Abstract
Mutational activation of the phosphatidylinositol 3-kinase (PI3K) pathway occurs in a wide variety of tumors, whereas activating Wnt pathway mutants are predominantly found in colon cancer. Because GSK3 is a key component of both pathways, it is widely assumed that active PI3K signaling feeds positively into the Wnt pathway by protein kinase B (PKB)-mediatefd inhibition of GSK3. In addition, PKB has been proposed to modulate the canonical Wnt signaling through direct stabilization and nuclear localization of β-catenin. Here, we show that compartmentalization by Axin of GSK3 prohibits cross-talk between the PI3K and Wnt pathways and that Wnt-mediated transcriptional activity is not modulated by activation of the PI3K/PKB pathway.
Introduction
Developmental signaling cascades typically transduce signals from the cell surface onto regulatory sequences of nuclear target genes. In the simplest model, signals transduced through different pathways are integrated at the level of the regulatory elements of individual genes. Such regulatory elements may be viewed as assemblies of cis-acting response elements that are tailored to create the unique expression pattern for each gene. However, numerous studies propose that signaling pathways may interact at any stage between the plasma membrane and the nucleus. One mechanism by which such cross-talk may occur involves the sharing of a common component between two different pathways. It is often tacitly assumed that such shared components are equally accessible to all pertinent pathways.
Glycogen synthase kinase 3-α and -β, collectively termed GSK3, are constitutively active serine/threonine kinases (1). GSK3 features in two signaling pathways that are of particular importance in cancer. GSK3 is a downstream component of the phosphoinositide 3-OH kinase (PI3K)2 pathway (2, 3). Growth signals, activated Ras proteins, or loss of the phosphatase and tensin homolog (PTEN) all activate PI3K, which in turn phosphorylates and activates protein kinase B (PKB) (3). Active PKB phosphorylates GSK3α on Ser-21 (4) and GSK3β on Ser-9 (5), in both cases leading to inhibition of the constitutive kinase activity. GSK3 is also a component of the Wnt cascade (6). GSK3 is bound by Axin (Axis inhibition protein) (7) and phosphorylates β-catenin, thus targeting it for ubiquitination and degradation by the proteasome. Wnt signaling is assumed to block GSK3-mediated β-catenin phosphorylation, leading to the accumulation and nuclear translocation of β-catenin (6). It remains unclear how the Wnt cascade controls the activity of the dedicated Axin1-bound GSK3 pool. A recent genetic experiment has demonstrated that removal of the inhibitory serines from the two GSK3 proteins has no effect on Wnt signaling (8).
Although an early study proposed that the two pathways do not cross-talk at the level of GSK3 (9), a multitude of papers have since appeared that are based on the premise that a single pool of GSK3 is targeted by both signals (see supplemental Table S1). Moreover, direct stabilization of β-catenin by the PI3K/PKB pathway has been claimed in several additional studies (see supplemental Table S1). Mutational activation of the Wnt pathway through loss of adenomatous polyposis coli protein (APC), Axin1/2, or through point mutations in β-catenin occurs in a limited diversity of cancers, most notably of the intestine (6), and it is characterized by stabilized β-catenin and constitutive transcriptional activity of β-catenin-TCF complexes in the nucleus. This can be readily read out by the constitutive activity of β-catenin/TCF reporters such as pTOPFlash (10). Mutational activation of the PI3K pathway occurs in a wide variety of tumors through mutational activation of any of the Ras genes, v-raf murine sarcoma viral oncogene homolog B1 (BRAF), PI3K, or loss of PTEN (3). If GSK3 would indeed represent a focal point of cross-talk between the two pathways, β-catenin/TCF-driven transcription would be activated in tumors harboring PI3K-activating mutations. This has major implications for our thinking on the molecular pathogenesis of cancer.
EXPERIMENTAL PROCEDURES
Q Descendants Migration Count in Caenorhabditis elegans
The final positions of the Q descendants was scored using a mec-7::gfp (muIs32) reporter transgene (11). All assays were performed at 20 °C. The daf-18(ok480) allele (provided by the C. elegans gene knock-out project at the Oklahoma Medical Research Foundation) was detected by PCR using the following primers: daf-18int-in (CAACGCAGTACATCTCGAAGCC) and daf-18int-out (CCAGCTGATACCGATGATGTTGAT).
