Abstract
Heavy metals are known to generate reactive oxygen species that lead to the oxidation and fragmentation of proteins, which become toxic when accumulated in the cell. In this study, we investigated the role of the proteasome during cadmium stress in the leaves of Arabidopsis thaliana plants. Using biochemical and proteomics approaches, we present the first evidence of an active proteasome pathway in plants. We identified and characterized the peptidases acting sequentially downstream from the proteasome in animal cells as follows: tripeptidyl-peptidase II, thimet oligopeptidase, and leucine aminopeptidase. We investigated the proteasome proteolytic pathway response in the leaves of 6-week-old A. thaliana plants grown hydroponically for 24, 48, and 144 h in the presence or absence of 50 μm cadmium. The gene expression and proteolytic activity of the proteasome and the different proteases of the pathway were found to be up-regulated in response to cadmium. In an in vitro assay, oxidized bovine serum albumin and lysozyme were more readily degraded in the presence of 20 S proteasome and tripeptidyl-peptidase II than their nonoxidized form, suggesting that oxidized proteins are preferentially degraded by the Arabidopsis 20 S proteasome pathway. These results show that, in response to cadmium, the 20 S proteasome proteolytic pathway is up-regulated at both RNA and activity levels in Arabidopsis leaves and may play a role in degrading oxidized proteins generated by the stress.
Introduction
Cadmium is a highly toxic and persistent environmental poison for plants, yeasts, and animals (1). Cadmium is released into the environment mainly through industrial wastes and is transferred to animals through the food chain (2). Cadmium interferes with many cellular functions mainly by complex formation with organic compounds such as proteins, lipids, and nucleotides leading to the inhibition of gene expression and metabolic activities. Cadmium toxicity involves production of active oxygen species and free radicals, although the mechanism is still obscure (3). Cadmium is not a transition metal and does not produce hydroxyl radicals through Fenton or Haber-Weiss reactions, but it induces the production of superoxide anion, nitric oxide, and hydrogen peroxide, enhances lipid peroxidation and depletion of cellular glutathione, and finally generates oxidative stress (3, 4). Active oxygen species can lead to oxidation of side chains of amino acid residues and formation of protein-protein covalent cross-linkage, which can lead to protein inactivation or denaturation (5–7). If they are not rapidly degraded, oxidatively modified proteins can undergo direct fragmentation or can form large aggregates due to covalent cross-linking and increased surface hydrophobicity, which lead to cell death (5, 8).
In animal cells, the proteasome has been shown to recognize and degrade mildly oxidized proteins in the cytosol, nucleus, and endoplasmic reticulum, thus minimizing their cytotoxicity (5). From in vitro studies, it was shown that the 20 S proteasome recognizes and degrades oxidized proteins, probably through a “by default” degradation mechanism (9), although the 26 S proteasome does not (10–12). This may be explained by the fact that a mild oxidative stress rapidly inactivates both the ubiquitin-activating/conjugating system and 26 S proteasome activity in intact cells, but it does not affect 20 S proteasome activity (10, 12, 13).
In mammals, a set of proteases has been identified acting sequentially downstream of the proteasome to peptide degradation and releasing free amino acids into the cytosol. The peptides issued from the proteasome range from 3 to 25 amino acids and have different features according to their size. The smallest products (2–6 amino acids) are directly degraded by aminopeptidases, mainly leucine aminopeptidases (LAPs),3 although larger peptides (>6 amino acids) are first cleaved by intermediate endopeptidases (14). The most important endopeptidases for these processes are the thimet oligopeptidase (TOP) (1) for peptides of 9–17 residues and tripeptidyl-peptidase II (TPPII) for longer products (15–25 amino acids) (15). The proteasome pathway (proteasome, TPPII, TOP, and aminopeptidases) constitutes the main protein degradation pathway in mammalian cells. It has been shown to play a central role in the degradation of many proteins in normal and disease states (16) and also in antigen processing for major histocompatibility complex class 1 presentation (17, 18). However, the specific involvement of the proteasome pathway in the degradation of oxidized protein has not been investigated so far.
In plants, the presence of the proteasome is now well established. It was first identified in pea (19) and then in all species investigated so far (20). Both the 20 S and 26 S proteasomes were characterized at molecular (21, 22) and biochemical levels (23) in Arabidopsis thaliana. As observed in animal and yeast cells, plant proteasome has been found to be up-regulated, at transcriptional or translational levels, in oxidative conditions such as carbon starvation (24) or cadmium stress (25–28), and its involvement in the response to oxidative stress has been postulated. In plants, the preferential involvement of the 20 S proteasome in the degradation of oxidized proteins was recently supported by in vivo experiments using A. thaliana mutants (29). The authors showed that the loss of function of some subunits of the 19 S regulatory particle of the 26 S proteasome resulted in a decrease of the 26 S accumulation at the benefit of the 20 S level, together with a higher tolerance to oxidative stress. The presence of TPPII, and many aminopeptidases, was also reported in plants (30–32), but no TOP-like protease has been identified to date. Thus, the following questions have been raised: (i) whether the proteasome pathway (including the 20 S proteasome and the downstream acting proteases) is similar in plants to that found in animals; (ii) whether this pathway is responsive to oxidative stress; and (iii) whether it is involved in the degradation of oxidized proteins.
To address these questions, proteomics and classical enzymatic approaches were used to investigate the presence of the following different peptidases of the proteasome pathway: proteasome, TPPII, TOP, and LAP in Arabidopsis leaves. The response of the proteasome pathway in the leaves of 6-week-old plants grown for 24, 48, and 144 h in the presence or absence of 50 μm cadmium was also determined. Oxidative stress was estimated through protein oxidation level. The changes in RNA and protein accumulation and activity of the different proteases of the pathway were analyzed. Oxidized or nonoxidized bovine serum albumin, lysozyme, and casein were used to test the capacity of the 20 S proteasome and TPPII to degrade oxidized substrates.
EXPERIMENTAL PROCEDURES
Growth Conditions
A. thaliana seeds (ecotype Columbia) were germinated in sand and then grown hydroponically in controlled conditions. The photoperiod was 14 h, with a photosynthetic photon flux density of 140–190 μmol photons m−2 s−1. The day/night temperatures were 23/18 °C, and the relative humidity was maintained close to 55%. The nutritive solution (pH 5.9) contained the following elements: 805 μm Ca(NO3)2, 2 mm KNO3, 60 μm K2HPO4, 695 μm KH2PO4, 1.1 mm MgSO4, 20 μm Na2EDTA, 20 μm FeSO4, 74 nm (NH4)6Mo7O24, 3.6 μm MnSO4, 3 μm ZnSO4, 9.25 μm H3BO3, and 785 nm CuSO4. The culture medium was renewed every 2 days and was continuously sparged with air. When seedlings were 6 weeks old, stress was induced by adding Cd(NO3)2 to the mineral solution at a 50 μm final concentration. No cadmium was added to control cultures. Cadmium-containing solutions were renewed every 2 days. At 0, 24, 48, and 144 h of cadmium exposure, leaves were harvested and quickly frozen in liquid N2, ground to a powder in liquid N2, and stored at −80 °C until analysis.
Preparation of Clarified Extracts
Five hundred mg of frozen leaf powder were homogenized in a mortar at 4 °C with four times their fresh weight of extraction medium (50 mm Tris-HCl (pH 7.5), 5 mm β-mercaptoethanol, and 0.3% (w/v) insoluble polyvinylpyrrolidone). The homogenate was centrifuged for 20 min at 16,000 × g, and the supernatant was used for protease purification and activity measurements.
Chromatography Steps
The soluble leaf protein extract (6 ml) was applied to a HiPrep 26/60 Sephacryl S-300 gel filtration column (Mr 10,000–1,500,000; Amersham Biosciences) equilibrated with 50 mm Tris-HCl (pH 7.5) and 150 mm NaCl. The column was calibrated with the following markers: thyroglobulin (670 kDa), bovine γ-globulin (158 kDa), chicken ovalbumin (44 kDa), equine myoglobin (17 kDa), and vitamin B12 (1.35 kDa). The fractions containing chymotrypsin-like AAF-AMC-like (TPPII), Mcc-PLGPK-Dnp-like (TOP) (1), and Leu-pNA-like activities were pooled and used for activity measurement in the presence or absence of various chemical agents to characterize the different proteases as described below.
Determination of Protein-bound Carbonyls
Carbonyl contents in soluble protein extracts, from 100 mg FW of each sample, were quantified by reaction with dinitrophenylhydrazine (DNPH) according to Ref. 33. Proteins were reacted with 200 μl of 10 mm DNPH (in 2 m HCl) or 2 m HCl (control) for 1 h at 25 °C. For each determination, two replicates and their respective blanks were used. Proteins were precipitated with 20% (w/v) trichloroacetic acid, centrifuged, and washed three times with ethanol/ethyl acetate (1:1 (v/v)). The protein pellets were finally dissolved in 6 m guanidine hydrochloride, 20 mm potassium phosphate buffer (pH 2.3), and the absorption at 370 nm was measured. Carbonyl content was calculated using a molar absorption coefficient of 22,000 m−1 cm−1. To determine the yield of protein recovery after the carbonyl derivation step with DNPH, proteins were quantified before and after the derivation procedure by reading the absorption of the samples at 280 nm (33). Protein amounts were calculated from a BSA standard curve prepared in the same conditions. Protein recovery was 88 ± 4%.
Proteolytic Activity Measurements
Global endopeptidase activity measurement was adapted from Brouquisse et al. (34). One hundred μl of clarified extract and 100 μl of azocasein (5 mg ml−1 in 200 mm MES-KOH (pH 6.0)) were incubated for 3 h at 37 °C. The reaction was stopped by the addition of 100 μl of 10% (v/v) trichloroacetic acid. After 10 min on ice, samples were centrifuged at 15,000 × g for 10 min; 250 μl of supernatant were then added to 750 μl of 1 m NaOH, and the absorbance was read at 440 nm. The extinction coefficient ϵ1% azocasein in 1 m NaOH = 37 liters cm−1 g−1 was used to calculate the azocasein degradation activity.
