Abstract
There is functional evidence that polycystin-2 (TRPP2) interacts with other members of the transient receptor potential family, including TRPC1 and TRPV4. Here we have used atomic force microscopy to study the structure of the TRPP2 homomer and the interaction between TRPP2 and TRPC1. The molecular volumes of both Myc-tagged TRPP2 and V5-tagged TRPC1 isolated from singly transfected tsA 201 cells indicated that they assembled as homotetramers. The molecular volume of the protein isolated from cells expressing both TRPP2 and TRPC1 was intermediate between the volumes of the two homomers, suggesting that a heteromer was being formed. The distribution of angles between pairs of anti-Myc antibodies bound to TRPP2 particles had a large peak close to 90° and a smaller peak close to 180°, consistent with the assembly of TRPP2 as a homotetramer. In contrast, the corresponding angle distributions for decoration of the TRPP2-TRPC1 heteromer by either anti-Myc or anti-V5 antibodies had predominant peaks close to 180°. This decoration pattern indicates a TRPP2:TRPC1 subunit stoichiometry of 2:2 and an alternating subunit arrangement.
Introduction
Autosomal dominant polycystic kidney disease is one of the commonest inherited human disorders (reviewed in Ref. 1). It has a population prevalence of over 1:1,000 in all ethnic groups and is a leading cause of end stage renal failure. Autosomal dominant polycystic kidney disease is characterized by the progressive loss of normal renal parenchyma secondary to the development of multiple fluid-filled cysts derived from renal tubular epithelial cells. It is caused by mutations in two genes, PKD1 and PKD2, whose protein products, polycystin-1 (2, 3) and polycystin-2 (or TRPP2) (4) form a Ca2+-permeable ion channel complex (5). This complex transduces extracellular mechanical stimuli via the renal primary cilium (6) and regulates multiple intracellular Ca2+-sensitive signaling pathways (5, 7). TRPP2 also appears to have a role, independent of polycystin-1, in regulating Ca2+ efflux from the endoplasmic reticulum (8, 9). In addition to its interaction with polycystin-1, TRPP2 is known to interact with other members of the TRP4 superfamily, such as TRPC1 (10) and TRPV4 (11), raising the question of the architecture of these heteromeric complexes.
TRP channel complexes have been assumed to be tetramers, initially on the basis of the resemblance of the primary structure of the TRP channel subunits to that of the Shaker K+ channel, which is known to be tetrameric (12). Moreover, a variety of structural and functional techniques have been used to demonstrate a tetrameric structure for a number of TRP channel family members, including TRPC1 (13), TRPC3 (14), TRPV1 (15, 16), TRPV5 and TRPV6 (17), and TRPM2 (18). Intriguingly, data have recently been presented indicating that TRPP2 exists in the plasma membrane as a trimer, which is then able to interact with polycystin-1 to form a heteromer with a 3:1 stoichiometry (19). It is puzzling that TRPP2 appears to behave differently from all other TRP channels, and the stoichiometry of the TRPP2 homomer must be regarded as controversial. The structure of the TRPP2-TRPC1 heteromer has also recently been examined using atomic force microscopy (AFM) (20). However, the images presented had some unusual features; for instance, the molecular volumes of the proteins were much larger than expected. It is fair to say, therefore, that the molecular architecture of this protein complex is also still unclear.
We have developed a method, based on AFM imaging, for determining the arrangement of subunits within multimeric proteins (13, 21–24). This method involves engineering specific epitope tags, including His6, onto each subunit and expressing the proteins in a suitable cell line (e.g. tsA 201). Crude membrane fractions from the transfected cells are solubilized in detergent, and the proteins are isolated through binding to Ni2+-agarose beads. The isolated proteins are then imaged by AFM, and their mean molecular volume is compared with the molecular volume expected for the protein, based on its molecular mass. In this way, assembled multimers can be distinguished from unassembled subunits. The proteins are incubated with antibodies to the tags, and the resulting multimer-antibody complexes are imaged by AFM. Multimers with two bound antibodies are identified, and the angles between the antibodies are measured. A frequency distribution of these angles then reveals the architecture of the multimer. Here, we have used this method to study the stoichiometry of the TRPP2 homomer and the interaction of TRPP2 with TRPC1. We show that TRPP2 assembles a homotetramer and that TRPP2 and TRPC1 form a heterotetramer with a 2:2 stoichiometry and an alternating subunit arrangement.
EXPERIMENTAL PROCEDURES
Cell Culture
tsA 201 cells (a subclone of human embryonic kidney-293 cells stably expressing the SV40 large T-antigen) were grown in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin in an atmosphere of 5% CO2, air.
Channel Constructs
DNA encoding human TRPP2 was subcloned into the pcDNA3.1/Myc-His vector (Invitrogen), which produces a protein tagged at its C terminus with Myc and His6 epitopes. Human TRPC1 DNA was subcloned into the pcDNA3.1/V5-His-TOPO vector (Invitrogen), which produces a protein tagged at its C terminus with V5 and His6 epitopes (13). In addition, the sequence encoding the His6 tag was deleted from this construct to enable the expression of a protein containing only the V5 epitope.
