Abstract
The Rho GTPase family member RhoE inhibits RhoA/ROCK signaling to promote actin stress fiber and focal adhesion disassembly. We have previously reported that RhoE also inhibits cell cycle progression and Ras-induced transformation, specifically preventing cyclin D1 translation. Here we investigate the molecular mechanisms underlying those observations. RhoE inhibits the phosphorylation of the translational repressor 4E-BP1 in response to extracellular stimuli. However, RhoE does not affect the activation of mTOR, the major kinase regulating 4E-BP1 phosphorylation, as indicated by the phosphorylation levels of the mTOR substrate S6K, the dynamics of mTOR/Raptor association, and the observation that RhoE, as opposed to rapamycin, does not impair cellular growth. Interestingly, RhoE prevents the release of the eukaryotic initiation factor eIF4E from 4E-BP1, inhibiting cap-dependent translation. Accordingly, RhoE also inhibits the expression and the transcriptional activity of the eIF4E target c-Myc. Consistent with its crucial role in cell proliferation, we show that eIF4E can rescue both cell cycle progression and Ras-induced transformation in RhoE-expressing cells, indicating that the inhibition of eIF4E function is critical to mediate the anti-proliferative effects of RhoE.
Introduction
Rnd proteins (Rnd1, Rnd2, and RhoE/Rnd3) constitute a separate subfamily within the Rho family of small GTPases that have attracted recent attention because of their atypical features (1). Despite their high level of sequence similarity with RhoA, -B, and -C, Rnd proteins are remarkably different at both biochemical and functional levels. For instance, Rnd proteins do not cycle between an active form and an inactive form. Instead, they are thought to be constitutively GTP bound because of their extremely low GTPase activity and their increased affinity for GTP over GDP. Moreover, both Rnd1 and RhoE/Rnd3 induce opposite effects to those induced by RhoA on the actin cytoskeleton, promoting the disassembly of actin stress fibers and the loss of focal adhesions (2, 3).
The best characterized member of the Rnd family, RhoE/Rnd3, has been shown to antagonize RhoA cytoskeletal functions through two different mechanisms (4, 5). On the one hand, RhoE binds to the N terminus of ROCK I and inhibits its downstream activity (6), thus blocking the major RhoA-dependent pathway promoting actomyosin contractility. On the other hand, RhoE has been shown to bind to p190RhoGAP and to enhance its GTPase activity toward RhoA, thereby reducing RhoA-GTP levels (7). Altogether, these effects account for the so-called “round phenotype” elicited by Rnd proteins in different cell systems. It has been recently shown that RhoE function is regulated in part by ROCK I-dependent phosphorylation on multiple residues. Phosphorylated RhoE is predominantly located in the cytosol and is more stable. In agreement with these observations, higher levels of RhoE phosphorylation on a specific residue (serine 11) are associated with its increased ability to promote actin stress fiber disassembly (8). RhoE is also regulated at the transcriptional level, for example by p53 (9).
As well as their well-described role in actin cytoskeleton dynamics, Rho GTPases are key regulators of other cellular processes such as cell cycle progression (10). Using fibroblasts engineered to inducibly overexpress RhoE, we have shown that RhoE inhibits cell cycle progression and Ras-induced transformation (11). The anti-proliferative effects induced by RhoE are not caused by its cytoskeletal functions and do not involve alterations in RhoA/ROCK signaling. RhoE expression induces G1 arrest and prevents cyclin D1 expression, without affecting the activation of the ERK,3 Rac, and PI3K/Akt pathways. Interestingly, RhoE specifically prevents cyclin D1 biosynthesis but not its transcription, indicating that RhoE might impinge on translational regulators controlling cyclin D1 translation (11). Similarly, RhoE overexpression also induces G1 arrest and prevents cyclin D1 expression at a post-transcriptional level in glioblastoma cells (12).
Initiation of protein translation in mammalian cells requires the formation of the eIF4F translation initiation complex. This complex is comprised of three different proteins: the cap-binding protein, eIF4E, which binds to the cap structure (m7·GTP) found at the 5′-end of mRNAs, an ATP-dependent helicase, eIF4A, that helps to unwind highly structured 5′-UTRs (untranslated regions) and a large scaffolding protein, eIF4G, that provides the docking site for the 40 S ribosomal subunit (13). The activity of eIF4F is dependent on the availability and function of eIF4E. When eIF4E expression or activity is low, only “strong” mRNAs with simple 5′-UTRs are efficiently translated. In contrast, high levels of eIF4E activity favor the translation of a specific subset of genes characterized by complex and highly structured 5′-UTRs (14). Interestingly, these eIF4E-dependent genes, such as cyclin D1 and c-Myc, are mostly linked with growth-promoting, oncogenic functions (15, 16). In agreement with these observations, eIF4E has been defined as a bona fide oncogene (17), and its expression levels are substantially elevated in several types of human cancer including breast and colon, among others (18–20).
Although the activity of eIF4E can be regulated at multiple levels, including its expression and phosphorylation, eIF4E function is mainly controlled by its interaction with a family of translational repressors termed the 4E-binding proteins (4E-BPs). The best characterized member of this family, 4E-BP1, binds avidly to the eIF4E region that interacts with eIF4G, thereby effectively blocking the formation of the eIF4F complex and the onset of cap-dependent translation (21). 4E-BP1 interaction with eIF4E is relieved by its phosphorylation at several sites, which takes place in a hierarchical fashion under the appropriate conditions, including growth factor or hormone stimulation and nutrient availability. Upon a priming phosphorylation event on Thr-37/46, which is mTOR/Raptor-dependent, 4E-BP1 undergoes sequential phosphorylation at Ser-70 and Ser-65 leading to its dissociation from eIF4E and its functional inactivation as a translational repressor (22, 23).
Here, we have used RhoE-inducible cells to investigate whether RhoE affects translational regulators to inhibit cyclin D1 biosynthesis. We show that RhoE prevents 4E-BP1 phosphorylation at relevant sites induced by stimuli including phorbol esters and growth factors, leading to the stabilization of its interaction with eIF4E and the inhibition of cap-dependent translation. Accordingly, RhoE reduces the expression of c-Myc and attenuates its transcriptional activity. Interestingly, RhoE does not alter mTOR function to block 4E-BP1 phosphorylation because S6K signaling is not impaired in RhoE-expressing cells. We finally show that the inhibition of eIF4E function contributes to RhoE-mediated anti-proliferative effects, because eIF4E rescues both cell proliferation and Ras-induced transformation in RhoE transfectants.
EXPERIMENTAL PROCEDURES
Cell Culture and Cell Size Measurements
NIH 3T3 cells were grown in DMEM supplemented with 10% donor calf serum. COS-7 and HEK-293 cells were grown in DMEM supplemented with 10% fetal calf serum. RhoE-3T3 cells have been previously described (11) and were grown in histidine-free DMEM supplemented with 10% donor calf serum, 0.5 μg/ml tetracycline, 0.5 mm histidinol, and 2 μg/ml puromycin (Sigma). Cells were made quiescent by culturing them for 24 h in medium containing 0.5% fetal bovine serum. Platelet-derived growth factor (PDGF), 12-O-tetradecanoylphorbol-13-acetate (TPA), and rapamycin (all from Sigma) were added directly to the medium at the indicated concentration and cells were harvested at the time points indicated in the figure legends. For cell size measurements, serum-starved cells were stimulated with 10% FCS-containing medium for the indicated time points, collected by trypsinization, resuspended with phosphate-buffered saline and processed using a Casy Cell Counter (Scharfe System) according to the manufacturer's instructions. Results are presented as the mean ± S.D. of data from three independent experiments, each of which was conducted in duplicate.