For chymotrypsin-like activity measurement, Suc-Leu-Leu-Val-Tyr-7-amino-4-methylcoumarin (Suc-LLVY-AMC; 2 mm stock in dimethyl sulfoxide) was used as a substrate. The assay mixture contained 100 μl of 20 S proteasome fraction in 50 mm Tricine buffer (pH 8.1) and 100 μm of Suc-LLVY-AMC. After incubation at 37 °C for 1 h, the reaction was stopped by the addition of 100 μl of 10% (w/v) SDS and 700 μl of Tris-HCl 100 mm (pH 9.0). The AMC radical released was measured fluorometrically (λexcitation = 380 nm; λemission = 460 nm). Activity was calculated using AMC standard curve made in the same conditions.
For TPPII activity measurement, 100 μl of sample fraction were mixed with 50 mm Tris-HCl buffer (pH 7.6), 5 mm MgCl2, and 125 μm Ala-Ala-Phe-AMC (AAF-AMC) as substrate. After 1 h of incubation period at 37 °C, the reaction was stopped with the addition of 800 μl of 80 mm sodium acetate (pH 4.3). The AMC radical released was measured fluorometrically (λexcitation = 380 nm; λemission = 460 nm).
TOP activity was measured by mixing a 100-μl sample with 50 mm Tris-HCl buffer (pH 7.6), 5 mm MgCl2, 0.1 mm dithiothreitol, 0.05% Brij 35% (v/v), and 20 μm Mcc-Pro-Leu-Gly-Pro-d-Lys-OH-Dnp (Mcc-PLGPK-Dnp) as substrate. After 1 h at 37 °C, the reaction was stopped by adding of 800 μl of 80 mm sodium acetate (pH 4.3). The product was measured fluorometrically (λexcitation = 345 nm; λemission = 405 nm).
For LAP activity measurement, 300 μl of fraction were mixed with 50 mm Tris-HCl buffer (pH 8) and 0.5 mm MnCl2. The reaction was measured at 25 °C and started by the addition of 1 mm l-leucine-p-nitroanilide (Leu-pNA) as substrate. The pNA released was measured spectrophotometrically at 405 nm. The extinction coefficient (ϵ), pNA = 10,500 liters cm−1 m−1, was used to calculate LAP-like activity.
Protease Effectors
Protease effector stock solutions were prepared in the following solvents: E64 (2 mm), Na2EDTA (0.2 m), butabindide (1 mm), AAF-chloromethyl ketone (AAF-CMK), and benzyloxycarbonyl-AAF CMK (5 mm) in water; phenylmethylsulfonyl fluoride (0.2 m), 1,10-phenanthroline (0.2 m), and pepstatin (2 mm) in ethanol; lactacystine (1 mm), carboxy-3-phenylpropyl -AAF-polyclonal antibody (2 mm), and benzyloxycarbonyl-Leu-Leu-leucinal (MG132, 2 mm) and bestatin (2 mm) in dimethyl sulfoxide (DMSO). Effectors were first preincubated for 20 min at room temperature with protein extracts prior to substrate addition, and activities were measured as described above. Control assays were carried out with the corresponding solvent.
Immunoprecipitation Experiments
Polyclonal antibodies raised against the maize 20 S proteasome (24) and tomato LAP (35) were used for immunoprecipitation experiments. Serum IgG fractions were purified using 1 ml of HiTrap protein A column (Amersham Biosciences) as recommended by the manufacturer. Immunoprecipitation protocol was adapted from Ref. 24. Briefly, 100 μl of either clarified extract or partially purified fractions were incubated 1 h at 4 °C with increasing volumes of purified immune or preimmune serum. Immune complexes were incubated 1 h at 4 °C with a 5-fold (immunoglobulin-G binding) excess of protein A-agarose (Affi-Gel, Bio-Rad) and then centrifuged for 5 min at 10,000 × g. The enzymatic activities (chymotrypsin-like or Leu-pNA-like) were measured in each supernatant fraction as described above.
Two-dimensional Gel Electrophoresis
Proteins were extracted according to Bestel-Corre et al. (36) and lyophilized. Proteins were dissolved in solubilization buffer for two-dimensional PAGE (37). Protein concentrations were determined using a modified Bradford assay using BSA as a standard. SDS-PAGE and two-dimensional PAGE were performed as described by Wang et al. (38). After electrophoresis, gels were stained with Coomassie Blue R-250 or electrophoretically transferred to nitrocellulose filters. Electro-transfer and protein blots procedures were performed as described by Gu et al. (35). Protein blots were incubated with a 1:500 dilution of a polyclonal serum made against the tomato LAP-A1 (35). Signals were analyzed and quantified using Quantity One software (Bio-Rad).
Identification of Proteases of the Proteasome Pathway
For in-gel digestion, 20 μg of proteins from the Mono Q fractions were by SDS-PAGE. After coloration with Coomassie Blue R-250, the gel was cut into 1.5-mm slices. Each band was further cut and washed twice in 100 μl of destaining solution (50 mm NH4HCO3/CH3CN (50:50, v/v)) at room temperature for 30 min before dehydration with 100 μl of pure CH3CN. The solution was then removed, and the gel pieces were dried in a speed vacuum and rehydrated in 100 μl of 7% H2O2, at room temperature for 15 min, in the dark. The oxidizing solution was removed, and the gel slices were rinsed in water and then dehydrated in CH3CN as described above. After complete drying, the bands were rehydrated in 20 μl of digestion buffer (150 mm Tris-HCl (pH 8.1), 10 mm CaCl2, 100 mm urea/CH3CN (95:5, v/v)), containing 150 ng sequencing grade modified trypsin (Promega, Madison, WI). After 15 min of incubation at 4 °C, 30 μl of digestion buffer was added, and the digestion reaction was carried out at 37 °C for 5 h, under permanent shaking. The digestion solution was then collected, and peptides were extracted from the gel by diffusion in 50 μl of 0.3 m urea, 90% (v/v) CH3CN for 30 min with intermittent sonication. Digestion and extraction solutions were pooled and dried under speed vacuum. Peptide mixtures were redissolved in 25 μl 5% (v/v) CH3CN, 0.2% (v/v) formic acid in water prior to LC-MS/MS analysis.
For nano-liquid chromatography (nano-LC)-electrospray ionization-MS/MS, chromatographic separation of digested proteins was accomplished by loading 0.15 μg of peptide mixture onto a 15-cm fused silica C18 column (75 μm inner diameter, 3 μm, 100 Å, and 360 μm outer diameter; Dionex). Peptide elution was achieved using the following linear gradients: (a) from 10 to 40% solvent B (90% (v/v) CH3CN, 0.1% (v/v) formic acid in water) for 40 min and (b) from 40 to 90% solvent B for 5 min. The remaining percentage of the elution solvent was made of solvent A (water/CH3CN (1:1, v/v)) containing 0.1% formic acid). Flow rate through the nano-LC column was 200–300 nl/min. For automatic LC-MS/MS analysis, the QTOF Ultima instrument was run in data-dependent acquisition mode with the following parameters: 1-s scan time and 0.1-s interscan delay for MS survey scans; 400–1400 and 50–2000 m/z mass ranges for the survey and the MS/MS scans, respectively; five components; MS/MS to MS switch after 5 s; switchback threshold, 30 counts/s; include charge states 2, 3, and 4 with the corresponding optimized collision energy profiles. The acquired data were post-processed to generate peak lists (.pkl) with PeptideAuto, which is part of proteinLynx from Masslynx 4.0. Each peak list was submitted to searches using MASCOT 2.0. MASCOT search parameters used with MS/MS data were as follows: data base = A. thaliana (nuclear, mitochondrial, and chloroplastic genome), enzyme = trypsin/protein, one missed cleavage allowed, a peptide tolerance = 0.25 Da, MS/MS tolerance = 0.25 Da, peptide charge = 2+/3+/4+, and variable modifications such as oxidized methionine under sulfone and sulfoxide and cysteic acid. The algorithm can efficiently choose the best amino acid sequence, from all possible amino acid combinations, to interpret the MS/MS spectrum according to the same chemical modifications defined in Mascot.
Extraction of Total RNA and Estimation of Relative Transcript Level by RT-PCR
Total RNA was isolated from 100 mg of plant tissue using the TRIzol reagent protocol (Invitrogen). DNase treatment was performed to remove contaminant genomic DNA using the RQ1 DNase-RNase-free (Promega), according to the manufacturer's instructions. Reverse transcription was performed using 5 μg of total RNA (first strand cDNA synthesis, Amersham Biosciences) as detailed in the manufacturer's instructions.
Semi-quantitative RT-PCR was performed using 0.5 μl of a 10× dilution of the RT reaction with 0.1 μm of each oligonucleotide primer, using the TITANIUMTM Taq polymerase (Clontech). After an initial denaturation step of 3 min at 95 °C, PCR was performed as follow: 30 s denaturation (94 °C), 20 s hybridization (55, 60, or 62 °C according to the primers), and 45 s elongation (68 °C); the number of PCR cycles was defined for each gene studied to obtain a detectable signal without reaching saturation (Table 1). Amplification products (10 μl) were loaded on a 1.3% (w/v) ethidium bromide-stained agarose gel. Oligonucleotide pairs used for amplification are listed in Table 1. Three independent experiments were analyzed. PCRs were performed in duplicate. Each set of reactions was stopped at three different numbers of cycles and analyzed on agarose gel. Signals were analyzed and quantified using Quantity One software (Bio-Rad). The expression level of each gene was normalized according to the ACT2/8 level and the control conditions.
TABLE 1.
