Abstract
Pmel17 is a transmembrane protein that mediates the early steps in the formation of melanosomes, the subcellular organelles of melanocytes in which melanin pigments are synthesized and stored. In melanosome precursor organelles, proteolytic fragments of Pmel17 form insoluble, amyloid-like fibrils upon which melanins are deposited during melanosome maturation. The mechanism(s) by which Pmel17 becomes competent to form amyloid are not fully understood. To better understand how amyloid formation is regulated, we have defined the domains within Pmel17 that promote fibril formation in vitro. Using purified recombinant fragments of Pmel17, we show that two regions, an N-terminal domain of unknown structure and a downstream domain with homology to a polycystic kidney disease-1 repeat, efficiently form amyloid in vitro. Analyses of fibrils formed in melanocytes confirm that the polycystic kidney disease-1 domain forms at least part of the physiological amyloid core. Interestingly, this same domain is also required for the intracellular trafficking of Pmel17 to multivesicular compartments within which fibrils begin to form. Although a domain of imperfect repeats (RPT) is required for fibril formation in vivo and is a component of fibrils in melanosomes, RPT is not necessary for fibril formation in vitro and in isolation is unable to adopt an amyloid fold in a physiologically relevant time frame. These data define the structural core of Pmel17 amyloid, imply that the RPT domain plays a regulatory role in timing amyloid conversion, and suggest that fibril formation might be physically linked with multivesicular body sorting.
Introduction
Pmel17 (also known as gp100 or SILV) is a pigment cell-specific protein involved in the initial steps in the biogenesis of the lysosome-related organelle, the melanosome (1, 2). Within melanosome precursor organelles, Pmel17 forms a fibrillar matrix over which melanin pigments are deposited as they are synthesized in later stages of melanosome development. The function of the Pmel17 fibrils is not entirely clear, but several studies suggest that they play a cytoprotective role by sequestering toxic intermediates produced during melanin synthesis and/or by templating and accelerating melanin production (3, 4). Consistent with this notion, mutations in Pmel17 result in pigment dilution in a number of animal models, at least some of which are associated with poor health or viability of melanocytes (5–11). Moreover, Pmel17 fibrils from purified melanosomes bind amyloidogenic dyes such as Congo red and thioflavin S, and fibrils formed from purified recombinant Pmel17 isoforms resemble cross-β-sheet amyloid fibrils by a number of criteria (e.g. a cross-β-sheet x-ray fibril diffraction pattern), indicating that Pmel17 fibrils are a functional form of amyloid (4). Thus, understanding how Pmel17 fibrils form physiologically without promoting cytotoxicity might provide insight into pathologic amyloidogenic processes, such as those that occur in Alzheimer disease and the prion-dependent spongiform encephalopathies.
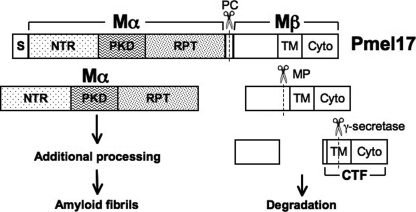
To understand how Pmel17 is converted to amyloid, it is essential to appreciate the cellular and structural features that govern this conversion (2). Pmel17 is synthesized as a type 1 integral membrane glycoprotein featuring a single transmembrane domain and a large lumenal domain exposed within the lumen of the endoplasmic reticulum (12). After terminal glycosylation in the Golgi apparatus, Pmel17 is eventually delivered to early endosomes (13), most likely indirectly after delivery to the plasma membrane and subsequent internalization; internalization is facilitated by interaction of a dileucine-based signal in the Pmel17 cytoplasmic domain with the clathrin adaptor AP-2 (14, 15). Within endosomes, Pmel17 partitions to membrane microdomains that invaginate to form intralumenal vesicles (ILVs)3 (1, 13). In association with the invaginating membranes and the ILVs, Pmel17 is proteolytically cleaved first by furin or a related proprotein convertase (16, 17) and then by a metalloproteinase (18), leading to the release of the amyloidogenic lumenal fragment, Mα, from the membrane-containing Mβ fragment and subsequent membrane-associated degradation products (Fig. 1). Once released from Mβ, Mα is only detected in subcellular fractions that are insoluble in nonionic detergents and that correspond to the melanosomal fibrils (16), consistent with its efficient formation of amyloid. The Mα fragment is further proteolyzed within maturing melanosomes (19–21), whereas the membrane-bound fragment that results from metalloproteinase cleavage of Mβ is a substrate for γ-secretase cleavage (18), likely facilitating the degradation of the remaining fragments (Fig. 1). This sequence of sorting and processing events is essential for fibril formation, as mutations that prevent either Pmel17 proteolytic cleavage or delivery to ILVs inhibit fibril formation (16–18, 22). Importantly, neither fibrils nor amorphous aggregates are observed in early biosynthetic compartments such as the endoplasmic reticulum or Golgi apparatus, indicating that the critical amyloidogenic switch occurs on ILVs within vacuolar early endosomes from which early stage melanosomes derive (13, 23). Within melanocytes, the fibrils begin to form in association with ILVs (23), suggesting that ILV contents might function to promote fibril formation. A similar role for ILVs has been implicated in the Alzheimer disease-associated proteolytic processing of amyloid precursor protein and the subsequent amyloid conversion of the resulting Aβ peptide (24–26). The membrane composition of ILVs, which is distinct from that of the endosomal limiting membrane (27–31), might potentially facilitate Pmel17 proteolytic processing and/or allow for a conformational switch that exposes the amyloidogenic core of Pmel17 to initiate fibril formation. To begin to dissect the nature and requirements of this conformational switch, it is critical to define the substructure of Pmel17 that serves as the core of its cross-β-sheet amyloid fibrils.
FIGURE 1.
Schematic representation of Pmel17 proteolytic processing. Shown is a scheme for the primary structure of human Pmel17 and its component domains. Full-length Pmel17 is cleaved first by furin or a related proprotein convertase (PC) into Mα and Mβ fragments. Mα is incorporated into amyloid fibrils and is further proteolytically processed during fibril maturation. Mβ is cleaved by a metalloproteinase (MP), and the resulting C-terminal fragment (CTF) is a target for γ-secretase cleavage, likely facilitating the degradation of Mβ-derived fragments in lysosomes and/or proteasomes. S, signal sequence; NTR, N-terminal region; PKD, polycystic kidney disease protein homology domain; RPT, repeat domain; TM, transmembrane domain; Cyto, cytoplasmic domain.
The functional properties of Pmel17 can be ascribed to individual subdomains that have been defined based on structural features deduced from its primary amino acid sequence (Fig. 1) (2). The fibrillogenic Mα fragment consists of three subdomains. An N-terminal region (NTR), homologous to a similarly placed domain in gpNmb (32), lacks homology to identifiable protein domains but contains two potential N-glycosylation sites and several cysteine residues that are likely engaged in disulfide bonds in the native protein. Following the NTR is a domain with significant homology to a repeated domain found in the extracellular region of the polycystic kidney disease (PKD) protein, polycystin-1, which has a β-pleated sheet-rich immunoglobulin-like fold (33, 34). The NTR and PKD domains are required for Pmel17 sorting onto ILVs within multivesicular endosomes (17, 22) and therefore for facilitating proprotein convertase cleavage. Thus, in the absence of these domains, fibrils do not form in vivo, at least in part because Pmel17 is not processed correctly and fails to reach the microenvironment conducive for fibril assembly. Immediately downstream of the PKD domain is a highly O-glycosylated RPT domain, consisting of a series of 10 imperfect direct repeats of 13 residues each rich in proline, serine, threonine, and glutamic acid residues (20, 35). Deletion of the RPT domain does not influence Pmel17 distribution within endosomes but ablates fibril formation in vivo (17, 22). Melanosome fibrils are reactive with antibodies to both the RPT and PKD domains (13, 20), suggesting that both might be critical for fibril formation. Because of the sorting requirement for the NTR and PKD domains, however, a direct assessment of the involvement of these domains in fibrillogenesis in vivo is not possible.
To define the subdomains that likely participate directly in fibril formation in vivo, we took an in vitro approach similar to that used by Fowler et al. (4). Using a variety of biophysical and spectroscopic approaches, we show that, contrary to a recent report (36), the unglycosylated RPT domain in isolation is very soluble, protease-sensitive, and unable to form amyloid fibrils in aqueous solution within the biologically relevant experimental time frames employed for the other Pmel17 domains. This finding suggests that the RPT domain likely plays a regulatory rather than a direct structural role in Pmel17 fibril formation. In striking contrast, both the NTR and the predicted β-sheet-rich PKD domain are amyloidogenic and contribute to the amyloid-like properties of Mα. Moreover, a fragment of the PKD domain cofractionates with melanosomal fibrils isolated from melanocytes. These results suggest that the PKD domain, perhaps in conjunction with the NTR, participates directly as the core of the Pmel17 amyloid fibril and that both domains have dual functions, contributing to both Pmel17 sorting and fibril formation.
EXPERIMENTAL PROCEDURES
Reagents
All reagents were obtained from Sigma or Thermo Fisher Scientific unless otherwise specified.
Antibodies
Affinity-purified polyclonal rabbit anti-peptide antibodies recognizing the N terminus (αPmel-N (16)) and C terminus of Pmel17 (αPep13h (1)) were described previously. Monoclonal antibodies recognizing the RPT (HMB45) and PKD (HMB50) domains of Pmel17 (20, 22) were purchased from LabVision/Thermo Fisher Scientific (Fremont, CA). I51 was a kind gift of P. Cresswell, N. Vigneron, and R. M. Leonhardt (Yale University, New Haven, CT) and was generated by Proteintech Group, Inc. (Chicago, IL), to a peptide with sequence AFTITDQVPFSVSVSGGC, corresponding to Pmel17 residues 206–220 (within the PKD domain) appended with a glycine-glycine linker and a cysteine, conjugated to keyhole limpet hemocyanin.