Expression Vectors and Transient Transfections
Cells were transfected using Lipofectamine2000 (Invitrogen) according to the manufacturer's instructions with the following expression plasmids: H-RasV12 (pcDNA3-H-RasV12, a gift of J. Downward), Flag-RhoE (pCMV5-Flag-RhoE, (6)), Flag-RhoAV14 (pCMV5-Flag-RhoAV14, a gift of G.O. Cory), Myc-eIF4E (pBabe-myc-eIF4E, a gift of G. Grech), mTOR (pcDNA1-HA-mTOR, a gift of I. Gout), Myc luciferase reporter (pM4-mintkluc, a gift of B. Lüscher) and the bicistronic luciferase/Renilla reporters (pRemcvF and pRhrvF, a gift of M. J. Coldwell).
Gel Electrophoresis and Immunoblotting
Cells were harvested in a buffer containing 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, and 1% (v/v) Triton X-100 plus protease and phosphatase inhibitors (2 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mm phenylmethylsulfonyl fluoride, 1 mm dithiothreitol, 1 mm NaF, and 0.2 mm Na3VO4). Protein content was measured by the Bradford procedure, using bovine serum albumin as a standard (24). Cell lysates or proteins from pull-down or immunoprecipitation experiments were electrophoresed in SDS-polyacrylamide gels. After electrophoresis, the proteins were transferred to Immobilon-P strips for 2 h at 60 V. The sheets were preincubated in TBS (20 mm Tris-HCl, pH 7.5, 150 mm NaCl), 0.05% Tween 20, and 5% defatted milk powder for 1 h at room temperature and then incubated for 1 h at room temperature in TBS, 0.05% Tween 20, 1% bovine serum albumin, and 0.5% defatted milk powder containing the appropriate antibodies: anti-HA (Covance Research, MMS-101R, 1:1000), anti-cyclin D1 (Santa Cruz Biotechnology, sc-8396, 1:1000), anti-Flag (Sigma, F3165, 1:2000), anti-β-tubulin (Sigma, T0198, 1:4000), anti-phospho-S6K T389 (Cell Signaling, 9206, 1:1000) mouse monoclonal antibodies, and anti-phospho-S6 ribosomal protein (Cell Signaling, 4856, 1:1000), anti-S6K (Cell Signaling, 9202, 1:1000), anti-eIF4E (Cell Signaling, 9742, 1:1000), anti-4E-BP1 (Cell Signaling, 9452, 1:1000), anti-phospho-4E-BP1 (S65) (Cell Signaling, 9451, 1:1000), anti-phospho-4E-BP1 (T37/46) (Cell Signaling, 9459, 1:1000), anti-c-Myc (Santa Cruz Biotechnology, sc-764, 1:1000), anti-mTOR (Santa Cruz Biotechnology, sc-8319, 1:1000), and anti-Raptor (a gift of D. Sabatini, 1:500) rabbit polyclonal antibodies. After washing in TBS, 0.05% Tween 20, the sheets were incubated with a peroxidase-coupled secondary antibody (1:2000 dilution, Amersham Biosciences) for 1 h at room temperature. After incubation, the sheets were washed twice in TBS, 0.05% Tween 20, and once in TBS. The peroxidase reaction was visualized by the enhanced chemiluminescence detection system.
Measurement of 4E-BP1 Binding to eIF4E
The capacity of eIF4E to bind to the cap structure was used to analyze the amount of 4E-BP1 bound to eIF4E, as described elsewhere. Cells (5–10 × 106) were washed with ice-cold phosphate-buffered saline, resuspended in cap-binding buffer (20 mm Hepes, pH 7.2, 1 mm EDTA, 100 mm KCl, 10% (v/v) glycerol, 7 mm 2-mercaptoethanol, 50 mm β-glycerophosphate, 50 mm NaF plus the protease and phosphatase inhibitors indicated above) and subjected to three consecutive freeze-thaw cycles. Cleared (10,000 × g) lysates were incubated for 45 min at 4 °C with m7·GTP Sepharose (Amersham Biosciences), and beads were washed four times in cap-binding buffer. Bound proteins were solubilized by the addition of 35 μl of Laemmli loading buffer and separated on 12.5% SDS-polyacrylamide gels. The amount of 4E-BP1 or eIF4E in the bound fraction was detected by Western blotting using specific antibodies.
Luciferase Reporter Assays
For luciferase assays, the activity of both Firefly and Renilla luciferase in cell lysates was measured using the Dual-luciferase reporter assay system (Promega). Assays were performed according to the manufacturer's recommendations. For cap-dependent translation analysis, NIH3T3 cells were seeded the day before transfection at a density of 1.5 × 105 cells per well on 6-well plates. Cells were transfected with 0.75 μg of the pRemcvF or the pRhrvF bicistronic vectors (25) together with the indicated expression plasmids. Cells were harvested 48 h later with passive-lysis buffer (Promega), and luciferase activities were measured. Cap-dependent translation levels in transiently transfected cells were calculated by normalizing Renilla levels to control (IRES-directed) firefly luciferase levels. For the data presented in Fig. 5B, the following average luciferase values were obtained: 49665 (control), 23752 (RhoE), and 60298 (eIF4E). Results are presented as the mean ± S.D. of data from three independent experiments, each of which was conducted in duplicate. For c-Myc reporter assays, cells (1.5 × 105) were transfected with 0.75 μg of Myc-luciferase reporter vector (pM4-mintkluc, a gift of Bernhard Lüscher) and the indicated expression vectors. 48 h after transfection, cells were harvested with passive lysis buffer (Promega), and luciferase activities were measured. Levels of reporter gene induction were obtained normalizing firefly luciferase levels to control Renilla luciferase levels. For the data presented in Fig. 6D, the following average luciferase values were obtained: 42293 (control) and 20065 (RhoE). Results are presented as the mean ± S.D. of data from three independent experiments, each of which was conducted in duplicate.
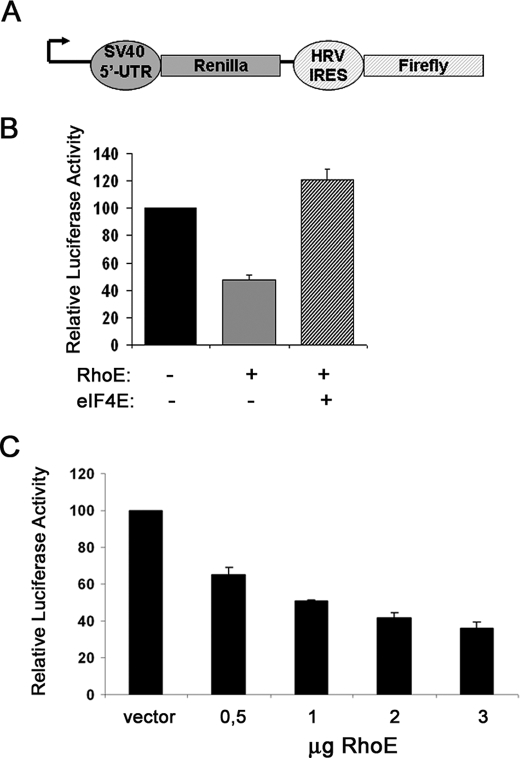
FIGURE 5.
