Abstract
We report characterization and the crystal structure of the Mycobacterium tuberculosis cytochrome P450 CYP125, a P450 implicated in metabolism of host cholesterol and essential for establishing infection in mice. CYP125 is purified in a high spin form and undergoes both type I and II spectral shifts with various azole drugs. The 1.4-Å structure of ligand-free CYP125 reveals a “letterbox” active site cavity of dimensions appropriate for entry of a polycyclic sterol. A mixture of hexa-coordinate and penta-coordinate states could be discerned, with water binding as the 6th heme-ligand linked to conformation of the I-helix Val267 residue. Structures in complex with androstenedione and the antitubercular drug econazole reveal that binding of hydrophobic ligands occurs within the active site cavity. Due to the funnel shape of the active site near the heme, neither approaches the heme iron. A model of the cholesterol CYP125 complex shows that the alkyl side chain extends toward the heme iron, predicting hydroxylation of cholesterol C27. The alkyl chain is in close contact to Val267, suggesting a substrate binding-induced low- to high-spin transition coupled to reorientation of the latter residue. Reconstitution of CYP125 activity with a redox partner system revealed exclusively cholesterol 27-hydroxylation, consistent with structure and modeling. This activity may enable catabolism of host cholesterol or generation of immunomodulatory compounds that enable persistence in the host. This study reveals structural and catalytic properties of a potential M. tuberculosis drug target enzyme, and the likely mode by which the host-derived substrate is bound and hydroxylated.
Introduction
The global threat to human health posed by the bacterium Mycobacterium tuberculosis (Mtb)5 was recognized by the World Health Organization some years ago (World Health Organization fact sheet on “Tuberculosis” located online at: www.who.int/mediacentre/factsheets/fs104/en), and it is estimated that one-third of the world's population is infected with the Mtb bacillus. Synergy with the HIV virus, failures in drug administration to patients, and the consequences of the development of drug and multidrug-resistant strains of Mtb have made the situation ever more perilous and it is widely acknowledged that novel intervention strategies are needed (1).
The determination of genome sequences of Mtb strains led to revelations relating to the protein repertoire of the pathogen, and highlighted the large number of enzymes involved in lipid metabolism (2, 3). Mtb has an extraordinary array of complex lipids, including unusual long chain, extensively substituted lipids (mycolipids) that form a waxy coat around the bacterium and are likely important in preventing antibiotic entry (4). Another interesting observation relating to lipid metabolizing enzymes is the large number (20) of Mtb cytochrome P450 (P450 or CYP) enzymes. P450s are heme-containing monooxygenases, well known for their roles in metabolism of fatty acids, steroids, and other lipophilic molecules (5). This suggests there may be critical roles for a number of these enzymes in Mtb lipid metabolism (6). Consistent with this theory, gene disruption and gene deletion studies have, to date, shown that Mtb CYP121 and CYP128 are essential genes for cell growth and viability (7, 8). These P450s have recently been proposed to have roles in C–C bond formation in a cyclic dipeptide and in hydroxylation of respiratory menaquinone, respectively (9, 10). Although physiological roles for many Mtb P450s remain unclear, Mtb CYP51B1 has been structurally and biophysically characterized, and catalyzes demethylation of various sterols (11, 12). This activity is consistent with that of eukaryotic CYP51 enzymes, suggesting that CYP51B1 has roles in host sterol metabolism. Importantly, it was demonstrated that various azole drugs (that inhibit fungal CYP51 by coordinating the heme iron) are also potent inhibitors of mycobacterial growth, thus suggesting that one or more Mtb P450s may be azole targets (13–15). Econazole and other azoles bind tightly to various Mtb P450s, including CYP121, CYP51B1, and CYP130 (10, 11, 13, 16, 17). Econazole is effective in clearing Mtb infection in a mouse model, and recent studies on Mtb CYP130 (a P450 whose gene is deleted in the vaccine strain Mycobacterium bovis BCG) revealed the binding mode of the drug to this P450 (16, 18).
Recently, a gene cluster in Rhodococcus sp. strain RHA1 was identified as being involved in catabolism of cholesterol (19). Several of these genes are conserved in Mtb, including the P450s CYP125 and CYP142 (20), suggesting that these have roles in cholesterol (or possibly other sterol) metabolism. Early studies of the protein interactions of the Mtb CYP125 with nitric oxide indicated that its ferrous-nitric oxide complex was relatively labile, and thus that CYP125 may be relatively resistant to macrophage-generated nitric oxide (21). Transcriptomic studies showed that Mtb H37Rv CYP125 is induced in macrophages, and it is reported to be essential for infection of mice; one of only 26 genes present in both categories (22). Furthermore, cholesterol, along with the phagosomal tryptophan-aspartate-containing coat protein, is crucial for Mtb entry into the macrophage and for establishment of intracellular infection by Mtb (23). In other work, genetic inactivation of the Mtb cholesterol oxidase (ChoD) resulted in attenuation of the choD mutant strain, implicating ChoD in Mtb pathogenesis (24). Also, recent studies implicated the actinobacterial mce4 gene locus (conserved in Mtb) with cholesterol/steroid uptake (25). Finally, it was shown that Mtb uses cholesterol as a source of carbon and energy for growth, suggesting that exploitation of host cholesterol may underlie persistence and survival in humans (26).
To investigate properties of the CYP125 P450 from the putative Mtb “cholesterol cluster,” we have purified Mtb CYP125 heterologously expressed in Escherichia coli and explored its thermodynamic and spectroscopic features, including its ligand-binding properties. We have determined the CYP125 crystal structure in a ligand-free state and in complex with econazole and androstenedione. Generation of a molecular model of the cholesterol complex indicated that cholesterol C25 and the terminal methyl (C26/27) carbons are exposed to the heme iron. Turnover studies demonstrated conclusively that CYP125 is a cholesterol 27-hydroxylase. Our data suggest a key role for CYP125 in Mtb cholesterol metabolism as a C27 hydroxylase, and thus its importance in infectivity and in persistence of Mtb in the human host.
EXPERIMENTAL PROCEDURES
CYPI25 Cloning, Expression, and Purification
CYP125 was cloned by PCR from a Mtb H37Rv cosmid library (from Institut Pasteur, Paris). The BAC clone containing CYP125 (Rv3545c) was prepared by standard protocols, and used as template DNA for the PCR using Pfu Turbo DNA Polymerase (Stratagene) and the oligonucleotide primers designed from the Mtb genomic sequence: upstream 5′-GGACAGCATATGTCGTGGAATCACCAGTCA-3′ and downstream 5′-CAGTGGGATAGATCTCCATTAGTGAGCAAC-3′. The bold letters in the upstream primer indicates an engineered NdeI restriction cloning site, including the initiation codon ATG. The underlined letters in the downstream primer indicate a BglII restriction cloning site. Amplification conditions were 95 °C for 2 min, 30 cycles of 95 °C for 50 s, 63 °C for 30 s, and 72 °C for 2 min, followed by a final polymerization step of 72 °C for 8 min. CYP125 was cloned into pET15b (Merck) using the NdeI and BamHI restriction sites and using the compatible cohesive ends between BglII on CYP125 and BamHI on the vector, allowing expression of the CYP125 gene from a T7lac promoter under isopropyl 1-thio-β-d-galactopyranoside induction, and producing a recombinant P450 protein with an N-terminal His6 tag.