Analysis of Cell-derived Detergent-soluble and -insoluble Fractions
Human HeLa (cervical carcinoma) and MNT-1 melanoma cells were grown, harvested, solubilized with Triton X-100, and fractionated into Triton X-100-soluble and -insoluble fractions as described previously (16). Proteins were separated by SDS-PAGE, transferred to Immobilon-P membranes (Millipore, Billerica, MA), and probed with antibodies directed against different Pmel17 subdomains. Bands were detected with alkaline phosphatase-conjugated secondary antibodies, enhanced chemifluorescence, and phosphorimaging analysis using a Storm 860 fluorescence imaging system, and ImageQuant software (GE Healthcare).
Immunoblotting and Immunoelectron Microscopy Analyses of Partially Purified Fibril Fractions
Triton X-100-insoluble fibril-enriched fractions were purified from a dense membrane fraction of MNT-1 melanoma cells as described previously (16). Briefly, postnuclear supernatants from MNT-1 cell homogenates were layered on a cushion of 2 m sucrose and centrifuged at 11,000 × g. Dense membranes collected from the interface were isolated by centrifugation at 100,000 × g. These dense membranes were then solubilized with 1% (w/v) Triton X-100, and insoluble material was separated from detergent-soluble material by a second centrifugation step at 20,000 × g. For immunoblotting, equal cell equivalents from each fraction were fractionated by SDS-PAGE and analyzed as described above. For immunoelectron microscopy, detergent-insoluble pellets were resuspended in phosphate-buffered saline, adhered to Formvar-coated grids, labeled with the indicated antibodies and protein A-conjugated 10-nm gold particles, and analyzed on a Philips CM120 electron microscope (FEI, Eindoven, The Netherlands) after contrasting and embedding in a mixture of uranyl acetate and methlycellulose. Digital acquisitions were made with a numeric Keen View camera (Soft Imaging System, Muenster, Germany).
Cloning, Protein Expression, and Purification
cDNAs encoding fragments of human Pmel17, bordered by BspHI/XhoI (Mα, ΔPKD, ΔRPT, and NTR) or NcoI/XhoI (ΔNTR, PKD, and RPT) restriction sites, were generated from pCI-Pmel17 (37) by thermal cycling amplification, using the Expand High Fidelity PCR system (Roche Applied Science), and corresponded to the following residues of the full-length protein: Mα (Lys25–Val467), NTR (Lys25–Ser205), PKD (Ala201–Arg314), RPT (Ser303–Val467), ΔNTR (Mα lacking Lys25–Leu200), ΔPKD (Mα lacking Ala206–Leu292), and ΔRPT (Mα lacking Pro315–Asp440). The fragments were subcloned into pET28a(+) (Novagen-EMD Biosciences, Gibbstown, NJ) for C-terminal hexahistidine (His6) tag fusions lacking the N-terminal His6 or T7 tags. Full-length Pallidin was subcloned into the EcoRI/SalI sites of pET28a(+) for expression with the N-terminal His6 and T7 tags instead of the C-terminal tag. All sequences were verified by dideoxy sequencing (University of Pennsylvania Cell Center, Philadelphia). The Sup35 amyloid-competent subdomain fused to a C-terminal His7 tag (NM-His) has been described previously (38). BL21 Escherichia coli expressing these constructs were grown at room temperature to an A600 = 0.4–0.6 and induced overnight with 1 mm isopropyl 1-thio-β-d-galactopyranoside at room temperature, except for NM-His-expressing bacteria which were grown and induced 3–4 h at 37 °C. Bacteria were harvested by centrifugation, lysed by lysozyme treatment and probe sonication, and fractionated into soluble and insoluble pools by centrifugation. Inclusion bodies were obtained as described previously (39) with some modifications. The insoluble fraction, representing primarily inclusion bodies, was washed three times with phosphate-buffered saline, 2 m urea, 2% (w/v) Triton X-100, and once with phosphate-buffered saline and then solubilized with 6 m guanidine HCl, 0.1 m NaH2PO4/Na2HPO4, 10 mm imidazole, pH 8.0, and subjected to centrifugation for 1 h at 100,000 × g at 4 °C to obtain a supernatant containing solubilized inclusion body proteins. His-tagged proteins were affinity-purified from solubilized inclusion body fractions under denaturing conditions using HisSelect resin (Sigma), according to the manufacturer's instructions. In the case of the soluble RPT domain, protein was purified under denaturing conditions from whole cell lysates rather than inclusion body fractions. Purified proteins were precipitated with methanol for long term storage (40) and resuspended in 8 m urea, 50 mm Tris-HCl, pH 7.5, prior to renaturation in aqueous buffer. Protein concentration was determined by A280 based on the calculated extinction coefficient for each protein (39).
Renaturation of Purified Proteins
Refolding and/or fibrillization was initiated by diluting a concentrated stock of affinity-purified protein into “physiological” assay buffer (5 mm KH2PO4/K2HPO4, 150 mm NaCl, pH 7.4) and incubating for varying times at 37 °C with agitation at 225 rpm. The final concentration was 10 μm unless otherwise indicated.
Sedimentation Analyses
Renatured protein reactions were sedimented by centrifugation at 100,000 × g for 1 h at 4 °C. Proteins from either the supernatant (soluble protein) or the insoluble pellet were diluted into SDS sample buffer, boiled, and analyzed by SDS-PAGE followed by Coomassie Blue staining. The relative intensities of the relevant band in each fraction were quantified using ImageQuant software (GE Healthcare).
Thioflavin T Binding Assays
An aliquot of renatured protein was diluted into glycine-NaOH, pH 9.0, containing thioflavin T (ThioT) to a final concentration of 2 μm protein and 200 μm ThioT. As a control for background fluorescence, assay buffer alone was diluted into glycine-NaOH buffer containing ThioT. In assays using proteins at different concentrations, 400 μm ThioT was used to saturate binding. Fluorescence emission at 490 nm (excitation, 440 nm; cutoff, 475 nm) was measured immediately after mixing, using a SpectraMax Gemini fluorometer and SoftMax Pro 4.0 software (Molecular Devices, Sunnyvale, CA). All assays were performed in triplicate.
Congo Red Binding Assays
An aliquot of renatured protein was combined with Congo red (CR) in assay buffer to a final concentration of 1 μm protein and 10 μm CR. Absorbance was measured at 477 and 540 nm using an Ultrospec 2000 spectrophotometer (Amersham Biosciences). All measurements were normalized to assay buffer alone combined with CR. The ratio of moles CR bound/mol of protein was calculated using the equation R = A540/25,295 − A477/46,306, as described previously (41). All assays were performed in triplicate.
Electron Microscopy of Recombinant Proteins
For analysis of recombinant protein morphology, renatured protein was centrifuged at 100,000 × g for 1 h at 4 °C, and the protein pellets were resuspended in a small volume of assay buffer and adsorbed onto 300-mesh Formvar-coated copper grids (Electron Microscopy Sciences, Hatfield, PA). Samples were negatively stained with 1% aqueous uranyl acetate (Electron Microscopy Sciences), and visualized with a Tecnai G2 transmission electron microscope (FEI, Hillsboro, OR) coupled to a Gatan digital camera system (Pleasanton, CA).
Electron Tomography of MNT-1 Cells
Thick (350 nm) sections of high pressure frozen, freeze-substituted MNT-1 cells were cut on a Reichert Ultracut S microtome (Leica Microsystems, Vienna, Austria) and collected on Formvar-coated copper grids (75 mesh) for analysis by electron tomography as described previously (23). In brief, sections were randomly labeled on the two sides with 10 nm of protein A-conjugated gold particles and post-stained with 2% uranyl acetate in methanol for 4 min and lead citrate for 2 min. Finally, tilt series (two perpendicular series per tomogram, angular range from −60 to +60° with 1° increment) were recorded using Xplore3D (FEI Co.) on a 200-kV transmission electron microscope (Tecnai 20 LaB6, FEI Co.) and used for reconstructing tomograms.
X-ray Diffraction Analyses
Affinity-purified proteins in 8 m urea, 50 mm Tris-HCl, pH 7.5, were dialyzed against deionized H2O, and aggregates were lyophilized until dry. Powder diffraction of proteins in quartz capillary tubes was carried out as described previously (4).
Proteinase K Digestion and Automated Edman Protein Sequencing
Proteinase K was added to renatured protein in assay buffer and incubated at 37 °C. Reactions were terminated by addition of phenylmethylsulfonyl fluoride, followed by boiling in SDS sample buffer. Proteinase-resistant fragments were fractionated by SDS-PAGE on 12.5 or 15% Tris-Tricine gels. In some experiments, bands were directly visualized by Coomassie Blue staining. To identify the protease-resistant cores, fractionated proteins were transferred to Immobilon-P membranes and visualized by Coomassie Blue staining and then excised and subjected to automated Edman protein sequencing by the Texas A&M University Protein Chemistry Laboratory (Department of Biochemistry, College Station, TX). The deduced sequences were manually aligned with the predicted amino acid sequence of Pmel17.
