Abstract
β-Galactose residues on N-glycans have been implicated to be involved in growth regulation of cells. In the present study we compared the galactosylation of cell surface N-glycans of mouse Balb/3T3 cells between 30 and 100% densities and found the β-1,4-galactosylation of N-glycans increases predominantly in a 100-kDa protein band on lectin blot analysis in combination with digestions by diplococcal β-galactosidase and N-glycanase. When cells at 100% density were treated with jack bean β-galactosidase, the incorporation of 5-bromodeoxyuridine into the cells was stimulated in a dose-dependent manner, suggesting the involvement of the galactose residues in growth regulation of cells. A galactose-binding protein was isolated from the plasma membranes of cells at 100% density by affinity chromatography using an asialo-transferrin-Sepharose column and found to be galectin-3 as revealed by mass spectrometric analysis. The addition of recombinant galectin-3 into cells at 50% density inhibited the incorporation of 5-bromodeoxyuridine in a dose-dependent manner, but the inhibition was prevented with haptenic sugar. An immunocytochemical study showed that galectin-3 is present at the surface of cells at 100% density but not at 30% density where it locates inside the cells. Several glycoproteins bind to a galectin-3-immobilized column, a major of which was identified as vascular cell adhesion molecule (VCAM)-1. Immunocytochemical studies showed that some galectin-3 and VCAM-1 co-localize at the surface of cells at 100% density, indicating that the binding of galectin-3 secreted from cells to VCAM-1 is one of the pathways involved in the growth regulation of Balb/3T3 cells.
Introduction
Cell surface carbohydrates bound to proteins and lipids play important roles in many biological events including cell-to-cell and cell-to-substratum interactions (1–3). Our previous study showed that N-acetylglucosamine (GlcNAc)-terminated N-glycans expressed predominantly in mammalian brain tissues are involved in neural cell adhesion by binding to Na+/K+-ATPase β1-subunit, a GlcNAc-binding protein (4, 5). Similarly, a variety of lectins expressed at the cell surface have been shown to stimulate or inhibit cell growth and differentiation and induce apoptosis by binding to their respective carbohydrates at the cell surface (6–8). In the case of galectins, which belong to a family of calcium-independent soluble β-galactose-binding proteins, galectin-1 has been shown to be involved in T cell apoptosis (9) and growth inhibition of cells by interaction with α5β1-integrin (10), galectin-3 in pre-mRNA splicing (11), metastasis of hepatoma cells (12), apoptosis of Jurkat E6–1 cells (13), endocytosis (14, 15), cell adhesion and cell growth (16–18), and galectin-9 in metastasis of mammary carcinoma cells (19).
One of the interesting roles of the galactose residues of N-glycans at the cell surface is that they may be involved in the growth regulation of normal cells in vitro, called contact-dependent inhibition of growth, which mimics wound healing in vivo (20, 21). In relation to this, our previous study showed that remodeling of the galactosylation of N-glycans by introducing the β-1,4-galactosyltransferase II cDNA or β-1,4-galactosyltransferase V antisense cDNA into B16-F10 mouse melanoma cells results in the suppression of tumor growth in vivo.2 A similar result has been observed in human glioma cells by introducing the β-1,4-galactosyltransferase V antisense cDNA (22). However, the molecular mechanisms to explain how such gene introduction suppresses tumor growth remain to be elucidated. In the present study we used Balb/3T3 cells in culture to examine whether the galactose residues of N-glycans at the cell surface are involved in growth regulation.
EXPERIMENTAL PROCEDURES
Cell Culture
Mouse fibroblast Balb/3T3 A31-1-1 (JCRB0601) was obtained from Human Science Research Resources Bank (Osaka, Japan). Cells were cultured in 100-mm plastic dishes containing 10 ml of Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, streptomycin (100 μg/ml) and penicillin (100 microunits/ ml) at 37 °C under humidified 5% CO2, 95% air. Cell numbers were counted with a Coulter cell counter every day after passing 4 × 104 cells into a 100 mm dish, and cell density was determined by taking the number of cells at confluence as 100%. In growth curve experiments, cells were plated in 6-well plates at a density of 2 × 104 cells per well. Cell numbers were counted in triplicate for each clone by Coulter counter.
Chemicals
Horseradish peroxidase (HRP)3-conjugated Ricinus communis agglutinin-I (RCA-I), leukoagglutinating-PHA (L-PHA), jack bean β-galactosidase and the Konica Immunostain HRP-1000 kit were obtained from Seikagaku Kogyo Co. (Tokyo, Japan). Human transferrin and diplococcal β-galactosidase were from Sigma. Goat anti-mouse galectin-3 antibody, rabbit anti-mouse vascular cell adhesion molecule-1 (VCAM-1) antibody, protein G-agarose, and HRP-conjugated rabbit anti-goat IgG antibody were from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit anti-mouse Na+/K+-ATPase β1-subunit antibody was from Millipore Co. (Tokyo, Japan). Phospho-p44/42 mitogen-activated protein kinase (MAPK) antibody and HRP-conjugated horse anti-mouse IgG antibody were from Cell Signaling Technology (Beverly, MA). Cy3-Conjugated donkey anti-goat IgG (H+L) antibody was from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA). Alexa Fluor® 488 donkey anti-rabbit IgG was from Molecular Probes, Inc. (Eugene, OR). CNBr-activated Sepharose 4B and enhanced chemiluminescence detection reagent were from GE Healthcare. Vectashield mounting medium with 4′,6-diamino-2-phenylindole was from Vector Laboratories (Burlingame, CA). Iodoacetamide, o-phenylenediamine, α-cyano-4-hydroxycinnamic acid, anti-bromodeoxyuridine (BrdUrd) antibody, anti-MAPK antibody, and d-mannose were from Sigma. d-Galactose, d-glucose, trifluoroacetic acid, isopropyl-β-d-thiogalactopyranoside, and dithiothreitol were from Nacalai Tesque Inc. (Kyoto, Japan).
Cell Proliferation Assay
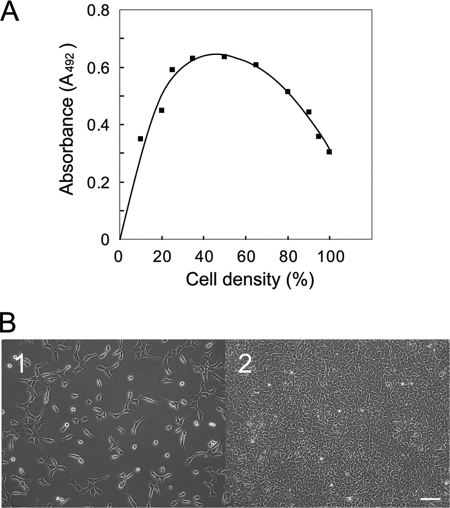
Cells at different densities were cultured in the presence of 10 μm BrdUrd for 2 h, and then amounts of BrdUrd incorporated into cells were assayed to determine growth activity of cells. To examine the effect of β-galactosidase or galectin-3 for cell growth, cells at 100 or 50% density in Dulbecco's modified Eagle's medium containing 2.5% fetal calf serum were incubated with different concentrations of jack bean β-galactosidase or recombinant galectin-3 for 18 h, then with BrdUrd for another 2 h. To determine the amounts of BrdUrd incorporated into cells, fixed cells were incubated with anti-BrdUrd antibody followed by HRP-conjugated secondary antibody. The antibody-BrdUrd complexes were visualized with o-phenylenediamine according to the manufacturer's instructions. The absorbance of each well at 492 nm was determined by an automated microtiter plate spectrophotometer (ImmunoMini NJ-2300, Nalge Nunc International, Tokyo). The relative incorporation of BrdUrd into cells at different densities was determined and is shown in Fig. 1A.
