Abstract
The laminins are a family of heterotrimeric basement membrane proteins that play roles in cellular adhesion, migration, and tissue morphogenesis. Through in silico analysis of the laminin-encoding genes, we identified a novel family of alternate splice isoforms derived from the 5′-end of the LAMA3 and LAMA5 genes. These isoforms resemble the netrins in that they contain a laminin N-terminal domain followed by a short stretch of laminin-type epidermal growth factor-like repeats. We suggest the terms LaNt (laminin N terminus) α3 and LaNt α5, for the predicted protein products of these mRNAs. RT-PCR confirmed the presence of these transcripts at the mRNA level. Moreover, they exhibit differential, tissue-specific, expression profiles. To confirm the existence of LaNt α3 protein, we generated an antibody to a unique domain within the putative polypeptide. This antibody recognizes a protein at the predicted molecular mass of 64 kDa by immunoblotting. Furthermore, immunofluorescence analyses revealed a basement membrane staining in epithelial tissue for LaNt α3 and LaNt α3 localized along the substratum-associated surface of cultured keratinocytes. We have also tested the functionality LaNt α3 through RNAi-mediated knockdown. Keratinocytes exhibiting specific knockdown of LaNt α3 displayed impaired adhesion, stress resistance, and reduced ability to close scratch wounds in vitro.
Introduction
The identification of a lower than initially predicted number of genes in mouse and human genomes has dramatically increased the focus on alternate splicing as an important mechanism of providing the necessary temporally and spatially restricted changes required in gene and protein expression as an organism both develops and responds to injury (1). Indeed, according to recent reports, 40–60% of human genes are alternatively spliced (2). One area where alternative splicing is thought to be of particular importance is in the extracellular matrix (ECM)4 (3, 4).
ECM proteins provide substrates for a range of functions including, but not limited to, structural roles in tissues such as skin, cartilage, and bone, in cell signaling, and in the apparently opposing roles of promoting cellular adhesion and migration (4, 5). Many ECM genes, including those that encode fibronectin, tropoelastin, the collagens, and proteoglycans, are large with multiple exons (4, 6–9). Alternative splicing of such genes results in the inclusion or exclusion of particular exons with the functional consequence of varying the domain composition of the ECM protein (8, 10). In addition, through alternate promoter or first exon usage or conversely through inclusion of an alternate exon or read through of an intron/exon boundary resulting in the inclusion of an in-frame stop codon, “truncated” forms of ECM proteins have been described (11–13). For example, splice site readthrough of the fibronectin exon III-Ia introduces a novel 3′-end containing a termination codon and leads to generation of a 70-kDa protein termed migration-stimulating factor (MSF) (12).
In this study, we have utilized an in silico approach to identify a family of transcripts derived by alternative splicing from the laminin extracellular matrix family. The laminins are a major family of basement membrane proteins with roles in maintenance of tissue integrity, in signaling, and development (14, 15). To date, 12 laminin-encoding genes have been identified, which, based on sequence identity, are subdivided into 5 α, 4 β, and 3 γ chains encoded by LAMA1–5, LAMB1–4, and LAMC1–3, respectively (16). Within the laminin family, the most striking alternative splicing occurs from the LAMA3 gene, where two major transcripts have been identified; LAMA3A, which consists of exons 39–76 and is initiated from an internal site in intron 38, encoding the N-terminally truncated laminin α3a protein (Fig. 1A) (13, 17). In contrast, LAMA3B contains exons 1–38 and 40–76 (i.e. skips exon 39), and encodes a full-length laminin termed α3b (Fig. 1B). Further alternate or minor isoforms have been identified in LAMC2 (18, 19), LAMA4 (20), LAMB3 (21), and LAMA2 (22).
FIGURE 1.
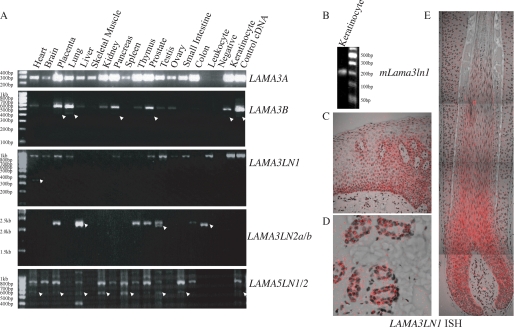
LAMA3 and LAMA5 transcripts and protein architecture. In A, the structure of previously characterized transcripts and proteins derived from the LAMA3 and LAMA5 genes as well as isoforms identified by in silico analyses are depicted diagrammatically. In the case of transcripts, white, gray, and black rectangles represent constitutive exons, alternate exons, and 5′ splice site readthrough exons, respectively. In the case of protein structure, gray rectangles indicate laminin-type epithelial growth factor-like repeats. LN, laminin N-terminal domain; G, laminin globular domain; 4a/Lβ, globular domains. B, comparison of 5′ splices site boundaries to consensus for readthrough sites of identified transcripts (for consensus; M is A or C, R is A or G). C, comparison of the 3′-end of the human and mouse LAMA3LN1 transcripts. In-frame stop codons are underlined, polyadenylation signal sites are capitalized.