RESULTS
Melanocyte-derived Fibrils Are Enriched in Proteolytic Fragments of Both the PKD and RPT Domains
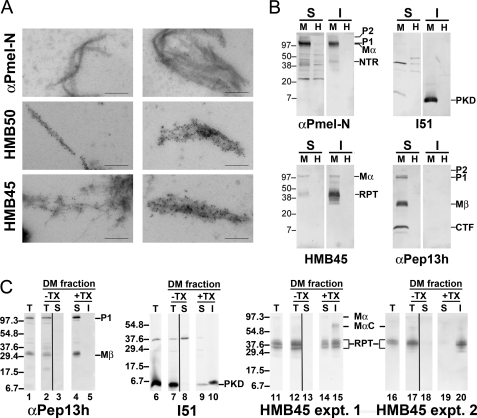
Melanosome fibrils are reactive by immunofluorescence and immunoelectron microscopy with antibodies to both the PKD and RPT domains (13, 16, 20), but due to the properties of the antibodies used thus far, only RPT-containing fragments, which are heavily O-glycosylated in vivo and thus protease-resistant (35), have been identified by biochemical analysis of detergent-insoluble fibril-enriched fractions isolated from melanosome-containing subcellular fractions (16, 20–22, 35). To determine whether other Pmel17 fragments are included in the fibrils, we assayed for immunoreactivity to a panel of anti-Pmel17 antibodies within an isolated fibril-enriched fraction from a human melanocytic cell line, MNT-1, in which there are numerous stage II melanosomes (13).
To first test whether isolated Pmel17 fibrils react with antibodies to distinct Pmel17 domains, a dense membrane fraction, enriched in stage II melanosomes, was isolated from postnuclear supernatants of MNT-1 cell homogenates as described previously (16). The fraction was treated with Triton X-100 to solubilize the membranes and then subfractionated into a detergent-soluble fraction, containing most cytoplasmic and membrane proteins, and a detergent-insoluble fraction in which Pmel17 fibrils and melanin granules are enriched (16, 22). Immunoelectron microscopy analysis of the detergent-insoluble fractions shows that the melanosomal fibrils are densely labeled with antibodies specific to both the PKD (HMB50) and RPT (HMB45) domains but not to the N terminus (αPmel-N) (Fig. 2A). This indicates that both the PKD and RPT domains are enriched within the fibrils either as part of a larger fragment or as separated domains.
FIGURE 2.
Melanosome-derived fibrils are enriched in PKD and RPT domain-containing fragments. A, dense membrane fraction from MNT-1 melanoma cell homogenates was solubilized with Triton X-100, and the insoluble fraction, enriched in melanin and melanosome fibrils, was labeled with the indicated antibodies and protein A-conjugated gold particles, and then analyzed by electron microscopy. Note that both thin immature fibrils and mature fibril sheets are recognized by antibodies HMB50 (to the PKD domain) and HMB45 (to the RPT domain), but not αPmel-N (to the N-terminal peptide), indicating that they are enriched in PKD and RPT-containing fragments but lack the N terminus. Scale bars, 200 nm. B, human MNT-1 melanoma (M) and nonmelanocytic HeLa (H) cells were lysed with 1% Triton X-100 and fractionated into Triton X-100-soluble (S) and -insoluble (I) cell fractions, the latter enriched in melanosome fibrils. Fractions were subjected to SDS-PAGE followed by immunoblotting using antibodies raised against the NTR (αPmel-N), PKD (I51), RPT (HMB45), and the cytoplasmic (αPep13h) domains of Pmel17. Note that the fragments reactive with antibodies to the RPT and PKD domains in the insoluble fractions of MNT-1 cells migrate with different molecular weights, indicating that these domains are proteolytically separated within Triton X-100-insoluble fibrils. C, MNT-1 melanoma cell homogenates (T for total; lanes 1, 6, 11, and 16) were fractionated by differential sedimentation on a 2 m sucrose cushion, and a dense membrane (DM) fraction was isolated as in A (T for total; lanes 2, 7, 12, and 17). Membranes in this fraction were collected by centrifugation at 100,000 × g for 1 h; supernatants (S) were also collected (lanes 3, 8, 13, and 18). The membranes were then treated with 1% Triton X-100 (TX), and the detergent-soluble (S; lanes 4, 9, 14, and 19) and -insoluble (I; lanes 5, 10, 15, and 20) fractions were separated by sedimentation at 20,000 × g for 20 min. Equal cell equivalents of each fraction were further fractionated by SDS-PAGE and analyzed by immunoblotting with the indicated antibodies. For HMB45 immunoblots, results are shown from two different experiments, each of which was representative of at least two separate repetitions.
The absence of NTR reactivity within the fibril fractions could be due either to degradation of this domain during fibril maturation or to sequestration of the epitope recognized by αPmel-N by immersion of this domain within the core of Pmel17 fibrils. To distinguish these possibilities and to better characterize the nature of the PKD and RPT domain-containing species, we analyzed fibril-enriched fractions by immunoblotting using antibodies directed against each of the subdomains of Mα. MNT-1 cells were lysed in Triton X-100, and cell lysates were fractionated into a soluble fraction and a fibril-enriched Triton X-100-insoluble fraction. To ensure specificity of detection by the anti-Pmel17 antibodies, nonmelanocytic HeLa cells, which do not express Pmel17, were fractionated and treated in the same way. Triton X-100-soluble and -insoluble fractions were denatured by boiling in SDS, which at least partially solubilizes the fibrils, and analyzed by SDS-PAGE and immunoblotting with antibodies to the N and C termini, to the RPT domain, and to a peptide spanning residues 206–220, corresponding to the N terminus of the PKD domain (Fig. 2B). Transfer conditions were optimized to detect lower molecular weight fragments that might arise from proteolytic processing of the higher molecular weight precursors. As shown previously, the Triton X-100-soluble fractions of MNT-1 cells are enriched in the immature precursor P1 form of full-length Pmel17 (reactive with αPmel-N and αPep13h, antibodies to the N and C termini, respectively), the membrane-associated Mβ and C-terminal fragments (products of proprotein convertase and metalloproteinase cleavage and reactive with αPep13h), and the full-length Golgi-modified P2 form (detected with αPmel-N and the RPT domain-reactive antibody, HMB45; see Ref. 20, for identification of these bands). By contrast, most of the fibrillogenic full-length Mα fragment (detected by αPmel-N and HMB45) is present within the fibril-enriched Triton X-100-insoluble fraction (note that P1 and Mα migrate similarly on the high percentage polyacrylamide gels used in these assays; their identity within each fraction is deduced from historical data; see Refs, 1, 16, 20). Nevertheless, as shown previously (19–22), the most prevalent bands detected by HMB45 in the Triton X-100-insoluble fraction are proteolytic digestion products that harbor the RPT domain and migrate with Mr 35,000–45,000. Importantly, antibodies directed against the NTR (αPmel-N) and PKD (I51) domains also recognize proteolytic Pmel17 fragments in the detergent-insoluble fraction of MNT-1 cells (Mr 45,000 and 7,000, respectively) but not from strictly analogous fractions from HeLa cells, indicating that the melanosome fibrils contain fragments derived from each of these domains. These fragments migrate with discrete molecular weights, indicating that they were separated by proteolytic processing within melanosomes or melanosome precursors. The proteolytic fragments are the predominant species detected by antibodies to the PKD and RPT domains, indicating that they are enriched relative to full-length Mα within the fibrils (note that in separate experiments not shown using different transfer conditions, P1 and Mα were detectable using I51 but were still less prevalent than the ∼7-kDa band). By contrast, despite using transfer conditions that favor detection of smaller fragments, the NTR-containing fragment detected by αPmel-N is much less intense than full-length Mα, indicating that it is not enriched. This result indicates that the lack of reactivity by αPmel-N in our immunoelectron microscopy analyses is not due to epitope sequestration but rather to proteolytic loss of the Pmel17 N terminus. Collectively, these data show that each of the three Mα lumenal domains are liberated from each other in melanosomes by proteolytic cleavage and that the PKD and RPT domains are both components of assembled fibrils. We cannot exclude the possibility that NTR fragments lacking the N terminus are also components of the fibrils.
To extend these analyses and determine whether either the PKD- or RPT-derived fragments could be dissociated from fibrils, we isolated melanosome-enriched subcellular fractions from MNT-1 cell homogenates, treated these fractions with Triton X-100, and similarly assayed for the presence of the PKD- and RPT-derived fragments in Triton X-100-soluble and -insoluble fractions by immunoblotting (Fig. 2C). We reasoned that the process of subcellular fractionation might release fragments that were not integral components of the fibrils, consistent with the ability of amyloidogenic dyes to label purified melanosomes but not melanosomes in situ (4). Consistent with results from whole cell lysates, P1 and Mβ (detected with αPep13h) were recovered exclusively in the detergent-soluble fraction (Fig. 2C, compare lanes 4 and 5), and the PKD domain-derived fragment with Mr 7,000 was always detected nearly exclusively in the detergent-insoluble fraction (compare lanes 9 and 10; note that an ∼38-kDa contaminant was recovered from the cytoplasmic fraction in lane 8 and not from the dense membrane fraction in lane 7). Interestingly, whereas in some experiments the HMB45-reactive Mr 35,000–45,000 RPT domain fragments were also recovered exclusively in detergent-insoluble pellets from the melanosome-enriched fraction (e.g. experiment 2, Fig. 2C, compare lanes 19 and 20), in other experiments these fragments were recovered nearly equally within the detergent-soluble supernatants (e.g. experiment 1, Fig. 2C compare lanes 14 and 15). The nature of the conditions that distinguish these two outcomes is not clear but appears to correlate with the degree of pigmentation within the starting cell population. Regardless, the latter result suggests that during preparation of the fibril fractions, the RPT domain fragments can be released from the fibrils and thus are unlikely to serve as the fibril core.