Protein was produced in E. coli HMS174 (DE3) (typically 15–20 liters, grown in 2×YT medium) by isopropyl 1-thio-β-d-galactopyranoside (0.15 mm) induction in the presence of the heme precursor δ aminolevulinic acid (0.1 mm) at OD600 = 0.6, with temperature then reduced from 37 to 23 °C and culture continued for 24 h. Thereafter, cells were harvested by centrifugation (9,000 × g, 4 °C, 20 min), resuspended in 50 mm potassium phosphate, 250 mm KCl, 10% glycerol, pH 8.0 (buffer A), containing protease inhibitors (Complete EDTA-free protease-free inhibitor tablets, Roche) at 4 °C, and re-centrifuged as before. The pellet was then resuspended in a minimal volume of buffer A (all buffers contained standard protease inhibitors), and the cells were broken by a combination of sonication and French pressure treatment, as described previously (17, 27). The disrupted cell extract was centrifuged (40,000 × g) for 30 min to remove particulate material and then loaded onto a nickel-nitrilotriacetic acid resin column (Qiagen). The column was washed twice in buffer A, containing 30 mm then 75 mm imidazole, and eluted using 200 mm imidazole in the same buffer. The CYP125-containing fractions were pooled and dialyzed versus 50 mm Tris, 1 mm EDTA, pH 7.2 (buffer B), prior to further fractionation using a Resource-Q column on an AKTA purifier (GE Healthcare). CYP125 was bound to the column in buffer B and eluted in a gradient of 0–500 mm KCl in buffer B. The most intensely red CYP125-containing fractions were retained, pooled, and concentrated to a final volume of <1 ml (using a Vivaspin 30 concentrator, Generon) prior to a final gel filtration step using a Sephacryl S-200 column (1.6 × 70 cm) with 10 mm Tris, pH 7.5. CYP125 purity was determined by SDS-PAGE and UV-visible spectroscopy. The most pure fractions were retained, concentrated as previously (to ∼500 μm), and used directly for crystallogenesis, or dialyzed into 50 mm potassium phosphate, pH 7.5 (buffer C), containing 50% glycerol and stored at −80 °C.
Ligand Binding and Thermodynamic Studies
Optical titrations for determination of azole ligand binding constants (Kd values) were done as previously described (11). Pure CYP125 (typically 2–5 μm) was suspended in buffer C in a 1-cm path length quartz cuvette and a spectrum for the ligand-free form recorded (250–800 nm) at 25 °C on a Cary UV-50 Bio scanning spectrophotometer (Varian, UK). Azole ligands (clotrimazole, econazole, fluconazole, miconazole, ketoconazole, voriconazole, 2-phenylimidazole, and 4-phenylimidazole) were titrated from concentrated stocks in dimethyl sulfoxide solvent (apart from the phenylimidazoles, which were prepared in 60% ethanol) until apparent saturation of the optical change was observed. Induced optical change versus ligand concentration data were fitted using Equation 1, which provides the most accurate estimation of Kd values for the tight binding azole drugs, as we have described in previous studies of the Mtb CYP121 and CYP51B1 P450s (8, 17). Data were fitted using Origin software (OriginLab, Northampton, MA).
 |
In Equation 1, Aobs is the observed absorbance change at ligand concentration S, Amax is the absorbance change at ligand saturation, Et is the CYP125 concentration, and Kd the dissociation constant for the CYP125-ligand complex.
Binding of the sterols cholesterol, testosterone, progesterone, and epiandrosterone was done by addition of small volumes of stock solutions of the sterols (suspended in EtOH) to CYP125 in buffer C, with spectral measurements taken before and after sterol addition. Other spectral measurements reporting on the sodium dithionite-dependent reduction, binding of CO to the ferrous enzyme form, and nitric oxide to the ferric form (for enzyme quantification and establishment of typical P450-type features of CYP125) were done using a Cary 50 UV-visible spectrophotometer, either aerobically or under anaerobic conditions in a glove box (Belle Technology, Portesham, UK) for ferrous enzymes (8, 28).
CYP125 redox titrations were performed in a Belle Technology glove box under nitrogen atmosphere, as described previously (29). Protein solution (approximately 9 μm in 5 ml of 100 mm potassium phosphate, 10% glycerol, pH 7.0) was titrated electrochemically by the method of Dutton (30) using sodium dithionite as reductant and ferricyanide as oxidant. Mediators were added to facilitate electrical communication between enzyme and electrode (2 μm phenazine methosulfate, 7 μm 2-hydroxy-1,4-naphthoquinone, 0.3 μm methyl viologen, and 1 μm benzyl viologen, to mediate in the range from +100 to −480 mV) (31). Spectra (250–800 nm) were recorded using a Cary UV-50 Bio UV-visible scanning spectrophotometer. The electrochemical potential of the solution was measured using a Mettler Toledo SevenEasy meter coupled to a Pt/Calomel electrode (ThermoRussell Ltd.) at 25 °C. The electrode was calibrated using the Fe3+/Fe2+ EDTA couple as a standard (+108 mV). A factor of +244 mV was used to correct relative to the standard hydrogen electrode. Redox titrations were performed in both reductive and oxidative directions to ensure that the redox processes were fully reversible and hysteretic effects were not observed. Absorption change versus applied potential data were fitted to the Nernst function (using Origin software) to derive the midpoint potential for the CYP125 heme iron Fe3+/Fe2+ couple (29).
Spectroscopic Studies
Electron paramagnetic resonance (EPR) was done on ligand-free and imidazole (10 mm)-bound ferric CYP125 (220 μm) in buffer C. EPR spectra were recorded on a Bruker ER-300D series electromagnet and microwave source interfaced with a Bruker EMX control unit and fitted with an ESR-9 liquid helium flow cryostat (Oxford Instruments), and a dual-mode microwave cavity from Bruker (ER-4116DM). Spectra were recorded at 10 K with a microwave power of 2.08 milliwatts and a modulation amplitude of 10 G. Resonance Raman was done using 15-milliwatt, 406.7 nm radiation from a Coherent Innova 300 krypton ion laser, and acquired using a Renishaw micro-Raman system 1000 spectrophotometer.
