Abstract
Accumulating evidence indicates that TBP (TATA-binding protein)-like protein (TLP) contributes to the regulation of stress-mediated cell cycle checkpoint and apoptotic pathways, although its physiological target genes have remained elusive. In the present study, we have demonstrated that human TAp63 is one of the direct transcriptional target genes of TLP. Enforced expression of TLP results in the transcriptional induction of the endogenous TAp63, but not of the other p53 family members such as TAp73 and p53. Consistent with these results, small interference RNA-mediated knockdown led to a significant down-regulation of the endogenous TAp63. Luciferase reporter assay and chromatin immunoprecipitation analysis revealed that the genomic region located at positions −487 to −29, where +1 represents the transcriptional initiation site of TAp63, is required for TLP-dependent transcriptional activation of TAp63 and also TLP is efficiently recruited onto this region. Additionally, cells treated with anti-cancer drug etoposide underwent apoptosis in association with the transcriptional enhancement of TAp63 in a p53-independent manner, and the knockdown of the endogenous TLP reduced etoposide-induced apoptosis through repression of TAp63 expression. Taken together, our present study identifies a TLP-TAp63 pathway that is further implicated in stress-induced apoptosis.
Introduction
Transcriptional regulation involves the functional integration of diverse factors and is a critical regulatory step for cellular events that include growth, differentiation, and death. These cellular activities often occur simultaneously due to the action of regulatory factors with broad targets. A representative example of such a factor is the tumor suppressor p53 and its family members, including p63 and p73, which contribute to tumor suppression, cell cycle checkpoint, DNA repair, and apoptosis (1). p63 acts as a pro-apoptotic transcription factor (2, 3) and, like p53 and p73, is expressed as multiple isoforms (4). They include the trans-activating (TA)3 isoform of p63, termed TAp63, and an NH2-terminal activation domain-deficient isoform, ΔNp63, that acts as a dominant negative factor over p53, TAp63, and TAp73 (2). p63 has been clearly implicated in a variety of developmental processes (5), whereas its anticipated role as a tumor suppressor is unclear, mainly because of its low frequency of the somatic mutations in human tumors (6, 7). However, a study focused on long term effects of p63 mutations in mice showed that mice bearing mutations in both p63 and p53 develop a more aggressive tumor, indicating the presence of a tumor suppressive activity of p63 (8). This is consistent with earlier studies indicating that p63 is required for p53-dependent apoptotic response (9) and that, in response to certain DNA damage insults, p63 activates an overlapping set of p53-target genes implicated in cell cycle arrest and apoptosis (10). Although extensive studies of p63 in human tumors have suggested that deregulated expression of TAp63 and ΔNp63 contributes to tumor development and progression (4), the precise molecular mechanisms behind the transcriptional regulation of TAp63 remain to be unclear.
TATA-binding protein (TBP) is a general transcription factor that plays a central role in the regulation of pre-initiation complex formation by eukaryotic RNA polymerases (11, 12). Eukaryotic cells also contain multiple TBP paralogs implicated in transcriptional regulation during cell growth, differentiation, and development (11, 12). TBP-like protein (TLP) (13), also known as TBP-related factor 2 (14, 15), TLF (16), or TRP (17), is one of the TBP paralogs common to Metazoa and has been implicated by genetic studies in various developmental processes, including spermiogenesis in mice (11, 12). Although TLP fails to bind to TATA box (11, 12), it stimulates transcription from several TATA-less promoters (18). Mammalian TLP, unlike TBP, does not associate with TAFs to form a transcription factor IID-type complex but, instead, associates with TFIIA in cells (14, 19). It was reported earlier (20) that mammalian TLP activates transcription from the TATA-less neurofibromatosis type 1 (NF1) promoter through site-specific binding, but represses the TATA-containing c-fos promoter, thus leading to the prediction of an anti-oncogenic ability of TLP as well as the potential for direct binding to other target genes. Shimada et al. (21) reported that chicken TLP represses the G2/M transition and, through its nuclear translocation, mediates apoptosis induction in a p53-independent manner. Hence, TLP is proposed to have both checkpoint and anti-oncogenic functions, although its physiological role and also the precise molecular mechanisms behind TLP-mediated apoptosis as well as cell cycle checkpoint remain to be elusive.
Here, we have analyzed mammalian TLP function in relation to TAp63 expression and show that TLP enhances the promoter activity of TAp63 and thus leads to apoptosis. Further observations suggest that this novel TLP-TAp63 pathway increases the sensitivity to anti-cancer drug etoposide.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
Chicken DT40 cells were grown in RPMI 1640 medium (Invitrogen) as previously described (21). DT40-TLP−/− cells derived from parental DT40 cells lack TLP (21). Human cervical carcinoma-derived HeLa and human hepatocellular carcinoma-derived HepG2 cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen), penicillin (100 IU/ml), and streptomycin (100 μg/ml). Human hepatocellular carcinoma-derived Hep3B cells were grown in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum and antibiotics. HepG2 and HeLa cells carry wild-type p53. Hep3B and DT40 cells lack p53. Where indicated, cells were exposed to etoposide (at a final concentration of 50 μm). For transfection, DT40 cells were transiently transfected with the indicated expression plasmids by electroporation. HeLa cells were transiently transfected with the indicated combinations of the expression plasmids using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer's instructions.