Cells and Cell Culture
HEK293T cells were maintained in RPMI 1640 medium (Invitrogen) supplemented with 5% fetal calf serum. All cancer cell lines used in this study are listed in Table 1. The prostate cancer cell lines LNCaP and PC3 were the kind gifts of Dr. J. Trapman and were cultured in RPMI 1640 medium with 10% fetal calf serum. The breast cancer cell lines EVSA-T and SK-BR-5/7 were the kind gifts of Dr. N. DeVleeschouwer (Institute Jules Brodet, Brussels, Belgium), Dr. H. S. Smith (California Pacific Medical Center, San Francisco), and Dr. E. Stockert (Sloan-Kettering Institute for Cancer Research, New York), respectively. The SUM series cell lines were generated in the Ethier laboratory (available on line). The cell line OCUB-F was obtained from Riken Gene Bank (Tsukuba, Japan). All other cell lines were obtained from American Type Culture Collection (Manassas, VA). All cell lines were grown according to the suppliers' recommendations and are unique and monoclonal as shown by extensive analysis of nearly 150 polymorphic microsatellite markers. Wnt3A-producing L-cells were the kind gift from R. Nusse, and Wnt3A/control condition media were generated by using the standard protocol provided by ATCC.
TABLE 1.
Constitutive activation of PI3K/PKB pathway does not activate Wnt target transcription
None of the tested cell lines with an active PI3K/PKB and Ras pathway displayed constitutive Wnt activity, except for DU4475 that harbors a mutated APC protein. The TCF/β-catenin transcription activity in all cancer cell lines was tested with the standard pTOPFlash assay. In the case when 20 mm LiCl responses were tested, LiCl was added 16 h after transfection, and the cells were lysed 24 h later. (See also supplemental Table S2.) ND means not done.
| Cell lines | Affected gene | Constitutive TCF/β-catenin signaling activity | Activation after LiCl treatment |
|---|---|---|---|
| Breast cell lines (13) | |||
| BT20 | PIK3CA | −(12) | ND |
| BT549 | PTEN | −(12) | + |
| CAMA-1 | PTEN | −(12) | ND |
| EVSA-T | PTEN | −(12) | ND |
| Hs578T | HRAS | −(12) | ND |
| MCF-7 | PIK3CA | −(12) | ND |
| MDA-MB-144VI | KRAS | −(12) | ND |
| MDA-MB-231 | BRAF, KRAS | −(12) | ND |
| MDA-MB-435s | BRAF | −(12) | ND |
| MDA-MB-453 | PTEN, PIK3CA | −(12) | ND |
| OCUB-F | PIK3CA | −(12) | ND |
| SK-BR-5 | PIK3CA | −(12) | ND |
| T47D | PIK3CA | −(12) | ND |
| DU4475 | BRAF, APC | +(12) | ND |
| BT483 | PIK3CA | − | + |
| HCC1937 | PTEN | − | − |
| MDA-MB-361 | PIK3CA | − | − |
| MDA-MB-415 | PTEN | − | + |
| MDA-MB-468 | PTEN | − | + |
| SK-BR-7 | KRAS, NRAS | − | + |
| SUM159-PT | PIK3CA, HRAS | − | + |
| SUM185PE | PI3KCA | − | + |
| UACC893 | PI3KCA | − | − |
| ZR-75-1 | PTEN | − | − |
| Prostate cancer cell lines (14) | |||
| LNCaP | PTEN | − | + |
| PC3 | PTEN | − | + |
Reporter Gene Assays
TCF reporter constructs were both modified to contain 10 optimized and 10 mutated TCF-binding sites as described previously. cDNA of Dkk1 (Dickkopf 1) (a kind gift from C. Niehrs), mutated β-catenin (S33A) (kind gift from K. Kinzler), and Wnt1 and Wnt3a (both kind gifts from R. Nusse) were subjected to PCR amplification and then cloned into pcDNA4TO. HCC1937, MDA-MB-361, MDA-MB-468, SUM159, SUM185, UACC893, and LNCaP were transfected with FuGENE HD transfection reagent (Roche Applied Science); BT483, BT549, MDA-MB-415, SK-BR-7, PC3, and 293T were transfected with polyethyleneimine (Polysciences); and ZR-75-1 was transfected with electroporation. The amount of DNA in each transfection was kept constant (250 ng/well in 24-well plate for FuGENE HD or polyethyleneimine transfection and 1.1 μg/well in 6-well plates for electroporation) by the addition of an appropriate amount of empty expression vector, pCDNA4TO, and the ratio of reporter plasmid (TOP10 or FOP10) with internal control plasmid (pCMV-Renilla) was kept constant at 10:1. 16 h after TOPFlash transfection, the cells were treated with condition medium or inhibitors (wortmannin and Akt inhibitor IV and Akt inhibitor VIII were both from Sigma), and the measurements were performed 12–16 h later. All experiments were performed in duplicate or triplicate and repeated at least twice.