CYP125 Crystallization, Structure Elucidation, and Molecular Modeling
CYP125 was concentrated to 13 mg/ml. Sitting drops were prepared by mixing 0.1 μl of CYP125 with 0.1 μl of mother liquor and incubating at 4 °C. Crystallization conditions were refined to two different conditions, both consisting of MgCl2 with 0.1 m HEPES, pH 7.0 or 7.5, and PEG 6000 (20%) or PEG 3350 (25%), respectively. The PEG 6000 conditions mainly generated crystals belonging to the C2221 space group, whereas crystals generated using PEG 3350 belonged to the P212121 space group. Ligands 4-androstene-3,17-dione (52 mm) and econazole (33 mm) were prepared in ethanol and diluted 1/10 in mother liquor prior to soaking single crystals for 15 min. Single crystals were cooled to 100 K after addition of 10% PEG 200 as cryoprotectant, and data were collected at ESRF and Diamond beamlines. The CYP125 structure was solved by molecular replacement using the P450terp structure as the search model. Full details are in the supplemental data section. Data and final refinement statistics for the CYP125 crystal structures are in supplemental Table S1.
Molecular modeling of the interaction of cholesterol with CYP125 was based on a soft-restrained molecular dynamics (MD) approach previously described for P450s (32). Briefly, cholesterol was positioned in the ligand-free structure of CYP125, close to the positioning of androstenedione in the androstenedione-bound CYP125 structure, in 4 different orientations, so that no steric clashes with CYP125 residues could be observed and such that either the cholesterol tetracyclic moiety or its alkyl chain was pointing to the heme. All 4 positions were chosen so that the cholesterol molecule main axis was aligned with the entrance channel, to minimize the large conformational changes that would occur during the substrate motion in the channel. Up to 5 different dockings were performed from each starting orientation, using small adjustments of the conformation and coordinates. In the following described protocols, the side chains of residues located in a 10-Å sphere centered on cholesterol, as well as water molecules, were defined as the only mobile atoms, to preserve the tertiary structure of CYP125 as observed in the crystal structure. All MD simulations and energy minimization experiments were performed using the NAMD program (33) with Amber force field parameters (34). Topology and parameter files for cholesterol were obtained using the Antechamber program (35) with AM1-BCC charges (36). The cut off parameter for the computation of non-bonded interactions was set to 12 Å, and the electrostatic forces were “softened” by defining a relative dielectric constant of 2 for the system. Energy minimization (1000 steps, conjugate gradient) and MD simulations (200 ps) were initially performed in vacuo at 100 K to thermally equilibrate CYP125-cholesterol complexes. Then, a distance-dependent constraint whose force constant values ranged from 1.5 to 2 (kcal/mol)/Å2 was applied between the heme iron and the closest cholesterol carbons (3 to 4 atoms), and MD simulations were performed at 100 K for 1 ns. Equilibration of the docked ligand in the active site was done by releasing the constraint in a final MD run of 1 ns at 100 K. Final minimization (1000 steps, conjugate gradient) was performed to obtain the CYP125-cholesterol complexes. Comparison and selection of the docked cholesterol models was done by comparing the stabilization energy due to the CYP125-cholesterol interactions (supplemental Table S4) and the minimal distances between cholesterol heavy atoms and the iron atom of the heme. Minimal distances greater than 7 Å led to the dismissal of the docked model. The model considered for the “Results” and “Discussion” was obtained from a starting position corresponding to orientation C (as represented in Fig. 7).
FIGURE 7.
Production of 27-hydroxycholesterol from CYP125-dependent oxidation of cholesterol. CYP125 was shown to produce exclusively 27-hydroxycholesterol in turnover assays with electrons provided by the E. coli flavodoxin reductase/flavodoxin system. A, HPLC separation (see “Experimental Procedures”) of reaction mixture containing [3H]cholesterol, showing substrate (retention time 17.36 min) and 27-hydroxycholesterol product (retention time 9.369 min). B, mass spectrum of the product peak (following GC separation) from the turnover reaction. Key peaks are indicated, including those with m/z values at 129.0, 417.4, 456.4, and 546.6. C, mass spectrum of the 27-hydroxycholesterol standard. Key peaks are at near identical m/z values as those for the reaction product in panel B, including the molecular ion at m/z = 546.5.
Reconstitution of Cholesterol Hydroxylase Activity of CYP125
Incubations with CYP125 and cholesterol were carried out in 1 ml of 50 mm potassium phosphate, pH 7.2, using 0.5 μm CYP125, 10 μm E. coli flavodoxin, 2.5 μm E. coli flavodoxin reductase, 2 nm [3H]cholesterol, and 1 mm NADPH with a NADPH regenerating system (glucose 6-phosphate and glucose-6-phosphate dehydrogenase) (37). The enzymatic reaction was initiated by the addition of NADPH and terminated by vortexing with 2 ml of CH2Cl2. The organic phase was isolated, evaporated, dissolved in acetonitrile, and subjected to HPLC as previously described (37).
To characterize the product of CYP125 activity by gas chromatography-mass spectrometry, the concentration of cholesterol in the enzyme assay was increased to 1 μm. After termination of the enzyme reaction, the substrate and product were extracted, converted into trimethylsilyl ethers, and injected into a VF-35MS capillary column (60 m × 0.32 mm × 0.25 μm) in a splitless mode at an injection temperature of 270 °C with a helium flow of 1.1 ml/min. The initial oven temperature was kept at 200 °C for 1 min, then increased to 280 °C (20 °C/min), ramped up to 310 °C (3 °C/min), and held for 14 min isothermally. The mass spectrometer (Agilent 5973N-MSD combined with an Agilent 6890 GC system) was operated in electron impact ionization (70 eV) at 230 °C. The retention time and mass spectrum of the trimethylsilyl CYP125 product was essentially identical to that of authentic 27-hydroxycholesterol (purchased from Steraloids, Newport RI), with the base peak at m/z 129 and prominent peaks at m/z 417, 456 and 546.
Materials
Bacterial growth medium (Tryptone, yeast extract) was from Melford Laboratories (Ipswich, Suffolk, UK). A 1-kb DNA ladder was from Promega. Azole drugs were from MP Biomedicals Inc. All other reagents were from Sigma and were of the highest grade available.
RESULTS
Genetic Context, Expression, and Production of M. tuberculosis CYP125
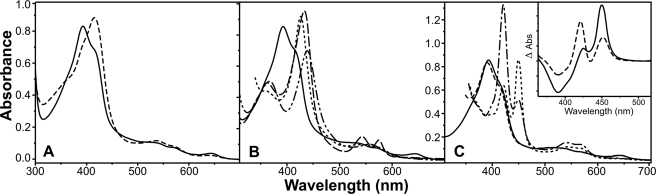
To define the biochemical and structural characteristics of CYP125, we expressed and purified the P450 from E. coli. Purified CYP125 was dark brown (not red) in color, and optical spectroscopy revealed an extensively high spin (HS, >80%) enzyme with heme Soret features at 393 (HS, major) and 416 nm (low spin, LS, shoulder) (Fig. 1A). The HS/LS ratio was affected by temperature, ionic strength, and pH, although the protein was predominantly HS under all conditions. In contrast, and despite apparent homogeneity by SDS-PAGE, certain fractions obtained during gel filtration purification had predominantly LS heme iron with Amax at 415 nm (Fig. 1A). Solvent treatments of HS CYP125 fractions did not result in extraction of potential substrates bound to the enzyme, but did demonstrate that the heme spin state could be readily modulated by organic solvents (e.g. methanol, see below).