Sets of PCR primer sequences used to amplify gene-specific regions and corresponding size of the amplified products
| Gene | AGI number | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) | Length | No. of cycles | |
|---|---|---|---|---|---|---|
| bp | ||||||
| Proteasome 20 S subunits | α7 | At2g27020 | AAGAAGGCGTTATCGAGGTG | AAAGACTCCAATGGTTCATGC | 317 | 23, 24, 25 |
| β1 | At4g31300 | TCTACGGGTTCTTCGACCAG | CGGGTCATTCATTACAACAGAA | 383 | 23, 24, 25 | |
| β2-1 | At3g27430 | TGATCTACATCAAGGCTCTGTG | TTTCGAAAGGACGAAGTCTCTC | 156 | 24, 25, 26 | |
| β2-2 | At5g40580 | TGAATCTCGTGTGAGAGACAGA | TTTCAAAACCGAATCATCCA | 184 | 24, 25, 26 | |
| β5-1 | At1g13060 | GTTTTCTAATCGTTCACTCCCTTT | TTTTTGCAGAAAAAGAGAATCTTATAG | 150 | 25, 26, 27 | |
| β5-2 | At3g26340 | CATCACTTATGTTTTGTTCATCAAT | TGTTATCTATTATGCAACGTGATCATT | 115 | 25, 26, 27 | |
| β6 | At3g60820 | GGTGCTCAAGGTTCTGGTTC | CAAGGATTCGAGTGATTCTCA | 431 | 24, 25, 26 | |
| β7 | At1g56450 | CTTGCTAGGCCAATTCTTCG | GATGTGGTTGCTTGGCTTCT | 259 | 24, 25, 26 | |
| Other proteases | TPPII | At4g20850 | CTAAATCAGTATCAGAACGCTTGG | CATGGCTAACCCTTTCTGGTATAG | 391 | 21, 22, 23 |
| NEURO | At1g67690 | GCTTTTGCCGTCTTTGACTCAG | GGTCAACACGTAGGCCCGAG | 307 | 30, 31, 32 | |
| TOP | At5g10540 | CGAACCATTTTCACATTCTCCAC | AATCGGAGAATTTGGGTGGAGG | 291 | 26, 27, 28 | |
| TOPorg | At5g65620 | CCATTTTTCCTACCACAACCAG | GGGAAAACTGAAGGAAAGGATTTG | 314 | 26, 27, 28 | |
| LAP1 | At2g24200 | GGAGTGGAGAGAAGCTGTGGAGG | CAGTCTCTCCCTCACGCTCTC | 297 | 23, 24, 25 | |
| LAP2 | At4g30920 | GGAGTGGAGAGAAGTTATGGAG | GCAAAAACACAACTAGTTCACATC | 314 | 21, 22, 23 | |
| LAP3 | At4g30910 | GGAGTGGAGAGAAGTTATGGAG | CTGAGAATGTTAAATAAGAACTGGG | 313 | 25, 26, 27 | |
| PIP2 | At2g14260 | CGTCGGACTCACACTTGC | ACATTCGGTTTCGCGTTAAC | 357 | 25, 26, 27 | |
| Constitutive genes | ACT2 | At3g18780 | GGTAACATTGTGCTCAGTGGTGG | AACGACCTTAATCTTCATGCTGC | 108 | 22, 23, 24 |
| ACT8 | At1g49240 |
Preparation of Oxidized Protein
To test the efficiency of the 20 S proteasome/TPPII couple in the degradation of oxidized proteins, oxidative treatment of 10–15 mg of either BSA, lysozyme, or α-casein was performed in vitro according to Ref. 24. Proteins were incubated in 20 mm sodium phosphate buffer (pH 7.2) in the presence or absence of 200 μm FeSO4 and 50 mm ascorbate in a final volume of 1 ml, at 37 °C, for 3 h. Five μl of either oxidized or nonoxidized proteins was submitted to DNPH derivatization to check their oxidation status, according to Basset et al. (24). Ascorbate and FeSO4 were eliminated using a 6-DG column (Bio-Rad). Primary amine residues were blocked by reductive methylation using the procedure of Ref. 39 with the following modifications: 0.2–0.5 μmol of protein were incubated with an excess of 100-fold formaldehyde and sodium cyanoborohydride at 25 °C for 23 h. These two reagents were then removed using a 6-DG column (Bio-Rad). After protein measurement, the samples were stored at −80 °C.
Degradation of Protein Substrates by the 20 S Proteasome/TPPII Mixture
After a Sephacryl chromatography step performed with the leaf protein extract, the 20 S proteasome and TPPII containing fractions were pooled (P1 pool) and used to monitor the degradation of the proteins oxidized in vitro. Twenty μg of methylated protein substrates were incubated in 0.1 m sodium phosphate buffer (pH 7.2), 5% glycerol, 1 mm dithiothreitol, and 35 μl of P1 pool (the protein concentration of P1 pool was 1 mg/ml), at 37 °C for 3, 4, and six h. Background changes of (i) the media, (ii) the protease mix, (iii) the substrates, and (iv) the protease inhibitors were monitored using a set of controls for each time point. Each measurement was made in duplicate.
The amount of newly formed α-amino groups was measured with fluorescamine (Sigma). Ten μl of the proteolytic reaction assay were harvested, and proteolysis was stopped with 10 μl of 10% trifluoroacetic acid. Then 100 μl of sodium phosphate buffer (pH 6.8) and 50 μl of fluorescamine (0.3 mg/ml in acetone) were added. The mixture was vortexed for 1 min, and water was added to a final volume of 1 ml. Fluorescence was measured using an excitation wavelength of 370 nm and an emission of 480 nm.
Other Assays
Proteins were quantified (40) using the Bio-Rad microassay reagent. Bovine γ-globulin was used as the standard. Native-PAGE was performed with 6% (w/v) polyacrylamide gels. Gels were fixed, stained with Coomassie Blue, and destained using standard methods. Western blot analyses were carried out as in Ref. 24 using polyclonal antibodies raised against either maize root 20 S proteasome or leaf tomato LAP.
RESULTS
Evidence for the Proteasome Pathway in A. thaliana
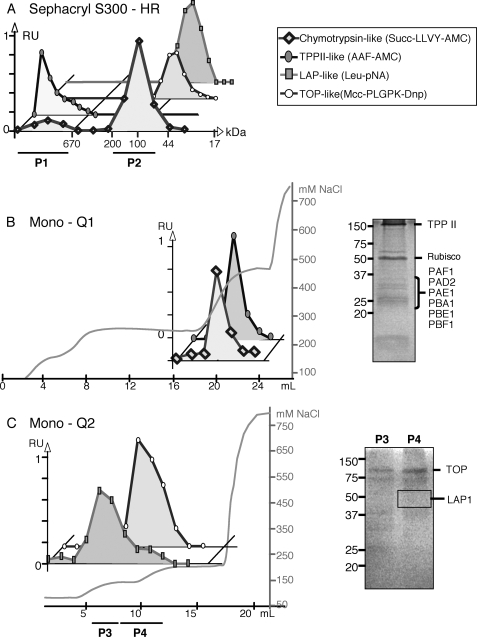
The presence of the different proteases involved in the proteasome pathway, including the 20 S proteasome, TPPII, TOP, and LAP, was investigated in the leaves of 6-week-old A. thaliana plants. A leaf protein extract was first submitted to a size-exclusion chromatography step, using a Sephacryl S-300 column. The pattern of the chromatogram obtained is presented in Fig. 1. Using specific substrates, the different peptidase activities were then assayed in the eluted fractions (Fig. 1A). The use of Suc-LLVY-AMC peptide has allowed the identification of two peaks of chymotrypsin-like activity (Fig. 1A). The first one, around 700 kDa, corresponds to the size of the 20 S proteasome, and its presence was confirmed by Western blot experiments using anti-plant 20 S proteasome antibodies (data not shown). The second major peak of chymotrypsin-like activity, which accounted for ∼80% of the total activity, was eluted with proteins having a molecular mass of around 100 kDa (no cross-reaction was observed with 20 S proteasome antibodies; data not shown). Thus we conclude that, in contrast to mammals and yeast, the 20 S proteasome is not responsible for the major part of the chymotrypsin-like activity.
FIGURE 1.
Partial purification and characterization of the proteases involved in the proteasome pathway from A. thaliana leaves. A, enzymatic activity elution pattern during S-300 Sephacryl chromatography. P1 refers to the fractions containing 20 S proteasome and TPPII activities; P2 refers to the fractions containing TOP and LAP activities. B, anion-exchange chromatography (Mono Q) of the proteins present in P1 pool (from the Sephacryl S-300-HR) and SDS-polyacrylamide gel of the fraction containing the maximum activity of proteasome 20 S and TPPII used for protein identification by mass spectrometry analysis. Rubisco, ribulose-bisphosphate carboxylase/oxygenase. C, anion-exchange chromatography step (Mono Q) of the proteins present in the P2 pool (from the Sephacryl S-300-HR) and SDS-polyacrylamide gels of the pools P3 and P4 used for protein identification by mass spectrometry analysis. Activities are expressed as relative units ranging from null to one. Salt concentration of elution buffer is expressed as millimolar NaCl. RU, relative units.
A TPPII-like activity, monitored using the AAF-AMC tripeptide as a substrate, was measured in the first peak corresponding to high molecular weight complexes (Fig. 1A). This is in agreement with a previous report showing that Arabidopsis TPPII is a large oligomeric 5–9-MDa complex (30). A TOP-like activity was measured using the specific fluorogenic substrate Mcc-PLGPK-Dnp. It was detected around the 80–100-kDa fractions, which are close to the 78-kDa size of mammalian TOP (41).
Leu-pNA was used as substrate for screening aminopeptidase activities with LAP or LAP-like activities. LAPs have been described in mammals and plants, as hexameric complexes composed of six 50–55-kDa subunits (42). The A. thaliana genome encodes three LAP proteins; only LAP1 has a putative cytosolic localization (43, 44). However, after screening the size-exclusion chromatography fractions, only one peak of Leu-pNA hydrolyzing activity was detected in the 80–100-kDa protein fractions (Fig. 1A). These data show that Leu-pNA hydrolyzing Arabidopsis peptidases distinct from the hexameric LAPs may be involved in degrading the peptides generated by the proteasome.
Two complementary approaches were used to further identify and characterize the peptidases of the proteasome pathway. The first strategy was a proteomics approach consisting of an additional protein purification step followed by an SDS-PAGE fractionation and identification by mass spectrometry. As a second approach, protease activities were characterized according to their sensitivity to known inhibitors.