Protein Analysis
The expression construct for human FLAG-tagged Axin1 was produced by PCR amplification of the coding region using the IMAGE clone ID 5809104 as a cDNA template, and this PCR product was cloned into pCDNA3. HEK293T cells were cultured to 80% confluence and starved at least 12 h before treatment with 10 mg/liter insulin (Invitrogen) for 10 min, 30 min, 1 h, and 4 h or with 200 nm wortmannin for 30 min, 1 h, and 4 h except control (0 min). Cells were washed with cold phosphate-buffered saline and lysed in cold lysis buffer containing 300 mm NaCl, 50 mm Tris (pH 7.5), 5 mm EDTA, 0.1% Nonidet P-40, 0.1 mm phenylmethylsulfonyl fluoride, 0.5 mm dithiothreitol, protease inhibitor mixture tablets (EDTA-free) (Roche Applied Science), and phosphatase inhibitor mixtures 1 and 2 (Sigma). A cycle of freeze-thawing was used for lysis. After clarification by centrifugation (14,000 rpm for 30 min at 4 °C), equal amounts of the total protein from each sample were incubated with anti-Axin1 antibody or mouse IgG immobilized on Protein G PLUS-agarose beads (Santa Cruz Biotechnology) at 4 °C overnight. After washing the immune complexes with cold lysis buffer six times, they were resuspended in 2× SDS sample buffer. Immunoblottings were done by reprobing the same blot after stripping (Pierce) using anti-phospho-GSK3β (S9) (Cell Signaling), anti-phospho-PKB (Ser-473) (Cell Signaling), anti-GSK3β (Cell Signaling), anti-PKB (Cell Signaling), and anti-Axin antibodies.
Generation of Axin-specific Antibodies
FLAG-tagged mouse Axin2/Conductin was a gift from Jürgen Behrens, and FLAG-tagged human Axin2 was cloned from IMAGE clone ID4053244 to pCDNA4TO vector by PCR. Antibodies directed against Axin were produced by immunizing BALB/c mice with the N-terminal part (amino acid 3–326) of mouse Axin1. Hybridomas were generated using standard procedures.
RESULTS AND DISCUSSION
When we screened a large panel of breast cancer cell lines for constitutive β-catenin/TCF reporter (pTOPFlash) activity, the hallmark of active Wnt signaling, we found only a single positive cell line to be responsive to Wnt (12). This cell line, DU4475, turned out to carry a mutation in the Wnt pathway tumor suppressor APC, a mutation typically found almost exclusively in colon cancers. We have since determined the mutational status of the three RAS genes (KRAS, HRAS, and NRAS), the BRAF kinase gene, the PTEN gene, and PI3K (PIK3CA) in a much larger panel of breast cancer cell lines (13). We conducted Tcf reporter assays for the entire breast cancer cell line panel. Moreover, we subjected two well defined prostate cancer cell lines, both of which harbor inactivating PTEN mutations (14). No TOPFlash activity was observed in any of the cell lines. LiCl is a well established inhibitor of GSK3 kinase activity and as such represents a pharmacological activator of the Wnt cascade at the level of GSK3 (15). Nine of 13 cell lines subjected to LiCl treatment responded by a potent activation of the TOPFlash reporter, indicating the presence of an intact intracellular Wnt pathway. Table 1 displays the identities of the mutations present in the individual cell lines, the presence of a constitutively active or inactive Wnt pathway as measured in the TOPFlash assay, as well as the inducibility of the Wnt pathway by LiCl which was tested in a subset of the cell lines. The underlying TOPFlash values are given in Table S2.