FIGURE 1.
Cell growth at different cell densities. A, shown is incorporation of BrdUrd into Balb/3T3 cells at different densities. Cells at different densities were cultured in the presence of BrdUrd for 2 h and then fixed. BrdUrd incorporated cells were detected with anti-BrdUrd antibody as described “Experimental Procedures.” B, Morphological appearances of Balb/3T3 cells at 30% with the highest BrdUrd incorporation (panel 1) and at 100% density with more than 50% inhibition of BrdUrd incorporation (panel 2), respectively, was used for further studies. The bar indicates 100 μm.
Lectin/Western Blot Analyses of Membrane Glycoproteins
Lectin blot analysis was performed as described previously (23). In brief, cells cultured were harvested and suspended in cold water to lysis for 15 min and then centrifuged (2,000 × g, 4 °C, 10 min) to remove nuclei. Membrane proteins were then precipitated with 50% acetone and recovered in pellets by centrifugation (2,000 × g, 4 °C, 20 min). Precipitates containing membrane glycoproteins were solubilized with SDS sample buffer and subjected to lectin blot analysis. For Western blot analysis cells were directly solubilized with 10 mm Tris-HCl buffer (pH 6.8) containing 1% SDS and protease inhibitors. Samples were subjected to SDS/polyacrylamide gel electrophoresis and then transferred onto polyvinylidene difluoride membranes. After blocking, the blots were incubated with HRP-conjugated RCA-I, anti-galectin-3 antibody, anti-VCAM-1 antibody, anti-MAPK antibody, or anti-phospho-MAPK antibody followed by HRP-labeled secondary antibody. Complexes were detected using a Konica Immunostain HRP-1000 kit or an enhanced chemiluminescence detection reagent.
Purification of Galactose-binding Protein by Asialo-transferrin Affinity Column Chromatography
Asialo-transferrin was prepared according to the method described previously (24) and immobilized to CNBr-activated Sepharose 4B. Balb/3T3 cells (2 × 108 cells) were homogenized in 10 mm Tris-HCl buffer (pH 6.8) containing 25 mm NaCl and protease inhibitors with a Dounce homogenizer and centrifuged at 4,000 × g at 4 °C for 10 min to remove nuclei. The supernatant was then centrifuged at 30,000 × g at 4 °C for 60 min, and resultant precipitates (crude plasma membranes) were solubilized with phosphate-buffered saline (PBS, pH 7.4) containing 1% Triton X-100, 1 mm CaCl2, and protease inhibitors at 4 °C for 2 h. Solubilized proteins were diluted 10 times with PBS containing 0.2 mm CaCl2, 0.1% CHAPS, and 0.02% NaN3 and were initially applied to a Sepharose column to eliminate those that bind non-specifically to Sepharose, and then the remaining proteins were applied to an asialo-transferrin-Sepharose column. The column was washed with buffer containing 0.5 m mannose, and then buffer containing 0.5 m glucose. A galactose-binding protein was eluted with buffer containing 0.5 m galactose. The pass-through and bound fractions were concentrated, subjected to SDS-PAGE, and stained with Coomassie Brilliant Blue (CBB). A stained band in the bound fraction was excised and used for protein identification.
Peptide Mass Fingerprinting by Matrix-assisted Laser Disruption Ionization/Time-of-flight Mass Spectrometry (MALDI- TOF/MS)
Peptide mass fingerprinting by MALDI-TOF/MS was performed to identify proteins by the method described previously (25). In brief, an excised protein band was reduced with 1.5 mg/ml dithiothreitol in 100 mm NH4HCO3 and then alkylated with 10 mg/ml iodoacetamide in 100 mm NH4HCO3. The protein was digested with 4 μg/ml sequencing grade modified trypsin (Promega, Tokyo) at 30 °C for 16 h. A peptide sample was mixed with 10 mg/ml α-cyano-4-hydroxycinnamic acid in 0.1% trifluoroacetic acid and spotted onto stainless steel targets for analysis with a MALDI-TOF mass spectrometer (Axima CFR, Shimadzu, Kyoto, Japan). The Mascot algorithm was used for protein identification.
Expression and Purification of Recombinant Galectin-3
The prokaryotic expression vector pIN-III-ompA2 was a gift from Dr. John L. Wang (Michigan State University). The galectin-3 cDNA was amplified by PCR using a cDNA library prepared from mouse NIH3T3 cells. PCR analysis was conducted using the 5′- and 3′-primers 5′-CGGAATTCATGGCAGACAGCTTTTCGCTTA-3′ and 5′-CGGGATCCGCTTAGATCATGGCGTGGTTAG-3′ for the mouse galectin-3 gene, designed on the basis of the mouse galectin-3 gene (GenBank™ accession no. NM 010705). The cDNA was digested with EcoRI and BamHI from pGEM®-T Easy (Promega) and inserted into pIN-III-ompA2 to generate the p-rgalectin-3 plasmid. An overnight culture of JM109 Escherichia coli (TOYOBO Bio-Chemical, Tokyo) transformed with a p-rgalectin-3 plasmid was diluted 1:500 and grown in 1 liter of Terrific broth containing 100 μg/ml ampicillin at 37 °C for 3 h. The cells were treated with 50 μm isopropyl-β-d-thiogalactopyranoside and incubated at 22 °C for 20 h. The cells were harvested by centrifugation, and the cell pellet was washed with 50 ml of 30 mm Tris-HCl buffer (pH 7.4) containing 20% sucrose and 1 mm EDTA. Then the cell pellet was incubated at 4 °C for 10 min in 50 ml of 5 mm MgSO4 solution containing a complete protease inhibitor mixture (Roche Diagnostics) and centrifuged at 10,000 × g, 4 °C for 10 min. A volume of 10×-concentrated PBS was added to the supernatant, and the mixture was applied to a 10-ml asialo-transferrin-Sepharose column as described above. The column was washed with PBS containing 0.1% CHAPS, 0.2 mm CaCl2, and 0.02% NaN3, and bound galectin-3 was eluted with buffer containing 0.5 m galactose. The eluted protein was desalted and concentrated by ultrafiltration using an Amicon Ultra-15 filter (Millipore, Billerica, MA).