Fractions eluted from the gel filtration column exhibiting chymotrypsin- and TPPII-like activities were pooled together (Fig. 1A, P1 pool) and applied on an anion-exchange Mono Q column. It appeared that both chymotrypsin- and TPPII-like activities eluted in the same fractions (around 300 mm NaCl, Fig. 1B). Proteins present in the fraction containing the maximum activities were separated by SDS-PAGE (Fig. 1B). After Coomassie Blue staining, bands present in the gel were excised and trypsin-digested, and peptide mixtures were submitted to nano-LC-ESI-MS/MS. This proteomics analysis led to the identification of several subunits of the 20 S proteasome (Table 2; Fig. 1B). The chymotrypsin activity of P1 pool from the Sephacryl S-300 and from the Mono Q1 was inhibited by 98 and 99%, respectively, when measured either in the presence of 10 μm lactacystin or after immunoprecipitation experiments with anti-20 S proteasome antibodies (Table 3). No inhibition was observed when chymotrypsin activity was measured in the presence of the TPPII inhibitor butabindide. These data clearly indicate that this activity was related to the 20 S proteasome.
TABLE 2.
Proteases of the proteasome pathway identified by mass spectrometry analysis
This table gives the protein acronym of proteases identified after SDS-PAGE in Mono Q1 and Mono Q2 fractions of Fig. 1; the gene designation; theoretical isoelectric point (pI); Mascot score of in-gel digestion protocol (Score column); deduced molecular mass; protein sequence coverage; and number of peptides assigned using the in-gel digestion protocol. The protease family was assigned by the peptidase MEROPS database.
| Proteins | Gene | pI | Score | Mass | Coverage | No. peptides |
|---|---|---|---|---|---|---|
| Da | % | |||||
| Proteases identified in Mono Q1 fractions | ||||||
| TPPII | At4g20850 | 5.92 | 584 | 152,273 | 7.1 | 9 |
| 20 S proteasome α-subunit D2 (PAD2) | At5g66140 | 8.73 | 84 | 27,307 | 9.2 | 2 |
| 20 S proteasome α-subunit F1 (PAF1) | At5g42790 | 4.99 | 73 | 30,457 | 10.1 | 2 |
| 20 S proteasome α-subunit E1 (PAE1) | At1g53850 | 4.70 | 75 | 25,931 | 14.4 | 3 |
| 20 S proteasome β-subunit F1 (PBF1) | At3g60820 | 6.95 | 51 | 24,628 | 9.9 | 2 |
| 20 S proteasome β-subunit E1 (PBE1) | At1g13060 | 5.77 | 75 | 25,264 | 12.2 | 2 |
| 20 S proteasome β-subunit A1 (PBA1) | At4g31300 | 5.24 | 84 | 25,280 | 4.7 | 2 |
| Proteases identified in Mono Q2 fractions | ||||||
| Peptidase M3 family protein/thimet oligopeptidase family protein (Thimet, TOPorg) | At5g65620 | 5.90 | 1479 | 88701 | 36.0 | 27 |
| Peptidase M3 family protein/thimet oligopeptidase family protein (Thimet, TOP) | At5g10540 | 5.45 | 736 | 78994 | 21.3 | 15 |
| Leucine aminopeptidase (LAP1) | At2g24200 | 5.66 | 175 | 54475 | 14.4 | 4 |
TABLE 3.
Effect of different effectors on proteolytic activities
Sephacryl S-300 fractions corresponding to pools 1 and 2 were used for inhibition measurement of the 20 S proteasome, TPPII, and TOP activities. Desalted clarified extracts were used for the immunoprecipitation of LAP activity. Activities were assayed as described under “Experimental Procedures.” The inhibitor and activator effects are expressed as percentage of the control (100%). Data correspond to the means of at least eight independent measurements ± S.D. The abbreviations used are as follows: Z, benzyloxycarbonyl; PMSF, phenylmethylsulfonyl fluoride; Cpp, carboxy-3-phenylpropyl; pAB, polyclonal antibody.
| Effector | Effector concentration | % of the control |
|---|---|---|
| 20 S proteasome (P1 pool) | ||
| Control | 100 | |
| Lactacystin | 10 μm | 11 ± 2 |
| Butabindide | 20 μm | 103 ± 20 |
| Anti-20 S antibody | 1 ± 4 | |
| Tripeptidyl peptidase II (P1 pool) | ||
| Control | 100 | |
| Butabindide | 20 μm | 1 ± 1 |
| AAF-CMK | 50 μm | 1 ± 1 |
| Z-AAF-CMK | 50 μm | 105 ± 9 |
| MG132 | 20 μm | 95 ± 6 |
| Bestatin | 20 μm | 96 ± 6 |
| PMSF | 2 mm | 1 ± 2 |
| E64 | 20 μm | 105 ± 8 |
| Pepstatin | 20 μm | 113 ± 8 |
| 1,10-Phenanthroline | 2 mm | 94 ± 8 |
| EDTA | 2 mm | 105 ± 8 |
| Thimet oligopeptidase (P2 pool) | ||
| Control | 100 | |
| Cpp-AAF-pAB | 20 μm | 21 ± 2 |
| PMSF | 2 mm | 96 ± 10 |
| E64 | 20 μm | 87 ± 1 |
| 1,10-Phenanthroline | 2 mm | 38 ± 1 |
| EDTA | 2 mm | 59 ± 5 |
| MnCl2 | 2 mm | 441 ± 22 |
| CoCl2 | 2 mm | 220 ± 11 |
| ZnCl2 | 2 mm | 2 ± 2 |
| Leucine aminopeptidase (crude extract) | ||
| Control | 100 | |
| Anti-LAP antibody | 83 ± 2 | |
TPPII was among the major proteins identified by SDS-PAGE and nano-LC-ESI-MS/MS (Fig. 1B; Table 2). Peptidase assays performed on either the Sephacryl S-300-P1 pool, or Mono Q1 fractions (Fig. 1), using two specific inhibitors of TPPII, butabindide and AAF-CMK, showed an inhibition close to 100% of this activity (Table 3). Moreover, benzyloxycarbonyl-AAF-CMK (a negative control for the serine protease inhibitor AAF-CMK) did not inhibit this activity. TPPII activity also showed a high sensitivity to the serine protease inhibitor phenylmethylsulfonyl fluoride, although it was not affected by proteasome (MG132), Cys protease (E64), or metallopeptidase (EDTA and 1,10-phenanthroline) inhibitors (Table 3). Aminopeptidase (bestatin) and Asp protease (pepstatin) inhibitors did not have any effect on this activity either (Table 3). These results are in accordance with previous descriptions of the Arabidopsis TPPII protease (30) and confirm the assumption that the AAF-AMC-like activity measured in these pools was due to TPPII.
The TOP- and LAP-like activities corresponding to proteins ranging from 75 to 140 kDa were pooled (Fig. 1A, P2 pool) and submitted to an anion-exchange chromatography step (Fig. 1C, Mono Q2). TOP activity was eluted with 135 mm NaCl. The corresponding fractions were pooled (P4) and submitted to SDS-PAGE (Fig. 1C). Subsequent proteomics analysis allowed the identification of two metallopeptidases, TOP (At5g10540) and TOPorg (At5g65620) (Table 2), which are annotated in the TAIR data base as M3/thimet oligopeptidase family proteins. In the Arabidopsis genome, another gene (At1g67690) presenting sequence similarities to animal thimet oligopeptidase family is annotated M3 peptidase similar to the cytosolic neurolysin, and named TOP-like (TOPL) in this work. Among the three M3/thimet oligopeptidase family proteins identified in A. thaliana, two are classified as cytosolic peptidases (TOP and TOPL), and the third one (TOPorg), presenting a transit peptide, is thought to have a mitochondrial or chloroplastic localization. The absence of TOPL peptides in the proteomic analysis (Table 2) suggests that TOPL is probably less abundant than TOP in the cytosol. The relative abundance of TOP and TOPorg compared with TOPL was also correlated with the levels of these RNAs in Arabidopsis plants. Compared with TOP and TOPorg RNAs, five additional PCR cycles were necessary to detect a TOPL RNAs using RT-PCR gene-specific primers (supplemental Fig. S1). This is a likely explanation of why no peptide was found to match with the product of the TOPL gene when the proteins of the Mono Q2 (Fig. 1C) were analyzed by mass spectrometry.
The sensitivity of TOP-like activity to a set of protease inhibitors (Table 3) confirmed the expectation, based on sequence comparisons, that these proteases are members of the TOP family. TOP-like activity was inhibited by the metalloprotease inhibitors (EDTA and 1,10-phenanthroline) (Table 3) suggesting they belong to this family. Moreover, the use of the TOP selective inhibitor, carboxy-3-phenylpropyl-AAF-polyclonal antibody (14), in the peptidase assay led to an 80% inhibition of the activity. Taken together, these results strongly suggest that the protease activity was effectively due to TOP enzyme. In addition, we were able to demonstrate that TOP is activated by MnCl2 (4.4-fold) and CoCl2 (2.2-fold), but 98% is inhibited by ZnCl2 (Table 3). Moreover, the inhibition of the TOP activity by EDTA was reversed and stimulated by the addition of MnCl2, but not by MgCl2 or ZnCl2. This suggests that the Arabidopsis TOP could be a manganese-dependent metalloprotease.