RPT Domain of Pmel17 Confers Solubility to Mα in Vitro
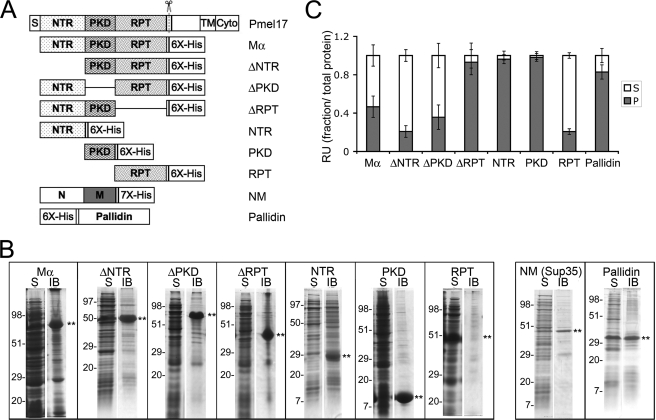
As indicated above, both the PKD and RPT domains are components of melanosome fibrils (Fig. 2). Previous studies have indicated that upon deletion of the RPT domain, Pmel17 is unable to form fibrils in cells (17, 22). Similarly, deletion of either the NTR or PKD domains results in loss of fibril formation, but because these domains are required for localization of Pmel17 to ILVs of endosomes and subsequent proteolytic activation, it is not known whether these domains also play a structural role in fibril formation. To directly assess how each of these subdomains participates in fibril formation, we took advantage of an in vitro approach used previously to demonstrate that a recombinant form of Mα, synthesized in bacteria, forms amyloid fibrils upon dilution out of denaturant (4). To delimit the region within Mα that forms the core of the amyloid fibrils, we similarly analyzed the in vitro amyloidogenic behavior of C-terminally hexahistidine (His6)-tagged constructs consisting of full-length Mα, Mα with deletions in the NTR, PKD, or RPT domains, or each domain in isolation (Fig. 3A). As a positive amyloidogenic control, analyses included a His-tagged form of the prion-competent subdomain, NM, of the well studied yeast prion protein Sup35 (40). As a negative control, analyses included a His-tagged isolated subunit (Pallidin) of the obligate multisubunit protein complex, BLOC-1 (42, 43). Pallidin is expected to be misfolded in the absence of its partner subunits but is not anticipated to efficiently form amyloid.
FIGURE 3.
Solubility of recombinant Pmel17 His-tagged constructs. A, schematic diagram of full-length Pmel17 and C-terminally His-tagged recombinant lumenal domain fragments. Δ denotes that the indicated domain has been deleted from full-length Mα. His6, hexahistidine tag on recombinant proteins. Also shown is the His7-tagged prion-forming subdomain (NM) of the yeast prion protein Sup35, used as a positive amyloid control, and the His6-tagged full-length Pallidin, used as a negative control. B, partitioning into soluble and insoluble bacterial fractions. BL21 E. coli expressing the different proteins indicated in A were harvested and processed as described under “Experimental Procedures.” The soluble (S) and insoluble IB fractions were separated by SDS-PAGE and analyzed by Coomassie Blue staining. Asterisks denote the position of the induced protein, and the migration of molecular weight standards is indicated to the left of each pair of lanes. Note that all of the Pmel17-derived proteins are found predominantly in the IB fraction with the exception of the RPT, which is found predominantly in the soluble fraction. C, sedimentation. Each of the recombinant proteins was solubilized from inclusion bodies in guanidine HCl and affinity-purified by His-bind chromatography. Affinity-purified protein was diluted out of the denaturant into physiological buffer and allowed to refold overnight with agitation at 37 °C. Aliquots were fractionated into a soluble supernatant (S) and insoluble pellet (P) by centrifugation at 100,000 × g for 1 h at 4 °C. Total, supernatant, and pellet fractions were analyzed by SDS-PAGE, stained with Coomassie Blue, and image scanned; the relative amount of protein in each fraction, assessed as signal intensity, was determined using ImageQuant software. The mean fraction of protein in the supernatant and pellet fractions relative to the total is plotted ± S.D. n = 3 for Pallidin, n ≥ 4 all others. RU, relative unit.
Each of the recombinant proteins was expressed in E. coli. Upon cell lysis, most of the Pmel17 constructs and the NM-positive control cofractionated predominantly with the insoluble inclusion body (IB) fraction (Fig. 3B). The sole exception was the isolated RPT domain, which partitioned primarily with the soluble fraction (Fig. 3B), inconsistent with the typical behavior of amyloidogenic polypeptides. The negative control Pallidin partitioned equally with the soluble and inclusion body fractions (Fig. 3B). The insolubility of Pmel17 fragments containing the NTR or PKD domains is consistent with the potential ability of these domains to form amyloid fibrils (44).
Because most of the constructs studied fractionated with IBs, we solubilized and purified each protein from the IB fraction under denaturing conditions, and we then diluted out the denaturant with a physiological buffer to initiate refolding and/or fibrillogenesis for 16 h. Most of the constructs treated in this manner were partially or totally insoluble and were pelleted by sedimentation at 100,000 × g for 1 h (Fig. 3C); the RPT domain was a notable exception, remaining largely in the supernatant. Moreover, whereas renatured Mα remained partially soluble under these conditions, deletion of the RPT domain caused a nearly complete shift into the pellet fraction. By contrast, the isolated NTR and PKD domains partitioned predominantly in the pellet fraction, and deletion of each of these domains from Mα resulted in a greater partitioning to the supernatant. These results indicate that the RPT domain confers partial solubility to Mα, whereas the NTR and PKD domains both contribute to its insolubility. These results are consistent with the potential of the NTR and PKD, but not the RPT, to form amyloid upon dilution out of the denaturant.
NTR and PKD Domains of Mα Have Amyloid-like Properties
Although insolubility in aqueous buffers is a property of amyloid, it is not unique to amyloid. To specifically assess amyloid formation, the following assays were performed with Pmel17 fragments and control proteins that were dissolved and monomerized in chaotropic solution and then allowed to refold or aggregate in buffer resembling physiological conditions.
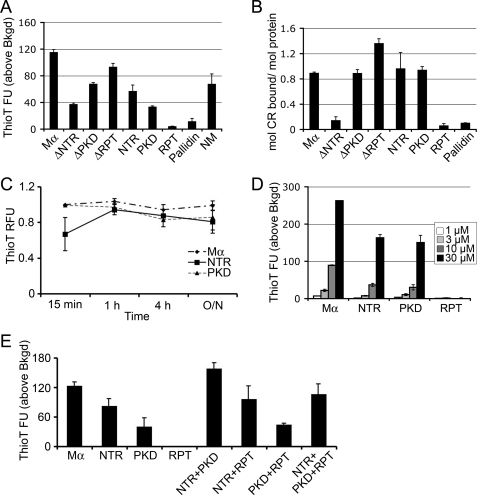
We first tested whether the isolated Mα subdomains or internal deletions of Mα, like full-length Mα, bind to amyloidophilic dyes. Addition of either the isolated NTR or PKD domain to solutions containing ThioT or CR leads to a large increase in fluorescence (ThioT) or absorption (CR). By contrast, the RPT domain does not alter the spectroscopic properties of either amyloidophilic dye significantly relative to background (Fig. 4, A and B). Consistent with these results, deletion of the RPT domain from Mα (ΔRPT) has minimal effect on dye binding, whereas deletion of the NTR (ΔNTR), and to a lesser extent the PKD domain (ΔPKD), results in a partial loss of dye binding. These data support the notion that in vitro the NTR and PKD domains, but not the RPT domain, have amyloidogenic potential.
FIGURE 4.
Amyloid dye binding properties of the NTR and PKD domains, but not the RPT domain, resemble those of Mα. A, ThioT fluorescence analysis of Mα subdomains. Affinity-purified proteins in denaturant were diluted into physiological assay buffer and incubated for 16 h at 37 °C with agitation to initiate refolding and/or fibrillogenesis. Aliquots were combined with ThioT, and fluorescence emission at 490 nm was measured upon excitation at 440 nm. Columns represent the mean fluorescent units (FU) above background (ThioT alone; average value, 20 fluorescent units) ± S.E. from at least three experiments. B, Congo red binding of Mα subdomains. Renatured proteins were prepared as in A and then combined with Congo red and analyzed by light spectroscopy. Plotted are the moles of CR bound/mol of protein, as determined according to Ref. 41. Bars represent mean ± S.E. from at least three experiments. C, time dependence of ThioT binding for Mα subdomains. Solubilized proteins were diluted out of denaturant and incubated for the indicated times, after which ThioT was added, and fluorescence emission was measured. ThioT fluorescence intensity was normalized relative to the maximum fluorescence intensity observed. Bars represent mean ± S.E. from at least two experiments done in triplicate. Values for each of the proteins in 8 m urea (“time 0”) were negligible relative to ThioT alone. RFU, relative fluorescent units. D, protein concentration dependence on ThioT binding and fluorescence. Increasing concentrations (as indicated) of protein prepared as in A were combined with ThioT, and fluorescence emission was measured and plotted in the bar graph. Bars represent mean ± S.D. from a representative experiment. E, ThioT fluorescence of Mα subdomains alone or in combination. Solubilized proteins were diluted out of denaturant and incubated at 37 °C overnight either alone or in combination as indicated (10 μm final concentration of each protein). ThioT was added at the end of the incubation, and fluorescence emission at 490 nm was measured. Columns represent ThioT fluorescence above background from a representative experiment performed in triplicate ± S.D. Note that the signal from each combination is roughly equivalent to the sum of the signals from each component, suggesting lack of significant synergy.
