Abstract
Cyclin E is a regulator of cyclin-dependent protein kinases (Cdks) and is involved in mediating the cell cycle transition from G1 to S phase. Here, we describe a novel function for cyclin E in the long term maintenance of checkpoint arrest in response to replication barriers. Exposure of cells to mitomycin C or UV irradiation, but not ionizing radiation, induces stabilization of cyclin E. Stabilization of cyclin E reduces the activity of Cdk2-cyclin A, resulting in a slowing of S phase progression and arrest. In addition, cyclin E is shown to be required for stabilization of Cdc6, which is required for activation of Chk1 and the replication checkpoint pathway. Furthermore, the stabilization of cyclin E in response to replication fork barriers depends on ATR, but not Nbs1 or Chk1. These results indicate that in addition to its well studied role in promoting cell cycle progression, cyclin E also has a role in regulating cell cycle arrest in response to DNA damage.
Introduction
Commitment to S phase and DNA replication is controlled by the cyclin-dependent protein kinase 2 (Cdk2)2 and its regulatory subunits cyclin E and cyclin A (1–3). Cyclin E and cyclin A have distinct roles in the initiation of DNA replication. Cyclin E accumulates in late G1 by the E2F-mediated gene transcription program, which in turn is activated by cyclin D-associated kinases via phosphorylation of the retinoblastoma protein. Upon entry into S phase, cyclin E is rapidly degraded by the ubiquitin-proteosome system by two different pathways employing distinct mechanisms. Cyclin E unbound to Cdk2 is targeted by the Cul3-based E3 ubiquitin ligase (4), whereas Cdk2-bound cyclin E is targeted by the SCFFbw7 ubiquitin ligase in a process that requires phosphorylation of cyclin E by both Cdk2 and GSK3β (5–12). A critical function of Cdk2-cyclin E is to promote replication licensing prior to initiation of S phase by phosphorylation of the prereplication complex (pre-RC) assembly factor Cdc6 (13, 14). This modification inhibits ubiquitylation and subsequent degradation of Cdc6 by the anaphase-promoting complex (APC)/cyclosome, thereby promoting pre-RC assembly. Interestingly, cyclin E also promotes pre-RC assembly in a Cdk2-independent fashion. Cyclin E interacts with the pre-RC complex members Cdt1 and Cdc6 on chromatin and facilitates loading of the minichromosome maintenance (MCM) complex (15). Cyclin A accumulates at the onset of S phase and is required for initiation of DNA replication in mammalian cells. In addition, Cdk2-cyclin A also prevents replicative reinitiation of the pre-RC via phosphorylation of Cdc6 (14, 16).
In normal replicating mammalian cells, cyclin E levels decline during S phase; however, in many human cancers cyclin E is overexpressed and deregulated as a function of the cell cycle (17–21), and this deregulation has been implicated as a causative factor in tumorigenesis (8, 22–24). Overexpression of cyclin E has been shown to induce both aneuploidy and polyploidy in mammalian cell lines (25, 26), and these events may represent the connection between deregulated cyclin E and cancer. Cyclin E overexpression accelerates entry into S phase, but somewhat paradoxically it also slows progression through S phase (25, 27–29). It has been shown that deregulation of cyclin E interferes with pre-RC assembly during early G1, and this defect leads possibly to impairment of replication initiation and/or fork elongation but does not affect the functions of cyclin E involved in the G1/S transition (30). Thus, this mechanism can potentially explain both the accelerated entry into S phase and the slower rate of DNA synthesis caused by cyclin E overexpression.
Cell cycle checkpoints are induced in response to DNA damage to allow additional time for lesions to be repaired and to carry out other aspects of the DNA damage response such as programmed cell death (31, 32). In response to the formation of double-strand breaks by ionizing radiation (IR), S phase checkpoints are mediated by two parallel pathways involving the upstream signaling kinases ATM and ATR and result in a rapid but transient inhibition of DNA synthesis (31, 33). The first of these pathways requires the activation of Chk1 and Chk2 kinases, both of which target the Cdc25A phosphatase for degradation resulting in an impairment of Cdk2 activation. The second pathway involves the MRN complex, Mdc1, and Smc1l however, how this pathway affects DNA replication is not known. Both of these pathways have also been implicated in the checkpoint responses to replication fork-blocking lesions such as DNA interstrand cross-links that are mediated by ATR (34, 35). However, interstrand cross-links cause an extended S phase arrest that can last for several days (36, 37) and that is not observed after exposure to IR, suggesting the possibility of additional checkpoint pathways that mediate long term arrest. Here, we show that in response to the cross-linking drug mitomycin C (MMC) or UV irradiation, but not to IR, cyclin E is strongly stabilized during S phase. This stabilization of cyclin E interferes with the activation of Cdk2-cyclin A and impedes DNA synthesis and S phase progression. Interestingly, the stabilization of cyclin E is not dependent on either Chk1 or Nbs1, but does depend upon ATR. In addition, cyclin E is required for the stabilization of Cdc6, which has been shown previously to be required for the activation of Chk1 (38–41). These findings suggest that ATR-mediated stabilization of cyclin E represents a novel mechanism that induces a sustained arrest in response to fork-blocking lesions.
EXPERIMENTAL PROCEDURES
Cell Culture, Synchronization, and BrdUrd Labeling
HeLa S3, HEK293T, and MRC5 cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. HeLa S3 cells were synchronized at G1/S phase by using a double-thymidine block as described previously (42). Briefly, HeLa S3 cells were cultured in Dulbecco's modified Eagle's medium with 2 mm thymidine for periods of 19 h and 16 h with an interval of 9 h in regular medium in between. After the second thymidine block period, cells were released into regular medium or were treated with MMC (Sigma), UV, or IR. Progression of the cell cycle was monitored by FACS analysis of propidium iodide (PI)-stained cells that were collected at the indicated time points. The modeling program FlowJo was used to calculate the peak of PI value which is the mode of the PI profile statistically.
Pulse labeling of cells with BrdUrd (10 μm) for 30 min was performed following the manufacturer's recommended protocol (Roche Applied Science in situ cell proliferation kit). Cells were trypsinized, fixed in 70% ethanol, incubated in blocking solution (4% bovine serum albumin in phosphate-buffered saline with 10% Triton X-100) for 30 min followed by incubation in 2 m HCl for 25 min. Cells were incubated with anti-BrdUrd for 45 min followed by a 20-min incubation with fluorescein isothiocyanate-conjugated secondary antibody.