RT-PCR
Total RNA was prepared from the indicated cells using RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's recommendations. For the RT-PCR, first strand cDNA was generated using SuperScript II reverse transcriptase (Invitrogen) and random primers. The resultant cDNA was subjected to the PCR-based amplification. The oligonucleotide primers used in this study were as follows: human TLP, 5′-CCTCTTCCCACGGATGTGAT-3′ (sense) and 5′-GAGTCCAATGTGCAGCAGT-3′ (reverse); mouse TLP, 5′-GCCATTTGAACTTAAGGA-3′ (forward) and 5′-TGTAAATTCTGGCAA-3′ (reverse); human TAp63, 5′-GTCCCAGAGCACACAGACAA-3′ (forward) and 5′-GAGGAGCCGTTCTGAATCTG-3′ (reverse); chicken TAp63, 5′-GAAACAGCCATGCCCAGTAT-3′ (forward) and 5′-CAAATGCGAGCTTCAAAACA-3′ (reverse); human TAp73, 5′-CGGGACGGACGCCGATG-3′ (forward) and 5′-GAAGGTCGAAGTAGGTGCTGTCTGG-3′ (reverse); human p53, 5′- ATTTGATGCTGTCCCCGGACGATATTGAAC-3′ (forward) and 5′-ACCCTTTTTGGACTTCCGGACGATATTGAAC-3′ (reverse); human p21waf1, 5′-GACACCACTGGAGGGTGACT-3′ (forward) and 5′-CCCTAGGCTGTGCTCACTTC-3′ (reverse); human 14-3-3σ, 5′-AGAGCGAAACCTGCTCTCAG-3′ (forward) and 5′-CTCCTTGATGAGGTGGCTGT-3′ (reverse); human Lamin A/C, 5′-CCGAGTCTGAAGAGGTGGTC-3′ (forward) and 5′-AGGTCACCCTCCTTCTTGGT-3′ (reverse); human BAX, 5′-TCTGACGCAACTTCAACAC-3′ (forward) and 5′-GAGGAGTCTCACCCAACCAC-3′ (reverse); human PUMA, 5′-GCCCAGACTGTGAATCCTGT-3′ (forward) and 5′-TCCTCCCTCTTCCGAGATTT-3′ (reverse); human NOXA, 5′-GCAAGAATGGAAGACCCTTG-3′ (forward) and 5′-GTGCTGAGTTGGCACTGAAA-3′ (reverse); human GAPDH, 5′-ACCTGACCTGCCGTCTAGAA-3′ (forward) and 5′-TCCACCACCCTGTTGCTGTA-3′ (reverse). The expression of β-actin or GAPDH was measured as an internal control. The PCR products were subjected to 1% agarose gel electrophoresis and visualized by ethidium bromide staining.
Real-time Quantitative RT-PCR
Total RNA was extracted from clinical samples using TRIzol reagent (Invitrogen) according to the manufacturer's instructions, and reverse transcription was performed with SuperScript II reverse transcriptase (Invitrogen). Real-time quantitative PCR (TaqMan PCR) using an ABI Prism 7700 sequence detection system (Perkin-Elmer Applied Biosystems, Foster City, CA) was carried out according to the manufacturer's protocol. All the reactions were performed in triplicate. The data were averaged from the values obtained in each reaction. The mRNA levels of each of the genes were standardized by β-actin.
siRNA
Hep3B cells transiently transfected with siRNA targeting TLP (TLP siRNA-1, 5′-UAACAGGGCCCAAUGUAAA-3′; TLP siRNA-2, 5′-GGAAGGAGCAAAUGUAAUU-3′), siRNA against TAp63 (TAp63 siRNA-1, 5′-CAGCUAUAUGUUCAGUUC-3′; TAp63 siRNA-2, 5′-GAUUGAGAUUAGCAUGGAC-3′) (Invitrogen) or with siRNA against Lamin A/C (Dharmacon, Chicago, IL) by using Lipofectamine RNAiMAX transfection reagent (Invitrogen) according to the manufacturer's instructions. Forty-eight hours after transfection, total RNA was prepared and processed for RT-PCR.
Immunoblotting
Cells were washed in ice-cold phosphate-buffered saline and lysed in an SDS-sample buffer. After brief sonication, whole cell lysates were boiled for 5 min, resolved by 12% SDS-PAGE, and electrotransferred onto Immobilon-P membranes (Millipore, Bedford, MA). The membranes were blocked with Tris-buffered saline containing 0.1% Tween 20 and 5% nonfat dry milk and then incubated with monoclonal anti-p21waf1 (Ab-1, Oncogene Research Products, Cambridge, MA), monoclonal anti-FLAG (M2, Sigma), monoclonal anti-Lamin B (Ab-1, Calbiochem), monoclonal anti-tubulin-α (Ab-2, Neomarkers, Fremont, CA), polyclonal anti-TAp63 (22), polyclonal anti-TLP (23), or with polyclonal anti-actin (20-33, Sigma) antibody for 1 h at room temperature followed by an incubation with the appropriate horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) for 1 h at room temperature. The chemiluminescence reaction was performed using ECL reagent (Amersham Biosciences, Piscataway, NJ).
Construction of Luciferase Reporter Plasmids
A luciferase reporter plasmid containing TAp63 promoter region encompassing from −2340 to +26, where +1 represents the transcriptional initiation site, was amplified by PCR-based strategy using genomic DNA prepared from human placenta as a template. Oligonucleotide primers used were as follows: F1, 5′-TTGGTAGAGCTCGAGGATAGCTTGAGTCCAGCAG-3′ (forward) and R1, 5′-AGATATCCCTTTCACATCCC-3′ (reverse); F2, 5′-GTGCATGTGTTTGAGGTAGG-3′ (forward) and R2, 5′-CTTAGAGCTAGCCCTTCAACTGTCTTTGATATCAACG-3′ (reverse). Underlined sequences indicate the positions of SacI restriction site in the forward primer F1 and NheI restriction site in the reverse primer R2. PCR products were gel-purified and subcloned into pGEM-T Easy plasmid (Promega, Southampton, UK) according to the manufacturer's protocol. The resultant plasmid DNA was digested with SacI plus HindIII or with HindIII plus NheI and then subcloned into SacI/NheI restriction sites of pGL3-basic plasmid (Promega) to give pGL3-TAp63(−2340). A series of the 5′ deletion mutants of pGL3-TAp63(−2340) were generated by using an Erase-A-Base system (Promega) according to the manufacturer's instructions.
Luciferase Reporter Assay
HeLa cells were transiently co-transfected with the constant amount of the indicated pGL3-TAp63 luciferase reporter constructs (100 ng), pRL-TK Renilla luciferase reporter plasmid (10 ng) together with or without 100 ng of the expression plasmid for FLAG-TLP. The total amount of the plasmid DNA per each transfection was kept constant (510 ng) with the empty plasmid (pCIneo). Forty-eight hours after transfection, cells were harvested and lysed, and both firefly and Renilla luciferase activities were measured by using a Dual-Luciferase reporter assay system (Promega). The firefly luminescence signal was normalized based on the Renilla luminescence signal.