Here, we describe the identification of multiple, short, alternate splice isoforms derived from the 5′-end of the genes (LAMA3 and LAMA5) encoding the α3 and α5 laminin subunits. Functional studies utilizing siRNA knockdown of one of these newly identified isoforms shows that it plays an important role in epidermal cell adhesion and in keratinocyte migration.
EXPERIMENTAL PROCEDURES
Antibodies
Exon9e of LAMA3 was amplified using primers; L3CprbFor 5′-GTA AGT TTC ATT TCA AGT TGG-3′ and L3CprbRev 5′-ATC CAA AAT TCAA AGA GAC TG-3′ and cloned into pCR2.1, sequence verified, and subcloned into pET4.1 (Novagen, Madison, WI). The pET41 construct was transformed into BL21(DE3)pLysS and protein expression induced by adding 1 mm isopropyl-β-d-thiogalactoside (Sigma-Aldrich) for 3 h at 37 °C. Proteins were extracted using Bugbuster (Novagen), and the fusion protein was bound to glutathione-Sepharose beads, washed extensively, and separated on a 4–12% NuPAGE bis/Tris gel. The band corresponding to GST-LAMA3exon9e was excised and used for rabbit polyclonal antibody production (Moravian Biotechnology, Czech Republic). Mouse monoclonal anti-β-actin was obtained from Sigma Aldrich; GB3, a mouse monoclonal against laminin γ2 and B1K, a mouse monoclonal against laminin β3, were obtained from Harlan Sera lab Ltd, England and Transduction Laboratories, Lexington, KY, respectively. Rabbit serum J18 against laminin 332 and mouse monoclonal RG13 against laminin α3 were described previously (23, 24). Secondary antibodies used were purchased from DakoCytomation, Denmark and Molecular Probes, Invitrogen.
MTcDNA Panel RT-PCR
Primer pairs were designed to specifically amplify unique regions of the human LAMA3A, LAMA3B, LAMA3LN1, LAMA3LN2, LAMA5LN1, LAMA5LN2, and mouse lama3ln1 transcripts (supplemental Table S1). RT-PCR with these primers were performed alongside those for G3PDH (BD Biosciences, Franklin Lakes, NJ) on human multiple tissue cDNA panels MTC1 and MTCII (BD Biosciences) and cDNA from cultured primary human keratinocytes, HaCaT cells, and mouse keratinocytes according to standard protocols (25–27).
In Situ Hybridization (ISH)
A 170-bp LAMA3LN1 specific probe (exon 9e) for ISH was generated by RT-PCR from HaCaT cDNA using primers: L3CprbFor 5′-GTA AGT TTC ATT TCA AGT TGG-3′ and L3CprbRev 5′-ATC CAA AAT TCAA AGA GAC TG-3′, Promega buffer, 15 mm MgCl2, and 1 unit of Taq (Promega) with conditions of: 1 cycle of 94 °C 2 min, 38 cycles of 94 °C 15 s; 55 °C 15 s; 72 °C 20 s, 1 cycle of 72 °C 10 min. The product size was confirmed on a 2% agarose gel and cloned into pCR2.1 (Invitrogen) and subcloned into pBluescript (Stratagene). ISH was performed as previously described (28).
Cell Culture
HaCaT cells and mouse epidermal keratinocytes (PAM cells) were maintained in Dulbecco's minimal Eagle's medium supplemented with 10% fetal calf serum (Sigma, DMEM), 10,000 units of penicillin and 10 μg/ml streptomycin (Sigma) (29, 30). Human epidermal keratinocytes, immortalized with human papilloma virus genes E6 and E7, were described previously (31). The cells were maintained in defined keratinocyte serum-free medium supplemented with a 1% penicillin/streptomycin mixture (Invitrogen). All cell cultures were maintained in a 37 °C, 5% CO2 environment. Proliferation assays were performed as previously described (32). Live cell images were generated using a Zeiss Axiovert 200 m (Carl Zeiss), digital camera (Hamamatsu), and Volocity 3DM software (Improvision, Coventry, UK).
siRNA
siRNA sequences were designed using a web-based design algorithm (33). siA 5′-UCA GCG AAG UCA UCA GGC U-3′, siB 5′-GUA UCU UAG CCU GUG UGU A-3′. For siRNA transfection HaCaT cells were seeded at ∼8.3 × 105 cells/well of 6-well plates (Invitrogen). 24 h after plating, 100 μl of DMEM were incubated with 15 μl of RNAifect (Qiagen) and 200 nmol of siRNA solution (Dharmacon Inc, Lafayette, CO) for 30 min at room temperature, then added dropwise to each well (2 ml total volume, siRNA final concentration, 100 μm).