Sucrose Gradient Centrifugation
Synchronized HeLa cells were collected at the indicated times and lysed in Nonidet P-40 buffer (50 mm Tris (pH 7.8), 120 mm NaCl, and 0.5% Nonidet P-40). Sucrose gradient analysis was performed as described previously (43). Briefly, cell lysates were loaded onto 5–30% sucrose gradients and spun at 28,000 rpm for 18 h in a Beckman SW 40i rotor. Fractions were collected and electrophoresed on SDS-polyacrylamide gels for immunoblot analysis. Protein bands were quantitated with Alpha Imager software (Alpha Innotech).
Antibodies and siRNAs
Antibodies used for immunoblot analysis, including cyclin E (sc-247), cyclin A (sc-751), ATR (sc-1887), Cdk2 (sc-163), proliferating cell nuclear antigen (sc-25280), Cdc6 (sc-9964), Nbs1 (sc-8580), MCM4 (sc-22779), p21 (sc-397-G), and HA (sc-7392) were purchased from Santa Cruz Biotechnology. Pin1 (3722), Chk1 (2345), and GSK3β (9315) antibodies were purchased from Cell Signaling. ORC2 (559260) and p27 (610241) antibodies were obtained from BD Pharmingen.
Nbs1 (L-009641-00), cyclin E (J-003213-10), Pin1 (J-003291-10), Chk1 (D-003255-06), and ATR (44) siRNAs were purchased from Dharmacon. An additional ATR siRNA was purchased from Sigma (SASI_HS01_00176271). Oligofectamine (Invitrogen) and the manufacturer's recommended protocol were used for siRNA transfections.
Ubiquitylation and Cdk2 IP Kinase Assays
For in vivo ubiquitylation assays, HeLa cells were transfected with pCDNA3/HA- ubiquitin using Lipofectamine 2000 (Invitrogen) followed by double-thymidine synchronization. Cells were collected and lysed in Nonidet P-40 buffer at the indicated time points. Anti-HA-conjugated agarose beads (sc-7392 AC; Santa Cruz Biotechnology) were used for HA IP.
IP kinase assays were performed as described previously (45). Briefly, Cdk2 or cyclin A was immunoprecipitated from cell lysates for 90 min at 4 °C. The precipitate was incubated with histone H1 (M2501; New England Biolabs) and [γ-32P]ATP in kinase buffer (50 mm Tris-HCl (pH 7.5), 20 mm EGTA, 10 mm MgCl, 1 mm dithiothreitol, 1 mm β-glycerophosphate, and 4 mm ATP) for 30 min at 25 °C. Proteins in the reaction mixtures were separated by SDS-PAGE and examined by Coomassie Blue staining and autoradiography.
RNA Preparation and Northern Blot Analysis
RNA was prepared from synchronized HeLa S3 cells using the RNeasy kit (Qiagen). pOTB7/cyclin E (clone ID 3841192) was purchased from the ATCC. A fragment produced by NcoI restriction digest of cyclin E cDNA was labeled by random priming and used for Northern blot analysis.
Chromatin Isolation
Chromatin isolation was performed by the method described previously (46). HeLa cells were collected at the time points indicated in the figure. Cells were suspended in buffer A (10 mm HEPES (pH 7.9), 10 mm KCl, 1.5 mm MgCl2, 0.34 m sucrose, 10% glycerol, and 1 mm dithiothreitol). Triton X-100 was added to a final concentration of 0.1%. Cells were incubated on ice for 8 min and then spun to collect the cytosolic fraction. The pellet was resuspended in buffer B (3 mm EDTA, 0.2 mm EGTA, and 1 mm dithiothreitol) and lysed on ice for 30 min. The supernatant (nucleoplasmic fraction) was combined with the cytosolic fraction as the soluble fraction. The pellet was dissolved in 1× SDS sample buffer as the chromatin fraction.
RESULTS
Agents That Introduce Replication Fork Barriers Stabilize Cyclin E by Inhibition of Ubiquitylation
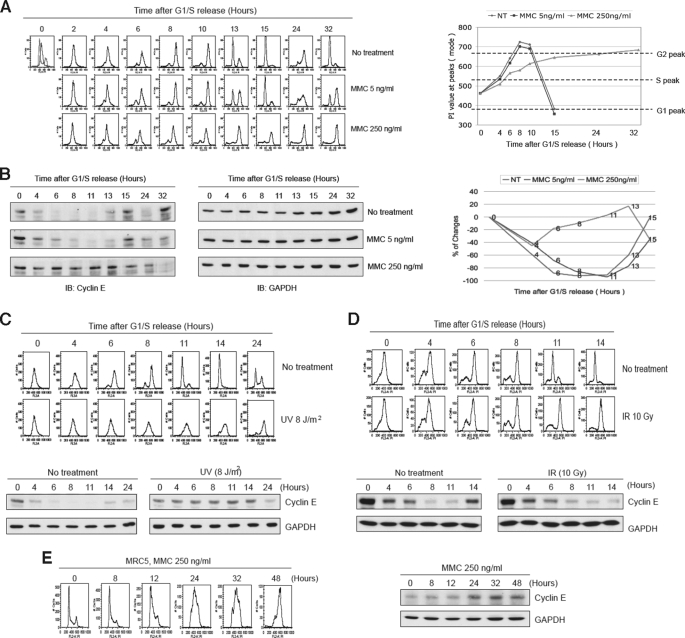
Cyclin E overexpression causes a prolonged S phase (25, 27–29), and recently it has been shown that cyclin E is a phosphorylation substrate of the checkpoint kinase ATR (47). These findings suggested the possibility that stabilization of cyclin E may mediate a long term arrest in the S phase of the cell cycle in response to replication stress. To investigate this hypothesis, we synchronized HeLa cells via a double-thymidine block and then released the cells into MMC. At 5 ng/ml MMC, cells exhibited a transient delay (note the 13 h time point), whereas at 250 ng/ml MMC a greatly decreased progression through S phase was observed (Fig. 1A). In untreated cells S phase was complete by 8 h, whereas in treated cells it was not fully complete by 24 h. Next we examined cyclin E levels, and we observed that it was slightly stabilized at 5 ng/ml MMC, but was strongly stabilized at the higher concentration of MMC (Fig. 1B). We performed similar experiments with synchronized HeLa cells using either UV or IR as the source of DNA damage (Fig. 1, C and D). Interestingly, UV, but not IR, also induced stabilization of cyclin E, suggesting that lesions that block replication forks induce the observed stabilization. As a further demonstration, we exposed unsynchronized primary human MRC5 fibroblast cells or HEK293T cells to MMC and observed stabilization of cyclin E for up to at least 48 h and a very prolonged S phase arrest (Fig. 1E and supplemental Fig. S1). To verify that cyclin E was responsible for the observed S phase delay, we overexpressed GFP-cyclin E and noted a prolongation of S phase (supplemental Fig. S2). Note that the exogenous GFP-cyclin E was strongly decreased compared with endogenous cyclin E, which is highly stabilized in the presence of drug or UV (Fig. 1, B and C).