ChIP Assay
Chromatin immunoprecipitation (ChIP) assay was performed according to the protocol provided by Upstate Biotechnology (Charlottesville, VA). In brief, HeLa cells were transiently transfected with the expression plasmid for FLAG-TLP. Forty-eight hours after transfection, cells were cross-linked with 1% formaldehyde in medium at 37 °C for 15 min. Cells were then washed in ice-cold phosphate-buffered saline and resuspended in 200 μl of SDS lysis buffer containing protease inhibitor mixture. The suspension was sonicated on ice and pre-cleared with protein A-agarose beads blocked with sonicated salmon sperm DNA (Upstate Biotechnology) for 30 min at 4 °C. The beads were removed, and the chromatin solution was immunoprecipitated with polyclonal anti-FLAG (Sigma) antibody at 4 °C, followed by incubation with protein A-agarose beads for an additional 1 h at 4 °C. The immune complexes were eluted with 100 μl of elution buffer (1% SDS and 0.1 m NaHCO3), and formaldehyde cross-links were reversed by heating at 65 °C for 6 h. Proteinase K was added to the reaction mixtures and incubated at 45 °C for 1 h. DNA of the immunoprecipitates and control input DNA were purified and then analyzed by standard PCR using human TAp63 promoter-specific primers.
Subcellular Fractionation
To prepare nuclear and cytoplasmic extracts, cells were lysed in 10 mm Tris-HCl, pH 7.5, 1 mm EDTA, 0.5% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, and a protease inhibitor mix (Sigma) and centrifuged at 5000 rpm for 10 min to collect soluble fractions, which were referred to as cytosolic extracts. Insoluble materials were washed in the lysis buffer and further dissolved in SDS-sample buffer to collect the nuclear extracts. The nuclear and cytoplasmic fractions were subjected to the immunoblot analysis using monoclonal anti-Lamin B (Ab-1, Oncogene Research Products) or monoclonal anti-tubulin-α (Ab-2, Neomarkers) antibody.
TUNEL Staining
Hep3B cells were grown on coverslips and transiently transfected with the indicated siRNAs. Forty-eight hours after transfection, cells were fixed in 4% paraformaldehyde and apoptotic cells were detected by using an in situ cell detection Kit (Roche Molecular Biochemicals, Mannheim, Germany) according to the manufacturer's protocol. The coverslips were mounted with 4′,6-diamidino-2-phenylindole-containing mounting medium (Vector Laboratories, Burlingame, CA) and observed under a Fluoview laser scanning confocal microscope (Olympus, Tokyo, Japan).
FACS Analysis
Transfected HepG2 cells were exposed to 50 μm of etoposide. Forty-eight hours after the treatment, floating and attached cells were collected, washed in ice-cold phosphate-buffered saline, and fixed in 70% ethanol at −20 °C. Following incubation with phosphate-buffered saline containing 40 μg/ml of propidium iodide and 200 μg/ml of RNase A for 1 h at room temperature in the dark, stained nuclei were analyzed by a FACScan machine (BD Biosciences, Mountain View, CA).
Statistical Analysis
The data obtained from real-time PCR were expressed as means ± S.E. of the mean.
RESULTS
TLP Has an Ability to Induce the Expression of TAp63
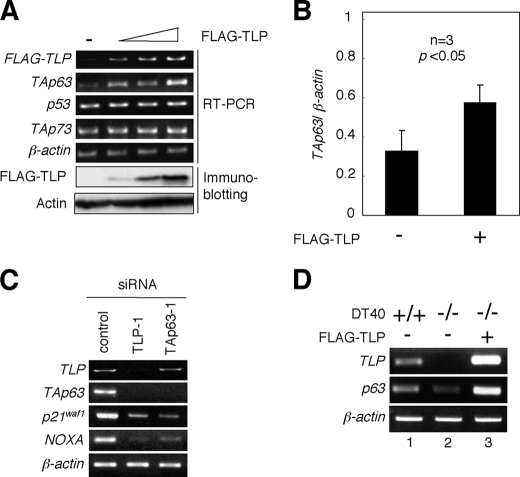
We have previously described that TLP induces both cell cycle arrest and apoptosis in chicken DT40 cells in a p53-independent manner (21). These results imply that the other p53 family members such as p73 or p63 could be closely involved in these cellular processes, because p53 family members play a dominant role in the regulation of cell fate determination. To examine the possible contribution of TLP to p53, TAp73, and TAp63 gene expression, human cervical carcinoma-derived HeLa cells were transiently transfected with FLAG-TLP expression plasmid. Intriguingly, enforced expression of FLAG-TLP resulted in a significant up-regulation of TAp63 but not of p53 and TAp73 (Fig. 1, A and B). Consistent with these results, siRNA-mediated knockdown of the endogenous TLP led to a remarkable down-regulation of the endogenous TAp63 as well as its direct transcriptional target genes such as p21waf1 and NOXA (Fig. 1C). p21waf1 and NOXA are involved in the induction of cell cycle arrest and apoptosis, respectively (24). Additionally, knocking down of the endogenous TAp63 had an undetectable effect on the expression level of the endogenous TLP, whereas the expression levels of the endogenous p21waf1 and NOXA dramatically decreased. In support with these results, TLP-deficient chicken DT40 cells expressed p63 at an extremely lower level as compared with wild-type DT40 cells, and ectopic expression of FLAG-TLP in TLP-deficient DT40 cells caused an increase in the expression level of p63 (Fig. 1D). Thus, it is likely that TAp63 is one of direct transcriptional target genes of TLP.
FIGURE 1.
Effects of ectopic expression of TLP on p53 family members. A, enforced expression of FLAG-TLP results in the up-regulation of TAp63 but not of p53 and TAp73. Human cervical carcinoma-derived HeLa cells were transiently transfected with or without the increasing amounts of FLAG-TLP expression plasmid (1.0, 1.5, or 2.0 μg). As a negative control, the empty plasmid (2.0 μg) was introduced into HeLa cells (−). Forty-eight hours after transfection, total RNA and whole cell lysates were prepared and analyzed by semi-quantitative RT-PCR using the indicated primers and immunoblotting with anti-FLAG antibody, respectively. For RT-PCR, β-actin was used as an internal control. For immunoblotting, actin was used as a loading control. B, quantitative real-time RT-PCR analysis. HeLa cells were transiently transfected with the constant amount of the empty plasmid or with the expression plasmid encoding FLAG-TLP (2.0 μg). Forty-eight hours after transfection, the expression levels of TAp63 were examined by quantitative real-time RT-PCR using β-actin as an internal control. Data represent -fold induction of TAp63 mRNA levels relative to those of β-actin mRNA. C, siRNA-mediated knockdown of the endogenous TLP. Human hepatocellular carcinoma-derived Hep3B cells were transiently transfected with 10 nm of control siRNA, siRNA against TLP (TLP-1) or with siRNA targeting TAp63 (TAp63–1). Forty-eight hours after transfection, total RNA was prepared and subjected to semi-quantitative RT-PCR. β-actin was used as an internal control. D, a significant correlation between the expression levels of TLP and TAp63. Chicken wild-type DT40 cells and TLP-deficient DT40 cells were transiently transfected with the empty plasmid (−) or with the expression plasmid for FLAG-TLP (+). Forty-eight hours after transfection, total RNA was prepared and processed for semi-quantitative RT-PCR. β-actin was used as an internal control.