Immunocytochemistry
For detection of galectin-3 at the cell surface, cells were incubated at 4 °C with goat anti-mouse galectin-3 antibody and rabbit anti-Na+/K+-ATPase β1-subunit antibody followed by Cy3-conjugated anti-goat IgG antibody and Alexa Fluor® 488-conjugated anti-rabbit IgG antibody and then fixed with PBS containing 2% paraformaldehyde. For detection of intercellular galectin-3, cells were fixed with PBS containing 4% paraformaldehyde and then treated with PBS containing 0.5% Triton X-100 for 5 min to permeabilize cells before incubation with antibodies. For detection of galectin-3 and VCAM-1 at the cell surface, cells at 100% density were fixed with PBS containing 4% paraformaldehyde and incubated with anti-galectin-3 antibody and rabbit anti-mouse VCAM-1 antibody followed by fluorescence-conjugated secondary antibodies. Fluorescent images were obtained under a laser-scanning microscope FV1000-D IX81 (Olympus, Tokyo).
Isolation of Counter-receptors for Galectin-3
Balb/3T3 cells at 100% density were solubilized with PBS containing 1% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycholate, and a protease inhibitor mixture at 4 °C for 16 h. Cell lysates were applied to a galectin-3-immobilized Sepharose column (6 mg protein/ml gel), and the column was washed with PBS containing 0.5 m NaCl and 0.1% CHAPS and then eluted with PBS containing 0.5 m galactose and 0.1% CHAPS. Proteins in both fractions were subjected to Western blot analysis and staining with CBB. The stained bands in the bound fraction were excised and identified by MALDI-TOF/MS.
Analysis of Complexes Formed with Galectin-3
Balb/3T3 cells at 100% density were lysed with PBS containing 0.1% Nonidet P-40 and a protease inhibitor mixture. The cell lysates were pretreated with 5 μg of normal goat IgG and protein G-agarose beads, and nonspecific immunocomplexes formed were precipitated by centrifugation at 15,000 × g for 15 min at 4 °C. Five micrograms of anti-galectin-3 antibody and then protein G-agarose beads were added to the supernatant. After overnight incubation, the immunocomplexes were isolated by centrifugation and washed three times with buffer. Bound proteins were subjected to Western blot analysis. In some experiments, 0.5 m mannose or 0.5 m galactose was included in the immunocomplexes.
Generation of Stable Gene Knockdown Cell Lines
Four or three sets of short hairpin RNA against mouse galectin-3 or VCAM-1 were inserted into pSilencer4.1-CMV hygro vector or pSilencer2.1-CMV puro vector (Ambion, Austin, TX). Target sequences for galectin-3 were as follows: 5′-AAGGATATCCGGGTGCATGGG-3′ (exon 3), 5′-AATCATGGGCACAGTGAAACC-3′ (exon 4), 5′-AACAACAGAAGAGTCATTGTG-3′ (exon 5), 5′-AAGCTGACCACTTCAAGGTTG-3′ (exon 6). Target sequences for VCAM-1 were as follows: 5′-AAAGGGACGATTCCGGCATTT-3′, 5′-AACCTTTACTCCCGTCATTGA-3′, 5′-AAATCCACGCTTGTGTTGAGC-3′. Stable transfectants were selected with 500 μg/ml hygromycin B (Calbiochem) or 10 μg/ml puromycin (Sigma). Negative control-short hairpin RNA plasmid (Ambion) was used as a control.
RESULTS
Galactosylation of Cell Surface N-Glycans in Balb/3T3 Cells at Different Densities
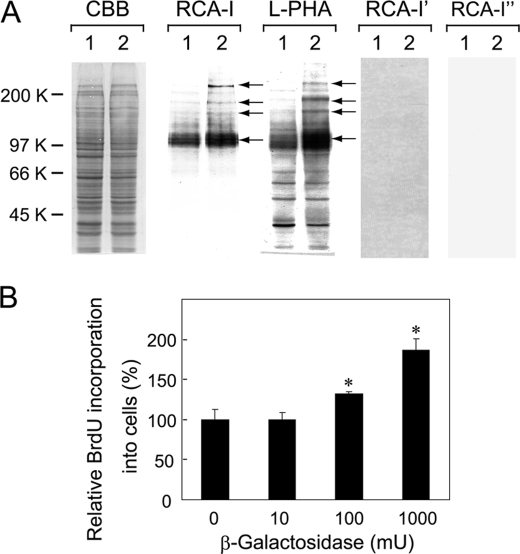
The relative amounts of BrdUrd incorporated into cells at different densities were determined to show the density-dependent inhibition of growth on cultured Balb/3T3 cells. The highest incorporation of BrdUrd into cells was detected at 30–50% densities, and the relative incorporation of BrdUrd into cells was gradually inhibited as cells become confluent (Fig. 1A). Then plasma membranes were prepared from mouse Balb/3T3 cells at 30 and 100% densities whose morphological appearances are shown in panels 1 and 2 of Fig. 1B, respectively. The membrane proteins were subjected to lectin blot analysis. The results showed that the binding of RCA-I, which preferentially binds β-1,4-galactosylated oligosaccharides (26), increases mainly in a 100-kDa protein band and slightly in 150-, 180-, and 220-kDa protein bands of cells at 100% density as compared with cells at 30% density (indicated with arrows, Fig. 2A, RCA-I). The blot was incubated with L-PHA, which preferentially binds highly branched N-glycans (27), and the binding increased markedly in a broad 100-kDa protein band and slightly in 150-, 180-, and 220-kDa protein bands of cells at 100% density as compared with cells at 30% density (indicated with arrows, Fig. 2A L-PHA). Binding of RCA-I to samples disappeared upon treatment of the blot with diplococcal β-galactosidase (Fig. 2A RCA-I'), which cleaves β-1,4-linkage (28), or N-glycanase (Fig. 2A RCA-I″). Similarly, L-PHA -binding to the samples also disappeared when the blots were treated with N-glycanase (data not shown). These results indicate that the galactosylation and high levels of N-glycan branching increase in particular proteins when cells become confluent.
FIGURE 2.
β-1,4-Galactosylation of surface glycoproteins at different cell densities. A, lectin blot analysis of membrane glycoprotein samples from Balb/3T3 cells is shown. Lanes 1 and 2 show samples from cells at 30 and 100% densities, respectively. The blots were incubated with CBB, HRP-conjugated RCA-I, or L-PHA. In some cases the blots were treated with diplococcal β-galactosidase and N-glycanase before incubation with RCA-I, shown as RCA-I′ and RCA-I″, respectively. Three independent experiments were conducted, and the representative results are shown. Arrows indicate protein bands with augmented lectin reactivities. B, shown is the effect of β-galactosidase on Balb/3T3 cell growth. Cells cultured at 100% density were treated with jack bean β-galactosidase (0, 10, 100, and 1000 milliunits against 1.5 × 104 cells/96-titer well, respectively) for 18 h and incubated with BrdUrd for another 2 h. Cell growth was assayed by determining the incorporation of BrdUrd into cells, and results are shown by taking the amount of BrdUrd incorporated into control cells as 100% with the mean ± S.D. of triplicate experiments. p values were obtained by Student's t test (*, p < 0.02 against control).