FIGURE 1.
Cyclin E is stabilized in response to replication fork barriers. A, cell cycle analysis of HeLa cells synchronized by a double-thymidine block and released into MMC. FACS analysis (left panel) and quantitation of PI staining (right panel) are shown. The highest value for the largest peak (mode) is represented. B, immunoblots showing cyclin E levels in cells presented in A. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a loading control. Far right panel shows quantitation of cyclin E bands. C and D, similar experiment as shown in A except that cells were treated with UV or IR and subsequently released into normal medium. Upper panels show FACS analysis, and lower panels show immunoblots. E, unsynchronized MRC5 primary human fibroblast cells were exposed to MMC and analyzed by FACS (left panel) and immunoblotting (right panel).
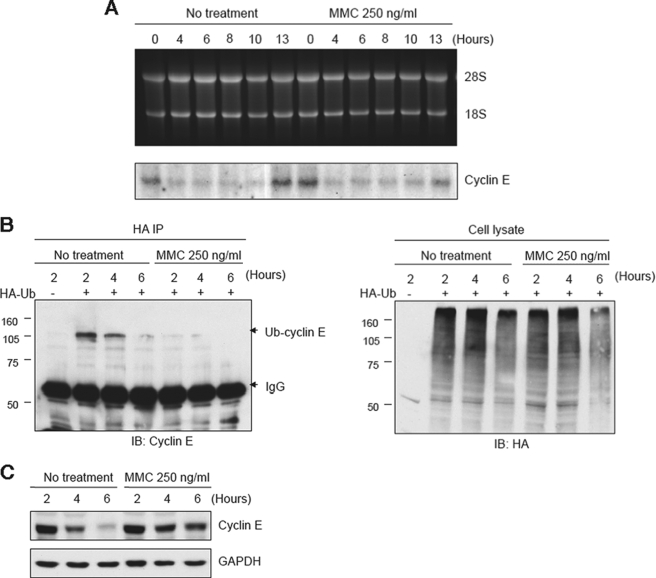
Next, we examined whether the stabilization of cyclin E occurred through a transcriptional or posttranslational mechanism. Synchronized HeLa cells were examined for levels of cyclin E transcripts by Northern blot analysis, and no increase was observed in the presence of MMC during the 4–10-h postrelease period when stabilization of cyclin E protein levels were observed (Fig. 2A). However, an examination of the degree of ubiquitylation of cyclin E showed that this modification was greatly reduced in the presence of MMC (Fig. 2B), and the stabilization of cyclin E in this experiment was confirmed by immunoblot analysis (Fig. 2C). These results demonstrate that exposure to MMC inhibits the normal ubiquitylation and degradation of cyclin E that occurs during S phase.
FIGURE 2.
MMC inhibits ubiquitylation of cyclin E. A, Northern blot showing that MMC does not affect transcript levels of cyclin E. HeLa cells were synchronized by double-thymidine block and analyzed at the indicated times after release into normal medium with or without MMC. B, immunoblot analysis showing that MMC prevents ubiquitylation of cyclin E. HeLa cells transiently expressing HA-ubiquitin (HA-Ub) were synchronized and released into medium with or without MMC. Lysates were subjected to IP with anti-HA and immunoblotted for cyclin E (left panel). Right panel shows a loading control. C, immunoblot analysis showing stabilization of cyclin E from the experiment in B.
MMC Alters the Association between Cyclin E and Factors Required for Its Ubiquitylation
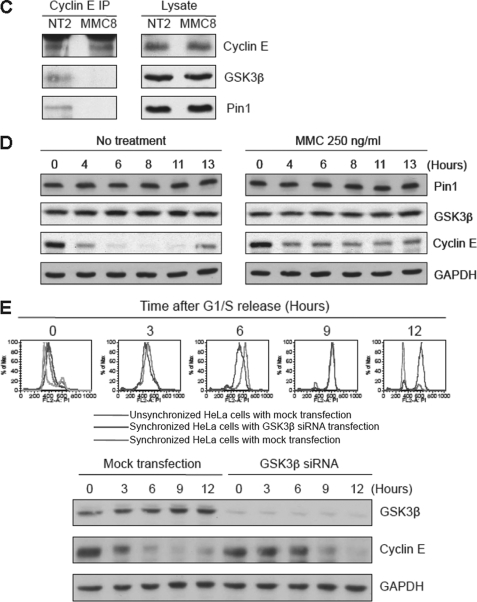
In addition to being a regulator of Cdk2, cyclin E is also a substrate of this kinase and GSK3β (5–12). Phosphorylation by these kinases is required for the ubiquitylation and ultimate degradation of cyclin E that occurs during S phase. In addition, the proline isomerase Pin1 is also required for ubiquitylation of cyclin E (12, 48). We therefore examined the association between these proteins and cyclin E by sucrose gradient sedimentation. As shown (Fig. 3A), at the time of release from synchronization the sedimentation profile of cyclin E partially overlaps with Cdk2 and GSK3β, but not with Pin1, in what appears to be a configuration intermediate between G1 and S phases. Two hours after release of untreated cells into S phase, the sedimentation profiles of all four proteins almost exactly overlap in a slower sedimenting complex (S phase configuration) that presumably mediates ubiquitylation of cyclin E. At 13 h after release, when cells have cycled into the G1 phase, cyclin E has returned to a G1 configuration with some overlap with Cdk2, but little overlap with GSK3β or Pin1. On the other hand, release of cells into MMC showed that cyclin E was maintained in the G1 configuration with only little or no overlap with GSK3β and Pin1 for many hours (Fig. 3B). This latter finding was also confirmed by co-IP experiments in which in the presence of MMC cyclin E no longer interacted with either GSK3β or Pin1 (Fig. 3C). Also, MMC did not affect the levels of either GSK3β or Pin1 (Fig. 3D). To confirm further that GSK3β is required for degradation of cyclin E during S phase, we depleted expression of GKS3β by siRNA and observed extended stabilization of cyclin E and slower progress through S phase (Fig. 3E). Taken together, these results indicate that treatment with MMC negatively affects the association between cyclin E and two factors that are required for its ubiquitylation, namely GSK3β and Pin1. Furthermore, they are consistent with the findings described above demonstrating that MMC induces stabilization of cyclin E via inhibition of ubiquitylation.