Transient Transfection of tsA 201 Cells
Transient transfections of tsA 201 cells with DNA were carried out using the CalPhosTM mammalian transfection kit (Clontech), according to the manufacturer's instructions. A total of 200 μg of DNA was used to transfect cells in five 162-cm2 culture flasks. When cells were doubly transfected, 100 μg of DNA for each construct was used. After transfection, cells were incubated for 24–48 h at 37 °C to allow protein expression. Protein expression and intracellular localization were checked using immunofluorescence analysis of small scale cultures. Cells were fixed, permeabilized, and incubated with appropriate primary antibodies (mouse monoclonal anti-Myc, anti-V5, and anti-His6, all from Invitrogen; mouse monoclonal anti-FLAG, from Sigma; and rabbit polyclonal anti-Myc, from Abcam), followed by either Cy3- or fluorescein isothiocyanate-conjugated goat secondary antibodies (Sigma). Cells were imaged by confocal laser-scanning microscopy.
Solubilization and Purification of His6-tagged Proteins
The solubilization/purification procedure was as described previously (24). Briefly, a crude membrane fraction prepared from the cells was solubilized in 1% (w/v) CHAPS, and the solubilized material was incubated with Ni2+-agarose beads (Probond, Invitrogen). The beads were washed extensively, and bound proteins were eluted with increasing concentrations of imidazole. Samples were analyzed by SDS-polyacrylamide gel electrophoresis, and proteins were detected by immunoblotting, using mouse monoclonal antibodies against Myc (TRPP2) or V5 (TRPC1).
AFM Imaging of TRP Proteins and TRP-Antibody Complexes
Isolated channels were imaged either alone or following overnight incubation at 4 °C with a 1:2 molar ratio (∼0.2 nm channel protein concentration) of anti-epitope tag monoclonal antibody (Invitrogen). Proteins were diluted to a final concentration of 0.04 nm, and 45 μl of the sample was allowed to adsorb to freshly cleaved, poly-l-lysine-coated mica disks. After a 10-min incubation, the sample was washed with Biotechnology Performance Certified-grade water (Sigma) and dried under nitrogen. Imaging was performed with a Veeco Digital Instruments Multimode AFM controlled by a Nanoscope IIIa controller. Samples were imaged in air, using tapping mode. The silicon cantilevers used had a drive frequency of ∼300 kHz and a specified spring constant of 40 newtons/m (Olympus). The applied imaging force was kept as low as possible (As/A0 ∼ 0.85).
The molecular volumes of the protein particles were determined from particle dimensions based on AFM images. After adsorption of the channels onto the mica support, the particles adopt the shape of a spherical cap. As described previously (24), the heights and radii were measured from multiple cross-sections of the same particle, and the molecular volume was calculated using the following equation,
where h is the particle height and r is the radius.
Molecular volume based on molecular mass was calculated using the equation,
where M0 is the molecular mass, N0 is Avogadro's number, V1 and V2 are the partial specific volumes of particle (0.74 cm3/g) and water (1 cm3/g), respectively, and d is the extent of protein hydration (taken as 0.4 g of water/g of protein).
RESULTS
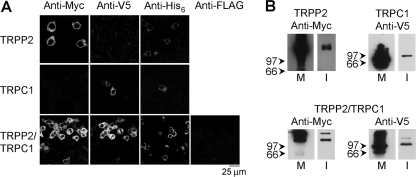
tsA 201 cells were transiently transfected with DNA encoding Myc/His6-tagged TRPP2, V5/His6-tagged TRPC1, or both constructs. Protein expression and localization was confirmed by immunofluorescence, using appropriate anti-tag antibodies. The staining signals with either anti-Myc or anti-His6 antibodies in cells transfected with DNA encoding TRPP2-Myc/His6 showed the expression of the channel (Fig. 1A, upper panels). In contrast, use of an anti-V5 antibody as a negative control produced only a background immunofluorescence signal. Conversely, cells transfected with TRPC1-V5/His6 gave positive immunofluorescence signals with anti-V5 and anti-His6 antibodies but not with anti-Myc (Fig. 1A, center panels). Cells transfected with both TRPP2-Myc/His6 and TRPC1-V5/His6 gave positive immunofluorescence signals with anti-Myc, anti-V5, and anti-His6 antibodies, indicating the presence of the two proteins (Fig. 1A, lower panels). As shown, the anti-Myc and anti-V5 signals in doubly labeled cell populations extensively overlapped, indicating that the majority of transfected cells expressed both TRPP2 and TRPC1. An anti-FLAG antibody gave only a background signal. The reticular staining patterns suggest that both TRPP2 and TRPC1 were localized predominantly in the endoplasmic reticulum, as has been reported previously (5, 9, 25).
FIGURE 1.
Expression and isolation of TRPP2 and TRPC1 homomers and TRPP2-TRPC1 heteromers. A, immunofluorescence detection of epitope-tagged proteins in transiently transfected tsA 201 cells. Cells were fixed, permeabilized, and incubated with appropriate anti-tag antibodies, as indicated, followed by fluorophore-conjugated secondary antibodies. For the TRPP2 and TRPC1 homomers, all primary antibodies were mouse monoclonals, and the secondary antibody was Cy3-conjugated. For the TRPP2-TRPC1 co-transfection, V5, His6, and FLAG epitopes were detected using mouse monoclonal primary antibodies, whereas Myc was detected using a rabbit polyclonal antibody, to allow double labeling of the same cell population with anti-Myc and anti-V5. Secondary antibodies were either Cy3-conjugated (Myc and FLAG) or fluorescein isothiocyanate-conjugated (V5 and His6). Cells were imaged by confocal laser-scanning microscopy. B, detection of proteins in membrane fractions from transfected cells (M) and after isolation (I) following elution from a Ni2+-agarose column. Samples were analyzed by SDS-PAGE and immunoblotting using mouse monoclonal anti-tag antibodies, followed by a horseradish peroxidase-conjugated goat anti-mouse secondary antibody. The total amount of protein in the initial membrane fraction was about 5 mg, of which 50 μg was loaded on the gels. The total yield of isolated proteins was about 500 ng, of which 30 ng was loaded on the gels. Immunoreactive bands were visualized using enhanced chemiluminescence. The arrowheads indicate molecular mass markers (kDa).