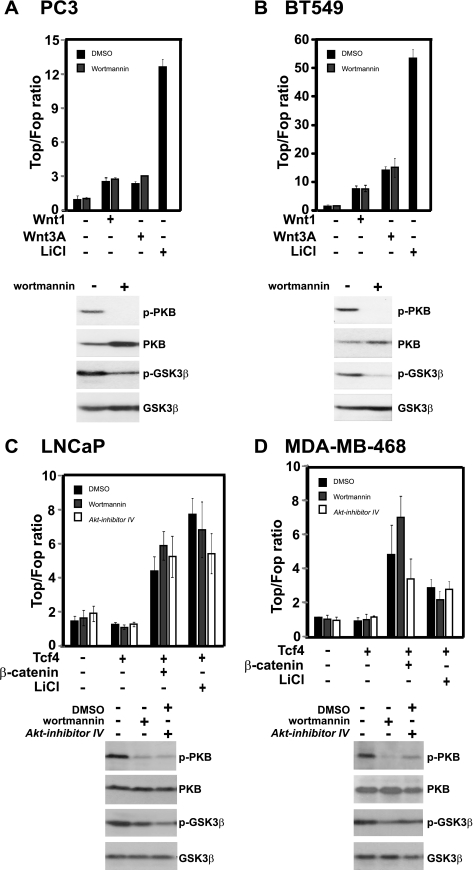
We next investigated the presence of a complete and functional Wnt pathway in the PTEN-mutant prostate cancer cell line PC3 and in the PTEN-mutant breast cancer cell line BT549 by transfection of Wnt1 or Wnt3A. Although modest responses were observed in the PC3 cells, the BT549 responded vigorously to the Wnt proteins (Fig. 1, A and B, top panels). We concluded that these two cell lines harbored fully functional Wnt pathways, which were not affected by the mutational status of the PI3K pathway but could readily be activated by Wnt proteins. To examine the effect of inhibition of the PI3K pathway on Wnt-induced transcriptional activity, we applied the pharmacological PI3K inhibitor wortmannin. Western blot analysis of phospho-PKB and phospho-GSK3β confirmed efficient inhibition of the PI3K pathway in these cell lines in response to wortmannin treatment (Fig. 1, A and B, bottom panels). TOPFlash reporter assays revealed no effect of wortmannin on Non-Wnt-stimulated cells (Fig. 1, A and B, top panels) or on Wnt1 or Wnt3A-stimulated cells (Fig. 1, A and B, top panels). In an independent experiment, we applied the PKB inhibitors AKT inhibitor IV and VIII. Although these inhibitors readily blocked phosphorylation of PKB and GSK3β (supplemental Fig. 1), no statistically significant differences were noted on LiCl-stimulated TOPFlash reporter activity in BT549 cells (supplemental Fig. 2). We extended these results by examining the effect of PI3K pathway inhibition on Wnt-induced transcription in two other prostate and breast cancer cell lines, LNCaP and MDA-MB-468, respectively. Both cell lines were transcriptionally induced in response to β-catenin/TCF4 expression or LiCl treatment (Fig. 1, C and D, top panels). However, addition of PI3K pathway inhibitors wortmannin and AKT inhibitor IV, at concentrations that efficiently blocked PKB and GSK3 phosphorylation (Fig. 1, C and D, bottom panels), had no significant effect on LiCl or TCF4/β-catenin induced transcriptional activation. Comparison of differences between TOPFlash activities was performed using two-tailed Student's t test (where equal variance between groups was assumed).