Identification of the Region within Human TAp63 Promoter Required for TLP-mediated Transactivation of TAp63
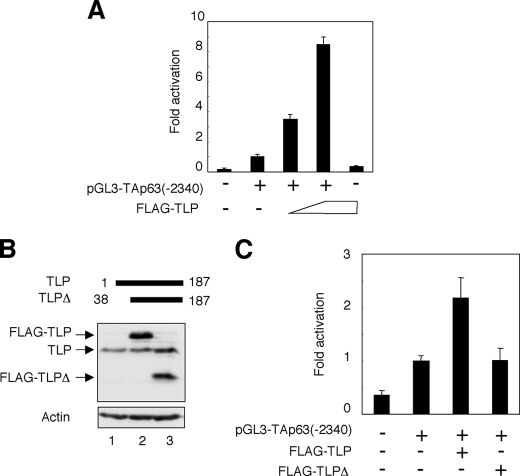
To identify the essential region within human TAp63 promoter required for TLP-dependent transcriptional activation of TAp63, we have generated the luciferase reporter plasmid bearing TAp63 genomic fragment spanning from positions −2340 to +26, where +1 represents the transcriptional initiation site, termed pGL3-TAp63(−2340). HeLa cells were transiently co-transfected with the constant amount of pGL3-TAp63(−2340), Renilla luciferase reporter plasmid together with or without the increasing amounts of the expression plasmid for FLAG-TLP. Forty-eight hours after transfection, cells were lysed, and their luciferase activities were measured. As shown in Fig. 2A, FLAG-TLP had an ability to enhance the luciferase activity driven by TAp63 promoter in a dose-dependent manner. Similar results were also obtained in p53-deficient human lung carcinoma-derived H1299 cells (data not shown). In addition, NH2-terminal deletion mutant of TLP (FLAG-TLPΔ) (Fig. 2B), which lacks a transcriptional activation function, failed to transactivate TAp63 promoter as examined by luciferase reporter assay (Fig. 2C). These observations suggest that the genomic region of TAp63 gene used in the luciferase reporter assay contains one or more TLP-responsive elements.
FIGURE 2.
NH2-terminal deletion mutant of TLP fails to transactivate TAp63 promoter. A, pGL3-TAp63(−2340) responds to TLP. HeLa cells were transiently co-transfected with the constant amount of the luciferase reporter plasmid termed pGL3-TAp63(−2340) (100 ng) and Renilla luciferase reporter plasmid (pRL-TK, 10 ng) along with or without the increasing amounts of the expression plasmid encoding FLAG-TLP (100 or 200 ng). Forty-eight hours after transfection, cells were lysed and their luciferase activities were examined. Firefly luminescence signal was normalized based on the Renilla luminescence signal. Results are shown as -fold induction of the firefly luciferase activity compared with control cells transfected with the empty plasmid. B, immunoblotting. HeLa cells were transiently transfected with the constant amount of the empty plasmid (lane 1), the expression plasmid for FLAG-TLP (lane 2), or with the expression plasmid encoding FLAG-TLPΔ (lane 3). Forty-eight hours after transfection, whole cell lysates were analyzed by immunoblotting with the anti-TLP antibody. Actin was used as a loading control. C, luciferase reporter assay. HeLa cells were transiently co-transfected with the constant amount of pGL3-TAp63(−2340) and Renilla luciferase reporter plasmid along with or without the expression plasmid for FLAG-TLP or FLAG-TLPΔ. Forty-eight hours after transfection, cells were lysed and their luciferase activities were determined as in A.
To further extend our study, a series of progressively 5′-truncated TAp63 promoter reporter constructs were generated and subjected to luciferase reporter assay. As clearly shown in Fig. 3A, deletion up to −731 and −28 resulted in a significant decrease in the luciferase activity driven by TAp63 promoter, indicating that there exist at least two independent genomic regions of TAp63 gene (−1101 to −732 and −487 to −29) required for TLP-dependent transactivation of TAp63. To examine whether TLP could be recruited onto the essential genomic region of TAp63 promoter, which we have identified, we performed a ChIP assay. For this purpose, we have designed four primer sets (#1, #2, #3, and #4) to amplify the indicated genomic regions of TAp63 promoter (Fig. 3B). HeLa cells were transiently transfected with the empty plasmid or with the expression plasmid for FLAG-TLP. Forty-eight hours after transfection, chromatin DNA was cross-linked and then processed for ChIP assay. The expression of the exogenous FLAG-TLP was examined by semi-quantitative RT-PCR (Fig. 3C, right panel). As clearly shown in Fig. 3C, genomic DNA extending from −506 to −257 was specifically amplified, suggesting that TLP is efficiently recruited onto the proximal promoter region of the human TAp63 gene (−487 to −29) and that the upstream sequences (−1101 to −732) might act to enhance the function of the TLP-bound downstream sequences.
FIGURE 3.
Identification of region(s) within human TAp63 promoter required for TLP-dependent transcriptional activation. A, luciferase reporter assays. HeLa cells were transiently co-transfected with the constant amount of the indicated luciferase reporter constructs bearing various length of human TAp63 promoter region (100 ng) and Renilla luciferase reporter plasmid (pRL-TK, 10 ng) together with the empty plasmid (pClneo) or with the expression plasmid for FLAG-TLP. Forty-eight hours after transfection, cells were lysed and their luciferase activities were measured as described in the legend for Fig. 2. Filled and open boxes indicate the putative TLP-responsive regions within TAp63 promoter. B, schematic drawing of TAp63 promoter region. Filled and open boxes indicate the putative TLP-responsive regions within TAp63 promoter. The positions of primer sets (#1, #2, #3, and #4) relative to the transcriptional initiation site (+1) used for ChIP assay are indicated. C, ChIP assay. HeLa cells were transiently transfected with the empty plasmid or with the expression plasmid for FLAG-TLP. Expression of FLAG-TLP was confirmed by semi-quantitative RT-PCR (right panels). Forty-eight hours after transfection, cells were cross-linked with formaldehyde and cross-linked chromatin was sonicated followed by immunoprecipitation with anti-FLAG antibody. Genomic DNA was purified from the immunoprecipitates and subjected to PCR using the indicated primer sets.