Effect of β-Galactosidase Treatment on Cell Growth
To examine the biological significance of the galactose residues of N-glycans at the cell surface, cells at 100% density were treated with jack bean β-galactosidase, which cleaves β-1,4- and β-1,3-linkages (29) and can cleave the sugars effectively from the cell surface at this cell density, and the incorporation of BrdUrd into cells was determined. A dose-dependent incorporation of BrdUrd into cells was observed by the β-galactosidase treatment (Fig. 2B), and RCA-I-binding disappeared, particularly from a 100-kDa protein band of the cell surface glycoproteins, and heat-inactivated enzyme did not affect cell growth (data not shown). Cells were also treated with N-glycanase under the same conditions as above; however, no significant incorporation of BrdUrd was observed. Lectin blot analysis of membrane glycoproteins prepared from cells treated with N-glycanase using concanavalin A did not show any significant change in lectin binding (data not shown), suggesting that N-glycanase does not act effectively on cell surface glycoproteins of cultured cells under the present conditions. The results suggest that β-galactose residues are important for growth inhibition of cells, probably by binding to a β-galactose-binding protein at the cell surface.
Isolation of a Galactose-binding Protein from Mouse Balb/3T3 Cells
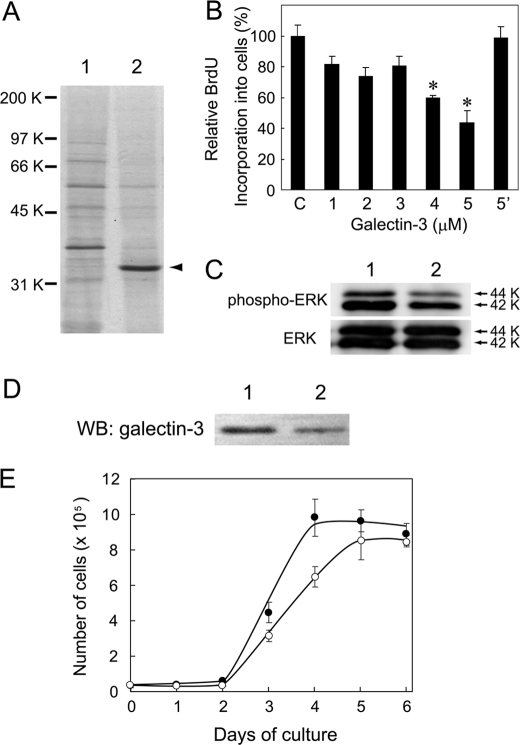
To isolate a galactose-binding protein, plasma membranes were prepared from Balb/3T3 cells at 100% density and solubilized with PBS containing 1% Triton X-100. Solubilized proteins were applied to an asialo-transferrin-Sepharose column. Proteins that passed through and those that bound and eluted from the column with PBS containing 0.5 m galactose and 0.1% CHAPS (bound fraction) were subjected to SDS-PAGE. When the gels were stained with CBB, a band with an approximate molecular mass of 33-kDa was obtained as a major protein in the bound fraction (lane 2 in Fig. 3A). The 33-kDa band was excised from the gel and digested with trypsin, and the products were subjected to MALDI-TOF/MS. Mascot analysis of the data revealed that the 33-kDa protein is mouse galectin-3.
FIGURE 3.
Isolation of a galactose-binding protein and its effect on cell growth. A, shown is isolation of a galactose-binding protein from Balb/3T3 cells by asialo-transferrin-Sepharose column chromatography. Plasma membranes were prepared from cells at 100% density, and proteins were solubilized and applied to the column. Lanes 1 and 2 indicate CBB-stained proteins that passed through and those that were bound and eluted with 0.5 m galactose from the column, respectively. A major 33-kDa protein band indicated with an arrowhead was isolated and identified to be galectin-3 using peptide mass finger printing. B, effect of galectin-3 on Balb/3T3 cell growth is shown. Cells cultured in Dulbecco's modified Eagle's medium containing 2.5% fetal calf serum at 50% density (7 × 103 cells/96-titer well) were treated with different amounts of recombinant galectin-3 for 18 h and then incubated with BrdUrd for another 2 h. Incorporated BrdUrd was detected by enzyme-linked immunosorbent assay. Galectin-3 showed dose-dependent inhibition of cell growth, which was nullified in the presence of 10 mm thiodigalactoside (data with 5 μm galectin-3 is shown in bar 5′ as a representative). The results are shown by taking the amount of BrdUrd incorporated into control cells as 100% with the mean ± S.D. of triplicate experiments. p values were obtained by Student's t test (*, p < 0.02 against control). C, shown is a Western blot analysis of MAPK proteins in Balb/3T3 cells. Cells at 50% density (7 × 103 cells/96-titer well) were treated with galectin-3 for 20 h (lane 2) or non-treated (lane 1), and soluble cellular proteins were subjected to Western blot analysis using anti-phospho-MAPK and anti-MAPK antibodies, respectively. The 42- and 44-kDa protein bands are ERK2 and ERK1, respectively. D, Western blot analysis (WB) of galectin-3 in its knockdown cells is shown. Cell lysates from mock-transfected (lane 1) and galectin-3-knockdown Balb/3T3 cells (lane 2) were subjected to Western blot analysis using an anti-galectin-3 antibody. E, a growth curve of galectin-3-knockdown cells (open circle) and mock-transfected cells (closed circle) is shown. Cells were cultured (2 × 104 cells/6-well plate) and counted with a Coulter counter every 24 h. The results are shown as the mean ± S.D. of triplicate experiments.
Effect of Galectin-3 on Cell Growth
To examine the biological roles of galectin-3 at the cell surface, recombinant galectin-3 was produced in E. coli and added into culture medium of Balb/3T3 cells at 50% density, and the incorporation of BrdUrd into the cells was determined. Because β-1,4-galactosylation of glycoproteins began to increase in cells at 50% density and because galectin-3 localized in cytoplasm of cells at 50% density (data not shown), cells at 50% density were used in the present experiment. The results showed that galectin-3 inhibits the incorporation of BrdUrd into cells in a dose-dependent manner (Fig. 3B) and that growth inhibition is abolished by inclusion of thiodigalactoside into medium (bar 5′ of Fig. 3B). The results indicate that the galectin-3 added inhibits the cell growth by binding to the cell surface galactose residues. The growth regulation with galectin-3 was also observed for normal canine kidney epithelial cells (supplemental Fig. S1).