Many pathological amyloidogenic peptides and proteins only form fibrils at high concentration and after extensive incubation times due to the slow kinetics of fibril nucleation. By contrast, Mα forms fibrils rapidly upon removal of denaturant (4). To determine whether this property can be ascribed to any of the subdomains, we tested the time and concentration dependence of ThioT binding by Mα subdomains. Like Mα, the isolated NTR and PKD domains display maximum dye binding within minutes (Fig. 4C). By comparison, the yeast prion protein NM requires 48 h of incubation before reaching maximum ThioT binding (supplemental Fig. S1A). In contrast to the PKD and NTR domains, the RPT domain is unable to bind ThioT above background even after 48 h (supplemental Fig. S1B). Moreover, whereas ThioT binding by Mα and the isolated NTR and PKD domains is directly proportional to protein concentration (Fig. 4D), the isolated RPT domain does not bind ThioT above background even at the highest concentrations used. Finally, ThioT bound equally well to the isolated PKD domain and full-length Mα over a pH range of 5.4–9.0, and binding to the NTR was half-maximal below neutral pH; however, ThioT did not bind to equivalent concentrations of the RPT domain significantly above background at any pH tested (supplemental Fig. S2). These data suggest that the NTR and PKD domains both contribute to Mα amyloid dye binding properties but that the RPT domain is not part of the amyloid core.
It was surprising to us that both NTR and PKD domains showed amyloidogenic potential by the dye binding assays, particularly given the lack of reactivity of melanocyte fibril fractions with the αPmel-N antibody (Fig. 2). To determine whether the NTR and PKD domains act independently of each other or synergistically and whether the RPT domain influences amyloidogenesis, we performed fibril assembly assays with each of the domains individually or in combination. As shown in Fig. 4E, the level of dye binding in samples containing both the NTR and PKD domains together is approximately the sum of the levels of dye binding by each domain alone, showing perhaps a modest synergistic effect. Moreover, the addition of the RPT domain does not influence dye binding by either the NTR or PKD domain and slightly inhibits the dye binding of the NTR + PKD combination. These data indicate that each of the NTR and PKD domains has independent amyloidogenic potential that is not dramatically influenced by the other domains in trans. Importantly, they also indicate that the RPT domain has no significant effect on the amyloid properties of either NTR, PKD, or both.
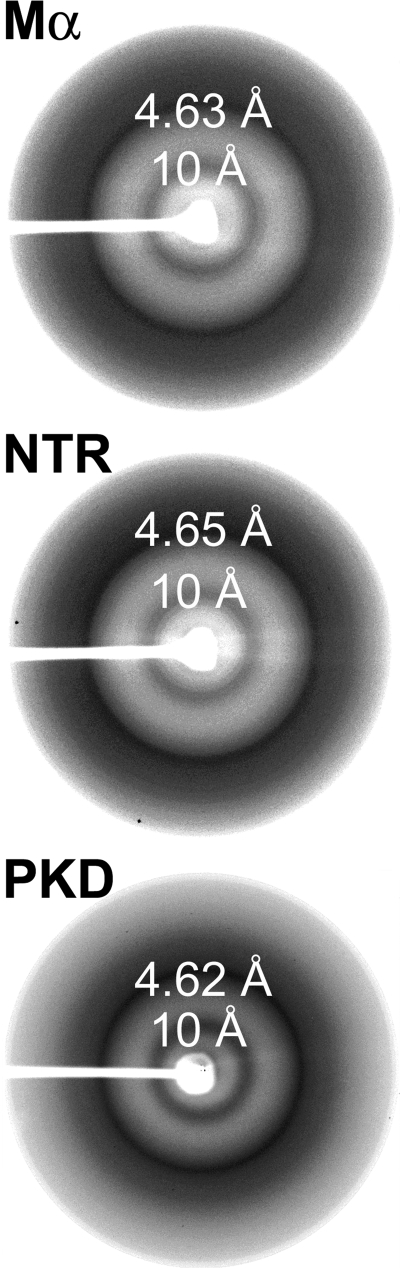
Mα fibrils exhibit additional biophysical and optical properties required for their classification as amyloid, including a characteristic cross-β-sheet-specific x-ray fiber diffraction pattern and a fibrillar three-dimensional morphology by electron microscopy. Consistent with their dye binding characteristics, the isolated NTR and PKD domains, like full-length Mα, both exhibit defined x-ray fiber reflections at 4.6 and 10 Å, indicative of the amyloid cross-β-sheet quaternary structure (Fig. 5). Electron microscopy analysis of Mα fibrils formed in vitro reveals that they have a short branched morphology (Fig. 6A, upper left panel), similar to the appearance of nascent fibrils that form in vivo in melanocytes as analyzed by electron tomography (Fig. 6B; see also supplemental movies 1–3 for tomographic reconstruction) (23). Electron microscopy analysis of the PKD domain also reveals a clear fibrillar morphology (Fig. 6A, lower left panel), but these fibrils are longer and thinner than those formed by full-length Mα, suggesting that other domains influence the quaternary assembly of the fibrils. Although the NTR did not form linear or branched fibrils, NTR aggregates were detected throughout the electron microscopy grids (Fig. 6A, upper right panel), perhaps reflecting higher order assembly of linear fibrils as is often observed for amyloidogenic proteins in vitro (45). In striking contrast to Mα, PKD domain, or NTR, neither fibrils nor frequent aggregates were detected upon analysis of the RPT domain (Fig. 6A, lower right panel), consistent with the results of the spectroscopic analyses. Occasional protein aggregates could be detected sparsely distributed on the grids, but these were rare, consistent with the high degree of solubility of RPT in aqueous buffers. These data support the notion that the PKD domain and NTR, but not the RPT domain, form amyloid fibrils in vitro.
FIGURE 5.
Amyloid-like x-ray diffraction patterns of the NTR and PKD domains. Resolubilized proteins in denaturant were dialyzed against deionized water, lyophilized, and analyzed by x-ray diffraction. The diffraction patterns of Mα, NTR, and PKD are shown. Note the reflections at 4.6 and 10 Å representing the regular spacing between strands within a β-sheet and between β-sheets, respectively.
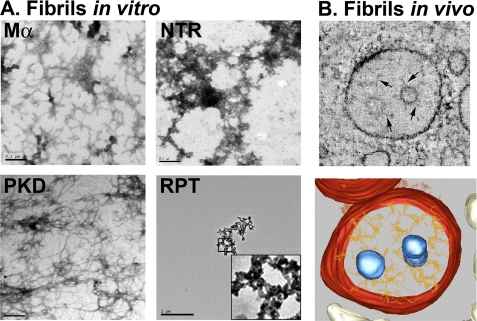
FIGURE 6.
Refolded PKD domain has fibrillar morphology, and refolded Mα resembles melanosome fibrils. A, affinity-purified proteins, as indicated, were renatured by dilution out of the denaturant into physiological assay buffer and incubated overnight at 37 °C with agitation. Samples were then centrifuged at 100,000 × g for 1 h at 4 °C, and pellets were resuspended in a small volume of assay buffer and mounted directly on coated grids, stained with uranyl acetate, and visualized by electron microscopy. Note the branched fibrillar structures apparent in the sample containing Mα and the long fibrillar structures in samples containing the PKD, whereas the NTR appears as aggregates. Fields containing the RPT domain were difficult to find; an isolated RPT aggregate is shown at low magnification (scale bar 1 μm as compared with 0.2 μm for the others) and magnified ×5 in the inset. B, electron tomography of early stage melanosomes. MNT-1 melanoma cells preserved by high pressure freezing were analyzed by electron tomography. Top, a slice from a single tomographic reconstruction showing branched fibrils emerging from internal membrane vesicles of a multivesicular endosome. Bottom, three-dimensional model of the same tomographic reconstruction. Note the similar branched morphology of the Mα fibrils formed in vitro (A, top left panel) and the protofibrils observed in cells (B, bottom panel).
NTR and PKD Domains Show Partial Resistance to Proteinase Digestion
Most amyloid fibrils are resistant to digestion by proteases due to the compact nature of the cross-β-sheet structure. Indeed, proteinase resistance has been used to define the amyloid core for amyloids such as Aβ and the fungal prion protein Ure2 (46, 47). To determine whether cores within Mα and its subdomains are protected from protease digestion, we incubated preformed fibrils in the presence of increasing concentrations of proteinase K and then separated the resulting digestion products by SDS-PAGE and visualized them by staining with Coomassie Brilliant Blue. With increasing concentrations of proteinase K, a number of digestion products are observed for Mα, NTR, and PKD (Fig. 7); the ladder-like nature of these digestion products is consistent with the notion that the NTR and PKD domains, like full-length Mα, self-assemble into a regular repetitive quaternary structure as is seen for other amyloids. The small resistant fragments, which migrate at ∼4.5–6 kDa, likely represent the cores of these repetitive structures. The smaller products of full-length Mα comigrate with those of both the NTR and PKD (Fig. 7, compare lanes 4, 8, and 11), suggesting that the protease-resistant cores of the NTR and PKD are similar, if not identical, to the protease-resistant fragments within Mα.
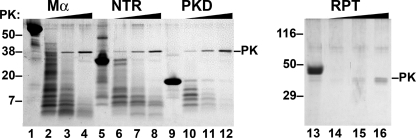
FIGURE 7.
NTR and PKD domains, but not the RPT, are resistant to proteinase digestion. Renatured proteins were treated with increasing concentrations (1, 3.33, and 10 μg/ml) of proteinase K (PK) for 30 min at 37 °C with agitation; digestion products were fractionated by SDS-PAGE and visualized by Coomassie Blue staining. The band corresponding to proteinase K is indicated to the right, and migration of molecular weight standards is indicated to the left. Note the absence of protease-resistant fragments of the RPT domain even at the lowest proteinase K concentration but the presence of resistant fragments for all other domains.
