Abstract
Cyp125 (Rv3545c), a cytochrome P450, is encoded as part of the cholesterol degradation gene cluster conserved among members of the Mycobacterium tuberculosis complex. This enzyme has been implicated in mycobacterial pathogenesis, and a homologue initiates cholesterol catabolism in the soil actinomycete Rhodococcus jostii RHA1. In Mycobacterium bovis BCG, cyp125 was up-regulated 7.1-fold with growth on cholesterol. A cyp125 deletion mutant of BCG did not grow on cholesterol and accumulated 4-cholesten-3-one when incubated in the presence of cholesterol. Wild-type BCG grew on this metabolite. By contrast, a parallel cyp125 deletion mutation of M. tuberculosis H37Rv did not affect growth on cholesterol. Purified Cyp125 from M. tuberculosis, heterologously produced in R. jostii RHA1, bound cholesterol and 4-cholesten-3-one with apparent dissociation constants of 0.20 ± 0.02 μm and 0.27 ± 0.05 μm, respectively. When reconstituted with KshB, the cognate reductase of the ketosteroid 9α-hydroxylase, Cyp125 catalyzed the hydroxylation of these steroids. MS and NMR analyses revealed that hydroxylation occurred at carbon 26 of the steroid side chain, allowing unambiguous classification of Cyp125 as a steroid C26-hydroxylase. This study establishes the catalytic function of Cyp125 and, in identifying an important difference in the catabolic potential of M. bovis and M. tuberculosis, suggests that Cyp125 may have an additional function in pathogenesis.
Introduction
Mycobacterium tuberculosis (Mtb)4 infects one-third of the human population and is the leading cause of lethal bacterial infections worldwide. Synergy of Mtb with the HIV virus and the emergence of extensively drug-resistant Mtb strains (XDR-TB) have further emphasized this pathogen as a major global health threat (1). However, understanding of many of its key physiological processes, particularly those contributing to intracellular survival, is limited. One element of Mtb physiology that may prove important for therapeutic development is its unusually high number of cytochromes P450 (P450s). P450s are heme-dependent mono-oxygenases that utilize reducing equivalents from NAD(P)H relayed via electron transfer proteins to activate heme-bound O2. Typical of actinomycete genomes, that of Mtb includes 20 genes encoding P450s (2). In these bacteria, P450s are typically involved in the initial catabolism of growth substrates and in secondary metabolite biosynthesis. Given the high number of P450s in Mtb and their susceptibility to azole drugs, this class of enzyme has been proposed as a promising target for antimycobacterial therapeutic development (2). However, the function of most mycobacterial P450s has yet to be determined.
The gene encoding one Mtb P450, cyp125 (Rv3545c), belongs to a large cluster of genes encoding cholesterol degradation (3). This cluster is conserved in several actinobacteria including the non-pathogenic soil bacterium, Rhodococcus jostii RHA1. The genes encode functions necessary for cholesterol import (4), as well as for the degradation of the steroid side chain and rings A and B (3). Several of the ring degradation enzymes from Mtb have been characterized biochemically (5–9). Animal infection studies of mutants deficient in cholesterol uptake and catabolism have indicated that cholesterol metabolism in Mtb plays an important role in infection, contributing to both dissemination in the host (7) and persistence (9, 10). These results are consistent with several mutant screens, which previously identified numerous genes in the cholesterol degradation pathway as having an impact on intracellular growth and survival in both mouse and macrophage models (11, 12), implicating cholesterol catabolism in Mtb pathogenicity.
Cyp125 has been implicated in the in vivo survival of Mtb, although its catalytic activity has not been demonstrated, and its physiological role remains unclear. The cyp125 gene is up-regulated during growth of Mtb in interferon γ-activated macrophages (13) and mutants lacking this gene exhibit in vivo growth defects and reduced pathology in mouse and macrophage infection models (12, 14, 15). Additionally, Cyp125 was found to be resistant to concentrations of nitric oxide that are physiologically relevant during the immune response (16). Biochemical data have established that the Cyp125 homologue of R. jostii RHA1 binds cholesterol, while molecular genetic data demonstrate that this enzyme initiates side chain degradation and is essential for cholesterol transformation in this organism and Rhodococcus rhodochrous RG32 (17). However, a mutant of Mtb lacking the igr locus, an operon encompassing cyp125 plus five additional genes, was reported to transform cholesterol (15). Nevertheless, the RHA1 and Mtb enzymes share ∼70% amino acid identity (3).
In this work, we examined the role of Cyp125 in the mycobacterial catabolism of cholesterol using a combination of in vivo and in vitro approaches. Mycobacterium bovis bacillus Calmette-Guérin (BCG) was used as a model for cholesterol catabolism in Mtb, as the cholesterol catabolic clusters share 99% nucleotide sequence identity and the igr operons, containing cyp125, are identical in sequence. The expression of cyp125 in BCG was investigated, and a deletion strain was prepared and characterized with respect to growth and metabolite accumulation. Cyp125 of Mtb H37Rv was heterologously produced and purified. The steroid binding properties of the enzyme were investigated. Finally, the activity of Cyp125 was reconstituted in vitro, and the reaction product was characterized. The findings shed new light on the initiation of cholesterol catabolism in mycobacteria.
MATERIALS AND METHODS
Chemicals and Reagents
Cholesterol, 4-cholesten-3-one, and 2-hydroxypropyl-β-cyclodextrin were purchased from Sigma-Aldrich. 26-Cholestenoic acid was a gift from Dr. Robert van der Geize. An EDTA-bridged β-cylodextrin dimer was a gift from Dr. José Vázquez Tato. Restriction enzymes and the Expand High Fidelity PCR System were purchased from New England Biolabs (Ipswich, MA) and Roche Applied Science (Laval, Quebec), respectively. All other reagents were of HPLC or analytical grade. Water for buffers was purified using a Barnstead Nanopure DiamondTM system (Dubuque, Iowa) to a resistivity of at least 18 mΩ.
Gene Expression
Reverse transcriptase quantitative-PCR (RT-qPCR) was performed as described previously (18) using 5′-6FAM-TCGCGCGTGAGGAC-3′ as the probe and 5′-TGATCCCGCGATTCAAGAA-3′ and 5′-GCATGACGAAGCGCTGAAC-3′ as the forward and reverse PCR primers, respectively. cDNA was synthesized using the ThermoScript RT-PCR System (Invitrogen, Carlsbad, CA) and random hexamers. The sigA gene was used as an internal standard in multiplex reactions and the Ct values were normalized (ΔCt) by subtracting those of the internal standard. Significant differences in ΔCt values were tested using a two-sample Student's t test assuming unequal variances. Relative fold differences were calculated as 2−ΔΔCt where ΔΔCt = ΔCt treatment − ΔCt control.