As carried out for TOP, the MS characterization of LAP was investigated in the 80–100-kDa protein fractions exhibiting LAP-like activity (Fig. 1A, P2 pool). These fractions were first loaded on a Mono Q column. Proteins exhibiting LAP-like activity were eluted from the Mono Q2 around 120 mm NaCl (Fig. 1C). The most active fractions were pooled (P3 pool) and subjected to SDS-PAGE fractionation. Mass spectrometry analysis allowed the identification of leucine aminopeptidase 1 (LAP1, At2g24200, see Table 2), corresponding to a 320–360-kDa hexameric enzyme (43, 45). AtLAP1 is cytosolic in location and is similar to the mammalian LAP that processes peptides from the proteasome. The co-localization of LAP activity and LAP1 peptides in the 80–100-kDa fractions was perplexing. Previous studies with the tomato LAP-A1 indicated that its hexameric structure was critical for LAP activity (46). Therefore, it was possible that the inactive LAP1 dimers were present in the 80–100-kDa fraction, and the Leu-pNA hydrolyzing activity (LAP-like activity) that was detected represented a novel aminopeptidase activity. Many different aminopeptidases are known to hydrolyze LAP substrates (32, 47). Given the possible complexity of Leu-pNA-hydrolyzing proteins in the leaf extracts, the use of aminopeptidase inhibitors with nonpurified protein fractions would not be useful. Therefore, LAP activity was determined in desalted leaf extracts in the presence or absence of anti-LAP antibodies. As reported in Table 3, LAP activity was found to represent 17% of the total Leu-pNA degrading activity in Arabidopsis leaves. This means that the major part of the Leu-pNA degrading activity is due to one or several nonhexameric LAPs. In A. thaliana, two other hexameric LAPs exist, LAP2 (At4g30920) and LAP3 (At4g30910), both predicted to be chloroplastic. On the basis of PCR analysis, LAP1 is the most abundantly expressed hexameric LAP in Arabidopsis leaves (supplemental Fig. S1), which may explain why only LAP1 was detected after MS analysis.
Cadmium Treatment Increases Protein Oxidation and Up-regulates the 20 S Proteasome Pathway in the Leaves of A. thaliana
As it has been hypothesized that the 20 S proteasome system in mammals could be involved in the degradation of oxidized proteins (8), the response of the proteasome pathway in response to cadmium stress was investigated. In addition, the capacity of the 20 S proteasome to degrade oxidized proteins was also determined.
To assess the impact of cadmium treatments on the proteasome pathway, A. thaliana plants were grown hydroponically and submitted to a 50 μm cadmium treatment. The leaves of 24-, 48-, and 144-h cadmium-treated and control plants were analyzed for protein content and oxidation level, as well as for proteolytic activities. No change in total protein content was observed in cadmium-treated leaves compared with controls (Table 4). However, the increase in carbonyl groups, measured after derivatization with DNPH, indicates that the protein oxidation state increased after 48 and 144 h of treatment with cadmium (Table 4). The total endopeptidase activities in leaf protein extracts also increased after 144 h of 50 μm cadmium addition (Table 4).
TABLE 4.
Protein and carbonyl contents and global endopeptidase activities in the leaves of Arabidopsis plants exposed to 50 μm Cd for 24, 48, and 144 h
Data are the mean ± S.D. of six (protein), two (carbonyl), and three (endopeptidase) independent experiments.
| Cadmium concentration (μm) | 24 h |
48 h |
144 h |
|||
|---|---|---|---|---|---|---|
| 0 | 50 | 0 | 50 | 0 | 50 | |
| Protein content (mg g−1 FW) | 19.9 ± 2.5 | 21.1 ± 2.8 | 18.8 ± 1.9 | 18.8 ± 1.8 | 20.0 ± 3.5 | 19.1 ± 2.6 |
| Carbonyl content (nmol mg−1 protein) | 3.04 ± 0.14 | 2.73 ± 0.07 | 2.83 ± 0.23 | 3.56 ± 0.07a | 2.94 ± 0.38 | 5.57 ± 1.04a |
| Endopeptidase activity (pg of azocasein min−1 mg−1 FW) | 5.3 ± 0.3 | 5.4 ± 0.5 | 4.8 ± 0.5 | 6.0 ± 1.1 | 5.3 ± 0.4 | 8.7 ± 0.9b |
a Significant difference between Cd-treated and control plants is <0.05 by Student's t test.
b Significant difference between Cd-treated and control plants is <0.01 by Student's t test.
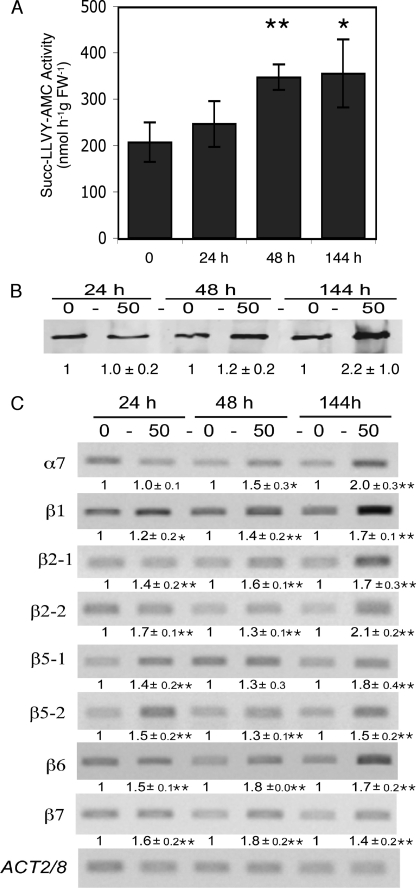
The 20 S proteasome activity was monitored during cadmium stress in the leaves of Arabidopsis plants harvested after 24, 48, or 144 h (6 days) of treatment. The chymotrypsin activity measured in the P1 pool after Sephacryl S-300 (Fig. 1A) was used to follow the 20 S proteasome activity. Our data indicated a 1.7-fold increase of the chymotrypsin activity of the 20 S proteasome after 48 and 144 h (Fig. 2A). Protein blot analysis also revealed an increase in 20 S proteasome content after 144 h of treatment (Fig. 2B).
FIGURE 2.
Changes in 20 S proteasome activity, amount, and transcript levels in the leaves of Arabidopsis plants grown for 24, 48, and 144 h in the presence of 50 μm cadmium. 20 S proteasome chymotrypsin-like activity (A), amount (B), and subunit transcript levels measured by semi-quantitative RT-PCR (C). Western blot (B) and luminescent (C) signals were quantified using the Quantity One software (Bio-Rad). Values represent the relative intensity of each signal normalized to control (0 cadmium) in B and to ACT 2/8 signal in C. Data represent the mean of three (activity), six (Western blot) and three (transcript) biological replicates. Asterisks indicate significant differences between cadmium-treated and control plants (**, <0.01; *, < 0.05, Student's t test).
To determine whether the increase of the 20 S proteasome activity in response to cadmium treatment was correlated with changes in proteasome subunit RNA levels, semi-quantitative RT-PCR experiments were performed. Specific primers were designed to monitor the RNAs of six proteasome subunits (Table 1). We chose three structural monogenic subunits as follows: α7 (PAG1, At2g27020), β6 (PBF1, At3g60820), and β7 (PBG1, At1g56450) and the three catalytic subunits β1, β2, and β5, respectively, encoded by either one gene, β1 (PBA1, At4g31300), or two genes, β21 (PBB1, At3g27430) and β22 (PBB2, At5g40580) and β51 (PBE1, At1g13060) and β52 (PBE2, At3g26340). The results, presented in Fig. 2C, showed an increase in transcript level for all the subunits analyzed from 24 h, except for α7, whose level increased only after 48 h. After 144 h, all the subunit transcripts presented a 1.4–2.1-fold increase. These data clearly indicate that RNAs encoding subunits of the 20 S proteasome were up-regulated in response to cadmium.
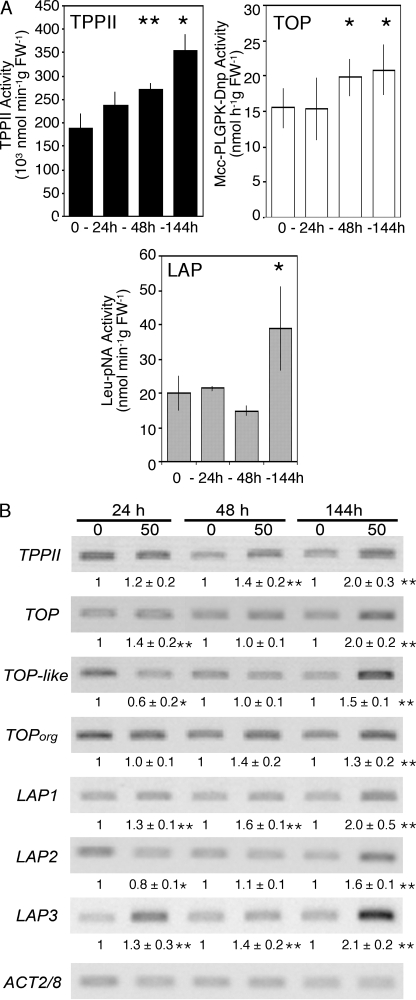
The expression and activity levels of TPPII, TOP, and LAP in response to cadmium treatment were also analyzed in Arabidopsis leaves. Specific primers were designed for each peptidase (Table 1), and the transcript analyses were performed using semi-quantitative RT-PCR (Fig. 3B). As for the 20 S proteasome, the TPPII activity from control and cadmium-treated leaves was measured in the P1 pool after the Sephacryl S-300 chromatographic step. From 24 h of treatment, TPPII presented a gradual increase in activity, reaching a 1.8-fold increase after 144 h (Fig. 3A). A similar increase was detected for its transcript level (At4g20850) (Fig. 3B).
FIGURE 3.
Changes in activity, amount, and transcript level of various proteases in the leaves of Arabidopsis plants grown for 24, 48, and 144 h in the presence of 50 μm cadmium. A, activity. TPPII and TOP + TOPL activities are expressed in nmol h−1 g FW−1; LAP activities are expressed in nmol min−1 g FW−1. Data represent the mean ± S.D. of three independent experiments. B, transcript levels measured by semi-quantitative RT-PCR. Luminescent signals were quantified using the Quantity One software (Bio-Rad). Values represent the relative intensity of each signal normalized to control (0 cadmium) and to ACT 2/8 signal. Data represent the means ± S.D. of three independent biological replicates. Asterisks indicate significant differences between cadmium-treated and control plants (**, <0.01, and *, < 0.05, Student's t test).