To define the regions within the PKD and NTR domains that were resistant to protease digestion, we used automated Edman sequencing to identify the N termini of the limiting digestion products of Mα, NTR, and PKD. The results (Table 1) identify two predominant protease-resistant peptides from within the NTR and one from the PKD domain, all of which are also predominant peptides derived by protease digestion of Mα. The major protease-resistant peptide from the NTR initiates at Ile131, and based on its Mr of 4,500, would be predicted to extend to residues 165–175; minor overlapping peptides initiating at Glu125 and Ser144 were also detected. These peptides are likely constrained in vivo because they span one to two cysteine residues that are predicted to be engaged in disulfide bonds; thus, the relevance of this fragment in vivo is not clear. A second NTR-derived peptide initiates at Ser171 and likely extends to the end of the NTR. The major protease-resistant peptide from the PKD domain initiates at Asp226 and a minor overlapping peptide initiates at Arg223. Interestingly, these residues fall within the predicted first β-strand of the PKD domain based on alignment with the PKD1 domain of polycystin-1 (33, 34). This region of Pmel17 is not predicted to be modified post-translationally within the eukaryotic secretory pathway. Moreover, the Mα- and PKD-resistant fragments obtained in vitro have a similar molecular weight to the PKD-derived fragment observed in fibril-enriched fractions obtained from melanocytic cells (see Fig. 2), which is not further digested by prolonged incubation with high levels of proteinase K (data not shown). These data further suggest that a core fragment within the PKD domain is the major amyloidogenic component within Mα.
TABLE 1.
N-terminal sequences of proteinase K-resistant peptide cores
Peptides from proteinase K digests of NTR, Mα, or PKD as shown in Fig. 7 were transferred to polyvinylidene difluoride membranes, excised, and submitted to automated Edman protein sequencing.
| Peptide prevalencea | Sequenceb | Initial yield | Position in Pmel17c |
|---|---|---|---|
| pmol | |||
| NTR (Mr ∼5,000) | |||
| 1° | XXPDGGPXXS | 15 | Ile131–Ser140 |
| 2° | SXG(T)GRAML | 5 | Ser171–Leu179 |
| Mα band 1 (Mr ∼7,000) | |||
| 1° | SIGTGRA | 33 | Ser171–Ala177 |
| 2° | ETDDAXI | 22 | Glu125–Ile131 |
| 3° | DGGNKHF | 24 | Asp226–Phe232 |
| 4° | IFPDGGX | 10 | Ile131–Pro137 |
| Mα band 2 (Mr ∼5,000) | |||
| 1° | IFPDGGPX | 74 | Ile131–Cys138 |
| 2° | S(I)GTGRAM | 56 | Ser171–Met178 |
| 3° | SQKR(S)FVY | 11 | Ser144–Tyr151 |
| PKD (Mr ∼7,000) | |||
| 1° | DGGNKHF | 20 | Asp226–Phe232 |
| 2° | RALD(G)(G)(N) | 3.8 | Arg223–Asn229 |
| 3° | DFGD(S)(S)(G) | 4.2 | Asp261–Gly267 |
a Prevalence of sequence is based on initial yield of each amino acid within the sequencing reaction. Peptides are denoted as primary (1°), secondary (2°), tertiary (3°), or quaternary (4°).
b Data are based on identity of the most prevalent amino acid at that position. X denotes the absence of a detected amino acid (often cysteine), and parentheses denote some degree of ambiguity.
c Data are based on alignment with the published amino acid sequence of human Pmel17, indicated as the three-letter amino acid code and its position relative to the translation start site. All peptide sequences were aligned without gaps or errors, conferring 100% confidence in their identity.
By contrast to Mα, PKD, and NTR, which exhibit proteinase K-resistant fragments, the RPT domain is fully digested even at the lowest concentration of proteinase K used, and no resistant fragment is detected (Fig. 7, compare lanes 13 and 14), suggesting that this soluble domain might have an extended structure. The NTR domain is more resistant to proteinase K digestion than the PKD or Mα (Fig. 7; note the higher protein abundance in lane 8 relative to lanes 4 and 12), perhaps because of its propensity to form large, hyperassembled insoluble aggregates upon dilution into aqueous solutions relative to the less aggregated/hyperassembled PKD and Mα fibrils (Fig. 6A).
DISCUSSION
Previous studies aimed at understanding the structural foundation for the amyloid-like fibrils within melanosomes have implicated a central role for the highly glycosylated and hydrophilic RPT domain of the melanosomal matrix protein Pmel17. This conclusion was based mainly on the findings that Pmel17 fibrils are immunoreactive with antibodies directed to the RPT domain (1, 13, 20, 21) and that deletion of the RPT domain results in loss of fibril formation in vivo (17, 22). Recently, McGlinchey et al. (36) additionally reported that a purified recombinant RPT domain, similar to that used here, was capable of forming amyloid-like fibrils upon very prolonged incubation (53 days) in acidic buffers. Our results are inconsistent with the RPT domain forming the core of melanosome fibrils in vivo. Rather, we propose that the amyloid core consists of the N-terminal region of the PKD domain, perhaps in combination with the NTR or a part thereof. We further propose that the RPT domain plays a regulatory role in fibril formation in vivo, perhaps by altering the kinetics of Pmel17 amyloid formation and ensuring that the fibrillogenic process takes place in the correct organelle at the correct time. These data have important implications for the mechanisms controlling the amyloid transformation of the Pmel17 Mα domain within the melanosome precursor organelles.
Our data indicate that the NTR and PKD domains both contribute to the amyloid properties of Mα. Upon dilution of denaturant, recombinant isolated Pmel17 fragments corresponding to both the NTR and PKD domains bind amyloidophilic dyes, form highly insoluble aggregates, display a cross-β-sheet structure by x-ray fiber diffraction, and have cores that are resistant to proteinase K digestion, all of which are hallmarks of amyloid. In addition, the fragments that are generated from both the NTR and PKD domains upon increasing proteinase K digestion appear in a ladder-like pattern, consistent with the formation of a compact quaternary structure as observed for other amyloid fibrils. Finally, the PKD domain forms isolated fibrils by electron microscopy analysis, and the NTR forms fibril aggregates similar to those formed by other amyloidogenic proteins in vitro. Based on its other properties, the NTR aggregates likely consist of fibrils that laterally associate to a higher degree than the PKD domain under our in vitro conditions. Such associations might not occur in vivo, as the native NTR within Pmel17 is N-glycosylated and likely constrained by intra- and/or inter-domain disulfide bonds; such modifications would potentially restrict the lateral aggregation that we observe in vitro. Importantly, the most prominent protease-resistant peptides within the NTR contain cysteine residues that likely participate in these disulfide bonds in vivo. It is therefore possible that the formation of amyloid by this peptide is only observed under our in vitro conditions with unmodified protein. By contrast, there is no evidence for post-translational modification of the PKD domain in cells, and therefore its conformation is likely to be similar to that of the recombinant protein. Therefore, our in vitro studies support a primary role for the PKD domain in amyloid formation in vivo, although we cannot exclude a secondary role for regions within the NTR. It is interesting that the PKD domain is predicted to be a β-sheet-rich structure (33, 34). If this prediction is correct, then perhaps only a minor conformational change would be necessary to generate the cross-β-sheet amyloid fold, consistent with a rapid transition from the nonfibrillar to fibrillar state in vivo (23). In this view, amyloidogenesis by the PKD domain might resemble amyloid formation by transthyretin or superoxide dismutase, which are thought to entail subtle “gain-of-interaction”' rearrangements of a pre-existing β-sheet-rich structure (48).
In contrast to recently published findings by McGlinchey et al. (36), our data indicate that the hexahistidine-tagged RPT domain is not able to form amyloid fibrils in vitro. We find that even after incubation for up to 48 h without chaotropic agents, the recombinant RPT domain is highly soluble in a variety of aqueous buffers (Fig. 3 and supplemental Fig. 2), and we detect no binding to amyloidogenic dyes and no formation of morphological fibrils (Figs. 4 and 6). Moreover, the high degree of sensitivity of the RPT domain in vitro to proteinase digestion (Fig. 7) suggests that it is likely to have a predominantly unstructured conformation, consistent with its hydrophilic and proline-rich composition. Finally, we find no influence of the RPT domain on the amyloidogenic potential of the PKD and NTR domains. Deletion of the RPT domain only results in decreased solubility and does not affect either Mα cross-β-sheet structure (data not shown) or dye binding by Mα or isolated domains (Fig. 4). Similarly, coincubation of RPT with PKD or NTR domains does not alter their amyloid dye binding properties or solubility (Figs. 3 and 4).
It should be noted that upon removal from aqueous solution by lyophilization, an x-ray diffraction pattern characteristic of amyloid was detected with recombinant RPT domains (supplemental Fig. S3), perhaps reflecting an intrinsic ability to form a cross-β-sheet structure in the absence of available hydrogen bonding to water. Such effects might explain some of the data from McGlinchey et al. (36) but appear not to reflect a physiological amyloid fibril given that the lyophilized material was readily resolubilized in nondenaturing aqueous buffers. By contrast, the PKD domain and NTR were insoluble in similar buffers both prior and subsequent to lyophilization. Thus, only the PKD and NTR domains appear to form true amyloid fibrils under physiological conditions.