Bacterial Strains and Vector Construction
For production and purification of Cyp125H37Rv, Rv3545c was amplified by PCR using the primers 5′-GCAATAGCATATGCCCAGCCCCAATCTGCCG-3′ and 5′-GCCAAGCTTCTGTTCCGCAGTGGGATCGAAATC-3′. The amplicon was digested with NdeI and HindIII (cleavage sites underlined in primer sequences) and ligated into NdeI- and HindIII-digested pTipQC2, yielding the expression vector pTipCP125.
An identical allelic exchange construct was used to replace the cyp125 gene of BCG-Pasteur (BCG_3609c) and Mtb H37Rv (Rv3545c) with a γδres-sacB-hyg-γδres cassette comprising the sacB and hygromycin resistance genes flanked by res-sites of the γδ-resolvase. Upstream and downstream flanking DNA regions of cyp125 were amplified by PCR employing the oligonucleotide pairs cyp125-LL (5′-TTTTTTTTGCATAAATTGCCGATCAACCCGGCAATCAGATG-3′) and cyp125-LR (5′-TTTTTTTTGCATTTCTTGCGCTCGACAAAG TCTCCAGAACC-3′) introducing BstAPI restriction sites (underlined) for amplification of the upstream flanking region cyp125-L; cyp125-RL (5′-TTTTTTTTCCATAGATTGGATCTTTAACGCCGTAGCCGACC-3′) and cyp125-RR (5′-TTTTTTTTCCATCTTTTGGGTTGACAGCGGCTTACCGAAC- 3′) introducing Van91I restriction sites (underlined) for amplification of the downstream flanking region cyp125-R. Amplified cyp125-L and cyp125-R were digested with BstAPI and Van91I, respectively, and ligated with Van91I-digested p0004S vector arms,5 resulting in the knock-out construct pcyp125 which was linearized with PacI and cloned and packaged into the temperature-sensitive phage phAE1596 as described (19), yielding the knock-out phage phcyp125. Allelic exchange in BCG and H37Rv using the phage phcyp125 was achieved by specialized transduction as reported previously (19), resulting in the mutant, Δcyp125, in which nucleotides 506–1167 of the cyp125 gene (1,302 bp) are deleted and replaced by the γδres-sacB-hyg-γδres cassette (Fig. 1A). The mutant was verified by Southern analysis of BamHI-digested genomic DNA using radiolabeled cyp125-R as probe (Fig. 1B).
FIGURE 1.
Construction of mycobacterial Δcyp125 mutants. A, genetic organization of the cyp125 locus in WT strains H37Rv and BCG-Pasteur together with the corresponding Δcyp125 mutants. The size of the BamHI fragments as well as the location of the probe used for Southern analysis are indicated. γδres indicates the res-sites of the γδ-resolvase; hyg indicates the hygromycin resistance gene. B, Southern analysis of BamHI-digested genomic DNA from BCG, its Δcyp125 mutant, H37Rv, and its Δcyp125 mutant. Gene deletion was confirmed employing a [α-32P]dCTP-labeled probe hybridizing to the position indicated in A.
For complementation of the Δcyp125 mutant, the cyp125 gene from BCG was amplified by PCR using the oligonucleotide pair 5′-TCGCAGCTGCATAAAAGGAGATCACTCGTGTCGTGGAATCACCAGTCAGTGG-3′ (introducing a PvuII site (underlined) and a ribosome binding site (bold) and 5′-GCCAAGCTTCTGTTCCGCAGTGGGATCGAAATC-3′ (introducing a HindIII restriction site (underlined)). Amplicons were cloned via the primer-induced restriction sites (underlined) as a PvuII-HindIII fragment downstream of the constitutive hsp60 promoter into plasmid pMV361KanR, which allows single copy integration into the genome and complementation in trans, resulting in the complemented mutant strain Δcyp125 attBL5::pMV361::cyp125 (Δcyp125-C). An empty vector control strain (Δcyp125-V) consisted of Δcyp125 attBL5::pMV361.
In Vitro Growth Conditions of Mycobacterial Cultures
Strain BCG was grown in Middlebrook 7H9 basal medium (BD-Difco) plus 0.5% Tyloxapol. Steroids were added to the medium dissolved in the Tyloxapol prior to autoclaving. Cultures were incubated at 37 °C on a bottle roller at one rotation per minute. As the low aqueous solubility of cholesterol results in turbidity, optical density was not used as a quantitative measure of cell growth. Rather, growth was measured on the basis of total protein, determined by disrupting cells via hot alkaline lysis followed by the BCA protein assay (Pierce), with bovine serum albumin as the standard.
H37Rv strains were grown in a minimal medium (1 g/liter KH2PO4, 2.5 g/liter Na2HPO4, 0.5 g/liter (NH4)2SO4, 0.15 g/liter asparagine, 50 mg/liter ferric ammonium citrate, 0.5 g/liter MgSO4 × 7 H2O, 0.5 mg/liter CaCl2, 0.1 mg/liter ZnSO4, 0.05% (v/v) Tyloxapol) containing no additional carbon source or 0.5 mm cholesterol. Cholesterol was added from a stock solution dissolved in isopropanol to the culture tubes. After complete evaporation of the solvent, the liquid medium was added, heated to 80 °C, and a homogenous dispersion was produced by ultrasonication. Exponentially growing precultures were washed and starved in phosphate-buffered saline containing Tyloxapol (0.05%, v/v) for 48 h before inoculation of test cultures. Growth was monitored by measuring colony forming units (CFU) by plating serial dilutions of cultures onto Middlebrook 7H10 agar supplemented with 10% (v/v) OADC enrichment (Becton Dickinson Microbiology Systems, Spark, MD) and 0.5% (v/v) glycerol.
Metabolite Analysis
Gas chromatography-coupled mass spectrometry (GC-MS) was performed using an HP 6890 series GC system fitted with an HP-5MS 30 m × 250 μm column and an HP 5973 mass-selective detector (Hewlett-Packard, Palo Alto, CA). NMR spectra were recorded using a Varian Unity 500 MHz and Inova 600 MHz (Varian Inc., Mississauga, ON, Canada). Chemical shifts, δ, are reported in ppm relative to CHCl3 (1H: δ = 7.24) and CDCl3 (13C: δ = 77.0) as internal standards.