FIGURE 3.
Gradient sedimentation profiles of cyclin E and its binding partners in response to MMC. A, HeLa cells were synchronized and released into regular medium with or without MMC. Lysates were subsequently examined by sucrose gradient sedimentation (left panels). Quantitation of bands is shown in the right panels. L, loading of unfractionated samples. For reference, cell cycle distributions of these cells by FACS analysis are shown in the top panel. B, same as in A except cells were released into MMC. C, immunoblot analysis showing that MMC reduces the interaction between cyclin E and both GSK3β and Pin1 as determined in co-IP experiments. NT2, cells without drug released for 2 h; MMC8, cells treated with MMC (250 ng/ml) and released for 8 h. D, immunoblot showing that the levels of cyclin E, but not Pin1 or GSK3β, are altered in the presence of MMC. E, depletion of GSK3β results in stabilization of cyclin E during S phase. Upper panel shows FACS analysis of HeLa cell treated as indicated. Lower panel shows an immunoblot analysis.
Stabilization of Cyclin E Suppresses Cdk2 Activity
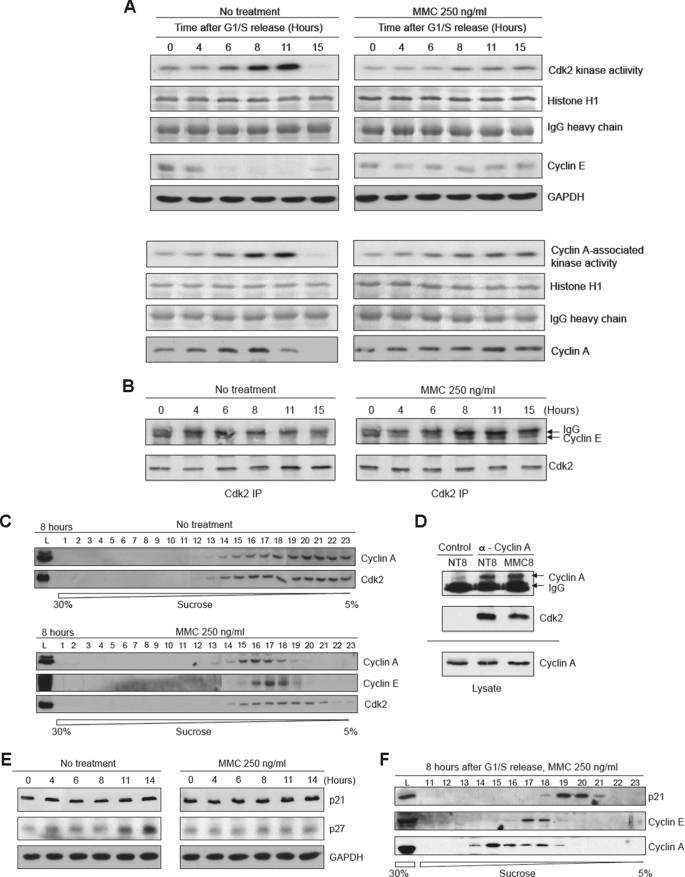
Ectopic overexpression of cyclin E is known to inhibit progression through S phase (25, 27–29). One possible explanation for this effect is that elevated levels of cyclin E may negatively affect Cdk2-cyclin A activity. To examine this hypothesis, HeLa cells were synchronized and released into medium with or without MMC. Approximately 6 h after release, untreated cells showed an increase in Cdk2 activity as determined in an IP-kinase assay using histone H1 as substrate (Fig. 4A, left upper panels). This increase in activity correlated with decreases in cyclin E levels. On the other hand, MMC-treated cells exhibited a distinctly lower level of Cdk2 kinase activity (Fig. 4A, right upper panels). We also measured cyclin A-associated kinase activity and found similarly that it was also reduced in the presence of MMC (Fig. 4A, lower panels). We also confirmed that stabilized cyclin E remained associated with Cdk2 as determined by a co-IP experiment (Fig. 4B). We next examined the sedimentation profile of cyclin A and found that 8 h after release the profiles for cyclin A and Cdk2 almost precisely overlapped in untreated cells. However, in the presence of MMC a portion of Cdk2 no longer co-sedimented with cyclin A (Fig. 4C). Furthermore, a co-IP experiment indicated that the interaction between cyclin A and Cdk2 was reduced by approximately half (Fig. 4D). These findings support the hypothesis that stabilization of cyclin E during S phase impairs the full activation of Cdk2-cyclin A.
FIGURE 4.
Cdk2 kinase activity is reduced in the presence of MMC. A, upper panels, analyses showing the results of IP-kinase assays of Cdk2 using histone H1 as the substrate. HeLa cells were synchronized and released into regular medium with or without MMC for the indicated times. The top rows show autoradiographs of phosphorylated histone H1. The next two rows shown Coomassie Blue staining of histone H1 and the IgG used for the IP. The bottom two rows show immunoblots of cyclin E and the loading control GAPDH. Lower panels, experiment as described above except that the IP was performed with an antibody to cyclin A. B, immunoblot analysis showing the co-IP of Cdk2 and cyclin E with or without MMC treatment. C, sucrose gradient sedimentation profiles of cyclin A and Cdk2 8 h after release from synchronization with or without MMC. D, immunoblot analysis showing co-IP between cyclin A and Cdk2 8 h after release from synchronization. NT8, nontreated cells at 8 h after release; MMC8, cells released into MMC for 8 h. Control indicates an IP with a nonspecific IgG. E, immunoblot analysis showing levels of p21 and p27 after release into regular medium with or without MMC. F, sucrose gradient sedimentation profiles of p21, cyclin E, and cyclin A at 8 h after release into MMC.
