the ribosomal E-site: A mutagenesis study. Nucleic Acids Res. 2005;33:6048–6056. doi: 10.1093/nar/gki910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nierhaus KH. Decoding errors and the involvement of the E-site. Biochimie. 2006;88:1013–1019. doi: 10.1016/j.biochi.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 30.O'C;onnor M, Willis NM, Bossi L, Gesteland RF, Atkins JF. Functional tRNAs with altered 3′-ends. EMBO J. 1993;12:2559–2566. doi: 10.1002/j.1460-2075.1993.tb05911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lill R, Lepier A, Schwagele F, Sprinzl M, Vogt H, Wintermeyer W. Specific recognition of the 3′-terminal adenosine of tRNAPhe in the exit site of Escherichia coli ribosomes. J. Mol. Biol. 1988;203:699–705. doi: 10.1016/0022-2836(88)90203-3. [DOI] [PubMed] [Google Scholar]
- 32.Sipley J, Goldman E. Increased ribosomal accuracy increases a programmed translational frameshift in Escherichia coli. Proc. Natl Acad. Sci. USA. 1993;90:2315–2319. doi: 10.1073/pnas.90.6.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harger JW, Meskauskas A, Dinman JD. An “integrated model” of programmed ribosomal frameshifting. Trends Biochem. Sci. 2002;27:448–454. doi: 10.1016/s0968-0004(02)02149-7. [DOI] [PubMed] [Google Scholar]
- 34.Pande S, Vimaladithan A, Zhao H, Farabaugh PJ. Pulling the ribosome out of frame by +1 at a programmed frameshift site by cognate binding of aminoacyl-tRNA. Mol. Cell. Biol. 1995;15:298–304. doi: 10.1128/mcb.15.1.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ivanov IP, Gurvich OL, Gesteland RF, Atkins JF. Recoding: site- or mRNA-specific alteration of genetic readout utilized for gene expression. In: Lapointe J, Barker-Gingras L, editors. Translation Mechanism. Austin, TX: Landes Bioscience; 2003. pp. 354–369. [Google Scholar]
- 36.Herr AJ, Wills NM, Nelson CC, Gesteland RF, Atkins JF. Factors that influence selection of coding resumption sites in translational bypassing: minimal conventional peptidyl-tRNA:mRNA pairing can suffice. J Biol Chem. 2004;279:11081–11087. doi: 10.1074/jbc.M311491200. [DOI] [PubMed] [Google Scholar]
- 37.Zaher HS, Green R. Quality control by the ribosome following peptide bond formation. Nature. 2009;457:161–166. doi: 10.1038/nature07582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inoue T, Shingaki R, Hirose S, Waki K, Mori H, Fukui K. Genome-wide screening of genes required for swarming motility in Escherichia coli K-12. J. Bacteriol. 2007;189:950–957. doi: 10.1128/JB.01294-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davids W, Zhang Z. The impact of horizontal gene transfer in shaping operons and protein interaction networks–direct evidence of preferential attachment. BMC Evol. Biol. 2008;8:23. doi: 10.1186/1471-2148-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu C, Parker J. A ribosomal frameshifting error during translation of the argI mRNA of Escherichia coli. Mol. Gen. Genet. 1994;243:434–441. doi: 10.1007/BF00280474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gurvich OL, Baranov PV, Zhou J, Hammer AW, Gesteland RF, Atkins JF. Sequences that direct significant levels of frameshifting are frequent in coding regions of Escherichia coli. EMBO J. 2003;22:5941–5950. doi: 10.1093/emboj/cdg561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.