Production and Purification of Cyp125
Cyp125 was heterologously produced in R. jostii RHA1 using the expression plasmid pTipCP125. Electrocompetent R. jostii RHA1 cells were transformed with pTipCP125 by electroporation using a Bio-Rad MicroPulser apparatus and Bio-Rad 0.1 cm GenePulser cuvettes. Transformed cells were grown on Luria-Bertani broth (LB) agar plates supplemented with 25 μg/ml chloramphenicol at 30 °C for 2 days. Colonies were used to inoculate 50 ml of LB supplemented with 25 μg/ml chloramphenicol, which was then incubated at 30 °C shaking at 200 rpm for 2 days. 10 ml of this preculture were used to inoculate 1 liter of the LB medium, incubated at 30 °C until the OD reached 0.5, and supplemented with thiostrepton to a final concentration of 50 μg/ml. The cells were allowed to grow for another 20 h before harvesting by centrifugation. Pellets were washed three times with 10 mm potassium phosphate, pH 7.0 containing 10% glycerol and frozen at −80 °C until use.
To purify Cyp125, the cell pellet from 3 liters of culture was suspended in 25 mm HEPES pH 7.5 supplemented with DNase I (Roche Applied Science). The suspended cells were subjected to 5 rounds of bead-beating for 3 min, alternating with 10 min of cooling on ice. The lysed cells were centrifuged for 7 min at 4,600 × g, and the supernatant was further clarified by ultracentrifugation at 10,000 × g for 45 min. The protein was purified by chromatography using an Äkta Explorer (Amersham Biosciences): the cell-free extract (∼30 ml) was passed through a syringe-driven 0.45-μm filter and loaded (in 10-ml aliquots) onto a 1 × 10 cm column of SourceTM15Q (GE Healthcare) resin equilibrated with 25 mm HEPES pH 7.5. After washing the column with 0.10 m NaCl, the enzyme was eluted with a linear gradient from 0.10–0.16 m NaCl over six column volumes at a flow rate of 3 ml/min. Red-colored fractions containing Cyp125, eluting at 0.14 m NaCl, were pooled, exchanged into 25 mm HEPES pH 7.5, concentrated to 10–15 mg/ml protein, flash frozen as beads in liquid nitrogen, and stored at −80 °C until use. Cyp125 concentrations were estimated by measuring thiolate-ligated heme iron using the extinction coefficient ϵ449–490 = 91 mm−1 cm−1 for the reduced CO-difference spectrum (20). KshB was purified as described previously (6).
Spectroscopic Analysis of Purified Cyp125
UV-vis absorption spectra were recorded using a Cary 5000 spectrophotometer equipped with a thermojacketed cuvette holder (Varian, Walnut Creek, CA). The CO-bound form of Cyp125 was generated by first incubating samples with ∼8 mm sodium dithionite for 10 min then slowly bubbling with CO for 30 s. The proportion of purified protein containing high-spin ferric heme iron was estimated by comparing the spectra of Cyp125 to linear combinations of the spectra of Cyp125 in a high spin state (generated by adding 12 μm 4-cholesten-3-one in 10% 2-hydroxypropyl-β-cyclodextrin (BCD) to 3.3 μm enzyme) and substrate-free P450Cam from Pseudomonas putida, generated as described previously (21)). The same values were obtained using low spin Cyp125 from R. jostii RHA1 as a standard (17). Substrate-induced spectral responses were recorded in 0.1 mm KPi pH 7.0 by titrating solutions of Cyp125 with 1.0 mm stock solutions of 4-cholesten-3-one or cholesterol in either 10% BCD or 10 mm of an EDTA-linked β-cyclodextrin dimer (22). Equilibrium dissociation constants were calculated using a quadratic equation as described for Cyp125 of R. jostii RHA1 (17). Non-linear least-squares fitting was done using the program R (23).
Activity Assay for Cyp125
Cyp125 activity was measured by monitoring substrate and product concentrations using a Waters 2695 Separations HPLC module equipped with a Waters 2996 photodiode array detector and a 250 × 4.60 mm C18 Prodigy 10u ODS-Prep column (Phenominex, Torrance, CA). Solvents used for HPLC analysis were 0.5% aqueous phosphoric acid (A) and methanol (B). Compounds were detected at 422 nm and eluted at a flow rate of 1 ml/min with a gradient from 80–90% B over 5 min, followed by isocratic elution at 90% B for 10 min, a gradient from 90–100% B for 5 min, and further isocratic elution for 15 min. Under these conditions, elution times for 4-cholesten-3-one and 4-cholesten-3-one-26-ol were 38.5 and 22.0 min, respectively. Concentrations of 4-cholesten-3-one and 4-cholesten-3-one-26-ol were calculated from their respective peak areas using a standard curve of 4-cholesten-3-one and assuming that the extinction coefficients for the two compounds are similar; the R2 value for the standard curve was >0.99. The standard assay was performed in air-saturated 0.10 m potassium phosphate at pH 7.0 containing 900 μm NADH, 50 μm 4-cholesten-3-one, 1.5 μm KshB, and 1.6 μm Cyp125. Stock solutions of 1 mm 4-cholesten-3-one were made in 10% BCD and stored at 4 °C; stock solutions of 180 mm NADH were made fresh daily in water and stored on ice. Assays were conducted in multiple tubes containing 200 μl of the standard assay mixture. At each time point, the reaction was quenched by the addition of 200 μl of methanol and vigorous mixing.
RESULTS
Up-regulation of cyp125 in Cholesterol-grown BCG
To investigate if cyp125 is up-regulated in BCG during growth on cholesterol, RT-qPCR was used to analyze cells growing on cholesterol versus on glucose, both sampled when at approximately half of final culture density. The cells grown on cholesterol exhibited a 7.1-fold increase in the level of normalized cyp125 transcript (n = 4, p < 0.05). These results correlate well with the expression ratio of 11 found for RHA1 cyp125 (ro04679) during growth on cholesterol versus pyruvate (3), as well as for expression ratios of other cholesterol catabolic genes in BCG grown on cholesterol versus glucose (2.1- and 4.6-fold for hsaC and kshA, respectively) (3).