Effect of Galectin-3 Treatment on MAPK Phosphorylation
Because the cell growth was inhibited by galectin-3, changes in the phosphorylation of MAPK molecules, which are involved in growth regulation of cells (30, 31), were examined by Western blot analysis. Cell lysates were prepared from galectin-3-treated cells and nontreated cells, and solubilized proteins were subjected to Western blot analysis using anti-phospho-MAPK and anti-MAPK antibodies. A remarkable decrease in the phosphorylation of the extracellular signal-regulated kinase 1 (ERK1) and ERK2 proteins (44 and 42 kDa, respectively) was observed in cells treated with galectin-3 for 20 h (lane 2 in the upper panel osf Fig. 3C) without a change in the amounts of ERK1 and ERK2 proteins (lower panel in Fig. 3C), indicating that growth regulation by galectin-3 is mediated by the inactivation of MAPK proteins.
Role of Galectin-3 in Growth Regulation of Cells
To examine the biological role of galectin-3 in growth regulation of cells, a Balb/3T3 cell clone with 50% knockdown of galectin-3 was obtained using short hairpin RNA as determined by Western blot analysis (Fig. 3D). The results showed that galectin-3-knockdown cells grow slower and saturate at lower density than mock-transfected cells (Fig. 3E).
Localization of Galectin-3 at Cell Surface
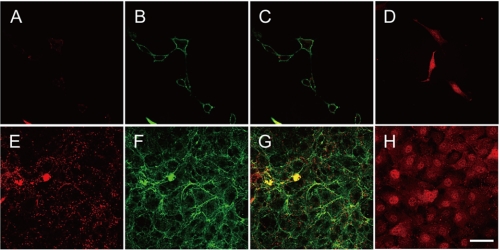
Galectin-3 has been shown to localize not only in the cytoplasm and nuclei but also at the cell surface (16, 17, 32, 33). To demonstrate the presence of galactin-3 on the surface of cells at 100% density, cells were initially incubated with the antibodies as described under “Experimental Procedures.” In cells at 30% density, galectin-3 was not detected at the cell surface (panel A), whereas in cells at 100% density, it was detected at the cell surface (panel E) (Fig. 4). To establish the cell surface localization of galectin-3, cells at 30 and 100% densities were incubated with anti-Na+/K+-ATPase β1-subunit antibody and visualized (panels B and F of Fig. 4, respectively) as Na+/K+-ATPase is a marker of plasma membranes. When panels E and F were overlaid, more than 50% of the fluorescence derived from galectin-3 was merged with that from Na+/K+-ATPase (panel G) but not when panels A and B were overlaid (panel C) (Fig. 4). Galectin-3 was also detected in cytoplasm and nuclei in cells both at 30 and 100% densities (panels D and H, respectively, of Fig. 4). These results indicate that galectin-3 is secreted to the outer surface of cells at 100% density.
FIGURE 4.
Localization of galectin-3 in Balb/3T3 cells. Cells at 30% density and 100% density were incubated initially with anti-galectin-3 antibody or anti-Na+/K+-ATPase β1-subunit antibody followed by secondary antibody and fixed (panels A–C and E–G). Cells at 30 and 100% densities were fixed, permeabilized with detergent, and then incubated with anti-galectin-3 antibody (panels D and H, respectively). Galectin-3 at the cell surface was detected with Cy3-conjugated secondary antibody (panels A, D, E, and H). Na+/K+-ATPase as a cell surface marker was detected with Alexa Fluor® 488-conjugated secondary antibody (panels B and F). A double fluorescent image for galectin-3 and Na+/K+-ATPase was obtained (panels C and G). The bar indicates 40 μm.
Interaction of Galectin-3 with VCAM-1
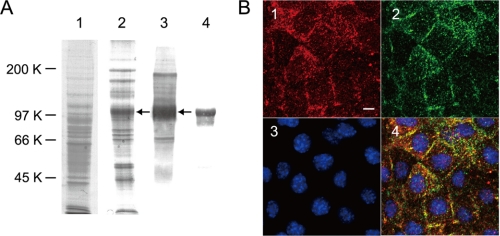
Galectin-3 has been shown to bind several cell surface glycoproteins including β1-integrin and epidermal growth factor receptor to exert a variety of functions (14, 15). To determine to which molecules galectin-3 binds significantly, solubilized proteins were applied to a galectin-3-immobilized column, and the bound proteins were eluted with PBS containing 0.5 m galactose and 0.1% CHAPS. Proteins in the pass-through and bound fractions were subjected to SDS-PAGE. When the gel was stained with CBB, several protein bands were detected in the bound fraction, and a band with an approximate molecular mass of 100 kDa was prominent in the bound fraction (indicated with an arrow in lane 2 of Fig. 5A). To confirm that these protein bands contained galactosylated N-glycans, the blot containing proteins in the bound fraction was subjected to lectin blot analysis using RCA-I. The results showed that the 100-kDa protein band is heavily galactosylated (data not shown) like that shown in Fig. 2A. When the blot containing the bound fraction was incubated with L-PHA that binds galactosylated tri- and tetra-antennary complex-type N-glycans, the 100-kDa protein band reacted mainly with the lectin (indicated with an arrow in lane 3 of Fig. 5A). Therefore, the 100-kDa band was excised from the gel, and trypsinized peptides were subjected to MALDI-TOF/MS. The Mascot analysis of the data revealed it to be mouse VCAM-1. Then the blot was incubated with anti-mouse VCAM-1 antibody, and a broad 100-kDa band reacted with the antibody (lane 4 in Fig. 5A). To confirm this association of galectin-3 with VCAM-1 further, the solubilized membrane proteins were incubated with anti-galectin-3 antibody and then immunoprecipitated. Analysis of the immunocomplexes revealed galectin-3 to be associated with VCAM-1 (lane 1), and this association could be inhibited partly with 0.5 m galactose (lane 2) but not with 0.5 m mannose (lane 3) (supplemental Fig. S2A), indicating that galectin-3 binds galactose residues expressed on VCAM-1. With a secondary antibody alone, there was no galectin-3 in immunoprecipitates (data not shown). Furthermore, when cell lysates were digested exhaustively with N-glycanase and then applied to a galectin-3-immobilized column, de-N-glycosylated VCAM-1 passed through the column as revealed by Western blot analysis (supplemental Fig. S2B), indicating that galectin-3 binds VCAM-1 via its N-glycans.
FIGURE 5.
Isolation of a counter-receptor for galectin-3 from Balb/3T3 cells and its cellular localization. A, shown is isolation of a counter-receptor for galectin-3. Proteins were solubilized from cells at 100% density and applied to a galectin-3-Sepharose column. Lanes 1 and 2 indicate CBB-stained proteins that passed through and those that bound and were eluted with 0.5 m galactose from the column, respectively. The blots containing proteins that bound to the column were incubated with L-PHA (lane 3) and anti-VCAM-1 antibody (lane 4). B, localization of galectin-3 and VCAM-1 on Balb/3T3 cells is shown. Cells at 100% density were fixed with PBS containing 4% paraformaldehyde. Fixed cells were incubated with anti-galectin-3 antibody or anti-VCAM-1 antibody followed by Cy3-conjugated (panel 1) or Alexa Fluor® 488-conjugated secondary antibody (panel 2). Nuclei were stained with 4′,6-diamino-2-phenylindole (panel 3). A triple staining image for galectin-3, VCAM-1, and 4′,6-diamino-2-phenylindole was obtained and shown in panel 4. The bar indicates 10 μm.