FIGURE 1.
Spectral analysis of CYP125 and its interactions with inhibitory ligands. A, the native HS (solid line) and fractionated LS (dashed line) forms of CYP125. Spectral maxima are at 393 and 415 nm, respectively. B, HS CYP125 (solid line), and its adducts with imidazole (dotted line, 10 mm), nitric oxide (dashed line), and cyanide (dot-dot-dash line, 20 mm). Maxima are 393, 426, 433, and 439 nm, respectively. C, HS CYP125 (solid line), its dithionite-reduced form (dashed line) and the Fe2+·CO complexes of the HS (dotted line) and LS (dot-dash line) forms. Maxima for both HS and LS Fe2+·CO complexes are at 450 and 422 nm. The inset shows difference spectra generated by subtracting spectra for the dithionite-treated CYP125 enzymes from the Fe2+·CO complexes for the HS (solid line) and LS (dashed line) forms. CYP125 concentration in all cases was 9.5 μm.
Ligand Binding Characteristics of CYP125
Addition of heme coordinating ligands resulted in occupancy of the 6th (distal) position on the heme iron, with Soret optical shifts seen for imidazole (maximum at 426 nm), cyanide (439 nm), and nitric oxide (433 nm) (Fig. 1B). A fundamental property of P450s is their binding of carbon monoxide (CO) to ferrous heme iron to give a spectral species with maximum near 450 nm. For CYP125 the Fe(II)·CO complex spectrum has two maxima at 450 (P450) and 422 nm (P420), suggesting protonation of the proximal cysteinate ligand (Cys377) to a thiol in the P420 form, as seen previously (11) (Fig. 1C). Consistent with this conclusion, higher buffer pH increased the P450:P420 ratio, with optimal P450 content achieved in 100 mm potassium phosphate, pH 9.0. The LS form of CYP125 showed lower stability of heme thiolate ligation in the Fe(II)·CO complex than did the major HS fraction, with a higher P420:P450 ratio observed (Fig. 1C).
Preceding studies have revealed high affinity and type II binding characteristics for the interactions of various azole drugs with other Mtb P450s (e.g. CYP121, CYP51B1, and CYP130) (11, 17, 21). Azoles typically directly coordinate to P450 heme iron to produce type II (red) shifts of the Soret band. For CYP125, unusual binding properties of various azoles were seen. Voriconazole did not induce a spectral shift, whereas fluconazole and ketoconazole produced small type II shifts, suggesting ∼20 and 35% heme iron coordination, respectively. In the case of econazole, previous work showed its binding induced a near complete HS conversion (21). Although we found this reproducible at ambient temperature, treatment of the HS CYP125 at low temperature (10 °C) with methanol or an ethanol/methanol mixture (10%) produced a form of CYP125 that displayed type II binding for econazole (Fig. 2A). For miconazole and clotrimazole, these azoles also bound to the HS form of CYP125 to produce type I shifts at low concentration (up to ∼0.5 μm), but type II shifts (to ∼422 nm) at higher drug concentrations (Fig. 2B). Kd values for azole binding were determined as described under “Experimental Procedures,” and were in the range ∼4–45 μm (supplemental Table S2). In addition, the LS CYP125 fractions obtained from gel filtration studies (see above) also displayed type II binding of these azoles.
FIGURE 2.
Optical properties of CYP125 complexes with azoles and cholesterol. A, spectra for HS CYP125 (solid line) and its type I (dotted line) and type II (dashed line, post-methanol treatment) complexes with econazole. Spectral maxima are at 393, 393, and 418 nm, respectively. B, titration of CYP125 with miconazole, showing a type II optical shift from 393 (solid line) to 421 nm (dashed line) (Kd = 4.6 ± 0.4 μm). Intermediate spectra are shown by thin dotted lines. C, type I (substrate-like) spectral shift on addition of cholesterol to CYP125 (from 415 to 393 nm). The starting (mainly LS) solvent-treated form of CYP125 (solid line) and the final HS form (dashed line) at the end of the cholesterol titration are shown, with intermediate spectra as dotted lines. CYP125 concentration in all cases was 3 μm.
In view of the likelihood that CYP125 binds sterols, optical binding studies of the interactions with various sterol-type molecules were done. The predominant HS state of the purified CYP125 precluded accurate attempts to establish further type I binding of most molecules. However, type I optical changes were induced by addition of androstenedione and cholesterol to the solvent-treated form (which exhibited increased LS heme content), whereas negligible spectral changes were induced by the addition of other steroids (e.g. testosterone, pregnenolone) (Fig. 2C). In parallel studies, no significant CYP125 optical perturbation was induced by addition of various fatty acids and terpenes, including palmitic acid and α-terpineol.
Spectroscopic and Thermodynamic Analysis of CYP125
To further probe the properties of CYP125, we undertook EPR, resonance Raman, and redox potentiometry studies, as described under “Experimental Procedures,” and previously (8). EPR of ligand-free CYP125 at 10 K was typical for a thiolate-coordinated, LS P450, with the major set of g values at gx = 2.40, gy = 2.25, and gz = 1.94 (supplemental Fig. S1). A very small signal from a HS species was detected at 10 K. Room temperature resonance Raman confirmed the ferric state of the CYP125 heme iron, with the main oxidation state marker band (ν4) at 1372 cm−1. The spin state marker band (ν3) showed features at 1487 (major) and 1500 cm−1, reflecting a dominant population of HS heme iron over the LS form. Binding of imidazole (10 mm) to CYP125 resulted in a LS form (see Fig. 1B) with ν3 at 1501 cm−1 predominant (supplemental Fig. S2). The redox potential for the Fe3+/Fe2+ transition of the CYP125 heme iron was −303 ± 5 mV (versus NHE), consistent with the mainly HS nature of the P450 (supplemental Fig. S3) (21). Full analyses of EPR, resonance Raman (supplemental Tables S2 and S3), and thermodynamic data are presented in the supplemental data.
Crystallization and Structural Determination of Ligand-free CYP125
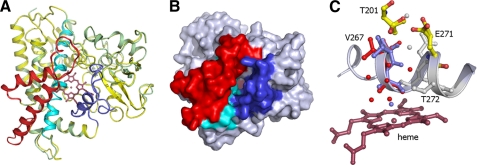
In view of the importance of CYP125 to Mtb viability in its host, we determined the crystal structure in both the presence and absence of ligands. The structure was solved to 1.4 Å by molecular replacement using the structure of the Pseudomonas sp. P450terp (CYP108A1) as the search model (38). CYP125 has a typical P450-fold with the heme cofactor sandwiched between a major α helical domain and a smaller domain with substantial β sheet content (Fig. 3A). An entrance to the active site is clearly defined by the B′ and F α-helices and their preceding loop regions (Val96–Lei117 and Met200–Ile221, respectively) in addition to contributions by the I-helix (Phe260–Thr272) and Trp414–Leu415 from the C-terminal loop region. The entire cavity is lined by hydrophobic residues and resembles a “letterbox” shape with the B′ and F helices defining the opposite sides (Fig. 3B). This putative substrate binding pocket becomes a funnel-like shape, with a progressive narrowing of the active site cavity on approach to the heme. The position and nature of the active site residues in the immediate vicinity of the heme group bear remarkable resemblance to the P450terp structure, despite the apparent lack of α-terpineol binding to CYP125.