P21Cip1 and p27Kip1 are well known inhibitors of Cdk2 (49, 50) and could be responsible for the prolonged S phase arrest observed in the presence of MMC. We therefore examined whether treatment with MMC causes an increase in the levels of either of these inhibitors. As shown (Fig. 4E), neither p21 nor p27 levels were increased upon release into MMC compared with untreated cells. Furthermore, p21 did not co-sediment with either cyclin E or cyclin A in the presence of MMC (Fig. 4F). Attempts to detect p27 in these gradients were unsuccessful. Nevertheless, these experiments suggest that neither p21 nor p27 is responsible for the decrease in Cdk2 activity or the prolonged arrest observed in the presence of replication fork-blocking agents.
Cyclin E and Cdc6 Are Maintained on Chromatin in the Presence of MMC
To examine the effect of MMC on DNA synthesis, we released synchronized cells and pulse-labeled them with BrdUrd at various times during the subsequent incubation. For untreated control cells, the fraction of BrdUrd-positive cells had decreased dramatically by 8 h after release, whereas for MMC-treated cells, a large fraction still incorporated BrdUrd at 15 h after release (Fig. 5 A). We also examined the degree of BrdUrd incorporation and found that it was still ongoing in the MMC-treated cells, although not to the extent observed in the control cells (supplemental Fig. S3). Thus, cells damaged by MMC treatment exhibit a prolonged and substantial rate of DNA synthesis, suggesting that ongoing DNA replication is not completely blocked. Some component of the observed BrdUrd incorporation in the MMC-treated cells may be due to DNA repair synthesis.
FIGURE 5.
Stabilized cyclin E and Cdc6 are maintained on chromatin in the presence of MMC. A, FACS analysis showing BrdUrd incorporation in HeLa cells released into regular medium with or without MMC (left panel). At each indicated time point cells were pulsed for 30 min with BrdUrd and then harvested for analysis. The percentage of BrdUrd-positive cells is quantitated as shown in the graph (right panel). NT, no treatment. B, cells treated as described in A were fractionated into soluble (S) and chromatin (P) fractions and subsequently analyzed by immunoblotting. C, immunoblot analysis of total lysates of samples shown in B. D, depletion of cyclin E prevents stabilization of Cdc6 in response to MMC. Cells depleted of cyclin E by siRNA were synchronized and released into MMC for the indicated times, and cellular lysates were examined by immunoblotting.
Next, we examined the cellular localization of cyclin E and other markers of DNA replication. Cyclin E binds to replication forks and is known to be required for the loading of the MCM complex in a kinase-independent manner (15). Cyclin E has also been shown to regulate fork progression negatively in Drosophila ovarian follicle cells (51). Release of synchronized cells into MMC caused the retention of cyclin E, Cdc6, and proliferating cell nuclear antigen on chromatin, consistent with the ongoing DNA synthesis shown above (Fig. 5, B and C). In untreated cells, cyclin E was only transiently associated with chromatin during early S phase. Interestingly, the degradation of Cdc6 that normally occurs upon mitotic exit (13, 52) is inhibited in the presence of MMC. MCM4, on the other hand, was not affected by MMC treatment, and this result is consistent with the findings of others that association of the MCM complex with chromatin is greatly decreased in mammalian cells at the onset of S phase (46).
The apparent stabilization of Cdc6 in the presence of MMC was intriguing because Cdc6 has been shown to be involved in the activation of Chk1 and in mediating the S-M cell cycle checkpoint in response to stalled replication forks (38–41, 53). To determine whether the apparent stabilization of Cdc6 was dependent upon cyclin E, we used siRNA to deplete cells of cyclin E. As shown (Fig. 5D), depletion of cyclin E in the presence of MMC resulted in a significant drop in Cdc6 levels. Taken together, these results indicate that replication fork barriers induce stabilization of replication forks with concomitant retention of cyclin E and Cdc6, which in turn causes a slowing of S phase progression and activation of the replication checkpoint.
ATR Regulates the Degradation of Cyclin E in Response to MMC
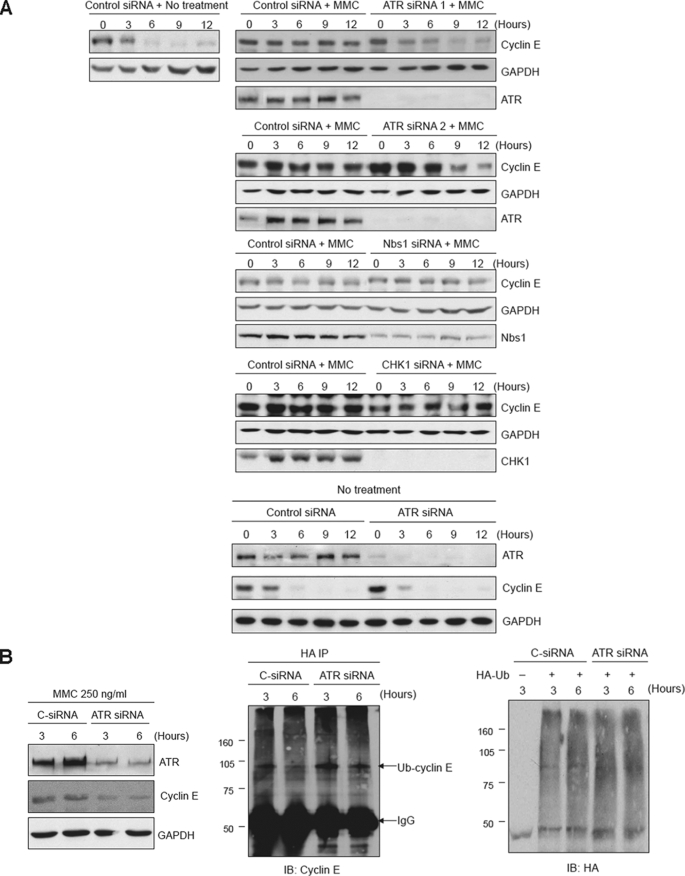
Cyclin E has recently been shown to be a phosphorylation substrate of ATR in response to DNA damage (47). To determine whether ATR plays a role in the DNA damage-mediated stabilization of cyclin E, we used siRNA to deplete HeLa cells of this checkpoint kinase. Release of synchronized cells into MMC showed that depletion of ATR significantly decreased the observed stabilization of cyclin E (Fig. 6 A). However, depletion of ATR in the absence of MMC did not induce stabilization of cyclin E (Fig. 6A). Interestingly, the downstream effectors of ATR-mediated checkpoints, Nbs1 and Chk1, were not required for stabilization of cyclin E (Fig. 6A), suggesting that ATR may directly regulate the degradation of cyclin E. To examine this possibility, ATR was depleted, and the ubiquitylation of cyclin E in the presence of MMC was examined in vivo. As shown (Fig. 6B), depletion of ATR resulted in an increase in ubiquitylation of cyclin E.