Cyp125 Is Essential for the Growth of BCG but Not H37Rv on Cholesterol
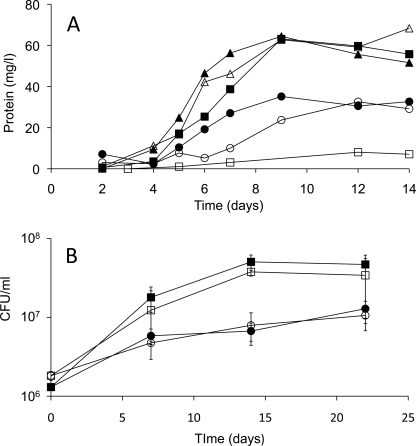
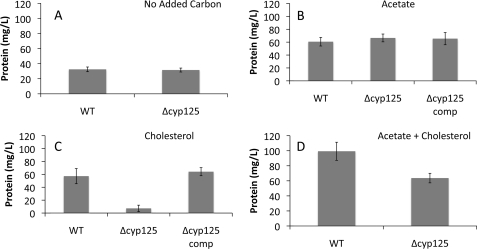
Growth of both BCG and H37Rv was robust on 0.5 mm cholesterol. For BCG, growth kinetics were very similar on cholesterol or 10 mm acetate (Figs. 2 and 3). To determine if Cyp125 is essential for growth on cholesterol, cyp125 deletion mutants were created in both BCG and H37Rv using a transduction phage method. Southern blot analysis confirmed the expected chromosomal deletion and disruption (Fig. 1). The BCG Δcyp125 strain grew normally on 10 mm acetate, with kinetics very similar to BCG (Fig. 2A). Both strains also showed some growth on the basal medium with no added carbon source, because of a low level of glutamate. As hypothesized, BCG Δcyp125 failed to grow on cholesterol. Moreover, the protein yield of the mutant was lower in the cholesterol-supplemented medium than in the no-carbon control, suggesting an inhibitory effect of cholesterol. To test this hypothesis, growth was tested on medium containing acetate plus cholesterol. While the WT exhibited a higher protein yield than on either cholesterol or acetate alone, the BCG Δcyp125 mutant strain exhibited a protein yield similar to that on acetate alone. Thus, cholesterol is not inhibitory to this mutant under all conditions. Finally, all growth phenotypes observed for the BCG Δcyp125 mutant could be complemented with cyp125 provided in trans while the empty vector control strain exhibited the same growth phenotypes as the mutant strain in medium supplemented with acetate or cholesterol (Fig. 3).
FIGURE 2.
Growth of mycobacterial Δcyp125 mutants. A, representative growth curves of WT BCG (solid symbols) and Δcyp125 mutant (open symbols) on basal medium containing no added carbon source (circles), 10 mm acetate (triangles), or 0.5 mm cholesterol (squares). B, growth of WT Mtb H37Rv (solid symbols) and Δcyp125 mutant (open symbols) on basal medium containing no added carbon source (circles) or 0.5 mm cholesterol (squares). Values represent mean of triplicates ± S.D.
FIGURE 3.
Final protein yields of wild-type (BCG), mutant (Δcyp125), and complementation (Δcyp125-C) strains. Cultures were incubated for 14 days in basal medium supplemented with no added substrate (A), 10 mm acetate (B), 0.5 mm cholesterol (C), or 10 mm acetate plus 0.5 mm cholesterol (D). Plotted values represent mean values of 3–6 replicates, and error bars indicate S.E.
Strain BCG was additionally able to grow on 4-cholesten-3-one. The growth phenotype of BCG Δcyp125 on 4-cholesten-3-one was the same as on cholesterol: the mutant was unable to grow on 4-cholesten-3-one, and 4-cholesten-3-one inhibited its growth on the basal medium (data not shown). Thus, Cyp125 is also necessary for metabolism of this compound. BCG failed to grow on ergosterol, β-sitosterol, cholic acid, or progesterone. The former three steroids did not inhibit growth on the basal medium in the manner of cholesterol, but progesterone did so.
In contrast to the BCG mutant, the H37Rv Δcyp125 mutant was not impaired in growth on cholesterol. The wild-type and mutant strains had nearly identical growth kinetics (Fig. 2B). This demonstrates a clear difference in the capacity of BCG and H37Rv to metabolize cholesterol.
BCG Δcyp125 Mutant Transforms Cholesterol
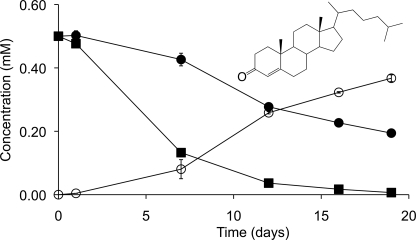
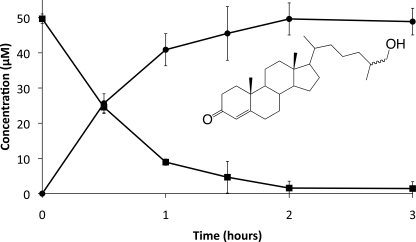
Cyp125 has been proposed to catalyze the obligate first step in cholesterol catabolism in R. jostii RHA1 (17). We hypothesized that the same would be true for Mtb and BCG, given the similarity of the cholesterol catabolic pathway in rhodococci and mycobacteria (3). Accordingly, we compared cholesterol depletion in cultures of BCG versus BCG Δcyp125 during growth in medium with acetate plus cholesterol. BCG completely removed cholesterol in 19 days and did not accumulate any detectable steroid metabolites (Fig. 4). By contrast, Δcyp125 exhibited slower cholesterol removal and stoichiometrically accumulated 4-cholesten-3-one. Thus, cells lacking Cyp125 transform cholesterol via the 3β-hydroxysteroid dehydrogenase (3β-HSD)-catalyzed oxidation of the C3 hydroxyl group and subsequent C5-C4 isomerization of the ring structure double bond (8). Subsequent transformation is blocked in cells lacking Cyp125 activity. 3β-HSD acts intracellularly (8), suggesting that cyp125 deletion does not disrupt cholesterol uptake, but appears to catalyze a reaction essential for further degradation of 4-cholesten-3-one. These results further indicate that neither cholesterol nor 4-cholesten-3-one is toxic to BCG under these conditions. Thus, BCG lacking Cyp125 can transform cholesterol but is unable to metabolize it sufficiently to support growth.
FIGURE 4.
Cholesterol transformation by BCG and Δcyp125 cultures. Cholesterol removal (solid symbols) and 4-cholesten-3-one (inset) accumulation (open symbols) is shown for BCG (squares) and Δcyp125 (circles) growing in medium with 10 mm acetate plus 0.5 mm cholesterol. Plotted values are the average of duplicate experiments, and error bars indicate S.E.