Co-localization of Galectin-3 with VCAM-1
To confirm the above observation, cells at 100% density were incubated with anti-galectin-3 and anti-VCAM-1 antibodies and examined immunocytochemically as described under “Experimental Procedures.” The cells were also incubated with 4′,6-diamino-2-phenylindole to stain cell nuclei (panel 3 in Fig. 5B). Most of the galectin-3 and VCAM-1 were detected at the cell surface as dots (panels 1 and 2, respectively, in Fig. 5B). When these panels were overlaid, more than 50% of the dots merged (panel 4 in Fig. 5B), indicating a fraction of galectin-3 molecules interacts with VCAM-1 molecules at the cell surface.
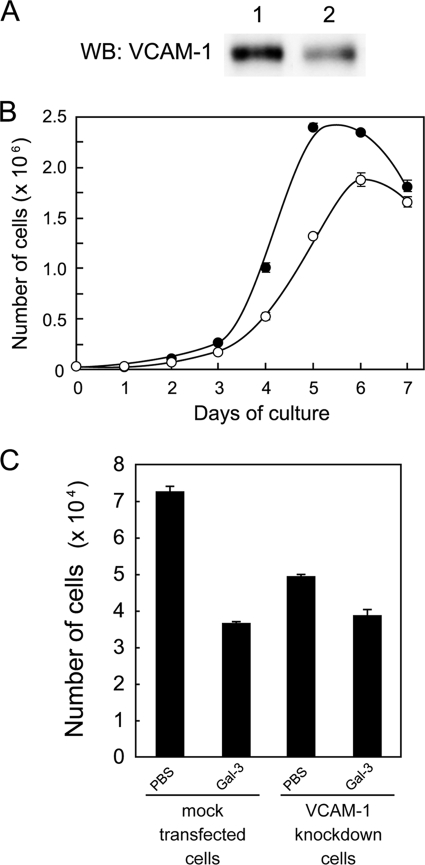
Role of VCAM-1 in Growth Regulation of Cells
To examine the role of VCAM-1 in growth regulation of cells, a Balb/3T3 cell clone with 60% knockdown of VCAM-1 against the control was obtained using short hairpin RNA as determined by Western blot analysis (Fig. 6A). The results showed that VCAM-1-knockdown cells grow slower and saturate at lower density than mock-transfected cells (Fig. 6B). When the effect of galectin-3 on cell growth was examined using these cells, growth of VCAM-1 knockdown cells was not inhibited strongly with galectin-3 when compared with that of mock-transfected cells (Fig. 6C), indicating that the decreased expression of VCAM-1 attenuates the growth inhibition of cells with galectin-3.
FIGURE 6.
Effect of VCAM-1 on growth regulation of cells. A, Western blot (WB) analysis of VCAM-1 in its knockdown Balb/3T3 cells is shown. Cell lysates from mock-transfected (lane 1) and VCAM-1-knockdown cells (lane 2) were analyzed by Western blot analysis with an anti-VCAM-1 antibody. B, shown is a growth curve of VCAM-1-knockdown cells (open circle) and mock-transfected cells (closed circle). Cells were cultured (2 × 104 cells/6-well plate), and their numbers were counted with a Coulter counter every 24 h. The results are shown as the mean ± S.D. of triplicate experiments. C, shown is the effect of galectin-3 on growth of VCAM-1-knockdown cells. VCAM-1-knockdown and mock-transfected cells cultured in Dulbecco's modified Eagle's medium containing 2.5% fetal calf serum (3 × 104 cells/24-well plate) were treated with 5 μm recombinant galectin-3 or PBS for 20 h and then counted by a Coulter counter. The results are shown as the mean ± S.D. of triplicate experiments.
DISCUSSION
The present study shows that the galactosylation and high level branching of N-glycans at the cell surface increase markedly in a 100-kDa protein when Balb/3T3 cells are grown to confluency as revealed by lectin blot analysis. An increase in binding of L-PHA, which binds highly branched N-glycans, in cells at higher densities indicates that enhanced galactosylation of N-glycans could be due to an increased number of branched sugar chains of N-glycans but not of increased galactosylation of GlcNAc residues of N-glycans as no significant change was observed in binding to PVL, which binds terminal GlcNAc residues (34), between cells at low and high densities (data not shown). Although high level branching of N-glycans is one of the characteristic features of malignantly transformed cells (35, 36), such N-glycans are also expressed in normal cells whose growth is arrested through contact-dependent inhibition (37), which suggests the importance of a galactose residue(s) on a particular branch(es) of N-glycans for growth regulation of cells. In support of this, the treatment of cells at 100% density with jack bean β-galactosidase results in a stimulation of BrdUrd incorporation into the cells. There are many factors that affect the β-1,4-galactosylation of N-glycans, and among them β-1,4-galactosyltransferases are the most effective factors to the increased binding of RCA-I to proteins. Because expression levels of the β-1,4-galactosyltransferase genes I, II, III, V, and VI increased when cells become confluent (supplemental Fig. S3), β-1,4-galactosyltransferases I, II, III, and/or V could be involved in the galactosylation of N-glycans in cells at 100% density as these β-1,4-galactosyltransferases can galactosylate N-glycans in vivo (38, 39).
Galectin-3 was isolated from the plasma membranes of cells at 100% density as a galactose-binding protein in the present study, and recombinant galectin-3 was added exogenously to cells at 50% density inhibits cell growth. The growth of normal canine kidney epithelial cells was also inhibited with galectin-3 (supplemental Fig. S1), indicating that galectin-3 is involved in growth regulation of several lines of cells. These results strongly indicate that the galactose residues on cell surface N-glycans are important for growth regulation of cells and act by binding to galectin-3. To establish the role of galectin-3 in growth regulation of cells, galectin-3-knockdown cells were obtained, which showed a slower growth rate and a lower saturation density of cells when compared with control cells. This is probably due to loss-of-functions of galectin-3 distributed widely within cells, particularly in nuclei and cytoplasm, and such loss-of-functions appeared strongly in these cell clones. Therefore, the reduced expression of endogenous galectin-3 in Balb/3T3 cells is not an appropriate way to establish the role of galectin-3 at the cell surface, and some other approach would be required. A similar result was observed in MDA-MB-435 breast cancer carcinoma cells using antisense cDNA of galectin-3 (40).