FIGURE 3.
Crystal structure of CYP125. A, overall topology of CYP125 structure with both crystal structures overlaid (colored green and yellow, respectively). The B′-helix, I-helix, and the FG helices are colored in blue, cyan, and red. B, solvent accessible surface of CYP125 with B′-helix, I-helix, and FG helices colored as in panel A. A large crevice is seen sandwiched between the B′-helix and the FG helices that allows access to the heme and presumably functions as the substrate binding site. C, detail of the CYP125 active site. The alternative positions for Val267 with associated waters are shown colored in blue (conformation A) and red (conformation B). Residues depicting multiple conformations that are possibly linked to proton transport to heme iron are shown in atom colored sticks. Residues or waters that do not display multiple conformations are colored gray.
A distinct crystal form (form 2) could be obtained that gave data until 1.7 Å and also contained one CYP125 monomer in the asymmetric unit. No significant changes were observed when comparing both crystal structures (Fig. 3A) with the notable exception of the environment and position of the I-helix residue Val267 that is located in the immediate vicinity of the heme distal pocket. In both crystal structures, the Val267 side chain is clearly defined as occupying two positions, but the relative occupancy of these positions is markedly different in both crystal structures (Fig. 3C). In one orientation (A), the Val267 carbonyl backbone oxygen is involved in I-helix H-bonding interactions, whereas the second orientation (B) positions this atom within the heme distal pocket. In conformation B, a water molecule occupies a position similar to that observed for the Val267 carbonyl backbone oxygen in conformation A. The relative occupancy of states A and B appears directly linked to the coordination state of the heme iron, with the Val267 A orientation linked to a hexa-coordinate LS state, whereas the B conformation gives rise to a penta-coordinate HS state. In state B, an indirect H-bonding interaction between the Val267 carbonyl backbone oxygen and the water molecule closest to the heme iron is observed. This could account for the observed link between heme iron coordination state and Val267 conformation, as reorientation of this residue affects the heme distal pocket H-bonding network and hence the extent to which water will ligate the heme. Thus, it is possible that upon substrate binding there is a reconfiguration of active site organization and that the structural rearrangement of Val267 is a trigger for aqua ligand displacement and concomitant P450 heme LS to HS conversion. This would link the conserved Thr272 (implicated in proton delivery) via the newly introduced water molecule (only observed in conformation B) to a network of hydrophilic residues (Thr201 and Glu271) and water molecules that could easily serve as a proton relay. It is also likely that CYP125 reduction itself is gated by a LS to HS transition, as seen for other P450s (39, 40).
Crystal Structures of CY125 Androstenedione and Econazole Complexes
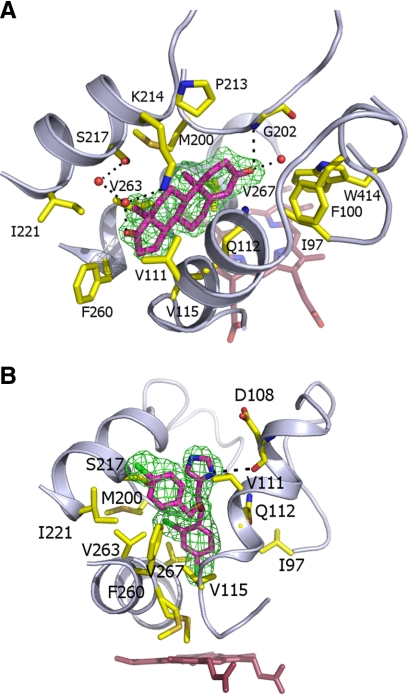
Soaking CYP125 crystals with both the steroid androstenedione and the azole econazole produced complexes that were solved to resolutions of 2.0 and 2.2 Å, respectively. In both cases, these molecules are bound within the observed letterbox cavity, with neither ligand able to penetrate the funnel-shaped access tunnel to the heme group (the closest atoms to the heme iron are at 12.9 and 9.3 Å for androstenedione and econazole, respectively). The binding mode for androstenedione (which lacks the alkyl side chain found in cholesterol) is not compatible with P450 oxidation, and the funnel-like nature of the active site clearly prevents the steroid moiety from reaching the direct vicinity of the heme iron (Fig. 4A). Binding of this ligand appears to introduce little change in the protein structure with ligand-protein interactions predominantly through hydrophobic packing of the steroid moiety between residues from the B′-helix and F-helix regions. In addition, a limited set of polar contacts are made between both hydrophilic substituents on the steroid moiety and residues Gly202, Lys214, and Ser217. Econazole binds in a similar hydrophobic region, and is again prevented from further migration into the active site by steric constraints (Fig. 4B). In contrast to androstenedione, econazole binding introduces a minor change in the position and conformation of Val267 due to the close contact made with the econazole chloride substituent that is closest to the heme. In similar fashion to the androstenedione-CYP125 structure, protein-ligand contacts are dominated by a series of hydrophobic interactions with the B′- and F-helix residues, in addition to a single polar contact between the azole moiety and Asp108. Ligand binding studies revealed the ability of econazole to coordinate heme iron only in an enzyme form obtained by solvent treatment at low temperature, and these data are consistent with conformational rearrangements of the enzyme induced by alteration of the chemical environment and ambient temperature, and that enable the ingress of econazole toward the heme in a proportion of the enzyme molecules.
FIGURE 4.
Structures of the CYP125 androstenedione and econazole complexes. A, structure of the steroid androstenedione in complex with the CYP125 hydrophobic active site. The ligand is in magenta colored sticks with the associated Fo − Fc map contoured at 3σ. B, structure of econazole in complex with the CYP125 active site. The ligand is in atom colored sticks (magenta carbons) with the associated Fo − Fc map contoured at 3σ. In both A and B, residues contacting the ligand are in atom colored sticks, whereas hydrogen bonding networks established with the ligand hydrophilic groups are shown in black dotted lines.
In addition to androstenedione and econazole, we sought to establish the binding mode of cholesterol to CYP125. However, crystal soaks with cholesterol persistently failed to reveal interpretable density for the cholesterol ligand, whereas co-crystallization attempts failed to generate crystals of suitable quality for diffraction studies. For this reason, we investigated the cholesterol docking mode using molecular modeling methods.