Crude membrane fractions prepared from cells expressing TRPP2-Myc/His6, TRPC1-V5/His6, or both proteins were solubilized in CHAPS detergent (1% (w/v)), and proteins were isolated through the binding of the His6 tags to Ni2+-agarose beads. Both the membrane fractions and the isolated proteins were subjected to SDS-PAGE and immunoblotting using either anti-Myc or anti-V5 antibodies. Anti-Myc labeled a band of molecular mass 110 kDa in fractions prepared from cells expressing TRPP2, and anti-V5 antibody labeled a band at 85 kDa in fractions from cells expressing TRPC1 (Fig. 1B). These bands are consistent with the expected sizes of the two subunits, with their epitope tags. Both immunoreactive bands were detected on immunoblots of fractions from cells expressing both proteins. Hence, the Ni2+-agarose beads successfully captured the appropriate proteins from the transfected cells.
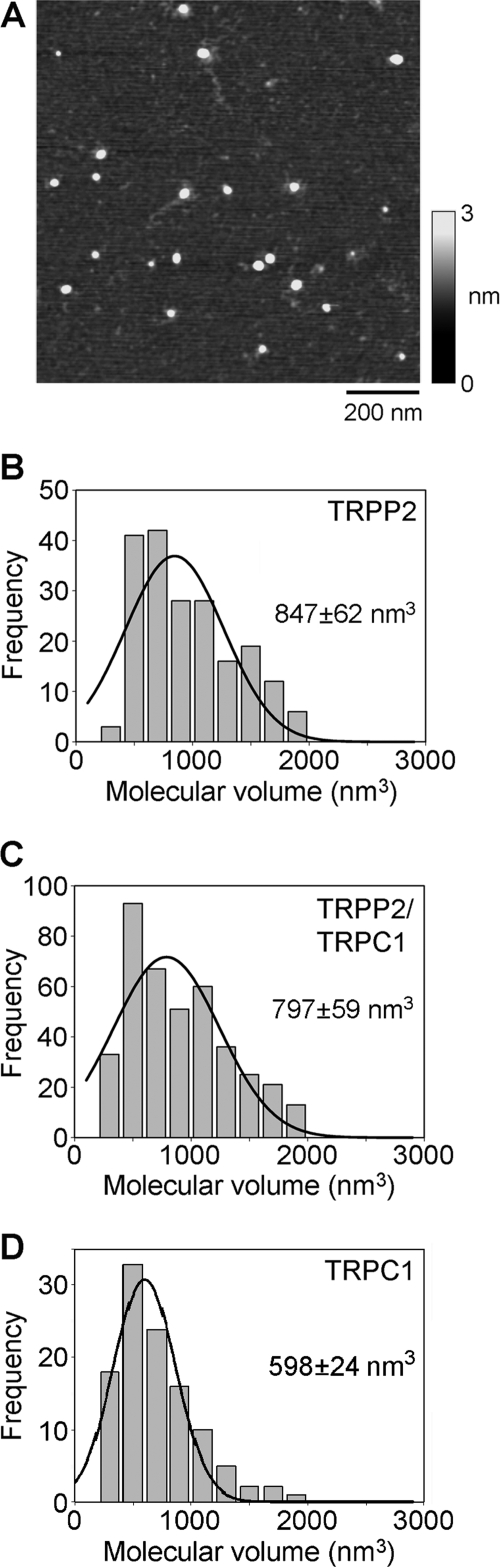
Proteins isolated from cells expressing TRPP2, TRPC1, and TRPP2-TRPC1 were imaged by AFM. A representative low magnification AFM image of particles isolated from TRPP2-expressing cells is shown in Fig. 2A. The image shows a homogenous spread of particles, indicating the presence of a single major protein species. Similar images were obtained from samples prepared from TRPC1-expressing and TRPP2-TRPC1-expressing cells (not shown). Frequency distributions of molecular volumes were produced for the three types of particles (Fig. 2, B–D). The mean ± S.E. molecular volumes were 847 ± 62 nm3 (n = 195) for TRPP2 (Fig. 2B), 797 ± 59 nm3 (n = 399) for TRPP2-TRPC1 (Fig. 2C), and 598 ± 24 nm3 (n = 111) for TRPC1 (Fig. 2D). The molecular volumes predicted for TRPP2 and TRPC1 subunits on the basis of their molecular masses are 209 and 161 nm3, respectively; hence, tetramers would have predicted molecular volumes of 836 nm3 (TRPP2) and 644 nm3 (TRPC1). The measured volumes, therefore, indicate a tetrameric structure in both cases, as already demonstrated for TRPC1 (13).
FIGURE 2.