Identification of the Putative TLP-responsive Element within the Proximal Region of TAp63 Promoter
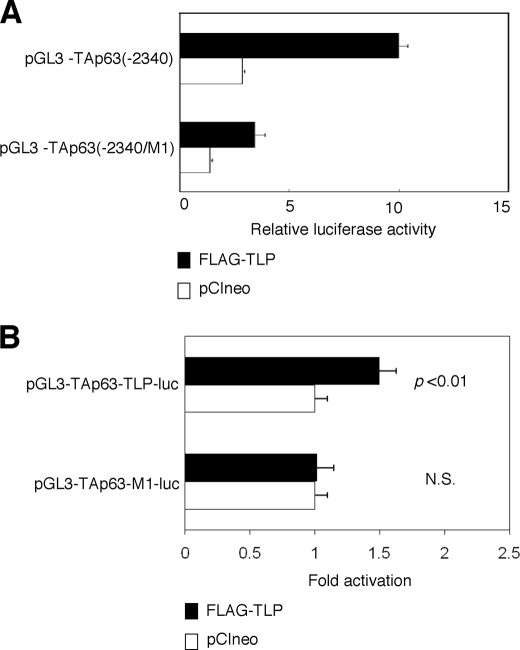
During the extensive search of the proximal region of human TAp63 promoter, we have found out the sequence element (5′-AGCTGGAGCA-3′), which was also included within one of the TLP-binding sequences of NF1 gene (5′-AGCTGAGAGCA-3′). Of note, this sequence element was well conserved among mouse, chicken, and dog TAp63 promoter regions (over 80% sequence identity). To address the functional significance of this sequence element in the regulation of TLP-dependent transcriptional enhancement of TAp63 gene, we have generated a luciferase reporter plasmid bearing mutant TAp63 promoter in which the putative TLP-responsive element (5′-AGCTGGAGCA-3′) was substituted to the mutant sequence (5′-CTAGTGAGCA-3′) (pGL3-TAp63(−2340/M1)). HeLa cells were transiently transfected with the constant amount of pGL3-TAp63(−2340) or pGL3-TAp63(−2340/M1) along with the expression plasmid for FLAG-TLP or with the empty plasmid. As shown in Fig. 4A, introduction of mutations into the putative TLP-responsive element decreased the luciferase activity mediated by exogenously expressed FLAG-TLP, suggesting that this sequence element might act as a TLP-responsive element for the human TAp63 gene.
FIGURE 4.
TLP-responsive element within human TAp63 promoter. A, introduction of the mutations into the putative TLP-responsive element. We have introduced the mutations (5′-CTAGTGAGCA-3′) into the putative TLP-responsive element (5′-AGCTGGAGCA-3′) within pGL3-TAp63(−2340) to give pGL3-TAp63(−2340/M1). HeLa cells were transiently co-transfected with the constant amount of Renilla luciferase reporter plasmid together with pGL3-TAp63(−2340) or with pGL3-TAp63(−2340) in the presence of the expression plasmid for FLAG-TLP (closed bars) or empty plasmid (open bars). Forty-eight hours after transfection, cells were lysed and their luciferase activities were measured. B, functional significance of the putative TLP-responsive element. We have generated the luciferase reporter construct bearing four tandem repeats of the putative TLP-responsive element (5′-AGCTGGAGCA-3′) fused to just upstream of TAp63 core promoter termed pGL3-TAp63-TLP-luc and its mutant carrying four tandem repeats of mutant form of the canonical TLP-responsive element (5′-CTAGTGAGCA-3′) fused to just upstream of TAp63 core promoter termed pGL3-TAp63-M1-luc. HeLa cells were transiently co-transfected with the constant amount of Renilla luciferase reporter plasmid along with the constant amount of pGL3-TAp63-TLP-luc or with pGL3-TAp63-M1-luc in the presence of FLAG-TLP expression plasmid or the empty plasmid. Eighteen hours after transfection, cells were lysed, and their luciferase activities were determined. Open and closed bars indicate the relative luciferase activity in cells transfected with the empty plasmid and the expression plasmid for FLAG-TLP, respectively. N.S., not significant.
To further confirm this issue, we have inserted four tandem repeats of the putative TLP-responsive element or mutant form of the putative TLP-responsive element into just upstream of TAp63 core promoter region to give pGL3-TAp63-TLP-luc and pGL3-TAp63-M1-luc, respectively. As shown in Fig. 4B, luciferase reporter assays demonstrated that pGL3-TAp63-TLP-luc but not pGL3-TAp63-M1-luc responds to the exogenously expressed FLAG-TLP, suggesting that the putative TLP-responsive element found in the present study plays an essential role in the regulation of TLP-dependent transactivation of TAp63.
Etoposide-mediated Induction of TLP and TAp63 in p53-deficient Cells
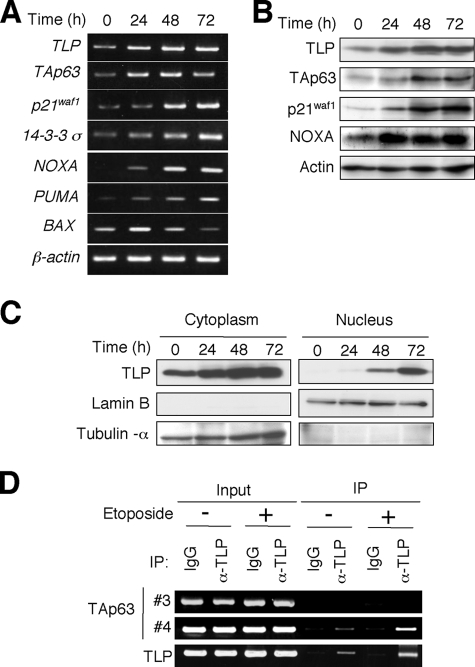
It has been shown that TAp63 is induced in human hepatocellular carcinoma-derived p53-deficient Hep3B cells in response to etoposide and also involved in the promotion of apoptosis (25, 26). We therefore investigated the effects of etoposide on the expression level of TAp63 and TLP in Hep3B cells. To this end, Hep3B cells were exposed to etoposide at a final concentration of 50 μm. At the indicated time points after etoposide treatment, total RNA and whole cell lysates were prepared and subjected to semi-quantitative RT-PCR and immunoblotting, respectively. As shown in Fig. 5A, the expression level of the endogenous TLP was induced in response to etoposide in association with the up-regulation of TAp63 as well as its transcriptional target genes such as p21waf1, 14-3-3σ, NOXA, PUMA, and BAX. Similarly, etoposide treatment led to a remarkable induction of the endogenous TLP protein as well as TAp63 protein and its target gene products, including p21waf1 and NOXA (Fig. 5B). Similar results were also obtained in human hepatocellular carcinoma-derived HepG2 cells (supplemental Fig. S1).