FIGURE 1.
Wnt ligands, β-catenin, and LiCl induce TCF/β-catenin-dependent transcription in PC3 (A), BT549 (B), LNCaP (C), and MDA-MB-468, (D) cell lines in a PI3K/Akt-independent manner. Cells were plated at 50–60% confluency in 24-well plates and transfected with pTOPFlash reporters, with or without Wnt1, Wnt3A, β-catenin, or TCF4 plasmids as indicated. 20 mm LiCl, 100 nm (BT509) or 250 nm (PC3, LNCaP, and MDA-MB-468) PI3K-inhibitor wortmannin, and 50 nm Akt inhibitor IV were added as indicated 16 h following transfection, and measurements were done after 24 h. The TOPFlash data (top panel) are representative of at least two independent experiments. Error bars represent ± S.D. Effective inhibition of the PI3K/PKB pathway in response to wortmannin and Akt inhibitor IV treatment is shown by Western blotting as indicated in the panels below the pertinent bar graphs.
We further confirmed our observations in an in vivo system, the nematode C. elegans. In the nematode C. elegans, canonical Wnt/β-catenin signaling regulates the left-right asymmetric migration of the Q descendants (16, 17). On the left side, egl-20 triggers the expression of the Hox gene mab-5, which directs the migration of the Q descendants (QL.d) toward the posterior (18). On the right side, mab-5 is not expressed, and as a result the QR descendants (QR.d) migrate in the default anterior direction. Mutations that disrupt Wnt/β-catenin signaling, such as mutations in mig-1/Fz, bar-1/β-catenin, or pop-1/Tcf, lead to loss of mab-5 expression in QL and anterior migration of the QL.d (17, 19, 20). Conversely, mutation of negative regulators of Wnt/β-catenin signaling, such as pry-1/Axin, leads to ectopic expression of mab-5 in QR and posterior migration of the QR.d (19, 21). The final positions of the Q descendants therefore provide a sensitive measure of Wnt/β-catenin pathway activity. We investigated whether mutation of the C. elegans PTEN ortholog daf-18 (22) affects Q descendant migration. The deletion allele daf-18(ok480) is expected to represent a strong loss of function or the null phenotype. We found that daf-18(ok480) has no effect on either QL.d or QR.d migration compared with wild type (Table 2; in each case, n = 50), indicating that loss of PTEN does not affect Wnt/β-catenin signaling in C. elegans.
TABLE 2.
Knock-out of daf-18 in C. elegans has no effect on the Wnt/β-catenin pathway
The mutation of the nematode C. elegans PTEN ortholog daf-18 has no effect on Q descendant migration. All assays were performed at 20 °C, and the final positions of the Q descendants were scored using a mec-7::gfp (muls32) reporter transgene.
| Genotype | Effect on Q descendant migration |
|---|---|
| Control (muls32(mec-7::gfp)) | 100% wild type, n = 50 |
| Daf-18(ok480); muls32,2 | 100% wild type, n = 50 |
| Daf-18(ok480); muls32,13 | 100% wild type, n = 50 |
| Daf-18(ok480); muls32,16 | 100% wild type, n = 50 |
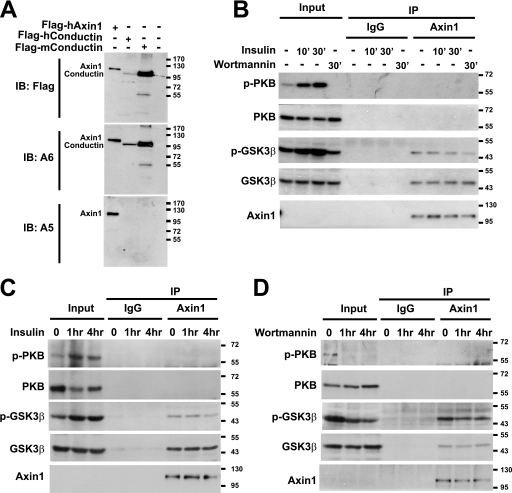
We then turned to a physiological model for insulin/PI3K signaling. HEK293T cells respond to insulin by activating PI3K, which phosphorylates and activates PKB/AKT (2), leading to inhibitory phosphorylation of GSK3β on Ser-9. As shown in Fig. 2, B and C, addition of insulin leads to a rapid phosphorylation of total cellular PKB and GSK3β (input, compare 1st 3 lanes). Inhibition of PI3K abolished PKB and GSK3β phosphorylation at 30 min (Fig. 2B, input, compare 1st with 4th lanes) and 1 and 4 h post-wortmannin treatment (Fig. 2D, compare lanes 1–3).