AFM imaging of TRPP2 and TRPC1 homomers and TRPP2-TRPC1 heteromers. A, low magnification AFM image of proteins isolated from TRPP2-expressing cells. A shade-height scale is shown at the right. B–D, frequency distributions of molecular volumes of proteins isolated from cells expressing TRPP2 (B), TRPP2-TRPC1 (C), and TRPC1 (D). The curves indicate the fitted Gaussian functions. The means of the distributions ± S.E. are indicated.
The size order of the three proteins is TRPP2 > TRPP2-TRPC1 > TRPC1. The observation that co-transfection with TRPP2 and TRPC1 produces particles of mean volume intermediate between the volumes of the two homomers is consistent with the possibility that TRPP2 and TRPC1 are interacting to form a heteromeric complex. However, given that the volume distributions are quite broad, a combination of TRPP2 and TRPC1 homomers might give a similar result. It should also be pointed out that measurement of molecular volumes by AFM is complicated by factors such as the convolution introduced by the geometry of the scanning tip. Hence, the measured volume is not by itself a reliable indicator of the stoichiometry of the channel.
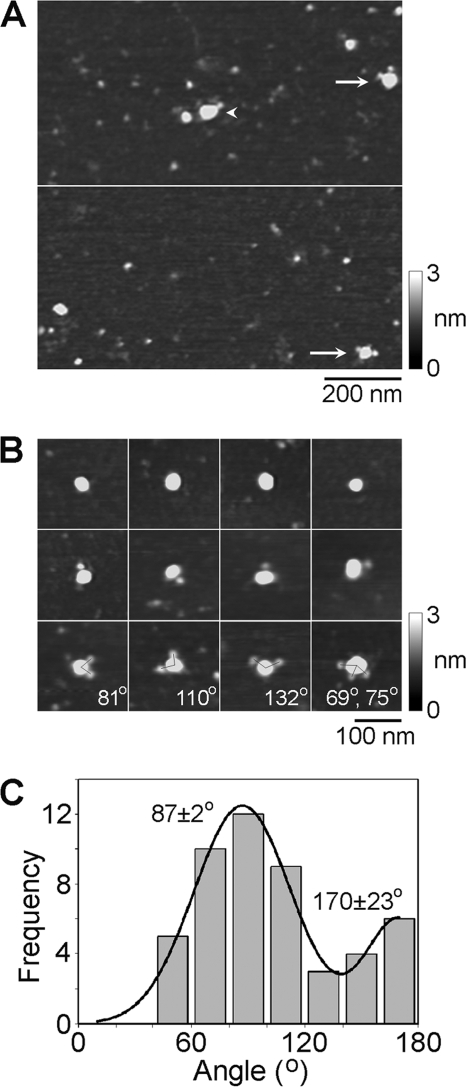
To conclusively establish the stoichiometry of the channels, they were imaged after antibody decoration. TRPP2 was imaged after incubation with the anti-Myc antibody (of the immunoglobulin G class), which should decorate the C-terminal Myc epitope tag present on each subunit. A low magnification AFM image of TRPP2-antibody complexes is shown in Fig. 3A. Several large particles can be seen, some of which have been decorated by either one (arrowhead) or two (arrows) smaller particles. The small particles had a molecular volume of around 260 nm3, close to the expected volume of 285 nm3 for an immunoglobulin G molecule, of molecular mass 150 kDa. Hence these particles represent anti-Myc antibodies bound to the Myc epitopes on the TRPP2 channel. A gallery of undecorated and singly and doubly decorated large particles is shown in Fig. 3B, along with an example of a triply decorated large particle. Of 888 large particles imaged, 520 (58.6%) were undecorated, 325 (36.6%) were singly decorated, 40 (4.5%) were doubly decorated, and three (0.3%) were triply decorated. No quadruply decorated channels were seen. This antibody decoration profile is similar to that predicted by the binomial distribution for a 12% occupancy of all potential binding sites (calculated assuming that the channel is a tetramer).
FIGURE 3.
Decoration of TRPP2 channels with anti-Myc antibodies. A, low magnification AFM images of a sample of isolated TRPP2 that had been incubated with anti-Myc antibodies. The arrowhead indicates a singly decorated TRPP2 particle; the arrows indicate doubly decorated particles. A shade-height scale is shown at the right. B, gallery of zoomed images of TRPP2 particles that are undecorated (top) or decorated by one (middle) or two (bottom) peripheral particles (antibodies). One triply decorated particle is also shown. Angles between pairs of antibodies are indicated. A shade-height scale is shown at the right. C, frequency distribution of angles between pairs of bound antibodies. The curve indicates the fitted Gaussian functions. The peaks of the distribution are indicated.
We identified TRPP2 channels that had been decorated by two or three antibodies and measured the angles between the bound antibodies. This was done in each case by joining the highest point on the central particle (the TRPP2 channel) to the highest points on the peripheral particles (the antibodies) by lines and then determining the angle between the two lines. The frequency distribution of angles obtained is shown in Fig. 3C. The angle distribution has two peaks: a large peak at 87 ± 2° and a smaller peak at 170 ± 23°. The ratio of the numbers of particles within the two peaks is 2.8:1. For a homotetramer, the expected result is a distribution with peaks at 90 and 180° in the ratio 2:1, close to the observed ratio. In contrast, a trimer would give a single angle peak at 120° (24), whereas a pentamer would give equally sized peaks at 72 and 144° (21). Hence, the antibody decoration profile confirms that TRPP2 assembles as a homotetramer, as we have previously shown for TRPC1, using a similar experimental approach (13).