FIGURE 5.
TLP and TAp63 are induced in response to DNA damage. A, etoposide treatment results in a significant induction of TLP and TAp63. Hep3B cells were treated with 50 μm of etoposide or left untreated. At the indicated time points after etoposide treatment, total RNA was prepared and subjected to semi-quantitative RT-PCR. B, immunoblotting. Hep3B cells were exposed to 50 μm of etoposide. At the indicated time points after exposure to etoposide, whole cell lysates were prepared and processed for immunoblotting with the indicated antibodies. C, etoposide-mediated nuclear accumulation of TLP. Hep3B cells were exposed to 50 μm etoposide. At the indicated time points after etoposide treatment, cells were biochemically fractionated into nuclear and cytoplasmic fractions followed by immunoblotting with anti-TLP antibody. Lamin B and tubulin-α were used as nuclear and cytoplasmic markers, respectively. D, ChIP assay. Hep3B cells were treated with 50 μm etoposide or left untreated. Forty-eight hours after etoposide treatment, cells were cross-linked with formaldehyde and cross-linked chromatin was sonicated followed by immunoprecipitation with normal goat IgG or with polyclonal anti-TLP antibody. Genomic DNA was purified from the immunoprecipitates and subjected to PCR using the indicated primer sets as described in the legend for Fig. 3.
Consistent with our previous observations (27), TLP accumulated in cell nucleus of Hep3B cells in response to etoposide as examined by immunoblotting (Fig. 5C). Similar results were also obtained in HepG2 cells (supplemental Fig. S1). Next, we sought to examine whether the endogenous TLP could be recruited onto TAp63 promoter in response to etoposide. For this purpose, Hep3B cells were treated with or without 50 μm etoposide. Forty-eight hours after etoposide treatment, cells were cross-linked and the immunoprecipitated genomic DNA was subjected to ChIP assay. As shown in Fig. 5D, the endogenous TLP was induced to be recruited onto TAp63 promoter region in response to etoposide. Because it has been shown that TLP has an ability to transactivate its own promoter (28), we examined the recruitment of TLP onto its promoter region as a positive control (bottom panel of Fig. 5D).
The Effect of Knocking Down of the Endogenous TLP or TAp63 on Etoposide-mediated Apoptosis
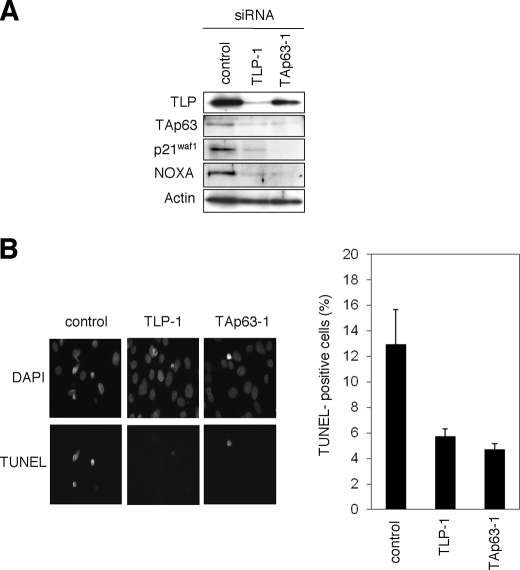
To examine the functional significance of the endogenous TLP and TAp63 in the regulation of etoposide-mediated apoptosis, Hep3B cells were transiently transfected with control siRNA, siRNA against TLP (TLP-1), or with siRNA targeting TAp63 (TAp63–1). Forty-eight hours after transfection, cells were exposed to etoposide for 48 h. As seen in Fig. 6A, knocking down of the endogenous TLP resulted in a significant down-regulation of the endogenous TAp63 as well as its target gene products such as p21waf1 and NOXA. Similarly, siRNA-mediated knockdown of the endogenous TAp63 caused a remarkable reduction in the expression level of p21waf1 and NOXA, suggesting that the TLP/TAp63 pathway contributes to the promotion of apoptosis mediated by etoposide. Consistent with this notion, TUNEL staining experiments demonstrated that knocking down of the endogenous TLP or TAp63 results in a significant reduction of number of TUNEL-positive cells as compared with that of control cells (Fig. 5B). Similar results were obtained in Hep3B cells transfected with the different set of siRNA (supplemental Fig. S2) and also obtained in HepG2 cells (supplemental Fig. S3). Taken together, our present findings strongly suggest that TLP has an ability to induce etoposide-mediated apoptosis through up-regulation of TAp63 expression.
FIGURE 6.
Effects of the endogenous TLP and TAp63 in the regulation of DNA damage response. A, siRNA-mediated knockdown of the endogenous TLP and TAp63. Hep3B cells were transiently transfected with 10 nm control siRNA, siRNA against TLP (TLP-1) or with siRNA targeting TAp63 (TAp63-1). Forty-eight hours after transfection, cells were exposed to 50 μm etoposide. Forty-eight hours after etoposide treatment, whole cell lysates were prepared and processed for immunoblotting with indicated antibodies. B, TUNEL staining. Hep3B cells were transiently transfected as in A. Forty-eight hours after transfection, cells were exposed to 50 μm etoposide. Forty-eight hours after etoposide treatment, cells were fixed in 4% parafolmaldeyde and subjected to TUNEL staining. Cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). The percentage of TUNEL-positive cells shown in each column represents the mean of three independent experiments (right panels).
DISCUSSION
Considering that TBP has an essential role in the regulation of basal transcription, TBP-related factors, including TLP might also participate in the transcriptional regulatory mechanisms. Previously, it has been shown that TBP is directly associated with tumor suppressor p53, and their complex formation contributes to the successful transcription (29). Although we have described that TLP has an intrinsic ability to prolong the G2 phase and to induce apoptosis in a p53-independent manner (21), our earlier study did not rule out the possible involvement of the other p53 family members such as TAp73 and TAp63 in this process. In the present study, we have found for the first time that, upon DNA damage mediated by anti-cancer drug etoposide, TLP is induced to accumulate in cell nucleus in association with a significant up-regulation of TAp63 as well as its direct target genes, suggesting that TLP acts as a transcriptional activator for pro-apoptotic TAp63.