TOP activity from control and cadmium-treated leaves was measured in the P2 pool after the Sephacryl S-300 chromatographic step. It presented a moderate increase (1.3-fold) only after 48 h. This increase was maintained up to 144 h (Fig. 3A). In semi-quantitative RT-PCR experiments, the transcript level of the three Arabidopsis thimet oligopeptidases, TOP (At5g10540), TOPL (At1g67690), TOPorg (At5g65620), was monitored. The most abundant cytosolic thimet oligopeptidase isoform, TOP, was induced after 144 h (with 1.4- and 2.0-fold augmentations, respectively), whereas the minor isoform, TOPL, first decreased at 24 h and then increased to reach 1.5 times the control level after 144 h (Fig. 3B). The transcript level of the organelle specific isoform, TOPorg, was slightly affected by cadmium treatment (Fig. 3B). It should be noted that the final increase of TOP activity measured may thus be overestimated because it corresponds to the activity of a mix of TOP, TOPL, and TOPorg.
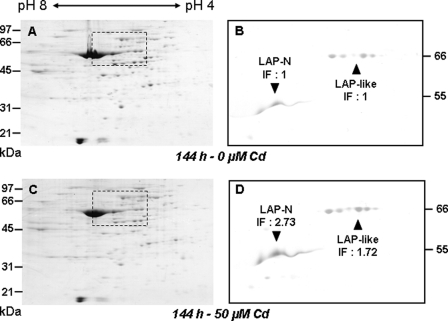
The activity of the hexameric LAP was measured in the desalted clarified extracts of control and cadmium-treated plants after immunoprecipitation with anti-LAP antibodies. In these extracts, LAP antibodies were found to precipitate only 11–18% of the total Leu-pNA degrading activity, which means that hexameric LAPs are not the major pNA-degrading peptidases in cadmium-treated as well as in control Arabidopsis leaf extracts. LAP activity did not change during the first 48 h of treatment and increased by a factor of 1.8 only after 144 h (Fig. 3A). This increase in activity was correlated with an increase in LAP amount. Indeed, immunoblot analysis of LAP proteins after two-dimensional PAGE showed that LAP and 66-kDa LAP-like proteins increased 2.7- and 1.7-fold, respectively, in 144-h cadmium-treated leaves compared with the untreated control leaves (Fig. 4). Among the three LAP genes (LAP1-At2g24200, LAP2-At4g30920, and LAP3-At4g30910), transcript levels were found to increase after 24 h of treatment for LAP1 and LAP3 (Fig. 3B), but the three genes presented a similar increase in transcript level after 144 h (2.0-, 1.6-, and 2.1-fold for LAP1, LAP2, and LAP3, respectively).
FIGURE 4.
Accumulation of LAP-A and 66-kDa LAP-like proteins in the leaves of Arabidopsis plants grown for 144 h in the presence of 50 μm cadmium. Total proteins (80 μg) were fractionated by two-dimensional PAGE and visualized by Coomassie Blue R-250 staining for protein loading control (A and C) or electroblotted and incubated with a 1:5000 (w/v) dilution of polyclonal LAP-A antiserum (B and D). LAP-A (▼) and LAP-like (▲) proteins masses are shown (in kDa). The pH range of the isoelectric focusing gels and the protein size markers are mentioned. Immunoblots B and D correspond to the dashed rectangles in Coomassie-stained gels A and C, respectively. Western blot signals were quantified using the Quantity One software (Bio-Rad). The increase factor (IF) in protein amounts in cadmium-treated versus control plants is indicated below each protein name.
It is noteworthy that all the different proteases of the 20 S proteasome pathway, including the 20 S proteasome, exhibited a similar increase in activity after 144 h (i.e. 1.3–1.8-fold increases). Transcript levels of the different proteases thought to belong to this pathway also increased in a proportional manner. These data indicate that the peptidases of the proteasome pathway are regulated at multiple levels, including transcript and protein level in response to cadmium, and suggest that the proteasome proteolytic pathway may be involved in the degradation of oxidized protein generated by the cadmium treatment.
Degradation of Oxidized Proteins by 20 S Proteasome and TPPII
Considering the increase in oxidized proteins in cadmium-treated Arabidopsis leaves (Table 4), and the induction of the different peptidases of the proteasome pathway (Figs. 2 and 3), we sought to determine whether oxidized proteins were preferential substrates for the 20 S proteasome pathway of A. thaliana. To test this hypothesis, BSA and lysozyme, two globular proteins whose structure can be significantly modified by oxidative treatment, were used as substrates. α-Casein, a linear protein, served as a control. These proteins were submitted to oxidative treatments and used as substrates for degradation by the 20 S proteasome and TPPII. The three proteins were first subjected to a mild metal-catalyzed oxidative treatment (in the presence of 50 mm ascorbate and 200 μm FeSO4). Their oxidation state was checked using anti-DNP antibodies after derivatization of the carbonyl groups with DNPH (supplemental Fig. S2A). Oxidized and nonoxidized proteins were then submitted to a reductive methylation (using formaldehyde and sodium cyanoborohydride) to block primary amine groups. In these conditions, the amount of primary amines, quantified by fluorometric assays using fluorescamine (a primary amine group reagent), was reduced by a factor of 80–100 (supplemental Fig. S2B).
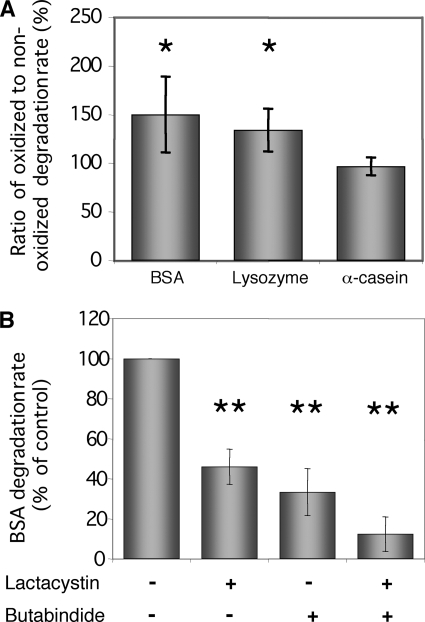
Methylated BSA, lysozyme, and α-casein were used as substrates for degradation by the 20 S proteasome and TPPII (P1 pool issued from the Sephacryl S-300 step, Fig. 1A). The degradation products of oxidized and nonoxidized proteins were quantified using fluorescamine. The data in Fig. 5A show the ratio of oxidized versus nonoxidized degraded substrate for each protein. BSA and lysozyme, but not α-casein, were better degraded under their oxidized form (1.5- and 1.3-fold, respectively). As expected, no enhanced degradation of oxidized α-casein was observed consistent with the facts that α-casein is a linear protein, whose structure is not further denaturated by oxidation (5).
FIGURE 5.
Degradation of oxidized protein by 20 S proteasome/TPPII. A, degradation rate of oxidized proteins are normalized to the degradation rate of nonoxidized proteins. The degradation rate of BSA, lysozyme, and α-casein was determined by fluorescent assays using fluorescamine as described under “Experimental Procedures.” Data represent the means ± S.D. of three independent biological replicates. B, effect of 20 S proteasome (10 μm lactacystin) and TPPII (20 μm butabindide) inhibitors on the degradation rate of BSA. Asterisks indicate significant differences between treated and control assays (**, <0.01, and *, <0.05, Student's t test).
To assess the contributions of the proteasome and TPPII to oxidized protein degradation, the degradation of BSA was tested in the presence or absence of proteasome and TPPII inhibitors. In the presence of either 10 μm lactacystin or 20 μm butabindide, the degradation of oxidized BSA was inhibited by 46 and 33%, respectively (Fig. 5B). These data indicated that both the 20 S proteasome and TPPII were able to degrade oxidized BSA. With both inhibitors, BSA degradation was almost completely inhibited (88%) (Fig. 5B). Similar results were obtained using lysozyme as substrate (data not shown). Based on inhibitor efficiency (Table 3), these data show that, in our assay, the degradation of BSA and lysozyme was fully due to the activity of the 20 S proteasome and TPPII.
DISCUSSION
Proteasome Proteolytic Pathway Occurs in A. thaliana
The proteasome pathway, as described in the cytosol of mammalian cells, may be defined by the more or less sequential action of the proteasome, TPPII, TOP, and LAPs (15). In this study, we first report the occurrence and the identification of the different proteases involved in the proteasome proteolytic pathway in the model plant A. thaliana. This characterization was done using classical enzymatic activity measurements with the help of specific inhibitors and effectors, by a proteomics approach, and by measuring gene expression at the transcript level. Besides the 20 S proteasome, which was previously identified in different plant species (19, 20), this work provides new insights into the functioning of peptidases previously identified in Arabidopsis-like TPPII (30) and LAP (48). In addition, we report the occurrence of proteases that have not been described in the plant kingdom until now, like TOP.
In animals, TPPII is the main exopeptidase capable of processing large proteasome products. Although not essential for the generation of antigenic peptides in antigen processing, it was supposed to be necessary for efficient protein turnover (49, 50). This protease was recently purified from Arabidopsis seedlings and characterized (30). In this study, we confirmed the presence of TPPII in the leaves of Arabidopsis plants and demonstrated its role in the catabolism of oxidatively damaged proteins.
In mammals, TOP appears among the most important elements of the proteasome proteolytic pathway (14). This peptidase, which belongs to the peptidase M3 family, is described as a zinc-dependent metallopeptidase and preferentially cleaves peptides 5–17 amino acids in length (14, 51). In A. thaliana, genome analysis reveals the existence of a small multigenic family composed of three genes encoding proteins similar to the mammalian TOP family. The proteins encoded by At5g10540 (TOP) and At5g65620 (TOPorg) are more similar to the mouse TOP (EC 3.4.24.15) (28% amino acids sequence identities), whereas TOPL (At1g67690) is more similar to the mouse neurolysin (EC 3.4.24.16) (34% amino acid sequence identity), a closely related M3 oligopeptidase. TOPorg is expressed as a preprotein with a transit peptide, which predicted a chloroplastic location (Predotar algorithm). S-300 Sephacryl fractionation of leaf crude extracts, activity characterization, and mass spectrometry analyses confirmed that Arabidopsis TOP and TOPorg are expressed as monomers of ∼80 kDa in healthy Arabidopsis leaves. The analysis of the influence of a set of compounds on TOP activity indicates that these proteases present similar inhibitor susceptibilities to mammalian TOPs.