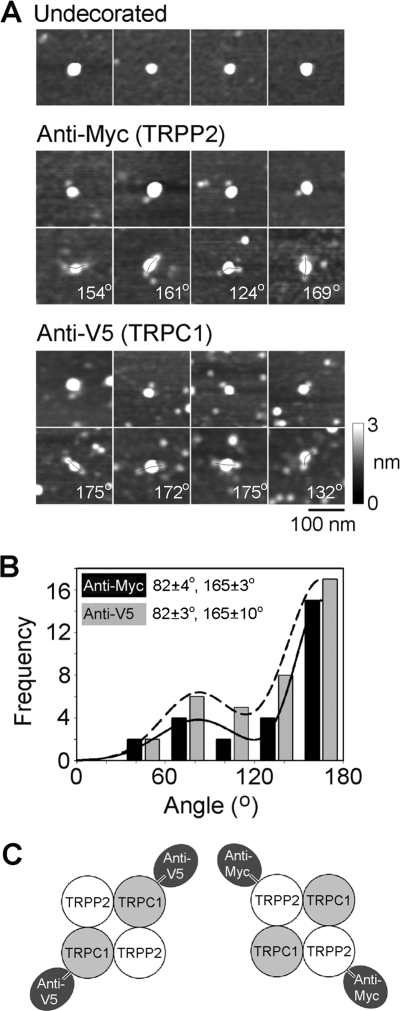
When proteins isolated from cells expressing both TRPP2-Myc/His6 and TRPC1-V5/His6 were incubated with anti-Myc or anti-V5 antibodies and then imaged by AFM, antibody-decorated large particles were again seen. A gallery of zoomed images of undecorated particles and particles that had been singly or doubly decorated by either anti-Myc or anti-V5 antibodies is shown in Fig. 4A. Of 1,267 large particles imaged, 27 (2.1%) were doubly decorated by anti-Myc antibodies. The extent of double decoration is therefore smaller than that seen with the TRPP2 homomer (above), indicating a reduced availability of antibody binding sites on each multimer. Similarly, of 745 large particles imaged, 38 (5.1%) were doubly decorated by anti-V5 antibodies, in comparison with 9.3% for the TRPC1 homomer under the same conditions (13). No triply or quadruply decorated particles were seen for either anti-Myc or anti-V5 antibodies.
FIGURE 4.
Decoration of TRPP2-TRPC1 channels with anti-Myc and anti-V5 antibodies. A, gallery of zoomed images of particles that are undecorated (top), decorated by one or two anti-Myc antibodies (middle), or decorated by one or two anti-V5 antibodies (bottom). Angles between pairs of antibodies are indicated. A shade-height scale is shown at the right. B, frequency distributions of angles between pairs of bound anti-Myc and anti-V5 antibodies. The curves indicate the fitted Gaussian functions. The peaks of the distributions are indicated. C, diagram illustrating the predominant subunit arrangement within the TRPP2-TRPC1 heteromer, as revealed by the antibody decoration patterns.
The angles between the pairs of bound antibodies were measured and used to construct frequency distributions. As for the TRPP2 homomer, both distributions had two peaks (Fig. 4B). However, in marked contrast to the results for the homomer, the smaller peak was at 82 ± 4° for anti-Myc and 82 ± 3° for anti-V5, whereas the larger peak was at 165 ± 3° for anti-Myc and 165 ± 10° for anti-V5. The ratios of the number of particles within the two peaks were 1:2.4 for anti-Myc and 1:1.9 for anti-V5. These distributions show a strong preference for decoration of the TRPP2-TRPC1 heteromer at around 180°, indicating a predominantly alternating subunit arrangement, as illustrated in Fig. 4C.
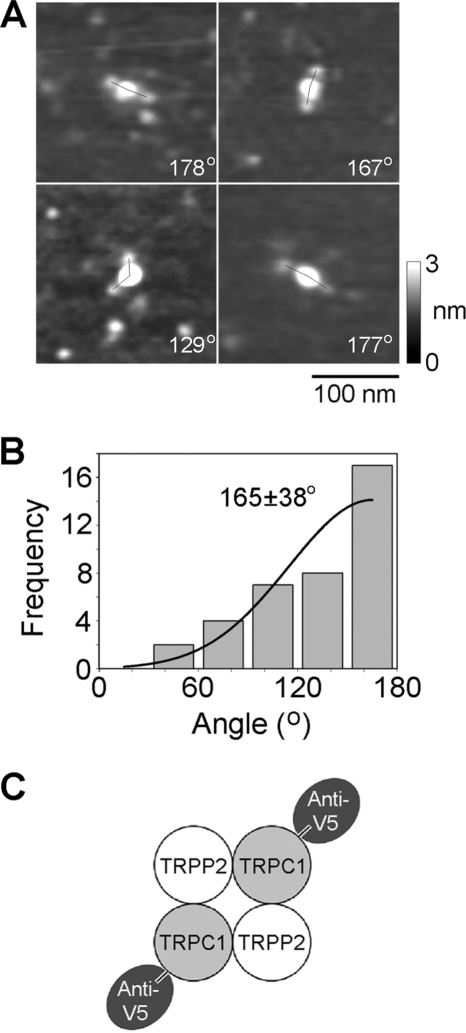
The TRPP2-TRPC1 heteromer showed small antibody decoration peaks at 90°, for both Myc and V5 epitope tags. These peaks could result from the isolation of minor populations of TRPP2 and TRPC1 homomers from the TRPP2-TRPC1-transfected cells. Alternatively, they could indicate that the architecture of the heteromer is not invariant and that some of the heteromers assemble with a TRPP2-TRPP2-TRPC1-TRPC1 arrangement. To distinguish between these two possibilities, we expressed a combination of TRPP2-Myc/His6 and TRPC1-V5 (lacking a His6 tag) in the tsA 201 cells. The normal isolation procedure was then used to produce protein complexes containing TRPP2-Myc/His6. Isolated proteins were then incubated with anti-V5 antibodies, which would decorate only heteromeric complexes containing TRPC-V5. A gallery of doubly decorated protein complexes is shown in Fig. 5A. All of the angles were now around 180°. Gaussian fitting showed that the frequency distribution of angles between pairs of bound antibodies (Fig. 5B) had only a single peak, at 165 ± 38° (n = 38); the peak at about 90° seen previously was not present. This result indicates that the heteromer assembles with the exclusive subunit arrangement TRPP2-TRPC1-TRPP2-TRPC1 and that the 90° peak was a result of the presence of a minority of homotetramers in the isolates from cells expressing both TRPP2-Myc/His6 and TRPC1-V5/His6.