According to our present results, ectopic expression of FLAG-TLP led to a significant induction of TAp63 but not of the other p53 family members such as p53 and TAp73, indicating that TLP acts as a specific transcriptional activator for TAp63. Consistent with these observations, siRNA-mediated knockdown of the endogenous TLP resulted in a remarkable down-regulation of TAp63. In addition, TLP had an undetectable effect on SV40 promoter (data not shown). Intriguingly, TLP-dependent transcriptional up-regulation was also observed in chicken DT40 cells, suggesting that the molecular mechanisms behind TLP-dependent transcriptional up-regulation of TAp63 are conserved among various species. By using luciferase reporter assays, we have identified the proximal and distal regions within human TAp63 promoter required for TLP-dependent transcriptional activation of TAp63. Among them, TLP was efficiently recruited onto the proximal region but not onto the distal region as examined by ChIP assay. Thus, we have focused our attention on the proximal region for further analysis. Chong et al. described that TLP has an ability to transactivate NF1 gene promoter, and they have identified the small independent two regions within NF1 promoter required for TLP-dependent transcriptional activation of NF1 gene (20). Based on their results, the above-mentioned two sequences directly bound to the purified TLP prepared from HeLa cells. During the extensive search of the proximal region of TAp63 promoter, we have found out the sequence element (5′-AGCTGGAGCA-3′), which was also included within one of the TLP-binding sequences of NF1 gene (5′-AGCTGAGAGCA-3′). Of note, this sequence element was well conserved among mouse, chicken, and dog TAp63 promoter regions (over 80% sequence identity). Introduction of mutations into this sequence element decreased the luciferase activity mediated by exogenously expressed FLAG-TLP, suggesting that this sequence element might act as a TLP-responsive element for human TAp63 gene.
As mentioned above, the distal region of TAp63 promoter had an indirect effect on TLP-dependent transcriptional regulation of TAp63. Consistent with these results, pGL3-TAp63(−2340/M1) in which mutations were introduced into the proximal TLP-responsive element retained an ability to respond to the exogenously expressed FLAG-TLP but to the lesser degree. This might be due to the presence of the additional regulatory sequence(s) within TAp63 promoter, including the distal region. When the four putative TLP-responsive elements were fused to core SV40 promoter, luciferase activity was undetectable in response to exogenously expressed FLAG-TLP (data not shown). In contrast, we have detected TLP-dependent luciferase activity driven by pGL3-TAp63-TLP-luc. Indeed, our luciferase reporter construct termed pGL3-TAp63(−487/+26) contained the proximal region and core TAp63 promoter region, suggesting that there could exist a functional relationship between the proximal and core promoter regions with respect to TLP-dependent transcriptional regulation of TAp63. In support of this notion, both TLP and TBP have been shown to be recruited onto NF1 genomic region containing TLP-responsive element and core promoter sequence (28). Further experiments should be required to adequately address this issue.
Another finding of our present study was that TLP is induced to accumulate in cell nucleus in response to etoposide, and contributes to etoposide-mediated apoptosis through the up-regulation of TAp63. Based on our present results, etoposide treatment promoted the efficient recruitment of the endogenous TLP onto TAp63 proximal promoter region and resulted in a strong induction of TAp63 as well as its direct target genes implicated in apoptosis such as PUMA, NOXA, and BAX. siRNA-mediated knockdown of the endogenous TLP led to a significant down-regulation of TAp63 as well as pro-apoptotic NOXA. Additionally, knocking down of the endogenous TLP resulted in a remarkable inhibition of etoposide-mediated apoptosis as examined by TUNEL assay. Similar results were also obtained in TAp63-knocked down cells. In a sharp contrast, Lantner et al. found that CD74 stimulation leads to the activation of pro-survival NF-κB and then activated form of NF-κB transactivates TAp63 followed by TAp63-dependent up-regulation of anti-apoptotic Bcl-2 (30). Their observations indicated that NF-κB is one of transcriptional activators for TAp63; however, the up-regulation of TAp63 contributes to cell survival in mature B cells. The differential biological outcomes of the up-regulation of TAp63 might be due to the cellular contexts employed in the experiments. To our knowledge, our present result is a first finding showing that TLP acts as a transcriptional activator for pro-apoptotic TAp63 and participates in the regulation of DNA damage response. Thus, the TLP/TAp63 pro-apoptotic pathway is a novel one in response to DNA damage.
From the clinical point of view, several lines of evidence indicated that the altered expression of p63 isoforms is observed in human tumor tissues (4), although loss of function mutations in p63 are rarely detected in various human tumors (6, 7). For example, the dysregulation of the oncogenic ΔNp63, which is expressed from a different promoter than that used for expression of TAp63, was detectable, especially in squamous cell carcinomas (4, 31). In support with this notion, ΔNp63 contributes to cell survival through a dominant negative effect toward wild-type p53, TAp63, and TAp73 (4, 31). In contrast, the possible involvement of TAp63 in tumor generation might be attributed to its low expression levels in human primary tumors (32, 33). In this connection, higher expression levels of TAp63 correlated with better prognoses of patients with bladder carcinoma (32). Thus, it is likely that TLP-TAp63 pathway might play an important role in certain tumor suppression. In accordance with this notion, it has been shown that TLP also induces the expression of NF1, a representative tumor suppressor gene (20). Given that etoposide stimulates nuclear accumulation of TLP and up-regulation of the TAp63, TLP-mediated TAp63 expression followed by induction of apoptosis might help to suppress tumor generation.
Acknowledgments
We are grateful to Drs. Robert G. Roeder, Jeong H. Park, Tomoe Ichikawa, Miki Ohira, Yohko Nakamura, Hajime Kageyama, Cheng Hong, Kyoung-ae Park, and Tomoko Mabuchi for valuable discussion.
This work was supported by a Grant-in-Aid from the Ministry of Health, Labor and Welfare for Third Term Comprehensive Control Research for Cancer (to A. N.), a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to T. T. and A. N.), a Grant-in-Aid for Scientific Research from Japan Society for the Promotion of Science (to A. N.), and grants from Uehara Memorial Foundation and Futaba Corporation (to T. T. and A. N.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- TA
- trans-activating isoform
- ChIP
- chromatin immunoprecipitation
- FACS
- fluorescence-activated cell sorter
- NF1
- neurofibromatosis type 1
- siRNA
- small interfering RNA
- TBP
- TATA-binding protein
- TLP
- TBP-like protein
- TUNEL
- terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling
- RT
- reverse transcription.