Production and Purification of Cyp125 from H37Rv
The cyp125 gene of Mtb was heterologously expressed in R. jostii RHA1 using a pTip vector. As this gene is 100% identical in nucleotide sequence to that of BCG, the proteins from these two species are not distinguished in the following text. Cyp125 production was confirmed by CO-difference spectra (results not shown). Initial attempts to purify this protein resulted in the production of two proteins of subtly different sizes, as determined by gel filtration chromatography and SDS-PAGE analysis. The two proteins exhibited very similar spectroscopic properties, both as isolated and upon formation of a ferrous-CO complex. MALDI-TOF mass spectrometry analyses indicated that the respective molecular masses of these proteins were 46.4 and 48.2 kDa. Based on this result, the annotated gene sequence was re-examined, and a second potential start codon preceded by a Shine-Dalgarno sequence (AAGGAG) was found 48 nucleotides downstream of the originally annotated one. The theoretical molecular masses of the proteins produced using the upstream and downstream start codons, 48.4 and 46.6 kDa, respectively, are consistent with the masses of proteins detected during purification attempts using the originally annotated gene. A sequence alignment with the closest homologues of cyp125H37Rv as identified by Swiss-Prot (24) revealed that the N-terminal 16 amino acids predicted using the original start codon were absent in all other sequences examined. Accordingly, the gene sequence was re-annotated as 1254- bp long and encompassing residues 3985397–3984144 of the H37Rv genome. The re-annotated gene was cloned and expressed in RHA1 using pTipCP125 for all subsequent experiments. This protein had a molecular mass of 46.4 kDa.
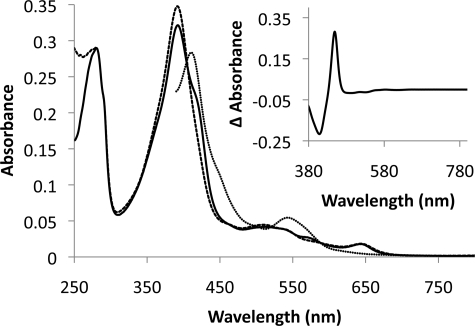
Purification of Cyp125 by anion exchange chromatography yielded ∼4 mg of purified protein per liter of harvested culture. SDS-PAGE analysis indicated that Cyp125 constituted >99% of the protein in the purified sample. The spectroscopic properties of the protein as isolated were atypical of bacterial P450s, as the protein was purified with the heme iron primarily in a high spin state; the UV spectrum of the purified material exhibited peaks at 280 and 392 nm, with a pronounced shoulder at 422 nm. This is consistent with the spectroscopic properties of Cyp125 found by Ouellet et al. (16). As isolated, Cyp125 had an absorbance ratio A392/A280 of 1.11 and was estimated to be 70% in the high spin state. Upon incubation with 4-cholesten-3-one, the shoulder disappeared and the A392/A280 increased to 1.20 (Fig. 5). Reduction with dithionite and exposure to carbon monoxide resulted in a reduced CO-difference spectrum with a peak at 449 nm, indicating an intact thiolate ligand to the heme iron (Fig. 5, inset).
FIGURE 5.
Spectra of Cyp125. The spectra of 3.4 μm Cyp125 as isolated with oxidized heme iron (solid line), upon addition of 10 μm 4-cholesten-3-one (dashed line) and subsequent reduction (dotted line) are shown. The reduced CO-difference spectrum is shown in the inset. Spectra were collected at 25 °C in 0.1 mm potassium phosphate buffer, pH 7.0.
Cyp125 of Mtb Binds C-27 Steroids
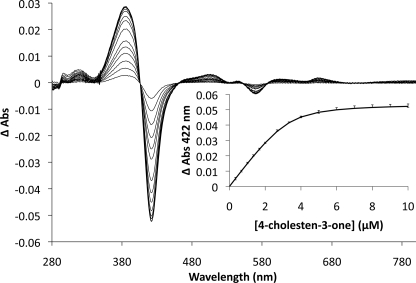
The binding of steroid ligands to Cyp125 was investigated by examining the relative absorbance at 392 (high spin heme iron) and 422 (low spin heme iron); ligand binding adjacent to the heme iron causes a shift in the iron from a hexa-coordinated low spin state to a penta-coordinated high spin state in P450 enzymes. Because of the low aqueous solubility of the potential steroid ligands, they were added to the enzyme as a complex with BCD (25). Upon addition of 4-cholesten-3-one, Cyp125 exhibited a type I difference spectrum (Fig. 6). No change in the Cyp125 spectrum was observed with BCD alone, indicating that the observed change in iron spin state was due to the steroid molecule binding in the active site. Cyp125 was also found by the same method to bind cholesterol, but not 5-cholestene-26-oic acid 3β-ol, a proposed product of the Cyp125 reaction (17). The Kdapp of Cyp125 for 4-cholesten-3-one was 0.27 ± 0.05 μm. Attempts to determine a Kd for cholesterol under these conditions were unsuccessful as a partial reversal of the spectral shift was observed at concentrations of BCD exceeded 0.02%. Such concentrations of BCD were attained well before the cytochrome was saturated with the steroid. To solve this problem, we solubilized 5 mm cholesterol in 25 mm of an EDTA-bridged β-cyclodextrin dimer (22). Using this reagent, we were able to obtain a binding curve for cholesterol with an Kdapp of 0.20 ± 0.02 μm. However, the binding reaction took much longer to equilibrate (∼15 min) than for 4-cholesten-3-one with BCD. With the dimer, 4-cholesten-3-one was not as soluble as cholesterol, and a similar problem of partial reversal of the spectral shift was observed using 4-cholesten-3-one with the dimer. Finally, using either steroid complexed with the dimer, a shoulder at 422 nm remained, even at saturation, which was not the case for binding studies performed using the 4-cholesten-3-one with BCD. Several additional solvent systems were tried for both steroids without success. Thus, a direct comparison of the binding affinity of Cyp125 for cholesterol versus 4-cholesten-3-one was not possible.
FIGURE 6.
Binding of 4-cholesten-3-one to Cyp125. Difference spectra of 3.3 μm Cyp125 upon the addition of 0.33 to 10 μm of 4-cholesten-3-one are overlaid. The resultant binding curve is inset with error bars representing the S.E. calculated from triplicate data; the solid line represents the best fit of a quadratic binding equation to the data as determined using the program R, with fitted parameters: KD = 0.27 ± 0.05 μm, [Cyp125] = 3.3 ± 0.2 μm, and ΔA422Max = 0.055 ± 0.001. 4-Cholesten-3-one was prepared as an aqueous solution in 10% 2-hydroxypropyl-β-cyclodextrin.