Galectin-3 localizes in the cytoplasm and nuclei and at the cell surface of a variety of cells. To establish the cell surface localization of galectin-3 in cells at 100% density, an immunocytochemical study was performed under the non-permeabilized condition. The results detected galectin-3 as dots on the surface of cells at 100% density but not at 30% density. Under the permeabilized condition, galectin-3 was in cytoplasm and nuclei, in which it is involved in pre-mRNA splicing, anti-apoptosis, and growth control as reported by Wang and co-workers (11, 41, 42). Because whole amounts of galectin-3 were not changed significantly by cell density as revealed by Western blot analysis (data not shown), the translocation of galectin-3 from the cytoplasm to the cell surface is a key issue of the growth regulation of Balb/3T3 cells. Galectin-3 has no signal sequence, but it can be secreted from cells through a non-classical pathway (43). Therefore, further studies are necessary to determine the molecular mechanism by which galectin-3 is secreted from the inside to the outside of cells at high density.
The effect of exogenously added galectin-3 on cell growth differs among cell types (18, 44–48). This could be due to differences in ligand glycoproteins to which galectin-3 binds, as galectin-3 binds fibronectin, laminin, integrin β1, and epidermal growth factor receptor expressed differentially among different cell types. In the present study using a galectin-3-immobilized column, a 100-kDa protein, identified to be mouse VCAM-1, was the major protein carrying galactose residues on its N-glycans. Furthermore, an immunocytochemical study showed that more than 50% of galectin-3 secreted to the cell surface co-localize with VCAM-1. Immunoprecipitation analysis also showed VCAM-1 to be co-precipitated with galectin-3. When VCAM-1 was treated with N-glycanase, de-N-glycosylated species passed through a galectin-3 column (lane 1), whereas partially de-N-glycosylated species bound to the column (lane 2) (supplemental Fig. S2B), indicating that galectin-3 binds to VCAM-1 via its N-glycans. Lectin blot analysis of proteins that bound to a galectin-3-immobilized column in the present study showed that VCAM-1 contains N-glycans with the β-1,6-GlcNAc-linked outer chain moiety, on which poly-N-acetyllactosamine is expressed preferentially (49), a preferred ligand for galectin-3 (50). Therefore, VCAM-1 could be a glycoprotein to which galectin-3 binds predominantly. Because the expression of VCAM-1 is slightly increased in cells at 100% density when compared with cells at 30% density (data not shown), it is necessary to conduct a more detailed analysis of cell density-dependent increase of the galactosylation and branching of N-glycans attached to VCAM-1.
To establish the role of VCAM-1 in growth regulation of cells, VCAM-1-knockdown cells were obtained, and they showed a slower growth rate and a lower saturation density of cells when compared with control cells. These results are not consistent with our present hypothesis but could be brought about by the loss-of-functions of VCAM-1, which possesses multiple functions such as interaction with α4β1 and α9β1 integrin molecules (51, 52). Because cross-linking of VCAM-1 with antibody has been shown to induce signals affecting ERK2 and protein-tyrosine phosphatase 1B activities (53, 54), VCAM-1 is also involved in growth regulation of cells. Quite interestingly, growth of the VCAM-1-knockdown cells was not inhibited strongly with galectin-3 compared with that of control cells, indicating that the expression of VCAM-1 is important for growth regulation of cells with galectin-3.
Finally, substantial evidence for our present hypothesis has to be obtained by some method other than analyzing galectin-3/VCAM-1 knockdown cells, and whether or not galectin-3 has the same biological effect in other cell types by binding to VCAM-1 expressed ubiquitously also remains to be elucidated.
Acknowledgment
We are grateful to Dr. John L Wang in Michigan State University for supplying the pIN-III-ompA2 plasmid.
This work was supported by Grants-in-aid for Scientific Research 10680696, 09240104, and 14580707 from the Ministry of Education, Science, Culture, and Sports of Japan, by the Research Promotion Fund from the Japan Science Technology Agency, and by Institutional Grants in 2006 and 2007 from Nagaoka University of Technology (to K. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
K. Shirane, M. Tagawa, T. Sato, S. Furukawa, M. Yu, Y. Kobayashi, K. Wada, and K. Furukawa, manuscript in preparation.
- HRP
- horseradish peroxidase
- BrdUrd
- 5-bromodeoxyuridine
- CBB
- Coomassie Brilliant Blue
- CHAPS
- 3-[(cholamidopropyl)dimethylammonio]-1-propane-sulfonate
- ERK
- extracellular signal-regulated kinase
- MALDI-TOF/MS
- matrix-assisted laser disruption ionization/time-of-flight mass spectrometry
- MAPK
- mitogen-activated protein kinase
- PBS
- phosphate-buffered saline
- RCA-I
- R. communis agglutinin-I
- VCAM-1
- vascular cell adhesion molecule-1
- CHAPS
- 3-[(cholamidopropyl)dimethylammonio]-1-propanesulfonate
- L-PHA
- leukoagglutinating PHA.
REFERENCES
- 1.Dwek R. A. (1995) Science 269, 1234–1235 [DOI] [PubMed] [Google Scholar]
- 2.Haltiwanger R. S., Lowe J. B. (2004) Annu. Rev. Biochem. 73, 491–537 [DOI] [PubMed] [Google Scholar]
- 3.Ohtsubo K., Marth J. D. (2006) Cell 126, 855–867 [DOI] [PubMed] [Google Scholar]
- 4.Kitamura N., Ikekita M., Hayakawa S., Funahashi H., Furukawa K. (2004) J. Neurosci. Res. 75, 384–390 [DOI] [PubMed] [Google Scholar]
- 5.Kitamura N., Ikekita M., Sato T., Akimoto Y., Hatanaka Y., Kawakami H., Inomata M., Furukawa K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2796–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hennet T., Chui D., Paulson J. C., Marth J. D. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 4504–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sasaki T., Hirabayashi J., Manya H., Kasai K., Endo T. (2004) Glycobiology 14, 357–363 [DOI] [PubMed] [Google Scholar]