McGlinchey et al. (36) concluded that neither Mα- nor PKD-containing fragments generated amyloid in vitro, but they based this conclusion entirely on an initial screen for single morphological fibers. In our hands, Mα aggregates extensively at pH 5.0, and although individual fibrils cannot be readily detected morphologically, Mα aggregates at low pH still bind amyloidophilic dyes such as thioflavin T (supplemental Fig. S2). At neutral pH, the fibrils that are detected are branched as they are in vivo, and it is not clear whether these branched structures would have been scored positively in their screen. In addition, the conditions under which McGlinchey et al. (36) observed RPT domain amyloid fibril formation are of questionable physiological relevance. Within melanocytes, Mα is incorporated into insoluble fibrils within minutes or at most a few hours, as judged by metabolic pulse/chase assays (1, 16) and by the detection of protofibrils during invagination of the limiting membrane of multivesicular endosomes (23). By contrast, the RPT domain does not form amyloid within 48 h in our hands, and it required many days (even with seeding) to form amyloid under the conditions of McGlinchey et al. (36). Moreover, although early stage melanosomes are indeed acidic as noted by McGlinchey et al. (36), as they mature melanosomes become less acidic and likely approach neutral pH (13), as required for optimal tyrosinase activity (49). Thus, the dissolution of RPT fibrils at neutral pH observed by McGlinchey et al. (36) is not consistent with the stability of the fibrils in later stage melanosomes in vivo (50–52). It is highly unlikely that the RPT domain in vivo is more prone to form amyloid, as it is highly modified by charged O-linked, sialylated glycans (20, 35) and therefore is even less likely to assemble into fibrils. This property of the RPT domain appears to be conserved in all Pmel17 homologues and has been suggested to be necessary for the formation of normal melanosome fibrils in vivo (35). By contrast, the number and sequence of the repeats are not conserved (2); this lack of sequence conservation makes it an even more improbable candidate for the core of the evolutionarily conserved functional amyloid fibrils.
Whereas the RPT domain is completely dispensable for fibril formation in vitro, it is required for fibril formation in vivo (17, 22) and is detected in association with the fibrils. We therefore propose that the RPT domain plays a regulatory function in vivo, rather than serving a structural role as the core of the fibrils. A comparable regulatory role for a repeat-rich domain has been observed for other amyloids, such as Sup35 and α-synuclein, in which repeated sequences have been found to influence the kinetics of fibril formation but are not completely sequestered in the amyloid core itself (53–59).
How the RPT domain might regulate Pmel17 amyloidogenesis is not entirely clear. One possibility is that the highly hydrophilic and heavily glycosylated domain serves to protect the amyloidogenic core of Pmel17 to prevent nonproductive, and potentially toxic, aggregation at the wrong stage of melanosome biogenesis. Alternatively, this heavily glycosylated domain might prevent higher order aggregation of individual Mα fibrils and ensure that Mα assembles within the organelle into ordered, functional arrays of fibrils rather than disordered plaques. It might also play a role in facilitating a conformational change that allows Pmel17 to adopt a fibrillogenic structure within endosomes. For example, it may prevent Mα from aggregating until reaching the endosome, where proteolytic maturation and a decreased pH might release the RPT domain and thus uncover the amyloidogenic domain within the NTR and/or PKD domains for the initiation of fibrillogenesis. Interestingly, deletion of the N-terminal repeats of α-synuclein by either mutation or protease digestion does not affect protofilament generation, but it results in thinner protofilaments that assemble less efficiently into protofibrils (58). Similarly, deletion of repeats within Sup35 retards fibril assembly (56). A similar function for the RPT domain in Pmel17 is consistent with previous studies showing that deletion of the RPT results in loss of mature fibrils but that unstructured aggregates can still be found in late endosomes (17, 22). Interestingly, a comparison between early melanosome fibrils observed in cells with those made by Mα in vitro reveals a striking resemblance in morphology (compare Fig. 6, A and B). In contrast, the fibrils obtained with the PKD domain are longer and thinner, underscoring the influence of additional domains within Mα during melanosome fibril formation and maturation.
That the results of our in vitro analyses reflect the nature of the fibrils formed in vivo is also supported by the identification of Pmel17 fragments that are found in fibril-enriched Triton X-100-insoluble fractions of pigmented human melanoma cells. Previous studies suggested that following release from the membrane-bound Mβ fragment in endosomes, the amyloidogenic Mα is further processed into smaller fragments within newly forming melanosomes (20–22). However, only full-length Mα- and RPT-containing fragments had been detected in fibril-enriched fractions due to a lack of suitable antibodies to other subdomains. We now show that these Pmel17 fibrillar fractions contain fragments corresponding to all three Mα subdomains. Importantly, distinct fragments are immunoreactive with antibodies to each subdomain, indicating that the three subdomains are cleaved from each other during the processing and maturation of fibrils, likely by lysosomal proteases that are present within melanosomes (60). The ∼7-kDa PKD domain-derived fragment within these fractions remains associated with the insoluble fibrillar fraction under all conditions, is similar in size to the limiting digestion product of the fibrils formed in vitro by the recombinant PKD domain, and is immunoreactive with an antibody generated to a peptide that corresponds to a region just N-terminal to this digestion product. We therefore propose that this fragment forms at least part of the core of the fibrils in vivo. Although the ∼40-kDa NTR fragment that we detected was present at very low levels relative to Mα and is thus likely substantially sub-stoichiometric with the PKD and RPT fragments, we cannot rule out that additional proteolytic fragments of the NTR that lack the N terminus (and that are thus nonreactive with the αPmel-N antibody) and that correspond to one of the limiting NTR digestion products in vitro might also be present in the fibril core. It is also possible, however, that post-translational modifications of this domain in vivo might mask its inherent ability to form amyloid. Regardless, the continued presence of fragments of at least two and possibly all three subdomains within the fibrils is suggestive of noncovalent interactions among them that likely contribute to the stability of the fibrils, their assembly into sheets (23), and/or their association with melanin intermediates (3, 4). Interestingly, release of the RPT domain from the insoluble fibrils in some experiments suggests that these interactions can be disrupted experimentally and provide further evidence against the RPT serving as the core of the fibrils.
Our data lead us to propose a model in which a dual role is assigned to the PKD domain, and perhaps the NTR, in Pmel17 trafficking and fibrillogenesis. Deletion of either of these domains results in missorting of Pmel17 to recycling endosomes and loss of fibril formation (17, 22). Our data here indicate that they also enable fibril formation by serving as the core of the Pmel17 amyloid fibrils. We speculate that these two roles are connected. Indeed, Pmel17 protofibrils in cells form in association with the ILVs of multivesicular compartments (23). Perhaps PKD domain-dependent association with components of the endosomal membrane that lead to incorporation of Pmel17 on the ILVs also results in a conformational change that facilitates the amyloid transformation of the PKD domain. Such a conformational change might reflect the interaction of a similar interface of the PKD domain both with ILV components and, subsequently, with another interface on adjacent PKD or NTR domains to effect fibril elongation. Such a model might have implications for the induction of amyloid formation under pathological conditions, for example by α-synuclein, Aβ, or prion proteins. Testing of such a model for Pmel17 will require definition of the residues involved in forming these interfaces, which is conceivable by combining our in vitro approach with in cellulo analyses of PKD domain-dependent sorting.
Acknowledgments
We are extremely grateful to Lawrence J. Dangott and the Protein Chemistry Laboratory of the Department of Biochemistry, Texas A&M University (College Station, TX) for automated Edman sequencing; to Willie J. C. Geerts and Arie Verkleij (Utrecht University, The Netherlands) and to the University of Pennsylvania Biomedical Imaging Facility for help with acquisition of the electron tomography and standard electron microscopy data, respectively; to Peter Cresswell, Natalie Vigneron, and Ralf M. Leonhardt (Yale University, New Haven, CT) for the I51 antibody and communication of unpublished results; and to Virginia M. Y. Lee (University of Pennsylvania) for advice and access to instrumentation.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AR048155 from the NIAMS (to M. S. M.), Grant R01 AG018917 from the NIA (to J. W. K.), and Director's New Innovator Award 1DP2OD002177-01 (to J. S.). This work was also supported by CNRS, Institut Curie, Fondation pour la Recherche Médicale, and Association pour la Recherche sur le Cancer (to G. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Movies 1–3.