Cyp125 of Mtb Transforms C-27 Steroids
Based on the binding results, we investigated the ability of Cyp125 to transform 4-cholesten-3-one and cholesterol. 4-Cholesten-3-one with BCD was used for quantitative Cyp125 activity analysis because of its ability to fully convert the P450 to a high spin state, short equilibration time for binding, and UV spectrum, which allowed for quantification by HPLC analysis. Bioinformatic analyses were performed to identify potential redox partners for Cyp125. These analyses indicated that the rhodococcal and mycobacterial cholesterol catabolic clusters contain more oxygenases than NAD(P)H-utilizing reductases. Moreover, the four steroid catabolic clusters identified in R. jostii RHA1 do not appear to each encode a full complement of reductases, suggesting that the reductase encoded by one cluster might be used by the oxygenases encoded by another cluster (3). Additionally, a kshB deletion mutant of Rhodococcus erythropolis SQ1 was unable to degrade the sterol side chain, prompting the authors to propose that KshB may function as part of the sterol 26-hydroxylase enzyme system (26). Accordingly, we attempted to reconstitute Cyp125 activity in vitro using KshB (Rv3571) as the reductase.
In assays containing Cyp125, KshB, and NADH, 4-cholesten-3-one was consumed, and a product was formed (Fig. 7). Assuming this product has a similar extinction coefficient as the substrate, it was formed in a 1:1 ratio with depleted 4-cholesten-3-one, and no other product was detected after incubation up to 24 h. Substrate turnover was dependent on NADH, KshB, and native Cyp125, as omitting any of these components or replacing Cyp125 with enzyme which had been heat inactivated by incubation at 65 °C for 30 min abolished substrate consumption and product formation. The reconstituted Cyp125 also transformed cholesterol (results not shown). The rate of the observed reaction using either steroid was very low, with a 4-cholesten-3-one specific activity of 0.01 unit/mg (Fig. 7). If Cyp125 was saturated with steroid under these conditions, this would correspond to an apparent kcat value of 31 h−1.
FIGURE 7.
Turnover of 4-cholesten-3-one by Cyp125. Depletion of 4-cholesten-3-one and accumulation of 26-hydroxy-4-cholesten-3-one (inset) are represented by squares and circles, respectively. Reactions were carried out in air-saturated 0.1 mm potassium phosphate, pH 7.0, containing 900 μm NADH, 50 μm 4-cholesten-3-one, 1.5 μm KshB, and 1.6 μm Cyp125.
In attempts to increase the rate of reaction, each of two potential ferredoxin components was added to the reaction mixture. Both Rv3503c and Rv3527, encoded in the cholesterol degradation gene cluster of Mtb, are predicted to encode small ferredoxins.7 The physiological role of neither protein has been determined. Each protein was heterologously produced in Escherichia coli. The absorption spectra of the purified proteins had broad absorption bands consistent with FeS clusters (results not shown). However, inclusion of either purified protein failed to increase the rate of Cyp125-catalyzed steroid turnover.
The rate of the Cyp125 reaction was also investigated with respect to oxygen concentration. There was a linear correlation between Cyp125 steroid consumption and oxygen concentration in buffer containing from 116 to 1160 μm O2 (supplemental Fig. S1). This indicates that the apparent KmO2 of Cyp125 was >1.2 mm. The estimated apparent kcat/KmO2 value of Cyp125 was 11 m−1 s−1, principally because of the low turnover number of the enzyme. A range of KmO2values have been reported for other cholesterol-degrading oxygenases of M. tuberculosis: >1.2 mm for KshAB (6), ∼100 μm for HsaAB,8 and ∼90 μm for HsaC (7).
Cyp125 Is a Steroid 26-Hydroxylase
The reaction products were identified by their mass fragmentation patterns as modified 4-cholesten-3-one or cholesterol bearing a hydroxyl group on the aliphatic side chain (supplemental Fig. S2). For example, the ion representing the total mass of each molecule (472 and 546 m/z for reactions with 4-cholesten-3-one and cholesterol, respectively) differed from the substrate molecules by the mass of one trimethylsianol-derivatized hydroxyl group (88 amu). Additionally, fragments representing the steroid nucleus (271 m/z for 4-cholesten-3-one missing C20-C27 and 255 m/z for cholesterol missing C20-C27 and the oxygen at C3) remain unaltered in both substrate-product pairs. Using a combination of 1H, HSQC, HMBC, and COSY NMR, the following chemical shifts for the hydroxylated 4-cholesten-3-one product were determined: 1H NMR (500 MHz, CDCl3): δ = 5.72 (s, 1H, 4), 3.50 (dd, 3J = 6.0, 10.0 Hz, 1H, 26x), 3.43 (dd, 3J = 6.0, 10.0 Hz, 1H, 26y), 2.40 (m, 4H, 2xy, 6xy), 2.01 (m, 2H, 1x, 12x), 1.83 (m, 2H, 7x, 16x), 1.69 (m, 1H, 1y), 1.61 (m, 2H, 15x, 25), 1.51 (m, 2H, 11x, 8), 1.44 (m, 1H, 11y), 1.40 (m, 4H, 20, 22x, 23x, 24x), 1.27 (m, 1H, 16y), 1.18 (s, 3H, 19xyz), 1.12 (m, 4H, 12y, 15y, 17, 23y), 1.02 (m, 4H, 7y, 14, 22y, 24y), 0.92 (m, 7H, 9, 21xyz, 27xyz), 0.71 (s, 3H, 18xyz); 13C NMR (125.8 MHz, CDCl3): δ = 199.7 (3), 171.7 (5), 68.2 (26), 55.9 (17), 55.7 (14), 53.6 (9), 42.3 (13), 39.4 (12), 38.5 (10), 36.0 (22), 35.8 (4), 35.6 (25), 35.5 (1, 20), 35.4 (8), 33.8 (2), 33.5 (24), 32.8 (6), 31.9 (7), 28.0 (16), 24.0 (15), 23.3 (23), 20.8 (11), 18.5 (21), 17.2 (19), 16.5 (27), 11.8 (18). Notably, five methyl groups were distinguishable in the spectrum for 4-cholesten-3-one, with the C26 and C27 protons represented by overlapping doublets centered at δ = 0.87 (data not shown). The spectrum of the reaction product did not contain this overlapping doublet, instead containing a signal at δ = 0.92 consistent with a downshifted methyl group and two protons on a hydroxylated carbon (δ = 3.50, 3.43). These results enabled the identification of the product as 26-hydroxy-4-cholesten-3-one. The stereochemistry at C25 was not able to be determined from this experiment.
DISCUSSION
The current study provides direct evidence that Cyp125 of Mtb catalyzes the C26 hydroxylation of cholesterol and 4-cholesten-3-one and that this is an obligate early step in the catabolism of these steroids by strains of the Mtb complex. Purified Cyp125 bound these compounds with submicromolar apparent dissociation constants and, when reconstituted with KshB, transformed them to the corresponding C26-hydroxysteroid with no evidence of further oxidation. A cyp125 deletion mutant of BCG failed to grow on cholesterol, but transformed it to 4-cholesten-3-one. It is unclear whether the physiological substrate of the mycobacterial Cyp125 is cholesterol, 4-cholesten-3-one or both. However, this work reveals that mycobacterial steroid ring degradation cannot proceed with substrates having a full-length aliphatic side chain.