Molecular Modeling of Cholesterol Binding to CY125A1
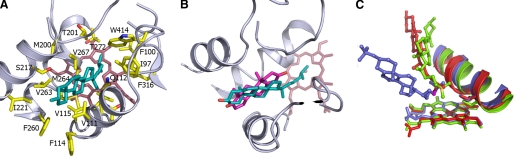
Cholesterol was docked using soft restrained dynamics docking (32) into the CYP125 active site, using the androstenedione binding pocket as the access channel. Several orientations were used as a starting point for docking (Fig. 5), with either the alcohol function on the tetracyclic moiety or the alkyl chain pointing to the heme. During molecular dynamics the backbone CYP125 coordinates were restrained to the conformation observed in the crystal structure. As described in the supplemental data, the final model was chosen considering the highest energy stabilization of the CYP125-cholesterol complex as well as the cholesterol-iron distances. The final model (Fig. 6A) exhibited the greatest stabilization energy among all the models obtained (more than 6 times higher than any others, see supplemental Table S4). The cholesterol is deeply buried in the CYP125 active site, with a calculated buried surface of 312 Å2, which corresponds to 86% of the total substrate surface. The tetracyclic portion of the cholesterol occupies the same region of the active site as seen in the androstenedione complex, but the molecule is “flipped” through 180° such that the hydroxyl group on ring A (a carbonyl in androstenedione) is orientated toward the mouth of the active site rather than being internalized. The cholesterol molecule is thus in a conformation where the alkyl chain extends toward the heme, and with the terminal methyl groups (C26/27) positioned close to the heme iron (at distances of 5.3 and 6.3 Å, respectively). This conformation of cholesterol is favored by stabilizing interactions between the hydrophobic cholesterol and several active site residues (Ile97, Leu117, Val267, Phe316, and Trp414), or residues along the active site access channel (Val111, Phe114, and Phe260). Despite the distinct binding modes, the cholesterol-CYP125 model obtained fits well with the CYP125-androstenedione crystal structure (Fig. 6B), as the tetracyclic portions of cholesterol and androstenedione can be readily superimposed, with methyl groups on the rings oriented in the same direction. The apparent rotation of the tetracyclic moiety between the androstenedione complex and the cholesterol model structures can be explained by the additional favorable binding energy associated with the burial of the cholesterol alkyl chain in the hydrophobic region leading to the heme (as opposed to burial and desolvation of the cholesterol alcohol when considering an androstenedione-like orientation). It is interesting to note that the terminal portion of the cholesterol side chain is in close contact with Val267, an interaction that may be important to promote conformational readjustment of the side chain to displace the distal water and trigger catalysis.
FIGURE 5.
Initial orientations of cholesterol for docking with CYP125. In all panels, heme is rendered in red sticks and Val111 and Ile221, residues that line the entrance of the androstenedione binding channel are represented in yellow sticks. Androstenedione is depicted in thin gray sticks to illustrate the position of the channel used for substrate docking (this compound is not present during the steered MD).
FIGURE 6.
Molecular model for cholesterol binding to CYP125. A, structure of cholesterol in complex with the CYP125 hydrophobic active site. The ligand is in cyan colored sticks, whereas residues contacting the cholesterol are shown in atom colored sticks. B, overlay of the cholesterol-CYP125 model with the androstenedione-CYP125 crystal structure. The tetracyclic moieties of both ligands bind in highly similar positions, although the cholesterol moiety (in cyan) is rotated by 180° with respect to the androstenedione (in magenta) conformation observed. This position allows the cholesterol alkyl chain to project down to the heme, whereas the single hydrophilic alcoholic group of cholesterol remains in contact with bulk solvent. The proximity of the terminal methyl groups to the heme iron suggests cholesterol C27 oxidation as an enzymatic activity for CYP125. C, overlay of the cholesterol-CYP125 model (in red) with the available cholesterol sulfate-CYP46A1 (in blue, PDB code 2Q9F) and vitamin D3-CYP2R1 (in green, PDB code 3G6C) structures. Carbon positions oxidized by CYP46A1 and CYP2R1 are shown in magenta (C24) and yellow (C25), respectively. For clarity, only the ligands, heme, and a portion of the I-helix are shown, including the side chain for the conserved threonine (Thr272) likely involved in oxygen binding/proton relay.
Experimental Validation of Cholesterol C27 Oxidation by CYP125
To establish that Mtb CYP125 actually catalyzed oxidation of cholesterol and determine the position(s) of oxidation, we reconstituted the P450 with a bacterial redox partner system (E. coli flavodoxin reductase and flavodoxin proteins and NADPH reductant) that has been well characterized and used widely to drive both prokaryotic and eukaryotic P450 catalysis (41, 42). Experiments were done using gas chromatography-mass spectrometry as performed previously for human CYP46A1 and as detailed under “Experimental Procedures” (37). A single product was formed using the E. coli redox system with CYP125. By comparison with authentic standards, this was shown to be 27-hydroxycholesterol, consistent with our predictions based on structural modeling of the mode of cholesterol association with CYP125 (Fig. 7).
DISCUSSION
The location of CYP125 in a gene cluster conserved from Rhodococcus to Mtb suggests a likely role in cholesterol metabolism (19). Cholesterol may be important for Mtb entry into macrophages, and for establishing infection. The fact that CYP125 is both induced in macrophages and reported as essential for establishing mouse infection is also indicative of a crucial role for this P450 (22, 23). CYP125 is retained in all Mtb strains and in some related actinobacteria, e.g. Nocardia and Streptomyces spp. The genetic context of CYP125 is conserved within these bacteria, and the surrounding acyl-CoA dehydrogenase genes (FADE28, FADE29, and FADA5, likely involved in lipid degradation) form an operon with CYP125. Gene knock-out studies on the CYP125 and associated FAD-containing intergenic region (igr) implicated this cluster of genes to have an important role in early mycobacterial infection (43). Despite genetic conservation in non-pathogens, many of the genes within the cholesterol operon are critical for Mtb pathogenesis. The Mtb cholesterol catabolic gene cluster is under the control of a TetR transcriptional repressor ktsR (Rv3574) likely to have an essential role in pathogenesis and lipid degradation. Genes in this cluster may metabolize diverse lipids, using the mce4 system involved in cholesterol/steroid uptake (44). Collectively these genetic studies and the presence of CYP125 in the cholesterol operon suggest a critical role in bacterial cholesterol metabolism, and in mycobacterial infection and pathogenesis. Our determination of the structure of CYP125 represents the first insight into active site architecture of this important P450, and explains unusual spectroscopic phenomena previously described (21).
Although type II azole binding has been demonstrated clearly for Mtb CYP51B1, CYP121, and CYP130 (11, 12, 16, 17), peculiar type I binding of econazole was reported for CYP125 (21). For the purified, HS form of CYP125 characterized here, this was shown to be the case for econazole. Moreover, clotrimazole and miconazole gave type I binding at low ligand concentrations, but type II binding (heme coordination) at higher concentrations. The phenomena observed for clotrimazole and miconazole suggest alternative binding modes and/or distinct conformers of the P450. On treatment of CYP125 with alcohol (10%) at 10 °C, we were able to produce a mixed spin species that gave type II binding with econazole. Higher concentrations of alcohol destabilized the protein, but also resulted in a further shift toward LS for the ligand-free enzyme. The crystal structure of the econazole-bound (Fig. 4B) CYP125 reveals narrowing of the active site “funnel” precluding further entry of econazole to coordinate the heme iron. The spectral studies are thus suggestive of different conformational states of the enzyme that are favored under different environmental conditions. EPR studies also suggest some heterogeneity in the thiolate-coordinated CYP125 species, which again may suggest the presence of different conformers in the enzyme population studied.