FIGURE 5.
Decoration of TRPP2-TRPC1 channels isolated from cells expressing TRPP2-Myc/His6 and TRPC1-V5 with anti-V5 antibodies. A, gallery of zoomed images of particles decorated by two anti-V5 antibodies. Angles between pairs of antibodies are indicated. A shade-height scale is shown at the right. B, frequency distributions of angles between pairs of bound anti-V5 antibodies. The curve indicates the fitted Gaussian function. The peak of the distribution is indicated. C, diagram illustrating the subunit arrangement within the TRPP2-TRPC1 heteromer, as revealed by the antibody decoration pattern.
DISCUSSION
Our results demonstrate that the TRPP2 homomer assembles as a homotetramer, as has been shown previously for a number of other TRP channel family members (13–18). This finding directly contradicts the conclusion of a recent report that TRPP2 in the plasma membrane is a trimer (19). The evidence presented in the previous report is 3-fold. First, TRPP2 migrated as three distinct bands on both blue native polyacrylamide gels and on conventional gels after chemical cross-linking; these bands were suggested to represent monomer, dimer, and trimer. Second, a coiled-coil domain present in the C-terminal region of TRPP2 crystallized as a trimer. Third, single-molecule fluorescence studies revealed that upon bleaching, the fluorescence of the molecules declined in a series of three steps. We suggest that is difficult to determine the precise molecular mass of proteins running close to the top of SDS-polyacrylamide gels (and above the largest molecular mass marker); indeed, the molecular mass of the largest cross-linked band was reported to be 400 kDa, between the values of 330 and 440 kDa expected for trimers and tetramers). Hence, the three bands seen could actually represent monomer, dimer, and tetramer. In addition, the behavior of a short (63-amino acid) peptide sequence in crystallographic studies might not reflect the behavior of the intact protein in the context of the plasma membrane. Finally, the possibility that in oocytes, exogenous TRPP2 associates with another endogenous protein to form a 3:1 complex cannot be excluded. In fact, the authors specifically mention endogenous TRPC1 as a possible binding partner. The formation of such a complex could account for the observed three-step bleaching pattern.
The ability of different TRP channels, and especially different members of the TRPC subfamily, to interact physically and functionally is well known (e.g. see Refs. 26 and 27). These interactions have been detected by co-immunoprecipitation and fluorescence resonance energy transfer and through the demonstration that co-expression of two different subunits produces channels with properties distinct from those formed after expression of either subunit alone (26, 27). None of these methods formally proves that the subunits form a heteromeric channel. It is equally possible that individual homomers interact together in such a way as to produce channels with novel properties. Such interactions have in fact been demonstrated for P2X receptors, which form signaling complexes not only with neighboring P2X receptors but also with Cys-loop receptors (28) and with channels belonging to the gap junction family (29). Within the P2X receptor family itself, there is considerable evidence for a functional interaction between different subunits. For instance, it has been suggested that the P2X4 and P2X7 subtypes form a heteromeric receptor (30). However, it has recently been demonstrated that the predominant assembly pathway of these two subunits, in at least two cell types, leads to the production of separate P2X4 and P2X7 homomers (31).
TRPP2 has been shown to interact with both TRPC1 (10) and TRPV4 (11). The earliest evidence for an interaction between TRPP2 and TRPC1 involved co-localization and co-immunoprecipitation of the two subunits (32). This interaction was mapped to two regions of TRPP2: the cytosolic C terminus and a larger region, including transmembrane regions 2–6 and the connecting loops. Functional evidence for a TRPP2-TRPC1 interaction includes the production of a channel with novel physiological and pharmacological characteristics after co-expression of the two subunits (10). The channels formed by co-expression of TRPP2 and TRPC1 had single-channel conductance and ion permeability properties distinct from those observed in cells expressing either TRPP2 or TRPC1 alone. Further, the channels had distinct sensitivities to stimulation via the M1-muscarinic receptor and to blockade by amiloride. TRPP2 and TRPC1 were shown to be co-localized at the primary cilium and to be co-immunoprecipitated from kidney cell membranes. In these cells, manipulation of the levels of the two proteins produced functional effects that again indicated an interaction between TRPP2 and TRPC1. These results were taken to indicate the generation of a heteromeric TRPP2-TRPC1 channel; however, the presence of such a heteromer was not directly demonstrated.