FIGURE 6.
ATR, but not Chk1 or Nbs1, is required for the stabilization of cyclin E in the presence of MMC. A, HeLa cells transfected with ATR, Chk1, Nbs1, or control siRNAs were synchronized and released into regular medium with MMC or without drug. Cyclin E levels were examined at the indicated times after release by immunoblot analysis. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a loading control. B, ATR prevents the ubiquitylation of cyclin E in response to MMC. Synchronized HeLa cells depleted of ATR by siRNA (left panel) were transfected with HA-ubiquitin and subsequently subjected to IP analysis by immunoblotting (center panel). Right panel shows the loading control.
DISCUSSION
Our findings described here demonstrate that in response to DNA damage, such as introduced by MMC or UV irradiation, cyclin E is stabilized by a mechanism that reduces its level of ubiquitylation and ultimate degradation. This phenomenon was observed in the cancer cell line HeLa, in HEK293T cells, and in primary human fibroblasts, suggesting that it is a general cellular response to stalled replication forks. Interestingly, production of double-strand breaks by IR did not induce stabilization of cyclin E, indicating that this response is specific to lesions that directly block replication forks. Overexpression of cyclin E has been shown in a number of contexts to inhibit the progression of cells through S phase in vivo (25, 27–29). A proposed explanation for this effect is that abnormally high levels of cyclin E interfere with the loading of the MCM complex in early G1 (30), and such a model has also been confirmed in vitro (16). However, this mechanism does not appear to explain our findings because we have used synchronization techniques to arrest cells at the G1/S boundary at a point in the cell cycle where pre-RC assembly would be completed. Rather, our results suggest that cyclin E may act to retard S phase progression by competing with cyclin A for binding to Cdk2 and thus inhibit origin firing, an apparent function of Cdk2-cyclin A (3, 16). Elevated levels of cyclin E have been shown to prevent cyclin A from activating replication origins in vitro (3, 16). Interestingly, even though cyclin E levels remained high in response to DNA damage, the overall activity of Cdk2 was decreased. This may be due to the activation of Chk1 and/or the possibility that the phosphorylated form of cyclin E may have decreased capacity to stimulate Cdk2. Cyclin E has also been shown to affect fork progression in Drosophila follicle cells directly, although the mechanism of this activity is unclear (51). Thus, cyclin E may retard S phase progression by both inhibiting replicon origin firing and replication fork progression.
The stabilization of cyclin E in response to DNA damage required ATR, and this effect was due to a requirement for ATR to inhibit the ubiquitylation of cyclin E. The mechanism of this inhibition may be mediated by direct ATR phosphorylation of cyclin E because cyclin E has been shown to be a substrate of ATR in response to DNA damage (47). Furthermore, the downstream ATR mediators Chk1 and Nbs1 were not required for cyclin E stabilization, indicating that they do not act upstream of cyclin E. Treatment with MMC prevented cyclin E from associating with factors such as GSKβ and Pin1 that are required for its ubiquitylation and appeared to maintain cyclin E in a complex that resembled a G1 configuration as opposed to an S phase configuration. Thus, ATR-mediated phosphorylation of cyclin E may cause disruption of the E3 ligase complex that ubiquitylates cyclin E during S phase, resulting in its stabilization. Interestingly, we have shown recently that degradation of cyclin E during recovery from the replication checkpoint requires the cell cycle regulator Artemis (54). This function of Artemis requires prior phosphorylation by ATR, indicating that ATR both regulates the stabilization of cyclin E during checkpoint arrest and its degradation during recovery from the checkpoint.
Concomitant with the stabilization of cyclin E, we also observed that DNA damage resulted in increased levels of Cdc6 protein. Cdc6 has been implicated in checkpoint activation in response to stress in fission yeast, Xenopus laevis extracts, and in mammalian cells (38–41, 55). In Schizosaccharomyces pombe, Cdc18/Cdc6 is required for the activation of the checkpoint kinase Cds1 and for the stabilization of stalled replication forks (40). In fact, more recently it has been shown that during S phase arrest, Cdc18/Cdc6 is stabilized on chromatin and serves as a receptor for the Rad3·Rad26 complex, the homologs of mammalian ATR and ATR-interacting protein (41). In vertebrate systems, Cdc6 has also been shown to be required for activation of Chk1 in response to stalled replication forks. Consistent with our findings with cyclin E, the function of Cdc6 in checkpoint regulation did not involve its role in loading of the MCM complex onto origins (39). The mechanism by which cyclin E stabilizes Cdc6 is unclear. In an unperturbed cell cycle, Cdk2-cyclin E phosphorylates Cdc6 during the G1 phase to prevent ubiquitylation of Cdc6 by the APC, which would otherwise result in its degradation. However, the APC is normally inactivated during S phase, and this also occurs in the presence of MMC because we observed that cyclin A, a substrate of the APC, increased after release from the G1/S synchronization block. Thus, it appears unlikely that cyclin E protects Cdc6 by preventing ubiquitylation by the APC in this context. A more plausible scenario is that because cyclin A is known to regulate Cdc6 negatively (16), the impairment of Cdk2-cyclin A activity by stabilized cyclin E might allow Cdc6 levels to rise, thus inducing activation of Chk1 and cell cycle arrest. Thus, this model (Fig. 7) puts cyclin E and Cdc6 upstream of Chk1 in response to stalled forks possibly by recruitment of ATR and ATR-interacting protein. However, it is clear that ATR can activate Chk1 in response to some forms of damage such as double-strand breaks induced by IR without stabilization of cyclin E (56, 57). Thus, the principal function of cyclin E may be to mediate a long term activation of the checkpoint in response to fork-blocking lesions. The repair of DNA interstrand cross-links is extremely slow compared with most other types of lesions such as double-strand breaks and has been estimated in human cells to occur at a rate of ∼10–12 adducts/hour (37). Thus, days are required to complete repair of even moderate levels of DNA damage necessitating a long term cell cycle arrest.