REFERENCES
- 1.Stiewe T. (2007) Nat. Rev. Cancer 7, 165–168 [DOI] [PubMed] [Google Scholar]
- 2.Yang A., Kaghad M., Wang Y., Gillett E., Fleming M. D., Dötsch V., Andrews N. C., Caput D., McKeon F. (1998) Mol. Cell 2, 305–316 [DOI] [PubMed] [Google Scholar]
- 3.Osada M., Ohba M., Kawahara C., Ishioka C., Kanamaru R., Katoh I., Ikawa Y., Nimura Y., Nakagawara A., Obinata M., Ikawa S. (1998) Nat. Med. 4, 839–843 [DOI] [PubMed] [Google Scholar]
- 4.Müller M., Schleithoff E. S., Stremmel W., Melino G., Krammer P. H., Schilling T. (2006) Drug Resist. Update 9, 288–306 [DOI] [PubMed] [Google Scholar]
- 5.Candi E., Dinsdale D., Rufini A., Salomoni P., Knight R. A., Mueller M., Krammer P. H., Melino G. (2007) Cell Cycle 6, 274–285 [DOI] [PubMed] [Google Scholar]
- 6.Hagiwara K., McMenamin M. G., Miura K., Harris C. C. (1999) Cancer Res. 59, 4165–4169 [PubMed] [Google Scholar]
- 7.Sunahara M., Shishikura T., Takahashi M., Todo S., Yamamoto N., Kimura H., Kato S., Ishioka C., Ikawa S., Ikawa Y., Nakagawara A. (1999) Oncogene 18, 3761–3765 [DOI] [PubMed] [Google Scholar]
- 8.Flores E. R., Sengupta S., Miller J. B., Newman J. J., Bronson R., Crowley D., Yang A., McKeon F., Jacks T. (2005) Cancer Cell 7, 363–373 [DOI] [PubMed] [Google Scholar]
- 9.Flores E. R., Tsai K. Y., Crowley D., Sengupta S., Yang A., McKeon F., Jacks T. (2002) Nature 416, 560–564 [DOI] [PubMed] [Google Scholar]
- 10.Levrero M., De Laurenzi V., Costanzo A., Gong J., Wang J. Y., Melino G. (2000) J. Cell Sci. 113, 1661–1670 [DOI] [PubMed] [Google Scholar]
- 11.Davidson I. (2003) Trends Biochem. Sci. 28, 391–398 [DOI] [PubMed] [Google Scholar]
- 12.Reina J. H., Hernandez N. (2007) Genes Dev. 21, 2855–2860 [DOI] [PubMed] [Google Scholar]
- 13.Ohbayashi T., Kishimoto T., Makino Y., Shimada M., Nakadai N., Aoki T., Kawata T., Niwa S., Tamura T. (1999) Biochem. Biophys. Res. Commun. 255, 137–142 [DOI] [PubMed] [Google Scholar]
- 14.Teichmann M., Wang Z., Martinez E., Tjernberg A., Zhang D., Vollmer F., Chait B. T., Roeder R. G. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 13720–13725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rabenstein M. D., Zhou S., Lis J. T., Tjian R. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 4791–4796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perletti L., Dantonel J. C., Davidson I. (1999) J. Biol. Chem. 274, 15301–15304 [DOI] [PubMed] [Google Scholar]
- 17.Moore P. A., Ozer J., Salunek M., Jan G., Zerby D., Campbell S., Lieberman P. M. (1999) Mol. Cell. Biol. 19, 7610–7620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohbayashi T., Shimada M., Nakadai T., Wada T., Handa H., Tamura T. A. (2003) Nucleic Acids Res. 31, 2127–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakadai T., Shimada M., Shima D., Handa H., Tamura T. A. (2004) J. Biol. Chem. 279, 7447–7455 [DOI] [PubMed] [Google Scholar]
- 20.Chong J. A., Moran M. M., Teichmann M., Kaczmarek J. S., Roeder R., Clapham D. E. (2005) Mol. Cell. Biol. 25, 2632–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimada M., Nakadai T., Tamura T. A. (2003) Mol. Cell. Biol. 23, 4107–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ichikawa T., Suenaga Y., Koda T., Ozaki T., Nakagawara A. (2008) Oncogene 27, 409–420 [DOI] [PubMed] [Google Scholar]
- 23.Ohbayashi T., Makino Y., Tamura T. A. (1999) Nucleic Acids Res. 27, 750–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ozaki T., Nakagawara A. (2005) Cancer Sci. 96, 729–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gressner O., Schilling T., Lorenz K., Schulze Schleithoff E., Koch A., Schulze-Bergkamen H., Lena A. M., Candi E., Terrinoni A., Catani M. V., Oren M., Melino G., Krammer P. H., Stremmel W., Müller M. (2005) EMBO J. 24, 2458–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petitjean A., Cavard C., Shi H., Tribollet V., Hainaut P., Caron de Fromentel C. (2005) Oncogene 24, 512–519 [DOI] [PubMed] [Google Scholar]
- 27.Park K. A., Tanaka Y., Suenaga Y., Tamura T. A. (2006) Mol. Cells 22, 203–209 [PubMed] [Google Scholar]
- 28.Bush S. D., Richard P., Manley J. L. (2008) Mol. Cell. Biol. 28, 83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin D. W., Muñoz R. M., Subler M. A., Deb S. (1993) J. Biol. Chem. 268, 13062–13067 [PubMed] [Google Scholar]
- 30.Lantner F., Starlets D., Gore Y., Flaishon L., Yamit-Hezi A., Dikstein R., Leng L., Bucala R., Machluf Y., Oren M., Shachar I. (2007) Blood 110, 4303–4311 [DOI] [PubMed] [Google Scholar]
- 31.Rocco J. W., Ellisen L. W. (2006) Cell Cycle 5, 936–940 [DOI] [PubMed] [Google Scholar]
- 32.Park B. J., Lee S. J., Kim J. I., Lee S. J., Lee C. H., Chang S. G., Park J. H., Chi S. G. (2000) Cancer Res. 60, 3370–3374 [PubMed] [Google Scholar]
- 33.Koster M. I., Lu S. L., White L. D., Wang X. J., Roop D. R. (2006) Cancer Res. 66, 3981–3986 [DOI] [PubMed] [Google Scholar]