The current findings extend those of a previous study demonstrating that in rhodococci, Cyp125 is involved in the formation of 26-cholestenoic acid and that this is an early step in cholesterol catabolism (17). In that study it was proposed that Cyp125 catalyzes the hydroxylation and possible further oxidation at C26 of cholesterol, while the current data indicate that Cyp125 of Mtb performs no further oxidation of the hydroxysteroid. Cholesterol catabolism appears to differ in the various actinobacteria in several respects. First, they do not share an obligatory order for side chain and ring structure modification. Thus, while the Δcyp125 mutants of BCG and R. rhodochrous RG32 transformed cholesterol to 4-cholesten-3-one, the corresponding mutant of RHA1 did not. This suggests that 3β-HSD from the different strains has different substrate specificities. A second difference is suggested by the inability of the BCG mutant to grow on or further transform 4-cholesten-3-one, whereas the RHA1 mutant could grow on this compound (17). It was suggested that this growth was caused by catabolism of the steroid rings. Nevertheless, it is unclear whether this reflects the different specificities of the ring-degrading enzymes in these two genera or the presence of multiple steroid degradation clusters in rhodococci (27).
This study also identified a significant difference in the potential for cholesterol metabolism between BCG and H37Rv. Whereas Cyp125 is essential for BCG to metabolize cholesterol beyond 4-cholesten-3-one, H37Rv appears to metabolize cholesterol normally without Cyp125. The most likely explanation is that H37Rv contains an enzyme that complements the loss of Cyp125. Bioinformatic analyses revealed eight genes predicted to encode oxidoreductases in H37Rv that do not occur in BCG-Pasteur (28), of which three are predicted to encode monooxygenases: Rv1256c, Rv3121, and Rv3618. The first two encode cytochromes P450 Cyp130 and Cyp141, respectively, while the third encodes a flavin-dependent monooxygenase related to those that hydroxylate alkyl moieties of cyclic terpenes. Both Rv1256c and Rv3618 were up-regulated during growth of Mtb in macrophages (11, 12), while transposon mutagenesis indicated Rv3618 to contribute to Mtb growth in IFNγ-activated macrophages (11, 12). As none of the three H37Rv genes have homologues in M. bovis, we predict that one of these three genes may complement cyp125 in Mtb, whereas cyp125 is likely to be essential for cholesterol metabolism in all M. bovis strains.
Characterization of the role of Cyp125 in Mtb confirms and extends recent studies on the respective physiological roles of FadA5 (9) and the igr locus (15) in Mtb H37Rv. The latter contains cyp125 and was concluded to be required for cholesterol metabolism, as cholesterol inhibited growth of an igr deletion mutant on glycerol. Although growth on cholesterol was not reported, the igr mutant transformed C4 of cholesterol to CO2 and incorporated C26 into mycobacterial lipids. This is consistent with the ability of the H37Rv Δcyp125 mutant to grow normally on cholesterol and further suggests that the effect of igr deletion on cholesterol metabolism is likely because of deletion of genes in the igr locus other than cyp125. Regardless of whether all the igr genes are involved in side chain degradation, these particular activities appear to be redundant in Mtb. Finally, FadA5, a thiolase involved in cholesterol side chain degradation, was found to be essential for growth on cholesterol (9). This indicates that not all steroid side chain-degrading activities in Mtb are redundant, and that side chain degradation is necessary to support growth on cholesterol.
The occurrence of a 26-hydroxylated steroid in the mycobacterial catabolism of cholesterol catabolism is potentially significant in pathogenesis, as hydroxysteroids are important signaling molecules in mammalian systems. Specifically, 25R-26-hydroxycholesterol (also called 27-hydroxycholesterol) plays a role in at least three processes: regulating cholesterol uptake and metabolism as an LXR ligand (30, 31); preventing accumulation of cholesterol in macrophages and regulating macrophage differentiation (32); and modulating the activity of human estrogen receptors (33). The current data do not indicate whether the reaction product of Cyp125 is a 25R- or 25S-hydroxysteroid: the initial attack could be at C26, C27, or both. Whereas most sterol hydroxylases characterized to date are stereoselective, a non-stereoselective 4-cholesten-3-one 26-hydroxylase was reported in Caenorhabditis elegans (34). Furthermore, several P450s catalyze the successive monooxygenation of terminal carbons to carboxylic acids. For example, the human sterol 27-hydroxylase (Cyp27) transforms cholesterol to 25R-26-hydroxycholesterol and, at a slower rate, to the carboxylic acid derivative (35, 36). Despite the uncertainty of the absolute configuration of the Cyp125 reaction product, the signaling roles of 27-hydroxycholesterol are particularly intriguing considering that cyp125 is not essential for Mtb growth on cholesterol but contributes to the pathogen's survival in animal models (15). Thus, Cyp125 may have a role in pathogenesis beyond cholesterol catabolism.
The ability of KshB to provide reducing equivalents for the Cyp125-catalyzed reaction is consistent with genetic evidence that KshB is the cognate reductase for Cyp125 in R. erythropolis SQ1 (26). Whereas the cumulative data do not conclusively demonstrate that KshB is the cognate reductase for Cyp125 in vivo, such a role for both KshA and Cyp125 would potentially be the first described example of an oxidoreductase exhibiting physiologically relevant activity toward two classes of oxygenase.
This study presents new insights into the initial steps of cholesterol degradation in mycobacterial species. In identifying Cyp125 as a steroid 26-hydroxylase, this work also suggests that Mtb contains additional C26-oxidase activity that transforms the 26-hydroxysteroid to the carboxylic acid derivative as well as a compensatory enzyme that can initiate side chain degradation. Further characterization of these essential early steps of Mtb cholesterol metabolism will lead to better understanding of the role of this process in mycobacterial pathogenesis.
Acknowledgments
We thank Mark Okon for skilled technical assistance and Dr. Robert van der Geize for critically reading the manuscript.