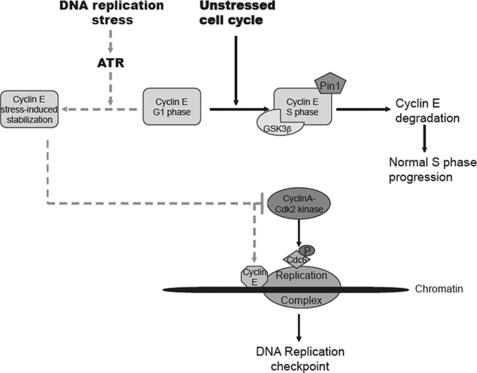
FIGURE 7.
Model for involvement of cyclin E in the replication checkpoint in response to replication fork barriers. In response to replication stress, ATR mediates stabilization of cyclin E resulting in inhibition of cyclin A-Cdk2 and retention of cyclin E and Cdc6 at stalled replication forks. These events contribute to the activation of the replication checkpoint.
Finally, our findings demonstrate that in response to fork-blocking lesions, cyclin E is stabilized resulting in the mediation of a long term replication checkpoint. The following considerations indicate that cyclin E is an integral component of this pathway: (i) stabilization of cyclin E was induced by MMC and UV, but not by IR, suggesting a specific response to stalled replication forks; (ii) DNA damage disrupted the interaction between cyclin E and proteins required for its ubiquitylation; (iii) cyclin E is a known substrate of the checkpoint kinase ATR; (iv) ATR was required for the stabilization of cyclin E by preventing its ubiquitylation; and (v) Cdc6, which has been implicated in checkpoint responses, required cyclin E for its stabilization. Taken together, these findings indicate that in addition to its well studied function in promoting cell cycle progression, cyclin E also has a role in regulating cell cycle arrest in response to DNA damage.
This work was supported, in whole or in part, by National Institutes of Health Grant CA097175 from the NCI. DNA sequencing resources were supported by the Cancer Center Support Grant CA16672.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- Cdk2
- cyclin-dependent protein kinase 2
- APC
- anaphase-promoting complex
- BrdUrd
- bromodeoxyuridine
- FACS
- fluorescence-activated cell sorter
- GSK3β
- glycogen synthase kinase 3β
- HA
- hemagglutinin
- IP
- immunoprecipitation
- IR
- ionizing radiation
- MCM
- minichromosome maintenance
- MMC
- mitomycin C
- PI
- propidium iodide
- pre-RC
- prereplication complex
- siRNA
- small interfering RNA
- ATM
- ataxia telangiectasia
- ATR
- ataxia telangiectasia and Rad3-related.
REFERENCES
- 1.Murray A. W. (2004) Cell 116, 221–234 [DOI] [PubMed] [Google Scholar]
- 2.Neganova I., Lako M. (2008) J. Anat. 213, 30–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woo R. A., Poon R. Y. (2003) Cell Cycle 2, 316–324 [PubMed] [Google Scholar]
- 4.Singer J. D., Gurian-West M., Clurman B., Roberts J. M. (1999) Genes Dev. 13, 2375–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clurman B. E., Sheaff R. J., Thress K., Groudine M., Roberts J. M. (1996) Genes Dev. 10, 1979–1990 [DOI] [PubMed] [Google Scholar]
- 6.Gupta-Rossi N., Le Bail O., Gonen H., Brou C., Logeat F., Six E., Ciechanover A., Israël A. (2001) J. Biol. Chem. 276, 34371–34378 [DOI] [PubMed] [Google Scholar]
- 7.Koepp D. M., Schaefer L. K., Ye X., Keyomarsi K., Chu C., Harper J. W., Elledge S. J. (2001) Science 294, 173–177 [DOI] [PubMed] [Google Scholar]
- 8.Moberg K. H., Bell D. W., Wahrer D. C., Haber D. A., Hariharan I. K. (2001) Nature 413, 311–316 [DOI] [PubMed] [Google Scholar]
- 9.Strohmaier H., Spruck C. H., Kaiser P., Won K. A., Sangfelt O., Reed S. I. (2001) Nature 413, 316–322 [DOI] [PubMed] [Google Scholar]
- 10.Welcker M., Singer J., Loeb K. R., Grim J., Bloecher A., Gurien-West M., Clurman B. E., Roberts J. M. (2003) Mol. Cell 12, 381–392 [DOI] [PubMed] [Google Scholar]
- 11.Won K. A., Reed S. I. (1996) EMBO J. 15, 4182–4193 [PMC free article] [PubMed] [Google Scholar]
- 12.van Drogen F., Sangfelt O., Malyukova A., Matskova L., Yeh E., Means A. R., Reed S. I. (2006) Mol. Cell 23, 37–48 [DOI] [PubMed] [Google Scholar]
- 13.Mailand N., Diffley J. F. (2005) Cell 122, 915–926 [DOI] [PubMed] [Google Scholar]
- 14.Petersen B. O., Lukas J., Sorensen C. S., Bartek J., Helin K. (1999) EMBO J. 18, 396–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geng Y., Lee Y. M., Welcker M., Swanger J., Zagozdzon A., Winer J. D., Roberts J. M., Kaldis P., Clurman B. E., Sicinski P. (2007) Mol. Cell 25, 127–139 [DOI] [PubMed] [Google Scholar]
- 16.Coverley D., Laman H., Laskey R. A. (2002) Nat. Cell Biol. 4, 523–528 [DOI] [PubMed] [Google Scholar]
- 17.Ekholm-Reed S., Spruck C. H., Sangfelt O., van Drogen F., Mueller-Holzner E., Widschwendter M., Zetterberg A., Reed S. I. (2004) Cancer Res. 64, 795–800 [DOI] [PubMed] [Google Scholar]
- 18.Erlandsson F., Wählby C., Ekholm-Reed S., Hellström A. C., Bengtsson E., Zetterberg A. (2003) Int. J. Cancer 104, 369–375 [DOI] [PubMed] [Google Scholar]
- 19.Keyomarsi K., Conte D., Jr., Toyofuku W., Fox M. P. (1995) Oncogene 11, 941–950 [PubMed] [Google Scholar]
- 20.Keyomarsi K., Tucker S. L., Bedrosian I. (2003) Nat. Med. 9, 152. [DOI] [PubMed] [Google Scholar]
- 21.Sandhu C., Slingerland J. (2000) Cancer Detect Prev. 24, 107–118 [PubMed] [Google Scholar]