In the proteasome pathway described in animal cells, aminopeptidases complete the total degradation of peptides into free amino acids. The mammalian LAP is involved in this process (14, 17). In plants, LAPs have been postulated to have functions in the salvage of C and N resources from the cotyledons of germinating seedlings, in the response to biotic and abiotic stresses, or for LAP-A to regulate jasmonic acid-mediated wound signaling (reviewed in Refs. 32, 44, 52). Our data show that although LAP1 subunits are detected in the Leu-pNA-hydrolyzing fraction, the hexameric LAP1 is not the dominant pNA-degrading peptidase in the cytosol and that other Leu-pNA-hydrolyzing enzymes are important in Arabidopsis leaves. Other candidate enzymes include three aminopeptidases of Arabidopsis encoded by M1-like peptidase genes (44), which are often referred to as LAPs in animals (31). Two of these M1-like peptidases have been characterized (53, 54), but peptides corresponding to these genes were not detected in the Leu-pNA fraction. Therefore, at present, there is no evidence supporting the role of the M1 family of peptidases in the proteasome degradative pathway. In the genome of A. thaliana, three genes encode leucine aminopeptidases, LAP1 (At2g24200), LAP2 (At4g30920), and LAP3 (At4g30910), which present 70–80% amino acid sequence identity with each other. Using gene-specific primers, we detected the transcripts of the three genes in the leaves of Arabidopsis (supplemental Fig. S1). Furthermore, LAP proteins were detected in healthy leaves after two-dimensional PAGE and immunoblot analysis with anti-LAP antibodies as reported previously (31).
Proteasome Proteolytic Pathway Is Responsive to Cadmium Stress
As reported in other plants species (25–27, 55–58), this study show that cadmium treatment causes an increase in oxidized proteins and endopeptidase activities in Arabidopsis leaves (Table 4). Recently, 20 S proteasome has been shown to be up-regulated in response to cadmium in sunflower, maize, and tomato leaves, as well as in Arabidopsis cells (25, 26, 28), suggesting that it could be involved in the degradation of oxidized proteins generated by cadmium-induced oxidative stress.
The data reported here demonstrate that the expression and activity of several proteasome pathway peptidases are up-regulated by cadmium stress. For example, 50 μm cadmium treatment led to an increase of transcripts level of three structural and three catalytic proteasome subunits, TPPII, TOP, TOPL, and LAP1–3 (Figs. 2 and 3). It also triggered a final 30–90% increase in the activity of the main elements of the pathway, i.e. 20 S proteasome, TPPII, TOP, and LAP-like peptidase. The increases in proteasome structural and catalytic subunit transcripts, followed by increases in proteasome quantity and chymotrypsin-like activity, are in good agreement with studies reporting that, in mammalian cells, the overexpression of one subunit of the 20 S proteasome leads to an up-regulation of the other subunits, an increase in proteasome content, and an enhancement of proteasome activities (59–61). The analysis of the regulation of these proteasome pathway-related peptidases was based on the similarity of the proteasome pathway between mammalian and plant systems. However, it does not exclude a priori the potential involvement in this pathway of other peptidases of which expression and activity were not taken into account in this study (for instance, nonhexameric LAPs).
The relatively low induction factor of the proteasome pathway in response to cadmium may give some doubt about the significance of this response. However, it must be kept in mind that in plants up to 50% of the total proteins are replaced every week (62) and that 26 S/20 S proteasome-dependent proteolysis accounts for the major part of protein turnover (20). Thus, even moderate changes in this central pathway (a 1.3–1.6-fold increase in the proteolytic capacity) are highly significant for cellular metabolism.
The role of the 20 S and 26 S proteasome in the response to oxidative stress and degradation of oxidized proteins appears to be complex as revealed from studies in both yeast and plants. In yeast, the degradation of abnormal proteins resulting from cadmium treatment was shown to be mediated, in part, by proteasome and up-regulation of genes coding for the limiting components of the ubiquitin pathway (63). Indeed, overexpression of a polyubiquitin gene conferred resistance to moderate oxidative stress in respiring yeast cells (64). In contrast, a recent study carried out with Arabidopsis showed that mutations of one of the subunits of the 19 S regulatory particle of the 26 S proteasome resulted in an enhanced accumulation of the 20 S versus 26 S proteasome and a higher tolerance to oxidative stress (29). Moreover, in contrast to the 26 S proteasome, the 20 S proteasome appears more resistant to oxidative stress as it maintains its activity even in the presence of H2O2 (13). In addition, Shringarpure et al. (65) demonstrated that, in vivo, oxidized proteins are degraded without ubiquitin conjugation. These data strongly support the involvement of the 20 S rather than the 26 S in the degradation of oxidized proteins, probably via a by default degradation mechanism (9).
This study indicated that the 20 S proteasome pathway is up-regulated at RNA and activity levels in response to cadmium in Arabidopsis leaves. This suggests that it is involved in the response to oxidative stress through the sequential degradation of oxidized proteins by the 20 S proteasome, TPPII, TOP, and finally LAPs. This suggestion is supported by the assay of oxidized versus nonoxidized protein degradation by the 20 S proteasome and TPPII (Fig. 5). The better degradation of oxidized BSA and lysozyme by the 20 S proteasome/TPPII couple, without any requirement for ATP, confirms the preferential degradation of oxidized proteins by the 20 S proteasome. Previous works reported that the degradation of oxidized proteins by the 20 S proteasome occurs via the recognition of hydrophobic amino acid residues that are exposed during oxidative rearrangement of secondary and tertiary protein structure (5, 66, 67). α-Casein, which is a structure-less protein, was used as a control for this and was found not to be better degraded when oxidized (Fig. 5). Therefore, these data further support the hypothesis that, in A. thaliana, the 20 S proteasome pathway takes part in the degradation of oxidized proteins resulting from a stress, such as cadmium stress.
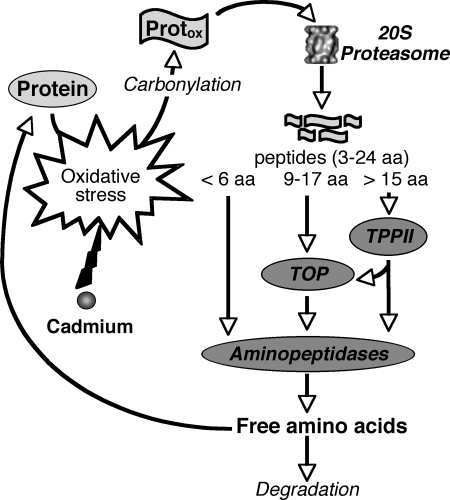
In summary, this work, together with previous knowledge acquired in mammalian and plant areas, led us to specify the function of the 20 S proteasome pathway during cadmium stress and more generally during oxidative stress. The general scheme of protein oxidation and degradation processes is summarized in Fig. 6. As a consequence of various oxidative stresses, oxygen radicals and activated oxygen species cause modifications to amino acids of proteins that result in the loss of protein function or enzymatic activity. To avoid the toxic effects of oxidized protein accumulation, the 20 S proteasome and the proteases acting downstream (TPPII, TOP, and aminopeptidases) sequentially degrade denaturated proteins into small peptides and amino acids. The free amino acids produced during this process are then either used for the synthesis of new proteins or degraded when they are present in oxidized form. Thus, the induction of the proteasome proteolytic pathway during cadmium stress could be part of the general response mechanism of plants to cope with daily and naturally occurring oxidative stresses.
FIGURE 6.
Putative function of the 20 S proteasome proteolytic pathway during cadmium and, more generally, oxidative stress. Results presented in this study strongly suggest that the 20 S proteasome, TPPII, TOP, and aminopeptidases participate in the degradation of oxidized proteins generated during stress to protect cells against toxicity. Free amino acids released from this pathway are used for the synthesis of new proteins, although oxidatively modified amino acids are degraded.
Acknowledgments
We are grateful to Dr. Maighred Gallagher-Gambarelli for critical reading of the manuscript. We thank Paul Legrand for help with the chromatographic steps and protease activity measurements.
This work was supported by the French “Program Inter-organismes CEA CNRS INRA INSERM de Toxicologie Nucléaire Environnementale.”

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- LAP
- leucine aminopeptidase
- TPPII
- tripeptidyl-peptidase II
- TOP
- thimet oligopeptidase
- Mcc
- 7-methoxycoumarin-3-carboxyl
- AMC
- 7-amino-4-methylcoumarin
- Suc
- succinimidyl
- RT
- reverse transcription
- BSA
- bovine serum albumin
- LC-MS/MS
- liquid chromatography/tandem mass spectrometry
- pNA
- p-nitroanilide
- MES
- 4-morpholineethanesulfonic acid
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- DNPH
- dinitrophenylhydrazine
- CMK
- chloromethyl ketone
- AAF
- Ala-Ala-Phe
- Dnp
- 2,4-dinitrophenyl
- FW
- fresh weight.