FIGURE 2.
PI3K/PKB pathway activity does not modulate Wnt/β-catenin signaling through GSK3β. A, mouse monoclonal A5 antibody is Axin1-specific, whereas A6 antibody recognizes both Axin1 and Conductin. 0.5 μg of DNA/well were transfected into HEK293T cells in a 6-well plate. All samples were lysed in 2× SDS sample buffer after 24 h, and Western blot analysis was performed using anti-FLAG, A6, and A5 antibodies, respectively. B–D, activation of PI3K/PKB pathway does not influence the phosphorylated or total amount of GSK3β protein that is bound to AXIN. HEK293T cells were cultured in 5% fetal calf serum/RPMI 1640 medium to 80% confluency and then starved in serum-free medium for at least 12 h before treatment with 10 mg/liter insulin for 10 and 30 min (B) and 1 and 4 h (C) or 200 nm wortmannin for 30 min (B) and 1 and 4 h (D). Cells were lysed and immunoprecipitated (IP) with either control mouse IgG or anti-Axin antibody. Resulting immunocomplexes were subjected to immunoblotting (IB) using the indicated antibodies.
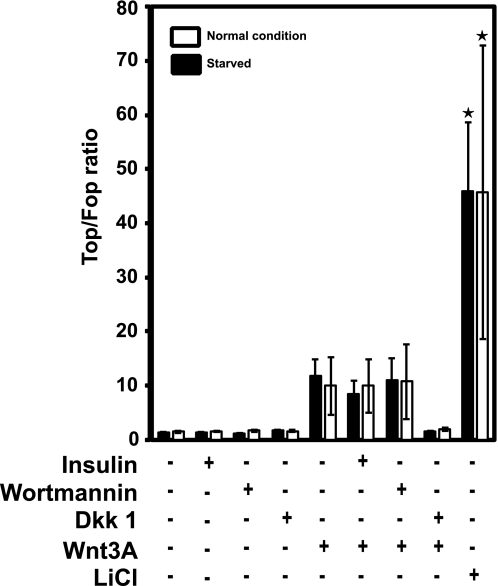
To study a potential effect of PI3K/PKB pathway activation on Wnt/β-catenin target gene expression in HEK293T cells, we used the pTOPFlash Wnt reporter assay. As shown in Fig. 3, Wnt-mediated transcriptional activity is not influenced by PI3K/PKB pathway activation but could readily be blocked by the soluble Wnt inhibitor Dkk1 in HEK293T cells.
FIGURE 3.
Tcf/β-catenin transcriptional activity is not modulated by activation of the PI3K/PKB pathway. HEK293T cells were plated at 30–40% confluency in 24-well plates and transfected with pTOPFlash reporters, Wnt3a and Dkk1 (Wnt inhibitor) plasmids as indicated. 16 h after transfection, half of the cells were starved with serum-free medium for at least 12 h before treatment with 20 mm LiCl, 10 mg/liter insulin, or 200 nm wortmannin as indicated. Measurement was done another 16–24 h later. Incubation periods of the nonstarved cells were the same for the starved cells. Data are from three independent duplicate experiments (except ★, n = 2). Error bars indicate mean ± S.D.