Recently, a detailed analysis of the channels generated by expression of TRPP2, TRPC1, and TRPP2 plus TRPC1 provided corroborative evidence for an interaction between the two proteins (20). This study also included an attempt to visualize the heteromer directly using AFM. Purified TRPP2 and TRPC1 were reconstituted into liposomes either singly or together, and the liposomes were used to produce supported lipid bilayers for AFM imaging. When TRPP2 alone was imaged, the particles observed had a mean molecular volume of 3663 nm3, approximately 4 times larger than the particles detected in the present study and, significantly, 4 times larger than expected for a TRPP2 homotetramer. In contrast, the TRPC1 homomer appeared to be much smaller (mean molecular volume 1053 nm3), but was still almost twice the size that we report (and almost twice the volume expected for a homotetramer). It was reported that the contribution of TRPP2 and TRPC1 monomers to the heterotetramers could be distinguished on the basis of particle heights within the imaged complexes. Tetrameric features were seen, composed of two large and two small particles. On the basis of these images, it was concluded that the heterotetramer has a stoichiometry of 2 TRPP2:2 TRPC1. Although not explicitly stated, the subunit arrangement implied by the geometry of the complexes was TRPP2-TRPP2-TRPC1-TRPC1.
The fact that the protein particles detected in this previous study (especially TRPP2) were so much larger than would be expected may give some cause for concern about what they actually represent. Furthermore, the generation of TRPP2-TRPC1 complexes after mixing of independently isolated proteins would necessitate the efficient disassembly of the two homotetramers and reassembly of heterotetramers. We feel that this process, if it occurs at all, is likely to be highly inefficient. For instance, in our previous studies of the architecture of 5-HT3 (21) and the GABAA receptor (22), we have seen no evidence for rearrangement of multisubunit protein complexes after their isolation.
In contrast to the results of Zhang et al. (20), the isolated channels imaged in our study have the expected molecular volumes. The multimeric structure of the channels was not directly visible in our AFM images, a phenomenon that we have reported previously for other multisubunit proteins of approximately the same size, such as the 5-HT3 receptor (21) and the GABAA receptor (22). Instead, we revealed the tetrameric structure of both the TRPP2 homomer and the TRPP2-TRPC1 heteromer using imaging of antibody-decorated complexes. The ratio of 90°/180° angles for pairs of antibodies bound to the TRPP2 homomer (2.8:1) is close to the predicted value of 2:1. In contrast, the decoration profiles for both epitope tags on the TRPP2-TRPC1 heteromer strongly favor the 180° angle, indicating that the subunit stoichiometry is 2:2 and that the subunit distribution around the channel pore is TRPP2-TRPC1-TRPP2-TRPC1.
In our experiments, we used equal amounts of DNA for the two constructs when expressing the TRPP2-TRPC1 heteromer. Although there is not necessarily a direct relationship between the amount of DNA used and the relative level of protein expression, our method probably favors the generation of heteromers containing equal numbers of the two subunits. The possibility remains that the subunit stoichiometry of the heteromer is not fixed but rather can be manipulated by changing the relative expression levels of the two subunits, as we have previously found for the P2X2-P2X6 receptor heteromer (23).
The method described here will allow us to study the interaction of TRPP2 with other proteins. For instance, we should be able to test whether TRPP2 and TRPV4 form a heteromer with the same subunit stoichiometry and arrangement as that reported here for TRPP2 and TRPC1. In addition, we should be able to determine the architecture of the complex formed between TRPP2 and polycystin-1 and to ascertain how clinically significant mutations in TRPP2 affect its ability to interact with its protein partners. We are optimistic that this approach will shed light on the etiology of autosomal dominant polycystic kidney disease.
Acknowledgments
We thank A. Stewart for help with the immunofluorescence experiments and S. Carnally and N. Barrera for helpful discussions.
Footnotes
- TRP
- transient receptor potential
- AFM
- atomic force microscopy
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate.