Both crystal structures of the ligand-free CYP125 reveal a clear active site crevice that is roughly rectangular in form and of dimensions well suited to the binding of cholesterol. The majority of this binding pocket is defined by the B′ and F helices, which, together with a section of the C-terminal loop and I-helix residues, also contribute to formation of the heme distal pocket. There are some important parallels in relation to the recently determined crystal structure of human CYP46A1, a cholesterol 24-hydroxylase (45; Protein Data Bank code 2Q9F) and of the vitamin D3-bound CYP2R1 (46) (PDB code 3C6G). An overlay of CYP125, CYP46A1, and CYP2R1 reveals that CYP125 and CYP2R1 share a common substrate binding pocket, whereas the sterol moiety of cholesterol in CYP46A1 is bound by a distinct region of the protein (Fig. 6C). In the cholesterol sulfate-CYP46A1 complex, the ligand C24 and C25 carbons are placed closest to the heme iron (both at distances of ∼5.7 Å), consistent with the preferred position of oxidation at C24, with the terminal methyl groups more distant. Similarly, the vitamin D3-CYP2R1 complex reveals the C25 and C26/27 carbons located at distances of 5.5 and 6.5 Å, respectively, from the heme iron, which again is in agreement with the observed oxidation at C25 (46). The cholesterol-CYP125 model predicts the C26/C27 cholesterol carbons to be close to the iron center, at a distance of ∼5.3 and ∼6.3 Å., and we therefore predicted that CYP125 would catalyze oxidation of cholesterol on one or both of the terminal methyl groups. This was proven to be the case in turnover studies, with CYP125 shown to form exclusively 27-hydroxycholesterol.
CONCLUSIONS
The CYP125 cytochrome P450 from M. tuberculosis was expressed, isolated, and structurally resolved. The P450 exhibits an obvious letterbox substrate access channel of dimensions appropriate for entry of the prospective substrate cholesterol. Complexes with androstenedione and econazole revealed ligand binding near the top of the active site cavity and exclusion for further ingress due to the narrowing of the active site funnel. Although solution state studies reveal econazole (and other azole drugs) are able to coordinate the heme iron under certain conditions, CYP125 clearly demonstrates lower type II binding affinity for a number of azole drugs compared with other Mtb P450s, e.g. CYP121 (17), consistent with the constricted nature of its heme access channel. Our model for the cholesterol-CYP125 interaction, and hence the catalytic activity, was obtained a priori and used to guide further experiments. This model indicates that the alkyl chain of this substrate can extend down the narrow binding funnel with the terminal methyl carbons of the chain presented to the heme iron to facilitate C27 oxidation, as confirmed by turnover studies. Given the likely role of CYP125 in catabolism of host cholesterol, this reaction is likely a primary event that enables the breakdown of the cholesterol side chain. However, the hydroxylation of cholesterol at the terminal position also has the potential to generate a product capable of modulating host cholesterol synthesis, competitively antagonizing estrogen receptor action, and inhibiting expression of nitric-oxide synthase (1). In this respect, it is tempting to speculate that CYP125 participates in cholesterol oxidation to generate a product that is further broken down to generate metabolic fuel for Mtb and/or is used directly to modulate host responses and thus facilitate persistence of the pathogen.
This work was supported in part by United Kingdom Biotechnology and Biological Sciences Research Council Grant C19757 and European Union Framework Programme VI project NM4TB.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” “Results,” Figs. S1–S3, and Tables S1–S4.
- Mtb
- bacterium Mycobacterium tuberculosis
- MD
- molecular dynamics
- HPLC
- high performance liquid chromatography
- HS
- high spin
- LS
- low spin
- EPR
- electron paramagnetic resonance.
REFERENCES
- 1.Umetani M., Domoto H., Gormley A. K., Yuhanna I. S., Cummins C. L., Javitt N. B., Korach K. S., Shaul P. W., Mangelsdorf D. J., et al. (2007) Nature Med. 13, 1185–1192 [DOI] [PubMed] [Google Scholar]
- 2.Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., 3rd, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., Davies R., Devlin K., Feltwell T., Gentles S., Hamlin N., Holroyd S., Hornsby T., Jagels K., Krogh A., McLean J., Moule S., Murphy L., Oliver K., Osborne J., Quail M. A., Rajandream M. A., Rogers J., Rutter S., Seeger K., Skelton J., Squares R., Squares S., Sulston J. E., Taylor K., Whitehead S., Barrell B. G. (1998) Nature 393, 537–544 [DOI] [PubMed] [Google Scholar]
- 3.Fleischmann R. D., Alland D., Eisen J. A., Carpenter L., White O., Peterson J., DeBoy R., Dodson R., Gwinn M., Haft D., Hickey E., Kolonay J. F., Nelson W. C., Umayam L. A., Ermolaeva M., Salzberg S. L., Delcher A., Utterback T., Weidman J., Khouri H., Gill J., Mikula A., Bishai W., Jacobs W. R., Jr., Venter J. C., Fraser C. M. (2002) J. Bacteriol. 184, 5479–5490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brennan P. J., Crick D. C. (2007) Curr. Top. Med. Chem. 7, 475–488 [DOI] [PubMed] [Google Scholar]
- 5.Denisov I. G., Makris T. M., Sligar S. G., Schlichting I. (2005) Chem. Rev. 105, 2253–2277 [DOI] [PubMed] [Google Scholar]
- 6.McLean K. J., Dunford A. J., Neeli R., Driscoll M. D., Munro A. W. (2007) Arch. Biochem. Biophys. 464, 228–240 [DOI] [PubMed] [Google Scholar]
- 7.Sassetti C. M., Boyd D. H., Rubin E. J. (2003) Mol. Microbiol. 48, 77–84 [DOI] [PubMed] [Google Scholar]