Axin is believed to be the central scaffolding component in the cytoplasmic destruction complex of the Wnt cascade, as it carries independent interaction domains for the Wnt pathway components APC, GSK3, CK1, Dsh, and β-catenin (6, 7). Moreover, it is by far the least abundant core component of the Wnt pathway (23). The pool of GSK3β that is dedicated to Wnt signaling is stably bound to Axin (6, 7). To be able to investigate whether the endogenous Axin-bound pool of GSK3β was targeted by insulin-activated PKB, we generated monoclonal antibodies against Axin (Fig. 2A). Fig. 2B (9th to 12th lanes) demonstrates the results obtained with immunoprecipitations of endogenous AXIN from the same lysates as used in input lanes. Endogenous GSK3β readily co-immunoprecipitated with AXIN (Fig. 2B, 9th to 12th lanes. This AXIN-GSK3 complex was robust, as it even resisted 3% Nonidet P-40/Triton X-100 (data not shown). From multiple experiments, we estimate that 3–5% of total cellular GSK3β resides within the AXIN complex.
When the blots were probed with anti-phospho-GSK3β antibodies, the phosphorylation level was found to remain unchanged upon both insulin signaling (Fig. 2, B and C) and treatment with wortmannin (Fig. 2, B and D). Furthermore, (phospho-) PKB was not co-precipitated. We conclude that, although insulin rapidly activated the PI3K pathway as evidenced by sequential phosphorylation of PKB and GSK3β, the small AXIN-bound pool of GSK3β is protected from activated PKB. Thus, activation or inhibition of PI3K/PKB does not target the Axin-bound GSK3β pool. These results were further confirmed in PC3 prostate cancer cells, which have previously been used to implicate a role for PI3K signaling in the Wnt pathway (supplemental Fig. 3) (24).
This result supports the hypothesis that different pools of GSK3 kinases exist in cells, participating separately in the PI3K/PKB pathway or the Wnt/β-catenin pathway as originally proposed by Frame and Cohen (25). The existence of Axin-GSK3 complexes appears to serve two functions. First, the Axin complex allows GSK3 to target β-catenin specifically by providing docking sites for both proteins (6). Second, when GSK3 is complexed to Axin, it is shielded from activated PKB. This molecular arrangement allows Wnt and PI3K inputs to have independent effects on the biological outputs of the cells that receive these signals.
These findings also help to clarify why mutations in canonical Wnt components (i.e. APC, Axin/conductin, or β-catenin) are mutually exclusive in colorectal cancer, although individual cases of colorectal cancer frequently combine an activating Wnt pathway mutation with a mutation in a component of the PI3K pathway (26). Similar observations are made for ovarian endometrioid adenocarcinoma, for example (27). If activating PI3K pathway mutations would activate the Wnt pathway, such mutations should not occur in combination with activating Wnt pathway mutations in cancer.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and Tables S1 and S2.
- PI3K
- phosphatidylinositol 3-kinase
- APC
- adenomatous polyposis coli protein
- PKB
- protein kinase B
- BRAF
- v-raf murine sarcoma viral oncogene homolog B1
- PTEN
- phosphatase and tensin homolog
- TCF
- T cell factor.