REFERENCES
- 1.Yoder B. K., Mulroy S., Eustace H., Boucher C., Sandford R. (2006) Expert Rev. Mol. Med. 8, 1–22 [DOI] [PubMed] [Google Scholar]
- 2.Hughes J., Ward C. J., Peral B., Aspinwall R., Clark K., San Millán J. L., Gamble V., Harris P. C. (1995) Nat. Genet. 10, 151–160 [DOI] [PubMed] [Google Scholar]
- 3.The International Polycystic Kidney Disease Consortium (1995) Cell 81, 289–298 [DOI] [PubMed] [Google Scholar]
- 4.Mochizuki T., Wu G., Hayashi T., Xenophontos S. L., Veldhuisen B., Saris J. J., Reynolds D. M., Cai Y., Gabow P. A., Pierides A., Kimberling W. J., Breuning M. H., Deltas C. C., Peters D. J., Somlo S. (1996) Science 272, 1339–1342 [DOI] [PubMed] [Google Scholar]
- 5.Hanaoka K., Qian F., Boletta A., Bhunia A. K., Piontek K., Tsiokas L., Sukhatme V. P., Guggino W. B., Germino G. G. (2000) Nature 408, 990–994 [DOI] [PubMed] [Google Scholar]
- 6.Nauli S. M., Alenghat F. J., Luo Y., Williams E., Vassilev P., Li X., Elia A. E., Lu W., Brown E. M., Quinn S. J., Ingber D. E., Zhou J. (2003) Nat. Genet. 33, 129–137 [DOI] [PubMed] [Google Scholar]
- 7.Delmas P., Nauli S. M., Li X., Coste B., Osorio N., Crest M., Brown D. A., Zhou J. (2004) FASEB J. 18, 740–742 [DOI] [PubMed] [Google Scholar]
- 8.Geng L., Boehmerle W., Maeda Y., Okuhara D. Y., Tian X., Yu Z., Choe C. U., Anyatonwu G. I., Ehrlich B. E., Somlo S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 15920–15925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wegierski T., Steffl D., Kopp C., Tauber R., Buchholz B., Nitschke R., Kuehn E. W., Walz G., Köttgen M. (2009) EMBO J. 28, 490–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bai C. X., Giamarchi A., Rodat-Despoix L., Padilla F., Downs T., Tsiokas L., Delmas P. (2008) EMBO Rep. 9, 472–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Köttgen M., Buchholz B., Garcia-Gonzalez M. A., Kotsis F., Fu X., Doerken M., Boehlke C., Steffl D., Tauber R., Wegierski T., Nitschke R., Suzuki M., Kramer-Zucker A., Germino G. G., Watnick T., Prenen J., Nilius B., Kuehn E. W., Walz G. (2008) J. Cell Biol. 182, 437–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacKinnon R. (1991) Nature 350, 232–235 [DOI] [PubMed] [Google Scholar]
- 13.Barrera N. P., Shaifta Y., McFadzean I., Ward J. P., Henderson R. M., Edwardson J. M. (2007) Biochem. Biophys. Res. Commun. 358, 1086–1090 [DOI] [PubMed] [Google Scholar]
- 14.Mio K., Ogura T., Kiyonaka S., Hiroaki Y., Tanimura Y., Fujiyoshi Y., Mori Y., Sato C. (2007) J. Mol. Biol. 367, 373–383 [DOI] [PubMed] [Google Scholar]
- 15.Kedei N., Szabo T., Lile J. D., Treanor J. J., Olah Z., Iadarola M. J., Blumberg P. M. (2001) J. Biol. Chem. 276, 28613–28619 [DOI] [PubMed] [Google Scholar]
- 16.Moiseenkova-Bell V. Y., Stanciu L. A., Serysheva I. I., Tobe B. J., Wensel T. G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7451–7455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoenderop J. G., Voets T., Hoefs S., Weidema F., Prenen J., Nilius B., Bindels R. J. (2003) EMBO J. 22, 776–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maruyama Y., Ogura T., Mio K., Kiyonaka S., Kato K., Mori Y., Sato C. (2007) J. Biol. Chem. 282, 36961–36970 [DOI] [PubMed] [Google Scholar]
- 19.Yu Y., Ulbrich M. H., Li M. H., Buraei Z., Chen X. Z., Ong A. C., Tong L., Isacoff E. Y., Yang J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 11558–11563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang P., Luo Y., Chasan B., González-Perrett S., Montalbetti N., Timpanaro G. A., Cantero M. del R., Ramos A. J., Goldmann W. H., Zhou J., Cantiello H. F. (2009) Hum. Mol. Genet. 18, 1238–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrera N. P., Betts J., You H., Henderson R. M., Martin I. L., Dunn S. M., Edwardson J. M. (2008) Mol. Pharmacol. 73, 960–967 [DOI] [PubMed] [Google Scholar]
- 22.Barrera N. P., Herbert P., Henderson R. M., Martin I. L., Edwardson J. M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12595–12600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrera N. P., Henderson R. M., Murrell-Lagnado R. D., Edwardson J. M. (2007) Biophys. J. 93, 505–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrera N. P., Ormond S. J., Henderson R. M., Murrell-Lagnado R. D., Edwardson J. M. (2005) J. Biol. Chem. 280, 10759–10765 [DOI] [PubMed] [Google Scholar]
- 25.Cai Y., Maeda Y., Cedzich A., Torres V. E., Wu G., Hayashi T., Mochizuki T., Park J. H., Witzgall R., Somlo S. (1999) J. Biol. Chem. 274, 28557–28565 [DOI] [PubMed] [Google Scholar]
- 26.Strübing C., Krapivinsky G., Krapivinsky L., Clapham D. E. (2001) Neuron 29, 645–655 [DOI] [PubMed] [Google Scholar]
- 27.Hofmann T., Schaefer M., Schultz G., Gudermann T. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7461–7466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khakh B. S., Zhou X., Sydes J., Galligan J. J., Lester H. A. (2000) Nature 406, 405–410 [DOI] [PubMed] [Google Scholar]
- 29.Pelegrin P., Surprenant A. (2006) EMBO J. 25, 5071–5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo C., Masin M., Qureshi O. S., Murrell-Lagnado R. D. (2007) Mol. Pharmacol. 72, 1447–1456 [DOI] [PubMed] [Google Scholar]
- 31.Boumechache M., Masin M., Edwardson J. M., Górecki D. C., Murrell-Lagnado R. D. (2009) J. Biol. Chem. 284, 13446–13454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsiokas L., Arnould T., Zhu C., Kim E., Walz G., Sukhatme V. P. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3934–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]