This work was supported in part by grants from the Canadian Institute for Health Research (to L. D. E.), the British Columbia Lung Association (to W. W. M.), and the Michael Smith Foundation for Health Research (MSFHR) Emerging Team programs.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
T. Hsu and W. R. Jacobs, Jr., unpublished results.
J. Kriakov and W. R. Jacobs, Jr., unpublished results.
R. van der Geize, personal communication.
C. Dresen and L. D. Eltis, unpublished observations.
- Mtb
- M. tuberculosis
- BCG
- M. bovis bacilli Calmette-Guérin
- P450
- cytochrome P450
- HPLC
- high-performance liquid chromatography
- RT-qPCR
- reverse-transcriptase quantitative-polymerase chain reaction
- GC-MS
- gas chromatography-coupled mass spectrometry
- BCD
- 2-hydroxypropyl-β-cyclodextrin
- WT
- wild type
- 3β-HSD
- 3β-hydroxysteroid dehydrogenase.
REFERENCES
- 1.World Health Organization (2008) Anti-tuberculosis Drug Resistance in the World, Report 4, pp. 14–21, World Health Organization, Geneva [Google Scholar]
- 2.Munro A. W., McLean K. J., Marshall K. R., Warman A. J., Lewis G., Roitel O., Sutcliffe M. J., Kemp C. A., Modi S., Scrutton N. S., Leys D. (2003) Biochem. Soc. Trans. 31, 625–630 [DOI] [PubMed] [Google Scholar]
- 3.van der Geize R., Yam K., Heuser T., Wilbrink M. H., Hara H., Anderton M. C., Sim E., Dijkhuizen L., Davies J. E., Mohn W. W., Eltis L. D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1947–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohn W. W., van der Geize R., Stewart G. R., Okamoto S., Liu J., Dijkhuizen L., Eltis L. D. (2008) J. Biol. Chem. 283, 35368–35374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lack N. A., Yam K., Lowe E. D., Horsman G. P., Owen R. L., Sim E., Eltis L. (2009) J. Biol. Chem., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capyk J. K., D'Angelo I., Strynadka N. C., Eltis L. D. (2009) J. Biol. Chem. 284, 9937–9946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yam K. C., D'Angelo I., Kalscheuer R., Zhu H., Wang J. X., Snieckus V., Ly L. H., Converse P. J., Jacobs W. R., Jr., Strynadka N., Eltis L. D. (2009) PLoS. Pathog. 5, e1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang X., Dubnau E., Smith I., Sampson N. S. (2007) Biochemistry 46, 9058–9067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nesbitt N. M., Yang X., Fontan P., Kolesnikova I., Smith I., Sampson N. S., Dubnau E. (2009) Infect. Immun. in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pandey A. K., Sassetti C. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4376–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rengarajan J., Bloom B. R., Rubin E. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8327–8332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sassetti C. M., Rubin E. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12989–12994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schnappinger D., Ehrt S., Voskuil M. I., Liu Y., Mangan J. A., Monahan I. M., Dolganov G., Efron B., Butcher P. D., Nathan C., Schoolnik G. K. (2003) J. Exp. Med. 198, 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang J. C., Harik N. S., Liao R. P., Sherman D. R. (2007) J. Infect. Dis. 196, 788–795 [DOI] [PubMed] [Google Scholar]
- 15.Chang J. C., Miner M. D., Pandey A. K., Gill W. P., Harik N. S., Sassetti C. M., Sherman D. R. (2009) J. Bacteriol. 191, 5232–5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ouellet H., Lang J., Couture M., Ortiz de Montellano P. R. (2009) Biochemistry 48, 863–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosloniec K., Wilbrink M. H., Capyk J. K., Mohn W. W., Ostendorf M., Van der Geize R., Dijkhuizen L., Eltis L. D. (2009) Mol. Microbiol. in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonçalves E. R., Hara H., Miyazawa D., Davies J. E., Eltis L. D., Mohn W. W. (2006) Appl. Environ. Microbiol. 72, 6183–6193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bardarov S., Bardarov S., Jr., Pavelka M. S., Jr., Sambandamurthy V., Larsen M., Tufariello J., Chan J., Hatfull G., Jacobs W. R., Jr. (2002) Microbiology 148, 3007–3017 [DOI] [PubMed] [Google Scholar]
- 20.Gunsalus I. C., Wagner G. C. (1978) Methods Enzymol. 52, 166–188 [DOI] [PubMed] [Google Scholar]
- 21.Eltis L. D., Karlson U., Timmis K. N. (1993) Eur. J. Biochem. 213, 211–216 [DOI] [PubMed] [Google Scholar]
- 22.Alcalde M. A., Antelo A., Jover A., Meijide F., Tato J. V. (2009) J. Incl. Phenom. Macrocycl. Chem. 63, 309–317 [Google Scholar]
- 23.The R Development Core Team (2008) R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 24.UniProt Consortium (2009) Nucleic Acids Res. 37, D169–D174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Souidi M., Parquet M., Lutton C. (1998) Clin. Chim. Acta 269, 201–217 [DOI] [PubMed] [Google Scholar]
- 26.van der Geize R., Hessels G. I., van Gerwen R., van der Meijden P., Dijkhuizen L. (2002) Mol. Microbiol. 45, 1007–1018 [DOI] [PubMed] [Google Scholar]
- 27.Yam K., Van der Geize R., Eltis L. (2010) in The Biology of Rhodococcus (Alvarez H. M. ed) Springer, New York, in press [Google Scholar]
- 28.Behr M. A., Wilson M. A., Gill W. P., Salamon H., Schoolnik G. K., Rane S., Small P. M. (1999) Science 284, 1520–1523 [DOI] [PubMed] [Google Scholar]
- 29.Deleted in proof
- 30.Chen W., Chen G., Head D. L., Mangelsdorf D. J., Russell D. W. (2007) Cell Metab. 5, 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Javitt N. B. (2002) J. Lipid Res. 43, 665–670 [PubMed] [Google Scholar]
- 32.Hansson M., Ellis E., Hunt M. C., Schmitz G., Babiker A. (2003) Biochim. Biophys. Acta 1593, 283–289 [DOI] [PubMed] [Google Scholar]
- 33.DuSell C. D., Umetani M., Shaul P. W., Mangelsdorf D. J., McDonnell D. P. (2008) Mol. Endocrinol. 22, 65–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Motola D. L., Cummins C. L., Rottiers V., Sharma K. K., Li T., Li Y., Suino-Powell K., Xu H. E., Auchus R. J., Antebi A., Mangelsdorf D. J. (2006) Cell 124, 1209–1223 [DOI] [PubMed] [Google Scholar]
- 35.Betsholtz I. H., Wikvall K. (1995) J. Steroid Biochem. Mol. Biol. 55, 115–119 [DOI] [PubMed] [Google Scholar]
- 36.Cali J. J., Russell D. W. (1991) J. Biol. Chem. 266, 7774–7778 [PubMed] [Google Scholar]