- 8.van Vliet S. J., Gringhuis S. I., Geijtenbeek T. B., van Kooyk Y. (2006) Nat. Immunol. 7, 1200–1208 [DOI] [PubMed] [Google Scholar]
- 9.Perillo N. L., Pace K. E., Seilhamer J. J., Baum L. G. (1995) Nature 378, 736–739 [DOI] [PubMed] [Google Scholar]
- 10.Fischer C., Sanchez-Ruderisch H., Welzel M., Wiedenmann B., Sakai T., André S., Gabius H. J., Khachigian L., Detjen K. M., Rosewicz S. (2005) J. Biol. Chem. 280, 37266–37277 [DOI] [PubMed] [Google Scholar]
- 11.Dagher S. F., Wang J. L., Patterson R. J. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 1213–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takenaka Y., Fukumori T., Raz A. (2004) Glycoconj. J. 19, 543–549 [DOI] [PubMed] [Google Scholar]
- 13.Yang R. Y., Hsu D. K., Liu F. T. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 6737–6742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furtak V., Hatcher F., Ochieng J. (2001) Biochem. Biophys. Res. Commun. 289, 845–850 [DOI] [PubMed] [Google Scholar]
- 15.Partridge E. A., Le Roy C., Di Guglielmo G. M., Pawling J., Cheung P., Granovsky M., Nabi I. R., Wrana J. L., Dennis J. W. (2004) Science 306, 120–124 [DOI] [PubMed] [Google Scholar]
- 16.Hughes R. C. (2001) Biochimie 83, 667–676 [DOI] [PubMed] [Google Scholar]
- 17.Ochieng J., Furtak V., Lukyanov P. (2004) Glycoconj. J. 19, 527–535 [DOI] [PubMed] [Google Scholar]
- 18.Sato S., Hughes R. C. (1992) J. Biol. Chem. 267, 6983–6990 [PubMed] [Google Scholar]
- 19.Irie A., Yamauchi A., Kontani K., Kihara M., Liu D., Shirato Y., Seki M., Nishi N., Nakamura T., Yokomise H., Hirashima M. (2005) Clin. Cancer Res. 11, 2962–2968 [DOI] [PubMed] [Google Scholar]
- 20.Gradl G., Faust D., Oesch F., Wieser R. J. (1995) Curr. Biol. 5, 526–535 [DOI] [PubMed] [Google Scholar]
- 21.Wieser R. J., Schütz S., Tschank G., Thomas H., Dienes H. P., Oesch F. (1990) J. Cell Biol. 111, 2681–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang J., Chen X., Shen J., Wei Y., Wu T., Yang Y., Wang H., Zong H., Yang J., Zhang S., Xie J., Kong X., Liu W., Gu J. (2006) J. Biol. Chem. 281, 9482–9489 [DOI] [PubMed] [Google Scholar]
- 23.Sato T., Furukawa K., Greenwalt D. E., Kobata A. (1993) J. Biochem. 114, 890–900 [DOI] [PubMed] [Google Scholar]
- 24.Furukawa K., Matsuta K., Takeuchi F., Kosuge E., Miyamoto T., Kobata A. (1990) Int. Immunol. 2, 105–112 [DOI] [PubMed] [Google Scholar]
- 25.Toda T., Sugimoto M., Omori A., Matsuzaki T., Furuichi Y., Kimura N. (2000) Electrophoresis 21, 1814–1822 [DOI] [PubMed] [Google Scholar]
- 26.Baenziger J. U., Fiete D. (1979) J. Biol. Chem. 254, 9795–9799 [PubMed] [Google Scholar]
- 27.Merkle R. K., Cummings R. D. (1987) Methods Enzymol. 138, 232–259 [DOI] [PubMed] [Google Scholar]
- 28.Paulson J. C., Prieels J. P., Glasgow L. R., Hill R. L. (1978) J. Biol. Chem. 253, 5617–5624 [PubMed] [Google Scholar]
- 29.Li S. C., Mazzotta M. Y., Chien S. F., Li Y. T. (1975) J. Biol. Chem. 250, 6786–6791 [PubMed] [Google Scholar]
- 30.Encinas M., Iglesias M., Llecha N., Comella J. X. (1999) J. Neurochem. 73, 1409–1421 [DOI] [PubMed] [Google Scholar]
- 31.Marshall C. J. (1995) Cell 80, 179–185 [DOI] [PubMed] [Google Scholar]
- 32.Théry C., Boussac M., Véron P., Ricciardi-Castagnoli P., Raposo G., Garin J., Amigorena S. (2001) J. Immunol. 166, 7309–7318 [DOI] [PubMed] [Google Scholar]
- 33.Wang J. L., Gray R. M., Haudek K. C., Patterson R. J. (2004) Biochim. Biophys. Acta 1673, 75–93 [DOI] [PubMed] [Google Scholar]
- 34.Endo T., Ohbayashi H., Kanazawa K., Kochibe N., Kobata A. (1992) J. Biol. Chem. 267, 707–713 [PubMed] [Google Scholar]
- 35.Dennis J. W., Laferté S., Waghorne C., Breitman M. L., Kerbel R. S. (1987) Science 236, 582–585 [DOI] [PubMed] [Google Scholar]
- 36.Santer U. V., Gilbert F., Glick M. C. (1984) Cancer Res. 44, 3730–3735 [PubMed] [Google Scholar]
- 37.Hahn T. J., Goochee C. F. (1992) J. Biol. Chem. 267, 23982–23987 [PubMed] [Google Scholar]
- 38.Furukawa K., Sato T. (1999) Biochim. Biophys. Acta 1473, 54–66 [DOI] [PubMed] [Google Scholar]
- 39.Guo S., Sato T., Shirane K., Furukawa K. (2001) Glycobiology 11, 813–820 [DOI] [PubMed] [Google Scholar]
- 40.Honjo Y., Nangia-Makker P., Inohara H., Raz A. (2001) Clin. Cancer Res. 7, 661–668 [PubMed] [Google Scholar]
- 41.Cowles E. A., Agrwal N., Anderson R. L., Wang J. L. (1990) J. Biol. Chem. 265, 17706–17712 [PubMed] [Google Scholar]
- 42.Moutsatsos I. K., Wade M., Schindler M., Wang J. L. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 6452–6456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moutsatsos I. K., Davis J. M., Wang J. L. (1986) J. Cell Biol. 102, 477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bullock S. L., Johnson T. M., Bao Q., Hughes R. C., Winyard P. J., Woolf A. S. (2001) J. Am. Soc. Nephrol. 12, 515–523 [DOI] [PubMed] [Google Scholar]
- 45.Inohara H., Akahani S., Raz A. (1998) Exp. Cell Res. 245, 294–302 [DOI] [PubMed] [Google Scholar]
- 46.John C. M., Leffler H., Kahl-Knutsson B., Svensson I., Jarvis G. A. (2003) Clin Cancer Res. 9, 2374–2383 [PubMed] [Google Scholar]
- 47.Kopitz J., von Reitzenstein C., André S., Kaltner H., Uhl J., Ehemann V., Cantz M., Gabius H. J. (2001) J. Biol. Chem. 276, 35917–35923 [DOI] [PubMed] [Google Scholar]
- 48.Maeda N., Kawada N., Seki S., Arakawa T., Ikeda K., Iwao H., Okuyama H., Hirabayashi J., Kasai K., Yoshizato K. (2003) J. Biol. Chem. 278, 18938–18944 [DOI] [PubMed] [Google Scholar]
- 49.Cummings R. D., Kornfeld S. (1982) J. Biol. Chem. 257, 11230–11234 [PubMed] [Google Scholar]
- 50.Frigeri L. G., Robertson M. W., Liu F. T. (1990) J. Biol. Chem. 265, 20763–20769 [PubMed] [Google Scholar]
- 51.Taooka Y., Chen J., Yednock T., Sheppard D. (1999) J. Cell Biol. 145, 413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vonderheide R. H., Springer T. A. (1992) J. Exp. Med. 175, 1433–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deem T. L., Cook-Mills J. M. (2004) Blood 104, 2385–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lazaar A. L., Krymskaya V. P., Das S. K. (2001) J. Immunol. 166, 155–161 [DOI] [PubMed] [Google Scholar]