RhoE inhibits cap-dependent translation. A, a schematic diagram representing the bicistronic reporter vectors used to measure cap-dependent translation. The Renilla and firefly luciferase genes are under the control of the 5′-untranslated region of SV40 and the human rhinovirus 2 internal ribosome entry site, respectively. B, graph represents the mean ± S.D. values of cap-dependent translation rates in cells transfected with 2 μg of the indicated plasmids, expressed as relative luciferase activity and obtained as indicated under “Experimental Procedures.” C, as in B, with the indicated amounts of transfected RhoE plasmid.
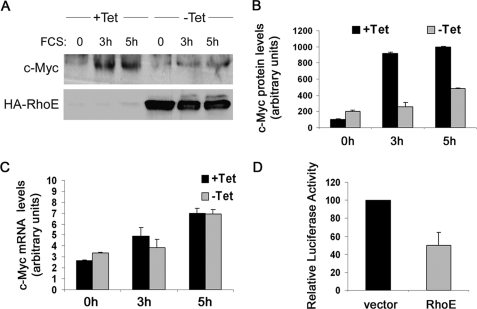
FIGURE 6.
RhoE inhibits c-Myc expression and its transcriptional activation. A, RhoE-3T3 cells were starved for 24 h in 0.5% FCS-containing medium in the presence (+Tet) or absence (−Tet) of tetracycline, stimulated for the indicated time with 10% FCS in the presence (+Tet) or absence (−Tet) of tetracycline and harvested. The expression levels of c-Myc and HA-RhoE were analyzed by Western blotting using specific antibodies. B, graph represents the mean ± S.D. value for quantified c-Myc protein levels in control (+Tet) and RhoE-expressing cells (−Tet) from three independent experiments. C, as in A, but the levels of c-Myc mRNA were measured by quantitative real-time PCR. The graph represents the mean ± S.D. values of normalized c-Myc mRNA levels in control (+Tet) and RhoE-expressing (−Tet) cells. D, representation of the mean ± S.D. values of c-Myc reporter activity in control vector and RhoE-transfected cells, from three independent experiments each conducted in duplicate.
Immunoprecipitations
Cells grown in 10-cm dishes were washed with ice-cold phosphate-buffered saline and harvested on ice in mTOR IP buffer (40 mm Hepes pH 7.5, 120 mm NaCl, 10 mm β-glycerophosphate, 50 mm NaF, 1 mm EDTA, and 0.3% (w/v) CHAPS, plus the protease and phosphatase inhibitors indicated above) and clarified by centrifugation (10,000 × g). Immunoprecipitations were carried out by incubating the cleared supernatants for 2 h at 4 °C with 2 μg of anti-mTOR (Santa Cruz Biotechnology, sc-1549) and then for 1 h at 4 °C with 15 μl of protein G-Sepharose (Amersham Biosciences). Immunoprecipitates were then washed four times in wash buffer (50 mm Hepes, pH 7.5, 40 mm NaCl, and 2 mm EDTA) and resuspended in Laemmli loading buffer. Immunoprecipitated proteins were run on 6% SDS-PAGE, and the amount of bound proteins was detected by Western blotting with specific antibodies.
Gene Expression Analysis
Total RNA was isolated using TRIzol® reagent (Invitrogen). 1 μg of RNA was reverse-transcribed into cDNA using pdN6 primers and the MMLV reverse transcriptase (Invitrogen). Subsequent real-time PCR reactions were performed in duplo in the LightCycler® 2.0 System (Roche Applied Science) using the SYBR Green detection methodology. c-Myc mRNA levels were measured using the two following sets of primers against murine c-Myc: 1) forward 5′-GCCCAGTGAGGATATCTGGA, reverse 5′- ATCGCAGATGAAGCTCTGGT and 2) forward 5′-AGTGCTGCATGAGGAGACAC, reverse 5′-GGTTTGCCTCTTCTCCACAG. Mouse Gapdh was measured as an internal control for normalization with the following primers: forward 5′-CAATGTGTCCGTCGTGGATCT, reverse 5′- GTCCTCAGTGTAGCCCAAGATG. Threshold cycle data were analyzed using the following formula: normalized gene expression level = (Etarget)CPtarget(sample)/(Eref)CPref(sample) (adapted from Ref. 26) to quantify the level of gene expression.
Focus Formation Assays
NIH 3T3 cells were seeded at a cell density of 2 × 105 cells per well in 6-well plates the day before transfection. Cells were transfected with the indicated vectors in 6-well plates in duplicate. After 24 h, cells were transferred to 10-cm plates, and the medium was substituted with 5% FCS-containing DMEM when cells reached confluency (typically, 2–3 days later). After 12–15 days, cells were stained with 0.5% (w/v) crystal violet in 70% ethanol, and the number of foci larger than 1 mm in diameter counted. Results are presented as the mean ± S.D. of data from three independent experiments, each of which was conducted in duplicate.
Clonogenic Assays
NIH 3T3 cells were seeded at a cell density of 2 × 105 cells per well in 6-well plates the day before transfection. Cells were transfected with the indicated vectors together with 0.2 μg of a plasmid encoding for Hygromycin resistance (pSV2-Hygro) in 6-well plates in duplicate. After 24 h, cells were transferred to 10-cm plates, and the medium was substituted with 10% DCS containing 0.5 μg/ml of hygromycin. After 7–10 days, antibiotic-resistant clones were stained with 0.5% (w/v) crystal violet in 70% ethanol and counted. Results are presented as the mean ± S.D. of data from three independent experiments, each of which was conducted in duplicate.