- ILV
- intralumenal vesicle
- CR
- Congo red
- IB
- inclusion body
- NTR
- N-terminal region
- PKD
- polycystic kidney disease-1 repeat
- RPT
- repeat domain
- ThioT
- thioflavin T
- Tricine
- N-[2-hydroxy-1,1- bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1.Berson J. F., Harper D. C., Tenza D., Raposo G., Marks M. S. (2001) Mol. Biol. Cell 12, 3451–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Theos A. C., Truschel S. T., Raposo G., Marks M. S. (2005) Pigment Cell Res. 18, 322–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakraborty A. K., Platt J. T., Kim K. K., Kwon B. S., Bennett D. C., Pawelek J. M. (1996) Eur. J. Biochem. 236, 180–188 [DOI] [PubMed] [Google Scholar]
- 4.Fowler D. M., Koulov A. V., Alory-Jost C., Marks M. S., Balch W. E., Kelly J. W. (2006) PLoS Biol. 4, e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunberg E., Andersson L., Cothran G., Sandberg K., Mikko S., Lindgren G. (2006) BMC Genet. 7, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark L. A., Wahl J. M., Rees C. A., Murphy K. E. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1376–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamilton H. (1940) Anat. Rec. 78, 525–548 [Google Scholar]
- 8.Kerje S., Sharma P., Gunnarsson U., Kim H., Bagchi S., Fredriksson R., Schütz K., Jensen P., von Heijne G., Okimoto R., Andersson L. (2004) Genetics 168, 1507–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quevedo W. C., Fleischmann R. D., Dyckman J. (1981) in Phenotypic Expression in Pigment Cells (Seiji M. ed) pp. 177–184, Tokyo University Press, Tokyo, Japan [Google Scholar]
- 10.Silvers W. K. (1979) The Coat Colors of Mice: A Model for Mammalian Gene Action and Interaction, Springer-Verlag, New York [Google Scholar]
- 11.Spanakis E., Lamina P., Bennett D. C. (1992) Development 114, 675–680 [DOI] [PubMed] [Google Scholar]
- 12.Kwon B. S., Chintamaneni C., Kozak C. A., Copeland N. G., Gilbert D. J., Jenkins N., Barton D., Francke U., Kobayashi Y., Kim K. K. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 9228–9232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raposo G., Tenza D., Murphy D. M., Berson J. F., Marks M. S. (2001) J. Cell Biol. 152, 809–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robila V., Ostankovitch M., Altrich-Vanlith M. L., Theos A. C., Drover S., Marks M. S., Restifo N., Engelhard V. H. (2008) J. Immunol. 181, 7843–7852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Theos A. C., Berson J. F., Theos S. C., Herman K. E., Harper D. C., Tenza D., Sviderskaya E. V., Lamoreux M. L., Bennett D. C., Raposo G., Marks M. S. (2006) Mol. Biol. Cell 17, 3598–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berson J. F., Theos A. C., Harper D. C., Tenza D., Raposo G., Marks M. S. (2003) J. Cell Biol. 161, 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Theos A. C., Truschel S. T., Tenza D., Hurbain I., Harper D. C., Berson J. F., Thomas P. C., Raposo G., Marks M. S. (2006) Dev. Cell 10, 343–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kummer M. P., Maruyama H., Huelsmann C., Baches S., Weggen S., Koo E. H. (2009) J. Biol. Chem. 284, 2296–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiamenti A. M., Vella F., Bonetti F., Pea M., Ferrari S., Martignoni G., Benedetti A., Suzuki H. (1996) Melanoma Res. 6, 291–298 [DOI] [PubMed] [Google Scholar]
- 20.Harper D. C., Theos A. C., Herman K. E., Tenza D., Raposo G., Marks M. S. (2008) J. Biol. Chem. 283, 2307–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kushimoto T., Basrur V., Valencia J., Matsunaga J., Vieira W. D., Ferrans V. J., Muller J., Appella E., Hearing V. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10698–10703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoashi T., Muller J., Vieira W. D., Rouzaud F., Kikuchi K., Tamaki K., Hearing V. J. (2006) J. Biol. Chem. 281, 21198–22208 [DOI] [PubMed] [Google Scholar]
- 23.Hurbain I., Geerts W. J., Boudier T., Marco S., Verkleij A. J., Marks M. S., Raposo G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19726–19731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ehehalt R., Keller P., Haass C., Thiele C., Simons K. (2003) J. Cell Biol. 160, 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajendran L., Honsho M., Zahn T. R., Keller P., Geiger K. D., Verkade P., Simons K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11172–11177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou L., Wang Z., Shen L., Bao G. B., Wang T., Kang J. H., Pei G. (2007) Cell Res. 17, 389–401 [DOI] [PubMed] [Google Scholar]
- 27.de Gassart A., Geminard C., Fevrier B., Raposo G., Vidal M. (2003) Blood 102, 4336–4344 [DOI] [PubMed] [Google Scholar]
- 28.Fivaz M., Vilbois F., Thurnheer S., Pasquali C., Abrami L., Bickel P. E., Parton R. G., van der Goot F. G. (2002) EMBO J. 21, 3989–4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi T., Stang E., Fang K. S., de Moerloose P., Parton R. G., Gruenberg J. (1998) Nature 392, 193–197 [DOI] [PubMed] [Google Scholar]
- 30.Möbius W., Ohno-Iwashita Y., van Donselaar E. G., Oorschot V. M., Shimada Y., Fujimoto T., Heijnen H. F., Geuze H. J., Slot J. W. (2002) J. Histochem. Cytochem. 50, 43–55 [DOI] [PubMed] [Google Scholar]
- 31.Möbius W., van Donselaar E., Ohno-Iwashita Y., Shimada Y., Heijnen H. F., Slot J. W., Geuze H. J. (2003) Traffic 4, 222–231 [DOI] [PubMed] [Google Scholar]
- 32.Weterman M. A., Ajubi N., van Dinter I. M., Degen W. G., van Muijen G. N., Ruitter D. J., Bloemers H. P. (1995) Int. J. Cancer 60, 73–81 [DOI] [PubMed] [Google Scholar]
- 33.Bycroft M., Bateman A., Clarke J., Hamill S. J., Sandford R., Thomas R. L., Chothia C. (1999) EMBO J. 18, 297–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hughes J., Ward C. J., Peral B., Aspinwall R., Clark K., San Millán J. L., Gamble V., Harris P. C. (1995) Nat. Genet. 10, 151–160 [DOI] [PubMed] [Google Scholar]
- 35.Valencia J. C., Rouzaud F., Julien S., Chen K. G., Passeron T., Yamaguchi Y., Abu-Asab M., Tsokos M., Costin G. E., Yamaguchi H., Jenkins L. M., Nagashima K., Appella E., Hearing V. J. (2007) J. Biol. Chem. 282, 11266–11280 [DOI] [PubMed] [Google Scholar]
- 36.McGlinchey R. P., Shewmaker F., McPhie P., Monterroso B., Thurber K., Wickner R. B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 13731–13736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nichols S. E., Harper D. C., Berson J. F., Marks M. S. (2003) J. Invest. Dermatol. 121, 821–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santoso A., Chien P., Osherovich L. Z., Weissman J. S. (2000) Cell 100, 277–288 [DOI] [PubMed] [Google Scholar]
- 39.Palmer I., Wingfield P. T. (2000) in Current Protocols in Protein Science (Coligan J. E., Dunn B. M., Ploegh H. L., Speicher D. W., Wingfield P. T. eds) pp. 6.3.1–6.3.15, John Wiley & Sons, Inc., New York [Google Scholar]
- 40.Chernoff Y. O., Uptain S. M., Lindquist S. L. (2002) Methods Enzymol. 351, 499–538 [DOI] [PubMed] [Google Scholar]
- 41.Klunk W. E., Pettegrew J. W., Abraham D. J. (1989) J. Histochem. Cytochem. 37, 1273–1281 [DOI] [PubMed] [Google Scholar]
- 42.Falcón-Pérez J. M., Starcevic M., Gautam R., Dell'Angelica E. C. (2002) J. Biol. Chem. 277, 28191–28199 [DOI] [PubMed] [Google Scholar]
- 43.Moriyama K., Bonifacino J. S. (2002) Traffic 3, 666–677 [DOI] [PubMed] [Google Scholar]
- 44.Wang Q., Johnson J. L., Agar N. Y., Agar J. N. (2008) PLoS Biol. 6, e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vitrenko Y. A., Gracheva E. O., Richmond J. E., Liebman S. W. (2007) J. Biol. Chem. 282, 1779–1787 [DOI] [PubMed] [Google Scholar]
- 46.Baxa U., Taylor K. L., Wall J. S., Simon M. N., Cheng N., Wickner R. B., Steven A. C. (2003) J. Biol. Chem. 278, 43717–43727 [DOI] [PubMed] [Google Scholar]
- 47.Kheterpal I., Williams A., Murphy C., Bledsoe B., Wetzel R. (2001) Biochemistry 40, 11757–11767 [DOI] [PubMed] [Google Scholar]
- 48.Nelson R., Eisenberg D. (2006) Adv. Protein Chem. 73, 235–282 [DOI] [PubMed] [Google Scholar]
- 49.Saeki H., Oikawa A. (1978) J. Cell Physiol. 94, 139–145 [DOI] [PubMed] [Google Scholar]
- 50.Birbeck M. S. (1963) Ann. N. Y. Acad. Sci. 100, 540–547 [DOI] [PubMed] [Google Scholar]
- 51.Maul G. G. (1969) J. Ultrastruct. Res. 26, 163–176 [DOI] [PubMed] [Google Scholar]
- 52.Moyer F. H. (1966) Am. Zool. 6, 43–66 [DOI] [PubMed] [Google Scholar]
- 53.Kochneva-Pervukhova N. V., Paushkin S. V., Kushnirov V. V., Cox B. S., Tuite M. F., Ter-Avanesyan M. D. (1998) EMBO J. 17, 5805–5810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koo H. J., Lee H. J., Im H. (2008) Biochem. Biophys. Res. Commun. 368, 772–778 [DOI] [PubMed] [Google Scholar]
- 55.Krishnan R., Lindquist S. L. (2005) Nature 435, 765–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu J. J., Lindquist S. (1999) Nature 400, 573–576 [DOI] [PubMed] [Google Scholar]
- 57.Parham S. N., Resende C. G., Tuite M. F. (2001) EMBO J. 20, 2111–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qin Z., Hu D., Han S., Hong D. P., Fink A. L. (2007) Biochemistry 46, 13322–13330 [DOI] [PubMed] [Google Scholar]
- 59.Toyama B. H., Kelly M. J., Gross J. D., Weissman J. S. (2007) Nature 449, 233–237 [DOI] [PubMed] [Google Scholar]
- 60.Diment S., Eidelman M., Rodriguez G. M., Orlow S. J. (1995) J. Biol. Chem. 270, 4213–4215 [DOI] [PubMed] [Google Scholar]