- 8.McLean K. J., Carroll P., Lewis D. G., Dunford A. J., Seward H. E., Neeli R., Cheesman M. R., Marsollier L., Douglas P., Smith W. E., Rosenkrands I., Cole S. T., Leys D., Parish T., Munro A. W. (2008) J. Biol. Chem. 283, 33406–33416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belin P., Le Du M. H., Fielding A., Lequin O., Jacquet M., Charbonnier J. B., Lecoq A., Thai R., Courçon M., Masson C., Dugave C., Genet R., Pernodet J. L., Gondry M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7426–7431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holsclaw C. M., Sogi K. M., Gilmore S. A., Schelle M. W., Leavell M. D., Bertozzi C. R., Leary J. A. (2008) ACS Chem. Biol. 3, 619–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McLean K. J., Warman A. J., Seward H. E., Marshall K. R., Girvan H. M., Cheesman M. R., Waterman M. R., Munro A. W. (2006) Biochemistry 45, 8427–8443 [DOI] [PubMed] [Google Scholar]
- 12.Bellamine A., Mangla A. T., Nes W. D., Waterman M. R. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 8937–8942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLean K. J., Marshall K. R., Richmond A., Hunter I. S., Fowler K., Kieser T., Gurcha S. S., Besra G. S., Munro A. W. (2002) Microbiology 148, 2937–2949 [DOI] [PubMed] [Google Scholar]
- 14.McLean K. J., Clift D., Lewis D. G., Sabri M., Balding P. R., Sutcliffe M. J., Leys D., Munro A. W. (2006) Trends Microbiol. 14, 220–228 [DOI] [PubMed] [Google Scholar]
- 15.Ahmad Z., Sharma S., Khuller G. K. (2006) FEMS Microbiol. Lett. 261, 181–186 [DOI] [PubMed] [Google Scholar]
- 16.Ouellet H., Podust L. M., de Montellano P. R. (2008) J. Biol. Chem. 283, 5069–5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLean K. J., Cheesman M. R., Rivers S. L., Richmond A., Leys D., Chapman S. K., Reid G. A., Price N. C., Kelly S. M., Clarkson J., Smith W. E., Munro A. W. (2002) J. Inorg. Biochem. 91, 527–541 [DOI] [PubMed] [Google Scholar]
- 18.Ahmad Z., Sharma S., Khuller G. K., Singh P., Faujdar J., Katoch V. M. (2006) Int. J. Antimicrob. Agents 28, 543–544 [DOI] [PubMed] [Google Scholar]
- 19.Van der Geize R., Yam K., Heuser T., Wilbrink M. H., Hara H., Anderton M. C., Sim E., Dijkhuizen L., Davies J. E., Mohn W. W., Eltis L. D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1947–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLean K. J., Munro A. W. (2008) Drug Metab. Rev. 40, 427–446 [DOI] [PubMed] [Google Scholar]
- 21.Ouellet H., Lang J., Couture M., Ortiz de Montellano P. R. (2009) Biochemistry 48, 863–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kendall S. L., Rison S. C., Movahedzadeh F., Frita R., Stoker N. G. (2004) Trends Microbiol. 12, 537–544 [DOI] [PubMed] [Google Scholar]
- 23.Pieters J. (2001) Microbes Infect. 3, 249–255 [DOI] [PubMed] [Google Scholar]
- 24.Brzostek A., Dziadek B., Rumijowska-Galewicz A., Pawelczyk J., Dziadek J. (2007) FEMS Microbiol. Lett. 275, 106–112 [DOI] [PubMed] [Google Scholar]
- 25.Mohn W. W., van der Geize R., Stewart G. R., Okamoto S., Liu J., Dijkhuizen L., Eltis L. D. (2008) J. Biol. Chem. 283, 35368–35374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pandey A. K., Sassetti C. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4376–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leys D., Mowat C. G., McLean K. J., Richmond A., Chapman S. K., Walkinshaw M. D., Munro A. W. (2003) J. Biol. Chem. 278, 5141–5147 [DOI] [PubMed] [Google Scholar]
- 28.Quaroni L. G., Seward H. E., McLean K. J., Girvan H. M., Ost T. W., Noble M. A., Kelly S. M., Price N. C., Cheesman M. R., Smith W. E., Munro A. W. (2004) Biochemistry 43, 16416–16431 [DOI] [PubMed] [Google Scholar]
- 29.Daff S. N., Chapman S. K., Turner K. L., Holt R. A., Govindaraj S., Poulos T. L., Munro A. W. (1997) Biochemistry 36, 13816–13823 [DOI] [PubMed] [Google Scholar]
- 30.Dutton P. L. (1978) Methods Enzymol. 54, 411–435 [DOI] [PubMed] [Google Scholar]
- 31.Lawson R. J., Leys D., Sutcliffe M. J., Kemp C. A., Cheesman M. R., Smith S. J., Clarkson J., Smith W. E., Haq I., Perkins J. B., Munro A. W. (2004) Biochemistry 43, 12410–12426 [DOI] [PubMed] [Google Scholar]
- 32.Lafite P., André F., Zeldin D. C., Dansette P. M., Mansuy D. (2007) Biochemistry 46, 10237–10247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K. (2005) J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cornell W. D., Cieplak P., Bayly C. I., Gould I. R., Merz K. M., Ferguson D. M., Spellmeyer D. C., Fox T., Caldwell J. W., Kollman P. A. (1995) J. Am. Chem. Soc. 117, 5179–5197 [Google Scholar]
- 35.Jakalian A., Jack D. B., Bayly C. I. (2002) J. Comput. Chem. 23, 1623–1641 [DOI] [PubMed] [Google Scholar]
- 36.Wang J., Wang W., Kollman P. A., Case D. A. (2006) J. Mol. Graph. Model. 25, 247–260 [DOI] [PubMed] [Google Scholar]
- 37.Mast N., Norcross R., Andersson U., Shou M., Nakayama K., Bjorkhem I., Pikuleva I. A. (2003) Biochemistry 42, 14284–14292 [DOI] [PubMed] [Google Scholar]
- 38.Hasemann C. A., Ravichandran K. G., Peterson J. A., Deisenhofer J. (1994) J. Mol. Biol. 236, 1169–1185 [DOI] [PubMed] [Google Scholar]
- 39.Sligar S. G., Gunsalus I. C. (1976) Proc. Natl. Acad. Sci. U.S.A. 73, 1078–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miles J. S., Munro A. W., Rospendowski B. N., Smith W. E., McKnight J., Thomson A. J. (1992) Biochem. J. 288, 503–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jenkins C. M., Waterman M. R. (1994) J. Biol. Chem. 269, 27401–27408 [PubMed] [Google Scholar]
- 42.McIver L., Leadbeater C., Campopiano D. J., Baxter R. L., Daff S. N., Chapman S. K., Munro A. W. (1998) Eur. J. Biochem. 257, 577–585 [DOI] [PubMed] [Google Scholar]
- 43.Chang J. C., Harik N. S., Liao R. P., Sherman D. R. (2007) J. Infect. Dis. 196, 788–795 [DOI] [PubMed] [Google Scholar]
- 44.Kendall S. L., Withers M., Soffair C. N., Moreland N. J., Gurcha S., Sidders B., Frita R., Ten Bokum A., Besra G. S., Lott J. S., Stoker N. G. (2007) Mol. Microbiol. 65, 684–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mast N., White M. A., Bjorkhem I., Johnson E. F., Stout C. D., Pikuleva I. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9546–9551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strushkevich N., Usanov S. A., Plotnikov A. N., Jones G., Park H. W. (2008) J. Mol. Biol. 380, 95–106 [DOI] [PubMed] [Google Scholar]