RESULTS
RhoE Prevents 4E-BP1 Phosphorylation in Response to Growth Factor Stimulation
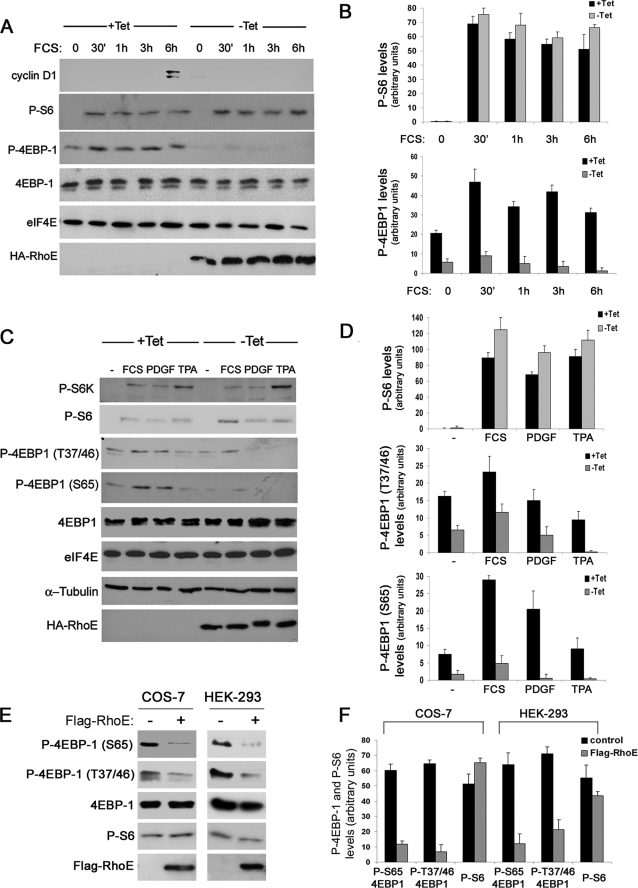
We have shown previously that RhoE blocks cyclin D1 biosynthesis and leads to cell cycle arrest in G1 (11). To understand the molecular mechanisms underlying the failure of RhoE-expressing cells to synthesize cyclin D1, we used RhoE-inducible cells to analyze the status of major signaling elements controlling protein translation. We focused on eIF4E, because this is a central regulator of translation initiation in mammalian cells (21). As expected, RhoE prevented serum-induced cyclin D1 expression, which is normally expressed at mid-G1 in control cells (Fig. 1 A). Although neither eIF4E nor its negative regulator 4E-BP1 expression levels were significantly altered upon RhoE expression, 4E-BP1 phosphorylation on serine 65 was strongly reduced in RhoE-expressing cells, whereas S6 phosphorylation remained unaffected (Fig. 1, A and B). The repressive function of 4E-BP1 is relieved by multiple phosphorylation events in response to extracellular stimuli. We next investigated whether RhoE was able to prevent 4E-BP1 phosphorylation induced by different signals at functionally relevant phosphorylation sites. Phosphorylation levels of both serine 65 and threonines 37/46, which have been shown to contribute to the disruption of 4E-BP1/eIF4E complexes, were similarly reduced by RhoE expression in response to stimuli including serum, PDGF, and the phorbol ester TPA (Fig. 1, C and D). PDGF induced a marked electrophoretic shift in HA-RhoE, consistent with its ability to induce endogenous RhoE phosphorylation on serine 11 (8). We next investigated whether RhoE could similarly inhibit 4E-BP1 phosphorylation in a different experimental system. For this purpose we transiently transfected COS-7 and HEK-293 cells with Flag-RhoE and monitored 4E-BP1 phosphorylation levels. In agreement with our previous results, RhoE expression inhibited 4E-BP1 phosphorylation, but not S6 phosphorylation, in both cell lines (Fig. 1, E and F). These observations indicate that RhoE inhibits 4E-BP1 phosphorylation in response to growth factor stimulation.
FIGURE 1.
RhoE inhibits growth factor-induced 4E-BP1 phosphorylation. A, RhoE-3T3 cells were starved for 24 h in 0.5% FCS-containing medium in the presence (+Tet) or absence (−Tet) of tetracycline and were stimulated with 10% FCS in the presence or absence of tetracycline and harvested at the indicated time points. The expression levels of the indicated proteins were analyzed by Western blotting with specific antibodies. B, representation of the mean ± S.D. levels of quantified P-S6 and p-4EBP1 in control (+Tet) and RhoE-expressing (−Tet) cells. C, RhoE-3T3 cells were starved as in A, stimulated with 10% FCS, PDGF (1 nm), or TPA (100 nm) for 30 min and harvested. The expression levels of the indicated proteins were analyzed by Western blotting with specific antibodies. D, representation of the mean ± S.D. levels of quantified P-S6 and p-4EBP1 (Ser-65 and Thr-37/46) in control (+Tet) and RhoE-expressing (−Tet) cells. E, COS-7 and HEK-293 cells were transfected with Flag-RhoE and harvested 48 h after transfection. The expression levels of the indicated proteins were analyzed by Western blotting with specific antibodies. F, representation of the mean ± S.D. levels of quantified P-S6 and p-4EBP1 (Ser-65 and Thr-37/46) in mock-transfected (control) and RhoE-transfected (Flag-RhoE) cells.
RhoE Does Not Affect mTOR/S6K Signaling
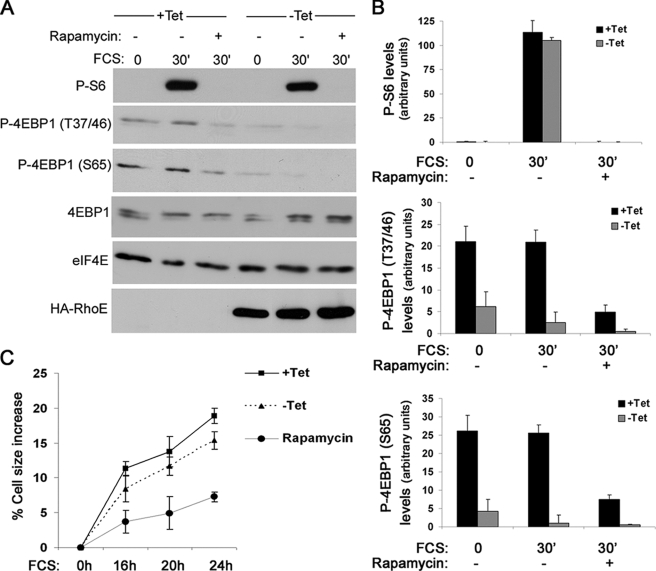
Because eIF4E function is mainly regulated by 4E-BP1, we next investigated how 4E-BP1 phosphorylation might be reduced in RhoE-expressing cells. 4E-BP1 phosphorylation in response to extracellular stimuli can be stimulated by the serine/threonine kinase mTOR, which is also responsible for activating S6K, coupling cap-dependent translation with ribosome biogenesis and general protein synthesis. We monitored the phosphorylation levels of S6K and S6, a substrate of S6K, as readout for mTOR/S6K activity. Unlike 4E-BP1, serum-induced S6 and S6K phosphorylation was not altered by RhoE expression (Figs. 1 and 2, A and B). Confirming our previous observations, phosphorylation of 4E-BP1 at the indicated sites was clearly reduced upon RhoE expression (Fig. 2, A and B). As expected, mTOR inhibition with rapamycin prevented both S6 and 4E-BP1 serum-stimulated phosphorylation (Fig. 2, A and B). These results suggest that RhoE specifically blocks 4E-BP1, but not S6K/S6 phosphorylation, and thus is unlikely to inhibit mTOR. As an alternative functional readout for mTOR/S6K activation, we measured the increase in cell size in response to growth factor stimulation, because this pathway but not 4E-BP1/eIF4E is thought to be responsible for driving general protein synthesis and thus cellular growth (27). Although RhoE-expressing cells appeared to be slightly smaller than control cells, their cell size increased upon serum stimulation similarly to that of control cells (Fig. 2C). In sharp contrast, mTOR inhibition with rapamycin strongly reduced the ability of cells to increase their size (Fig. 2C). Altogether, our data suggest that RhoE blocks 4E-BP1 phosphorylation without inhibiting the mTOR/S6K pathway.