REFERENCES
- 1.Kockeritz L., Doble B., Patel S., Woodgett J. R. (2006) Curr. Drug Targets 7, 1377–1388 [DOI] [PubMed] [Google Scholar]
- 2.Cully M., You H., Levine A. J., Mak T. W. (2006) Nat. Rev. Cancer 6, 184–192 [DOI] [PubMed] [Google Scholar]
- 3.Cross D. A., Alessi D. R., Cohen P., Andjelkovich M., Hemmings B. A. (1995) Nature 378, 785–789 [DOI] [PubMed] [Google Scholar]
- 4.Sutherland C., Cohen P. (1994) FEBS Lett. 338, 37–42 [DOI] [PubMed] [Google Scholar]
- 5.Sutherland C., Leighton I. A., Cohen P. (1993) Biochem. J. 296, 15–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clevers H. (2006) Cell 127, 469–480 [DOI] [PubMed] [Google Scholar]
- 7.Hart M. J., de los Santos R., Albert I. N., Rubinfeld B., Polakis P. (1998) Curr. Biol. 8, 573–581 [DOI] [PubMed] [Google Scholar]
- 8.McManus E. J., Sakamoto K., Armit L. J., Ronaldson L., Shpiro N., Marquez R., Alessi D. R. (2005) EMBO J. 24, 1571–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding V. W., Chen R. H., McCormick F. (2000) J. Biol. Chem. 275, 32475–32481 [DOI] [PubMed] [Google Scholar]
- 10.Korinek V., Barker N., Morin P. J., van Wichen D., de Weger R., Kinzler K. W., Vogelstein B., Clevers H. (1997) Science 275, 1784–1787 [DOI] [PubMed] [Google Scholar]
- 11.Ch'ng Q., Williams L., Lie Y. S., Sym M., Whangbo J., Kenyon C. (2003) Genetics 164, 1355–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van de Wetering M., Barker N., Harkes I. C., van der Heyden M., Dijk N. J., Hollestelle A., Klijn J. G., Clevers H., Schutte M. (2001) Cancer Res. 61, 278–284 [PubMed] [Google Scholar]
- 13.Hollestelle A., Elstrodt F., Nagel J. H., Kallemeijn W. W., Schutte M. (2007) Mol. Cancer Res. 5, 195–201 [DOI] [PubMed] [Google Scholar]
- 14.Vlietstra R. J., van Alewijk D. C., Hermans K. G., van Steenbrugge G. J., Trapman J. (1998) Cancer Res. 58, 2720–2723 [PubMed] [Google Scholar]
- 15.Stambolic V., Ruel L., Woodgett J. R. (1996) Curr. Biol. 6, 1664–1668 [DOI] [PubMed] [Google Scholar]
- 16.Silhankova M., Korswagen H. C. (2007) Curr. Opin. Genet. Dev. 17, 320–325 [DOI] [PubMed] [Google Scholar]
- 17.Korswagen H. C. (2002) BioEssays 24, 801–810 [DOI] [PubMed] [Google Scholar]
- 18.Salser S. J., Kenyon C. (1992) Nature 355, 255–258 [DOI] [PubMed] [Google Scholar]
- 19.Maloof J. N., Whangbo J., Harris J. M., Jongeward G. D., Kenyon C. (1999) Development 126, 37–49 [DOI] [PubMed] [Google Scholar]
- 20.Harris J., Honigberg L., Robinson N., Kenyon C. (1996) Development 122, 3117–3131 [DOI] [PubMed] [Google Scholar]
- 21.Korswagen H. C., Coudreuse D. Y., Betist M. C., van de Water S., Zivkovic D., Clevers H. C. (2002) Genes Dev. 16, 1291–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogg S., Ruvkun G. (1998) Mol. Cell 2, 887–893 [DOI] [PubMed] [Google Scholar]
- 23.Lee E., Salic A., Krüger R., Heinrich R., Kirschner M. W. (2003) PLoS Biol. 1, E10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Persad S., Troussard A. A., McPhee T. R., Mulholland D. J., Dedhar S. (2001) J. Cell Biol. 153, 1161–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frame S., Cohen P. (2001) Biochem. J. 359, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sjöblom T., Jones S., Wood L. D., Parsons D. W., Lin J., Barber T. D., Mandelker D., Leary R. J., Ptak J., Silliman N., Szabo S., Buckhaults P., Farrell C., Meeh P., Markowitz S. D., Willis J., Dawson D., Willson J. K., Gazdar A. F., Hartigan J., Wu L., Liu C., Parmigiani G., Park B. H., Bachman K. E., Papadopoulos N., Vogelstein B., Kinzler K. W., Velculescu V. E. (2006) Science 314, 268–274 [DOI] [PubMed] [Google Scholar]
- 27.Wu R., Hendrix-Lucas N., Kuick R., Zhai Y., Schwartz D. R., Akyol A., Hanash S., Misek D. E., Katabuchi H., Williams B. O., Fearon E. R., Cho K. R. (2007) Cancer Cell 11, 321–333 [DOI] [PubMed] [Google Scholar]