- 22.Bortner D. M., Rosenberg M. P. (1997) Mol. Cell. Biol. 17, 453–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajagopalan H., Jallepalli P. V., Rago C., Velculescu V. E., Kinzler K. W., Vogelstein B., Lengauer C. (2004) Nature 428, 77–81 [DOI] [PubMed] [Google Scholar]
- 24.Spruck C. H., Strohmaier H., Sangfelt O., Müller H. M., Hubalek M., Müller-Holzner E., Marth C., Widschwendter M., Reed S. I. (2002) Cancer Res. 62, 4535–4539 [PubMed] [Google Scholar]
- 25.Spruck C. H., Won K. A., Reed S. I. (1999) Nature 401, 297–300 [DOI] [PubMed] [Google Scholar]
- 26.Loeb K. R., Loeb L. A. (2000) Carcinogenesis 21, 379–385 [DOI] [PubMed] [Google Scholar]
- 27.Ohtsubo M., Roberts J. M. (1993) Science 259, 1908–1912 [DOI] [PubMed] [Google Scholar]
- 28.Resnitzky D., Gossen M., Bujard H., Reed S. I. (1994) Mol. Cell. Biol. 14, 1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wimmel A., Lucibello F. C., Sewing A., Adolph S., Müller R. (1994) Oncogene 9, 995–997 [PubMed] [Google Scholar]
- 30.Ekholm-Reed S., Méndez J., Tedesco D., Zetterberg A., Stillman B., Reed S. I. (2004) J. Cell Biol. 165, 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartek J., Lukas C., Lukas J. (2004) Nat. Rev. Mol. Cell Biol. 5, 792–804 [DOI] [PubMed] [Google Scholar]
- 32.Harper J. W., Elledge S. J. (2007) Mol. Cell 28, 739–745 [DOI] [PubMed] [Google Scholar]
- 33.Lambert S., Carr A. M. (2005) Biochimie 87, 591–602 [DOI] [PubMed] [Google Scholar]
- 34.Pichierri P., Rosselli F. (2004) EMBO J. 23, 1178–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mladenov E., Tsaneva I., Anachkova B. (2007) J. Cell. Physiol. 211, 468–476 [DOI] [PubMed] [Google Scholar]
- 36.Liu L., Akhter S., Bae J. B., Mukhopadhyay S. S., Richie C. T., Liu X., Legerski R. (2009) Cell Cycle 8, 628–638 [DOI] [PubMed] [Google Scholar]
- 37.Akkari Y. M., Bateman R. L., Reifsteck C. A., Olson S. B., Grompe M. (2000) Mol. Cell. Biol. 20, 8283–8289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clay-Farrace L., Pelizon C., Santamaria D., Pines J., Laskey R. A. (2003) EMBO J. 22, 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oehlmann M., Score A. J., Blow J. J. (2004) J. Cell Biol. 165, 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murakami H., Yanow S. K., Griffiths D., Nakanishi M., Nurse P. (2002) Nat. Cell Biol. 4, 384–388 [DOI] [PubMed] [Google Scholar]
- 41.Hermand D., Nurse P. (2007) Mol. Cell 26, 553–563 [DOI] [PubMed] [Google Scholar]
- 42.Whitfield M. L., Zheng L. X., Baldwin A., Ohta T., Hurt M. M., Marzluff W. F. (2000) Mol. Cell. Biol. 20, 4188–4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McDonald W. H., Ohi R., Smelkova N., Frendewey D., Gould K. L. (1999) Mol. Cell. Biol. 19, 5352–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geng L., Zhang X., Zheng S., Legerski R. J. (2007) Mol. Cell. Biol. 27, 2625–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blomberg I., Hoffmann I. (1999) Mol. Cell. Biol. 19, 6183–6194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Méndez J., Stillman B. (2000) Mol. Cell. Biol. 20, 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsuoka S., Ballif B. A., Smogorzewska A., McDonald E. R., 3rd, Hurov K. E., Luo J., Bakalarski C. E., Zhao Z., Solimini N., Lerenthal Y., Shiloh Y., Gygi S. P., Elledge S. J. (2007) Science 316, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 48.Yeh E. S., Lew B. O., Means A. R. (2006) J. Biol. Chem. 281, 241–251 [DOI] [PubMed] [Google Scholar]
- 49.Borriello A., Cucciolla V., Oliva A., Zappia V., Della Ragione F. (2007) Cell Cycle 6, 1053–1061 [DOI] [PubMed] [Google Scholar]
- 50.Abukhdeir A. M., Park B. H. (2008) Expert Rev. Mol. Med. 10, e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park E. A., Macalpine D. M., Orr-Weaver T. L. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 16739–16746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petersen B. O., Wagener C., Marinoni F., Kramer E. R., Melixetian M., Lazzerini Denchi E., Gieffers C., Matteucci C., Peters J. M., Helin K. (2000) Genes Dev. 14, 2330–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu L., Choi J. H., Yim H., Choi J. S., Park B. D., Cho S. J., Lee S. K. (2009) Int. J. Biochem. Cell Biol. 41, 1410–1420 [DOI] [PubMed] [Google Scholar]
- 54.Wang H., Zhang X., Geng L., Teng L., Legerski R. J. (2009) J. Biol. Chem. 284, 18236–18243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Borlado L. R., Méndez J. (2008) Carcinogenesis 29, 237–243 [DOI] [PubMed] [Google Scholar]
- 56.Zou L., Elledge S. J. (2003) Science 300, 1542–1548 [DOI] [PubMed] [Google Scholar]
- 57.Jazayeri A., Falck J., Lukas C., Bartek J., Smith G. C., Lukas J., Jackson S. P. (2006) Nat. Cell Biol. 8, 37–45 [DOI] [PubMed] [Google Scholar]