FIGURE 2.
RhoE does not affect mTOR/S6K signaling. A, RhoE-3T3 cells were starved for 24 h in 0. 5% FCS-containing medium in the presence (+Tet) or absence (−Tet) of tetracycline, stimulated for 30 min with 10% FCS alone or together with rapamycin (20 nm, preincubated for 30 min) in the presence or absence of tetracycline, and harvested. The expression levels of the indicated proteins were analyzed by Western blotting with specific antibodies. B, representation of the quantified P-S6 and p-4EBP1 (Ser-65 and Thr-37/46) levels in control (+Tet) and RhoE-expressing (−Tet) cells. C, RhoE-3T3 cells were starved for 24 h in 0.5% FCS-containing medium in the presence (+Tet) or absence (−Tet) of tetracycline and were stimulated with 10% FCS alone, in the presence (+Tet) or absence (−Tet) of tetracycline, or together with rapamycin (20 nm) in the presence of tetracycline (rapamycin). Cell size was measured at the indicated times and shown as the percentage in cell size increase relative to cell size at 0 h. Results represent the mean values from three independent experiments, each conducted in duplicate.
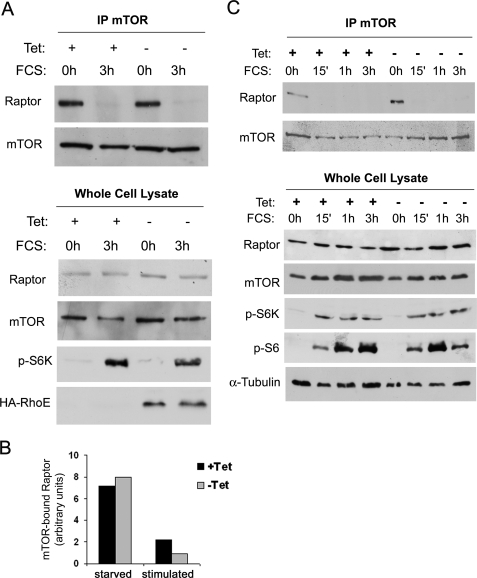
RhoE Does Not Alter the Dynamics of mTOR/Raptor Binding
mTOR signaling is regulated by its association to regulatory proteins such as Raptor and Rictor, which are crucial to couple mTOR to its downstream substrates and define different mTOR-containing complexes (28–31). Although it is believed that the mTOR/Raptor complex catalyzes phosphorylation of both S6K and 4E-BP1, it is possible that RhoE modulates the dynamics of these complexes to inhibit 4E-BP1, but not S6K, phosphorylation. We therefore co-immunoprecipitated mTOR in control and RhoE-expressing cells to investigate the impact of RhoE expression on the integrity and dynamics of these complexes. Rictor was associated with mTOR both in starved and stimulated conditions, and this interaction remained unaltered upon RhoE expression (data not shown). Interestingly, we consistently found that Raptor was strongly bound to mTOR in quiescent cells but this interaction disappeared upon mitogenic stimulation, irrespective of the presence or absence of RhoE (Fig. 3, A and B). We further confirmed this observation by performing a time course analysis of mTOR/Raptor interaction upon serum stimulation. Again, Raptor association with mTOR was significantly weakened upon mitogenic stimulation even at the earliest time point (15 min) irrespective of RhoE expression (Fig. 3C). Remarkably, the kinetics of mTOR/Raptor dissociation coincided with the onset of mTOR activity, as reflected by S6K and S6 phosphorylation (Fig. 3C). These results further indicate that RhoE does not inhibit 4E-BP1 by perturbing mTOR signaling.
FIGURE 3.
RhoE does not alter the dynamics of mTOR/Raptor binding. A, RhoE-3T3 cells were starved for 24 h in 0.5% FCS-containing medium in the presence (+Tet) or absence (−Tet) of tetracycline and stimulated for 3 h with 10% FCS in the presence (+Tet) or absence (−Tet) of tetracycline. Harvested cell lysates were subjected to immunoprecipitation with anti-mTOR antibody and the presence of mTOR and Raptor in the immunocomplex (top panel) and levels of indicated proteins in the input lysate (bottom panel) were analyzed by Western blotting with specific antibodies. B, graph represents the quantified levels of mTOR-bound Raptor in starved and stimulated control (+Tet) and RhoE-expressing cells (−Tet). C, as in A, but cells were stimulated for additional time points, as indicated.
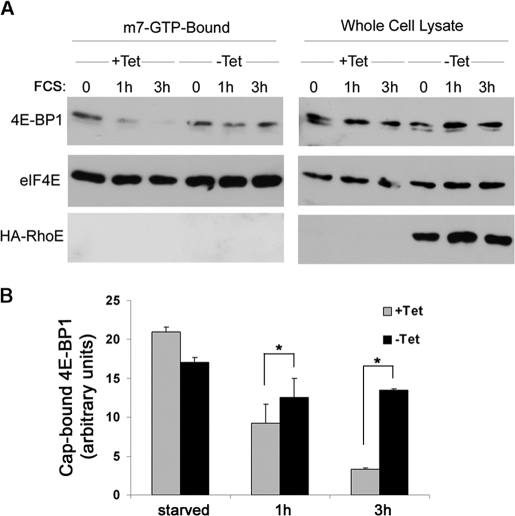
RhoE Inhibits 4E-BP1 Release from eIF4E in Response to Mitogenic Stimulation
Because phosphorylation of 4E-BP1 disrupts its inhibitory association with eIF4E, we analyzed whether RhoE expression could prevent this dissociation. To this end, we performed pull-downs with m7·GTP-Sepharose to assess the levels of 4E-BP1 bound to eIF4E in both starved and stimulated cells. As expected, eIF4E was recovered in the m7·GTP-Sepharose-bound fraction under all our experimental conditions (Fig. 4A). In contrast, whereas 4E-BP1 was present in the cap-bound fraction of resting cells, serum stimulation induced its dissociation from cap-bound eIF4E in control cells (Fig. 4, A and B). However, in RhoE-expressing cells the serum-induced dissociation of cap-bound 4E-BP1 was strongly reduced (Fig. 4, A and B). This observation indicates that RhoE inhibits eIF4E release from its negative regulator 4E-BP1 upon extracellular stimulation.
FIGURE 4.
RhoE inhibits 4E-BP1 release from eIF4E in response to mitogenic stimulation. A, RhoE-3T3 cells were starved for 24 h in 0.5% FCS-containing medium in the presence (+Tet) or absence (−Tet) of tetracycline and stimulated for the indicated time with 10% FCS in the presence (+Tet) or absence (−Tet) of tetracycline. Harvested cell lysates were pulled down with m7·GTP-Sepharose as indicated under “Experimental Procedures.” m7·GTP-Sepharose-bound proteins (left panel) and proteins in the input lysate (right panel) were analyzed by Western blotting with the indicated specific antibodies. B, graph represents the mean ± S.D. of quantified 4E-BP1/eIF4E ratio (cap-bound 4E-BP1) from three independent experiments. The differences in cap-bound 4E-BP1 levels between serum-stimulated control (+Tet) and RhoE-expressing cells (−Tet) are statistically significant (Student's t test: *, p < 0.05).