REFERENCES
- 1.Sanita di Toppi L., Gabbrielli R. (1999) Environ. Exp. Bot. 41, 105–130 [Google Scholar]
- 2.Wagner G. (1993) Adv. Agron. 51, 173–212 [Google Scholar]
- 3.Liu J., Qian S. Y., Guo Q., Jiang J., Waalkes M. P., Mason R. P., Kadiiska M. B. (2008) Free Radic. Biol. Med. 45, 475–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lafuente A., Cabaleiro T., Caride A., Romero A. (2008) Electron. J. Environ. Agric. Food Chem. 7, 3363–3371 [Google Scholar]
- 5.Davies K. J. (2001) Biochimie 83, 301–310 [DOI] [PubMed] [Google Scholar]
- 6.Davies K. J., Delsignore M. E. (1987) J. Biol. Chem. 262, 9908–9913 [PubMed] [Google Scholar]
- 7.Stadtman E. R. (1993) Annu. Rev. Biochem. 62, 797–821 [DOI] [PubMed] [Google Scholar]
- 8.Shringarpure R., Grune T., Davies K. J. (2001) Cell Mol. Life Sci. 58, 1442–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asher G., Reuven N., Shaul Y. (2006) BioEssays 28, 844–849 [DOI] [PubMed] [Google Scholar]
- 10.Shang F., Taylor A. (1995) Biochem. J. 307, 297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jahngen-Hodge J., Obin M. S., Gong X., Shang F., Nowell T. R., Jr., Gong J., Abasi H., Blumberg J., Taylor A. (1997) J. Biol. Chem. 272, 28218–28226 [DOI] [PubMed] [Google Scholar]
- 12.Obin M., Shang F., Gong X., Handelman G., Blumberg J., Taylor A. (1998) FASEB J. 12, 561–569 [DOI] [PubMed] [Google Scholar]
- 13.Reinheckel T., Sitte N., Ullrich O., Kuckelkorn U., Davies K. J., Grune T. (1998) Biochem. J. 335, 637–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saric T., Graef C. I., Goldberg A. L. (2004) J. Biol. Chem. 279, 46723–46732 [DOI] [PubMed] [Google Scholar]
- 15.Reits E., Neijssen J., Herberts C., Benckhuijsen W., Janssen L., Drijfhout J. W., Neefjes J. (2004) Immunity 20, 495–506 [DOI] [PubMed] [Google Scholar]
- 16.Lecker S. H., Goldberg A. L., Mitch W. E. (2006) J. Am. Soc. Nephrol. 17, 1807–1819 [DOI] [PubMed] [Google Scholar]
- 17.Rock K. L., York I. A., Goldberg A. L. (2004) Nat. Immunol. 5, 670–677 [DOI] [PubMed] [Google Scholar]
- 18.Kloetzel P. M., Ossendorp F. (2004) Curr. Opin. Immunol. 16, 76–81 [DOI] [PubMed] [Google Scholar]
- 19.Skoda B., Malek L. (1992) Plant Physiol. 99, 1515–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smalle J., Vierstra R. D. (2004) Annu. Rev. Plant Biol. 55, 555–590 [DOI] [PubMed] [Google Scholar]
- 21.Fu H., Doelling J. H., Arendt C. S., Hochstrasser M., Vierstra R. D. (1998) Genetics 149, 677–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu H., Doelling J. H., Rubin D. M., Vierstra R. D. (1999) Plant J. 18, 529–539 [DOI] [PubMed] [Google Scholar]
- 23.Yang P., Fu H., Walker J., Papa C. M., Smalle J., Ju Y. M., Vierstra R. D. (2004) J. Biol. Chem. 279, 6401–6413 [DOI] [PubMed] [Google Scholar]
- 24.Basset G., Raymond P., Malek L., Brouquisse R. (2002) Plant Physiol. 128, 1149–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Djebali W., Gallusci P., Polge C., Boulila L., Galtier N., Raymond P., Chaibi W., Brouquisse R. (2008) Planta 227, 625–639 [DOI] [PubMed] [Google Scholar]
- 26.Pena L., Pasquini L., Tomaro M., Gallego S. (2006) Plant Science 171, 531–537 [DOI] [PubMed] [Google Scholar]
- 27.Pena L. B., Pasquini L. A., Tomaro M. L., Gallego S. M. (2007) Phytochemistry 68, 1139–1146 [DOI] [PubMed] [Google Scholar]
- 28.Sarry J. E., Kuhn L., Ducruix C., Lafaye A., Junot C., Hugouvieux V., Jourdain A., Bastien O., Fievet J. B., Vailhen D., Amekraz B., Moulin C., Ezan E., Garin J., Bourguignon J. (2006) Proteomics 6, 2180–2198 [DOI] [PubMed] [Google Scholar]
- 29.Kurepa J., Toh-E A., Smalle J. A. (2008) Plant J. 53, 102–114 [DOI] [PubMed] [Google Scholar]
- 30.Book A. J., Yang P., Scalf M., Smith L. M., Vierstra R. D. (2005) Plant Physiol. 138, 1046–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chao W. S., Pautot V., Holzer F. M., Walling L. L. (2000) Planta 210, 563–573 [DOI] [PubMed] [Google Scholar]
- 32.Matsui M., Fowler J. H., Walling L. L. (2006) Biol. Chem. 387, 1535–1544 [DOI] [PubMed] [Google Scholar]
- 33.Reznick A. Z., Packer L. (1994) Methods Enzymol. 233, 357–363 [DOI] [PubMed] [Google Scholar]
- 34.Brouquisse R., Gaudillere J. P., Raymond P. (1998) Plant Physiol. 117, 1281–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu Y. Q., Pautot V., Holzer F. M., Walling L. L. (1996) Plant Physiol. 110, 1257–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bestel-Corre G., Dumas-Gaudot E., Poinsot V., Dieu M., Dierick J. F., van T. D., Remacle J., Gianinazzi-Pearson V., Gianinazzi S. (2002) Electrophoresis 23, 122–137 [DOI] [PubMed] [Google Scholar]
- 37.Hurkman W. J., Tanaka C. K. (1986) Plant Physiol. 81, 802–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang C. S., Walling L. L., Eckard K. J., Lord E. M. (1992) Plant Physiol. 99, 822–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu G., Liu Y., Kansal M. M., Sayre L. M. (1999) Chem. Res. Toxicol. 12, 855–861 [DOI] [PubMed] [Google Scholar]
- 40.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 41.Tisljar U., Barrett A. J. (1990) Biochem. J. 267, 531–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carpenter F. H., Vahl J. M. (1973) J. Biol. Chem. 248, 294–304 [PubMed] [Google Scholar]
- 43.Bartling D., Nosek J. (1994) Plant Sci. 99, 199–209 [Google Scholar]
- 44.Walling L. L. (2006) Curr. Opin. Plant Biol. 9, 227–233 [DOI] [PubMed] [Google Scholar]
- 45.Bolumar T., Sanz Y., Aristoy M. C., Toldrá F. (2003) Appl. Environ. Microbiol. 69, 227–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gu Y. Q., Holzer F. M., Walling L. L. (1999) Eur. J. Biochem. 263, 726–735 [DOI] [PubMed] [Google Scholar]
- 47.Walling L. L., Gu Y.-Q. (1996) in Aminopeptidases (Taylor A. ed) pp. 174–219, R. G. Landes Co., Austin, TX [Google Scholar]
- 48.Bartling D., Weiler E. W. (1992) Eur. J. Biochem. 205, 425–431 [DOI] [PubMed] [Google Scholar]
- 49.Saveanu L., Fruci D., van Endert P. M. (2002) Mol. Immunol. 39, 203–215 [DOI] [PubMed] [Google Scholar]
- 50.Tomkinson B. (1999) Trends Biochem. Sci. 24, 355–359 [DOI] [PubMed] [Google Scholar]
- 51.Barrett A. J., Brown M. A., Dando P. M., Knight C. G., McKie N., Rawlings N. D., Serizawa A. (1995) Methods Enzymol. 248, 529–556 [DOI] [PubMed] [Google Scholar]
- 52.Fowler J. H., Narvaez-Vasquez J. A., Aromdee D. N., Pautot V., Holzer F. M., Walling L. L. (2009) Plant Cell 21, 1239–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murphy A. S., Hoogner K. R., Peer W. A., Taiz L. (2002) Plant Physiol. 128, 935–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sánchez-Morán E., Jones G. H., Franklin F. C., Santos J. L. (2004) Plant Cell 16, 2895–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Balestrasse K. B., Benavides M. P., Gallego S. M., Tomaro M. L. (2003) Funct. Plant Biol. 30, 57–64 [DOI] [PubMed] [Google Scholar]
- 56.McCarthy I., Romero-Puertas M. C., Plama J. M., Sandalio L. M., Corpas F. J., Gomez M., Del Rio L. A. (2001) Plant Cell Environ. 24, 1065–1073 [Google Scholar]
- 57.Romero-Puertas M. C., Palma J. M., Gomez M., Del Rio L. A., Sandalio L. M. (2002) Plant Cell Environ. 25, 677–686 [Google Scholar]
- 58.Sandalio L. M., Dalurzo H. C., Gómez M., Romero-Puertas M. C., del Río L. A. (2001) J. Exp. Bot. 52, 2115–2126 [DOI] [PubMed] [Google Scholar]
- 59.Chondrogianni N., Stratford F. L., Trougakos I. P., Friguet B., Rivett A. J., Gonos E. S. (2003) J. Biol. Chem. 278, 28026–28037 [DOI] [PubMed] [Google Scholar]
- 60.Chondrogianni N., Tzavelas C., Pemberton A. J., Nezis I. P., Rivett A. J., Gonos E. S. (2005) J. Biol. Chem. 280, 11840–11850 [DOI] [PubMed] [Google Scholar]
- 61.Liu Y., Liu X., Zhang T., Luna C., Liton P. B., Gonzalez P. (2007) Mol. Vis. 13, 31–38 [PMC free article] [PubMed] [Google Scholar]
- 62.Vierstra R. D. (1996) Plant Mol. Biol. 32, 275–302 [DOI] [PubMed] [Google Scholar]
- 63.Jungmann J., Reins H. A., Schobert C., Jentsch S. (1993) Nature 361, 369–371 [DOI] [PubMed] [Google Scholar]
- 64.Cheng L., Watt R., Piper P. W. (1994) Mol. Gen. Genet. 243, 358–362 [DOI] [PubMed] [Google Scholar]
- 65.Shringarpure R., Grune T., Mehlhase J., Davies K. J. (2003) J. Biol. Chem. 278, 311–318 [DOI] [PubMed] [Google Scholar]
- 66.Grune T., Reinheckel T., Davies K. J. (1997) FASEB J. 11, 526–534 [PubMed] [Google Scholar]
- 67.Pacifici R. E., Kono Y., Davies K. J. (1993) J. Biol. Chem. 268, 15405–15411 [PubMed] [Google Scholar]