RhoE Inhibits Cap-dependent Translation
Because 4E-BP1 dissociation is crucial for eIF4E to promote translation initiation via the formation of the eIF4F complex (13), our observations strongly suggested that RhoE inhibits eIF4E function. To assess the impact of RhoE on eIF4E activity in vivo, we took advantage of an established method to measure cap-dependent translation rates using bicistronic reporter systems (25), as depicted in Fig. 5A. Using this assay, we measured the effect of RhoE on cap-dependent translation rates. RhoE expression led to a clear inhibition (∼50%) of cap-dependent translation rates in NIH3T3 cells, and eIF4E co-expression rescued cap-dependent translation rates in this assay (Fig. 5B). Dose response experiments confirmed that RhoE blocked cap-dependent over cap-independent translation (Fig. 5C), indicating that RhoE inhibits eIF4E activity in vivo.
RhoE Inhibits c-Myc Expression and Its Transcriptional Activity
We have shown that RhoE inhibits the expression of cyclin D1, a known target of cap-dependent translation. To determine if RhoE regulates the expression of other eIF4E targets, we investigated the expression of c-Myc, an established target of eIF4E-dependent translational regulation (32). To this end, we monitored the induction of c-Myc protein levels upon serum stimulation in control and RhoE-expressing cells. RhoE reduced serum-induced increase of c-Myc expression (Fig. 6, A and B) although it did not affect c-Myc mRNA levels (Fig. 6C). We also measured c-Myc-dependent transcriptional activity using a c-Myc-sensitive luciferase reporter vector (33). In agreement with the inhibition of c-Myc protein expression observed in RhoE-induced cells, RhoE similarly reduced c-Myc function (Fig. 6D). Altogether, our data are consistent with a model whereby RhoE-mediated inhibition of cap-dependent translation not only impairs cyclin D1 expression but also reduces c-Myc expression.
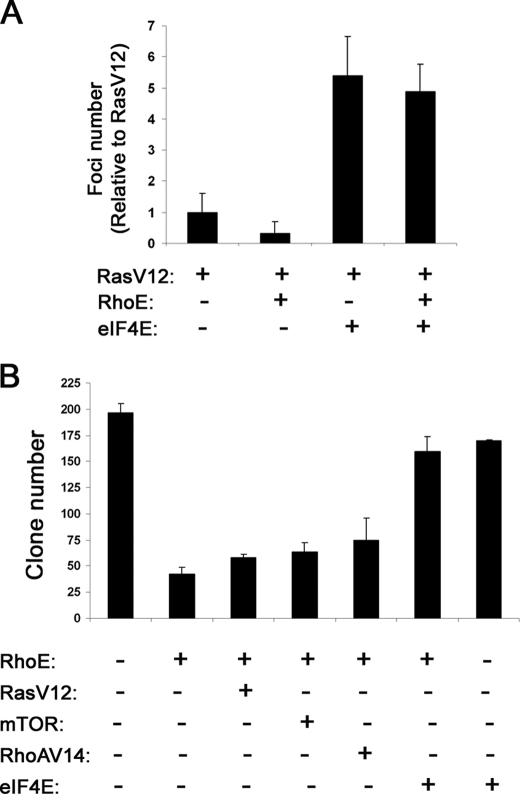
Inhibition of eIF4E Function Contributes to RhoE-mediated Cell Cycle Arrest
We have previously reported that RhoE can inhibit cell cycle progression and transformation (11). Because eIF4E activation is essential for cell proliferation, we next addressed whether RhoE-mediated inhibition of eIF4E function contributed to its anti-proliferative effects. First, because RhoE can inhibit Ras-mediated transformation in focus-formation assays in NIH3T3 cells (11) we investigated whether RhoE could also inhibit transformation induced by eIF4E. Because transfection of eIF4E alone results in low levels of transformation (data not shown), we used eIF4E in combination with very low, suboptimal levels of RasV12. These low levels of RasV12 induced very little transformation, which was nevertheless suppressed by RhoE co-expression (Fig. 7A). Interestingly, addition of eIF4E to suboptimal levels of RasV12 resulted in a dramatic increase in cell transformation, which could not be suppressed by RhoE co-transfection (Fig. 7A). This indicates that eIF4E, as opposed to other oncogenic signals including RasV12 and RafCAAX (11), is able to overcome the inhibition of cell transformation induced by RhoE. To extend these observations, we used clonogenic assays, in which we transfected NIH3T3 cells with RhoE alone or in combination with different constructs together with a plasmid conferring antibiotic resistance to select transfectants. Transfection of RhoE alone resulted in a low number of viable cell colonies, in agreement with its anti-proliferative ability (Fig. 7B) whereas co-expression of eIF4E dramatically increased colony numbers. Interestingly, neither mTOR, RhoA14, nor RasV12 were able to induce a similar response (Fig. 7B), indicating that eIF4E is specifically able to overcome RhoE-induced cell cycle arrest. Taken together, these results indicate that eIF4E, as opposed to other mitogenic signals, can override RhoE-induced inhibition of cell cycle progression and transformation.
FIGURE 7.
Inhibition of eIF4E function contributes to RhoE-mediated cell cycle arrest. A, NIH3T3 cells were transfected with the indicated plasmids and maintained in 5% serum for 12–15 days, replacing the medium every 2 days. After 12–15 days, cells were stained with crystal violet and the number of foci counted. The mean ± S.D. values from three independent experiments, each conducted in duplicate, are shown in the graph, representing the number of transformed foci relative to RasV12 transfectants. B, NIH3T3 cells were transfected with an empty vector or the indicated plasmids together with a plasmid conferring hygromycin resistance, exposed to hygromycin (0.5 μg/ml) for 7–10 days, stained with crystal violet, and the number of colonies counted. The graph represents the mean values ± S.D. from three independent experiments, each conduced in duplicate, showing the total number of cell colonies.
DISCUSSION
In this report, we have investigated the mechanistic basis for our previous observations on the role of RhoE in cyclin D1 expression and cell cycle regulation. We had previously reported that RhoE inhibited cyclin D1 biosynthesis without affecting its transcription (11). In agreement with our data, RhoE has been shown to inhibit cyclin D1 expression at the post-transcriptional level in U87 glioblastoma cells, also blocking cell cycle progression in this system (12). Because its ability to prevent cyclin D1 expression is likely to be causally related to its anti-proliferative effects, we sought to analyze whether RhoE could interfere with signaling pathways regulating protein translation.
A major determinant of protein translation initiation in mammalian cells is the formation of the eIF4F initiation complex, which is highly dependent on eIF4E availability. We thus analyzed the status of the 4E-BP1/eIF4E signaling axis using the RhoE-inducible cells we had generated (11). Interestingly, 4E-BP1 phosphorylation was strongly suppressed in RhoE-expressing cells. 4E-BP1 is hierarchically phosphorylated at multiple residues. Phosphorylation of threonines 37/46, which are thought to be mTOR-dependent, seems to prime 4E-BP1 for subsequent phosphorylation events at serines 65 and 70, among other sites (22, 23). These phosphorylated residues are collectively responsible for the conformational change that renders 4E-BP1 unable to bind eIF4E thus allowing eIF4E to recruit eIF4G and initiate protein translation (34). RhoE reduced 4E-BP1 phosphorylation at both threonines 37/46 and serine 65 in response to multiple stimuli. Interestingly, RhoE did not affect S6K phosphorylation nor that of its substrate S6, despite both 4E-BP1 and S6K being well-established mTOR targets. Indeed, inhibition of mTOR activation by rapamycin effectively reduced both S6 and 4E-BP1 phosphorylation levels in control and RhoE-expressing cells. This suggests that RhoE does not inhibit mTOR activation. Consistent with this, RhoE did not inhibit growth factor-induced cell size increase, another well characterized mTOR-regulated response. These results are in agreement with reports showing that eIF4E inhibition specifically affects cell proliferation and transformation, but not cell growth (27).
Further evidence that RhoE does not affect mTOR function is provided by our observation that it does not alter mTOR interaction with Raptor. mTOR activity is regulated by binding to its regulatory proteins Raptor and Rictor to form mTORC1 and mTORC2, respectively (35). mTORC1 (mTOR/Raptor) is thought to be mainly responsible for “classical” mTOR functions, such as S6K and 4E-BP1 phosphorylation, and is rapamycin-sensitive (28, 29). In contrast, mTORC2 (mTOR/Rictor) has been linked to actin cytoskeletal regulation and Akt phosphorylation on Ser-473 (30, 31, 36), and is rapamycin-insensitive, although long-term rapamycin treatment can block the formation of de novo mTOR/Rictor complexes (37). Interestingly, serum stimulation of 3T3 cells induced a rapid and sustained dissociation of mTOR and Raptor, in agreement with the fact that Raptor interaction with mTOR decreases in stimulated cells, inversely correlating with mTOR activation (29). Accordingly, the timing of mTOR/Raptor dissociation coincided with the onset of S6K phosphorylation. However, these changes in mTOR/Raptor binding, together with those of mTOR/Rictor (data not shown), are not altered by RhoE expression. Taken together, our results indicate that RhoE does not inhibit 4E-BP1 phosphorylation by altering mTOR signaling. In fact, other kinases have been reported to phosphorylate 4E-BP1 on some of the same residues as mTOR. For instance, cyclin-dependent kinase 1 (cdk1) can phosphorylate 4E-BP1 during mitosis (38). Although RhoE expression prevents mitosis entry and would therefore block cdk1 activation, the timing of the 4E-BP1 phosphorylation that is inhibited by RhoE upon mitogenic stimulation of quiescent cells does not support a role for cdk1 in our system. ERK has also been shown to induce 4E-BP1 phosphorylation on Ser-65 in response to phorbol ester stimulation (39). However, it is unlikely that RhoE regulates 4E-BP1 phosphorylation by ERK, because we have previously shown that RhoE does not inhibit ERK activation in these cells (11). It is possible that RhoE reduces the accessibility of 4E-BP1 to any of its upstream kinases, or titrates out an essential accessory protein participating in 4E-BP1 phosphorylation, perhaps through the regulation of the multiprotein eIF3-containing complex that has been shown to include many translation initiation regulators such as mTOR, Raptor, S6K, and 4E-BP1, among others (40). However, we have not been able to detect an interaction of RhoE with eIF4E, 4E-BP1, mTOR, or Raptor (data not shown). Alternatively, RhoE could regulate a still unidentified 4E-BP1-kinase or the dephosphorylation of 4E-BP1.
Our results indicate that RhoE effectively blocks the release of the translational repressor 4E-BP1 from eIF4E and consequently reduces cap-dependent translation rates, and this is likely to be responsible for inhibiting cyclin D1 biosynthesis (16). c-Myc is another major eIF4E target relevant to cell proliferation and transformation (15, 41). c-Myc can also be translated through an internal IRES (internal ribosome entry site) within its 5′-UTR (42), although this predominantly occurs under certain circumstances such as during mitosis or in apoptotic cells (43, 44), and is mostly associated with Myc2 isoform (15). Interestingly, RhoE expression was able to reduce c-Myc protein, but not mRNA, levels and its transcriptional activity upon mitogenic stimulation. Taken together, these results point to the modulation of 4E-BP1/eIF4E signaling as a major contributor to RhoE-mediated cell cycle arrest, because inhibiting eIF4E activation would attenuate both cyclin D1 and c-Myc expression. 4E-BP1/eIF4E regulation appears to be crucial for RhoE to promote cell cycle arrest, because eIF4E overexpression can rescue both Ras-driven transformation and clonogenic viability in RhoE-expressing cells. These results are specific to eIF4E, because expression of other growth-promoting agents including mTOR, RhoA-V14, or Ras-V12 does not induce similar effects in these same assays.
In summary, our results indicate that RhoE-mediated inhibition of the 4E-BP1/eIF4E axis is central to its effects on cell cycle progression and transformation, through the inhibition of eIF4E target genes including cyclin D1 and c-Myc. In this context, because eIF4E is overexpressed in many human tumors and RhoE expression has been found to be both up-regulated and down-regulated depending on the tumor type (45, 46), it will be interesting to investigate whether there is a correlation between RhoE and eIF4E expression in tumors and to assess the impact of RhoE as a modulator of the oncogenic function of eIF4E. These studies will help us to define more precisely the functions of RhoE in cancer cell biology.
Acknowledgments
We thank Julian Downward, Giles Cory, Mark Coldwell, Godfrey Grech, Ivan Gout, and Bernhard Lüscher for plasmids. We are also grateful to David M. Sabatini for the gift of Raptor and Rictor antibodies, Dos Sarbassov for technical advice on mTOR immunoprecipitations and mTORC analysis and Antònia Obrador for helpful advice on real time PCR analysis.
This work was supported by an AICR Project Grant (to A. J. R. and P. V.), a Ramón y Cajal fellowship from the Spanish “Ministerio de Educación y Ciencia” (to P. V.), and funding from the “Junta de Balears-AECC” (to P. V.).
- ERK
- extracellular signal-regulated kinase
- UTR
- untranslated region
- 4E-BP
- 4E-binding protein
- DMEM
- Dulbecco's modified Eagle's medium
- FCS
- fetal calf serum
- PDGF
- platelet-derived growth factor
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- TPA
- 12-O-tetradecanoylphorbol-13-acetate
- mTOR
- mammalian target of rapamycin.
REFERENCES
- 1.Chardin P. (2006) Nat. Rev. Mol. Cell Biol. 7, 54–62 [DOI] [PubMed] [Google Scholar]
- 2.Guasch R. M., Scambler P., Jones G. E., Ridley A. J. (1998) Mol. Cell. Biol. 18, 4761–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nobes C. D., Lauritzen I., Mattei M. G., Paris S., Hall A., Chardin P. (1998) J. Cell Biol. 141, 187–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chardin P. (2003) Curr. Biol. 13, R702–704 [DOI] [PubMed] [Google Scholar]
- 5.Riento K., Villalonga P., Garg R., Ridley A. (2005) Biochem. Soc. Trans. 33, 649–651 [DOI] [PubMed] [Google Scholar]
- 6.Riento K., Guasch R. M., Garg R., Jin B., Ridley A. J. (2003) Mol. Cell. Biol. 23, 4219–4229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wennerberg K., Forget M. A., Ellerbroek S. M., Arthur W. T., Burridge K., Settleman J., Der C. J., Hansen S. H. (2003) Curr. Biol. 13, 1106–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riento K., Totty N., Villalonga P., Garg R., Guasch R., Ridley A. J. (2005) EMBO J. 24, 1170–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ongusaha P. P., Kim H. G., Boswell S. A., Ridley A. J., Der C. J., Dotto G. P., Kim Y. B., Aaronson S. A., Lee S. W. (2006) Curr. Biol. 16, 2466–2472 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Villalonga P., Ridley A. J. (2006) Growth Factors 24, 159–164 [DOI] [PubMed] [Google Scholar]
- 11.Villalonga P., Guasch R. M., Riento K., Ridley A. J. (2004) Mol. Cell. Biol. 24, 7829–7840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poch E., Miñambres R., Mocholí E., Ivorra C., Pérez-Aragó A., Guerri C., Pérez-Roger I., Guasch R. M. (2007) Exp. Cell Res. 313, 719–731 [DOI] [PubMed] [Google Scholar]
- 13.Sonenberg N., Dever T. E. (2003) Curr. Opin. Struct. Biol. 13, 56–63 [DOI] [PubMed] [Google Scholar]
- 14.Koromilas A. E., Lazaris-Karatzas A., Sonenberg N. (1992) EMBO J. 11, 4153–4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carter P. S., Jarquin-Pardo M., De Benedetti A. (1999) Oncogene 18, 4326–4335 [DOI] [PubMed] [Google Scholar]
- 16.Rosenwald I. B., Lazaris-Karatzas A., Sonenberg N., Schmidt E. V. (1993) Mol. Cell. Biol. 13, 7358–7363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lazaris-Karatzas A., Sonenberg N. (1992) Mol. Cell. Biol. 12, 1234–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li B. D., Liu L., Dawson M., De Benedetti A. (1997) Cancer 79, 2385–2390 [PubMed] [Google Scholar]
- 19.Rosenwald I. B., Chen J. J., Wang S., Savas L., London I. M., Pullman J. (1999) Oncogene 18, 2507–2517 [DOI] [PubMed] [Google Scholar]
- 20.Bjornsti M. A., Houghton P. J. (2004) Cancer Cell 5, 519–523 [DOI] [PubMed] [Google Scholar]
- 21.Richter J. D., Sonenberg N. (2005) Nature 433, 477–480 [DOI] [PubMed] [Google Scholar]
- 22.Gingras A. C., Gygi S. P., Raught B., Polakiewicz R. D., Abraham R. T., Hoekstra M. F., Aebersold R., Sonenberg N. (1999) Genes Dev. 13, 1422–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gingras A. C., Raught B., Gygi S. P., Niedzwiecka A., Miron M., Burley S. K., Polakiewicz R. D., Wyslouch-Cieszynska A., Aebersold R., Sonenberg N. (2001) Genes Dev. 15, 2852–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 25.Stoneley M., Subkhankulova T., Le Quesne J. P., Coldwell M. J., Jopling C. L., Belsham G. J., Willis A. E. (2000) Nucleic Acids Res. 28, 687–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfaffl M. W. (2001) Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lynch M., Fitzgerald C., Johnston K. A., Wang S., Schmidt E. V. (2004) J. Biol. Chem. 279, 3327–3339 [DOI] [PubMed] [Google Scholar]
- 28.Hara K., Maruki Y., Long X., Yoshino K., Oshiro N., Hidayat S., Tokunaga C., Avruch J., Yonezawa K. (2002) Cell 110, 177–189 [DOI] [PubMed] [Google Scholar]
- 29.Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2002) Cell 110, 163–175 [DOI] [PubMed] [Google Scholar]
- 30.Sarbassov D. D., Ali S. M., Kim D. H., Guertin D. A., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2004) Curr. Biol. 14, 1296–1302 [DOI] [PubMed] [Google Scholar]
- 31.Jacinto E., Loewith R., Schmidt A., Lin S., Rüegg M. A., Hall A., Hall M. N. (2004) Nat. Cell Biol. 6, 1122–1128 [DOI] [PubMed] [Google Scholar]
- 32.De Benedetti A., Graff J. R. (2004) Oncogene 23, 3189–3199 [DOI] [PubMed] [Google Scholar]
- 33.Vervoorts J., Lüscher-Firzlaff J. M., Rottmann S., Lilischkis R., Walsemann G., Dohmann K., Austen M., Lüscher B. (2003) EMBO Rep. 4, 484–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mothe-Satney I., Yang D., Fadden P., Haystead T. A., Lawrence J. C., Jr. (2000) Mol. Cell. Biol. 20, 3558–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarbassov D. D., Ali S. M., Sabatini D. M. (2005) Curr. Opin. Cell Biol. 17, 596–603 [DOI] [PubMed] [Google Scholar]
- 36.Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 37.Sarbassov D. D., Ali S. M., Sengupta S., Sheen J. H., Hsu P. P., Bagley A. F., Markhard A. L., Sabatini D. M. (2006) Mol. Cell 22, 159–168 [DOI] [PubMed] [Google Scholar]
- 38.Heesom K. J., Gampel A., Mellor H., Denton R. M. (2001) Curr. Biol. 11, 1374–1379 [DOI] [PubMed] [Google Scholar]
- 39.Herbert T. P., Tee A. R., Proud C. G. (2002) J. Biol. Chem. 277, 11591–11596 [DOI] [PubMed] [Google Scholar]
- 40.Holz M. K., Ballif B. A., Gygi S. P., Blenis J. (2005) Cell 123, 569–580 [DOI] [PubMed] [Google Scholar]
- 41.Lin C. J., Cencic R., Mills J. R., Robert F., Pelletier J. (2008) Cancer Res. 68, 5326–5334 [DOI] [PubMed] [Google Scholar]
- 42.Stoneley M., Paulin F. E., Le Quesne J. P., Chappell S. A., Willis A. E. (1998) Oncogene 16, 423–428 [DOI] [PubMed] [Google Scholar]
- 43.Pyronnet S., Pradayrol L., Sonenberg N. (2000) Mol. Cell 5, 607–616 [DOI] [PubMed] [Google Scholar]
- 44.Stoneley M., Chappell S. A., Jopling C. L., Dickens M., MacFarlane M., Willis A. E. (2000) Mol. Cell. Biol. 20, 1162–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang C., Zhou F., Li N., Shi S., Feng X., Chen Z., Hang J., Qiu B., Li B., Chang S., Wan J., Shao K., Xing X., Tan X., Wang Z., Xiong M., He J. (2007) Ann. Surg. Oncol. 14, 2628–2635 [DOI] [PubMed] [Google Scholar]
- 46.Bektic J., Pfeil K., Berger A. P., Ramoner R., Pelzer A., Schäfer G., Kofler K., Bartsch G., Klocker H. (2005) Prostate 64, 332–340 [DOI] [PubMed] [Google Scholar]