Adhesion Assay
Cell-based adhesion assays were performed as described elsewhere (32, 34). HaCaT cells: 72-h post-transfection, cells were dissociated with trypsin, centrifuged, and washed with PBS. Cells were resuspended at 1.5 × 106 cells/ml and then plated onto 96-well dishes precoated with bovine serum albumin and allowed to adhere for 30, 60, or 360 min. Nonadherent cells were removed by PBS washes, and adherent cells were fixed with 3.7% formaldehyde and stained with crystal violet in 20% methanol (Sigma). It should be noted that by 360 min, >99% of cells had attached to substrate regardless of treatment. Crystal violet stain was eluted with 50% ethanol, 0.1 m sodium citrate, pH 4.2, and the absorbance at 540 nm was measured. The percentage of attached cells was calculated relative to the value obtained at the 360-min time point. To evaluate spreading, cells were plated at ∼7.5 × 105 cells/well in 6-well plates, and images taken from computer-selected random areas 2 h after seeding. The percentage of flattened cells in the adherent population was calculated.
Detachment Assay
A variation of the technique described by Yuen et al. (32, 35) was used. Cells were transfected with siRNA, and, 24 h later, they were trypsinized and replated. After an additional 48 h, the growth medium was removed, and wells were washed twice with PBS. They were then treated with 0.005% trypsin, 0.0004% EDTA in PBS (Invitrogen). Phase contrast images were taken every 5 min for 90 min, and the percentage of rounded cells in the population was determined.
Migration Assay
72 h after transfection, a scratch wound was created in a confluent monolayer of HaCaT cells with a 10-μl tip, washed twice with PBS, and the medium replaced with CO2-independent medium (Invitrogen) supplemented with 10% fetal calf serum. Images of the wound area were taken every 20 min for 36 h. At 1, 2, 3, 4, 5, 6, 9, 12, and 15 h post-scratching, the width of the wound was measured using Metamorph software (Universal Imaging Corp., Molecular Devices, Downingtown, PA).
SDS-PAGE/Western Blotting
Extracts of cells in monolayers were generated by scraping washed cells directly into urea/SDS sample buffer as previously described (36). ECM preparations of cultured cells were prepared as previously described (24). Briefly, cells were lysed by treatment with 20 mm NH4OH (Sigma) for 5 min, followed by extensive PBS washes to remove cellular debris. Microscopy confirmed complete removal of cells. ECM proteins were then solubilized in urea/SDS sample buffer. Conditioned medium of confluent or scratch-wounded cultured cells was collected, clarified by low speed centrifugation, and proteins precipitated using ammonium sulfate (1.5-4 m). Protein preparations were subject to SDS-PAGE/Western blotting as described elsewhere (36).
RESULTS
Immunofluorescence Analysis
Cells and tissues were prepared for indirect immunofluorescence as described previously (17, 23).
Identification of a Novel Family of Laminin Alternate Splice Isoforms
During the study of a rare variant of junctional epidermolysis bullosa (17), in silico analysis of the laminin α3-encoding gene was undertaken utilizing tools available on the University of Santa Cruz genome browser. Careful analyses of the 5′-end of the LAMA3 gene identified 2 ESTs containing readthrough of 5′ splice sites, raising the possibility that short, novel, splice isoforms are transcribed from this gene. IMAGE clone 1737146, isolated from a testis library (GenBankTM AI125611), was recognized as carrying an extension to the LAMA3 exon 9 (subsequently termed exon 9e) generated by readthrough of the exon 9-intron 9 splice boundary. This exon extension contained a stop codon and a polyadenylation signal, and the isolated EST carried a polyA tail and therefore appeared to be a genuine polyadenylated mRNA species, termed LAMA3LN1. Alternative splicing algorithms predicted that this transcript comprised exons 1–9 of LAMA3, encoding 487 amino acids of which the final 64 were encoded by exon 9e and were therefore unique to this peptide (Fig. 1A). RT-PCR products derived from human epithelial-containing tissues and keratinocytes confirmed the presence of this transcript at the mRNA level (Fig. 2A).
FIGURE 2.
mRNA expression profiling. A, cDNA from the indicated tissues and cultured cells were processed for sqRT-PCR using primers specific for LAMA3A, LAMA3B, LAMA3LN1, LAMA3LN2, and LAMA5LN1. Arrowheads denote presence of some minor products, which may represent additional isoforms. B, RT-PCR of mouse keratinocyte cDNA with primers specific for mLama3ln1 results in generation of a 220-bp product. C, D, and E show LAMA3LN1 in situ hybridization in human face skin (C and D) and in a human hair follicle (E). Reactive product is pseudocolored in red, whereas the tissue was counterstained with Mayer's hematoxylin (gray).
Analysis of the mouse and human sequences surrounding the exon 9/intron 9 splice boundary revealed perfect cross species conversation; however, in both there are three changes within the exonic sequence relative to the consensus splice site sequence suggesting a weak splice site (Fig. 1B) (37). Readthrough into mouse lama3 intron 9 also introduced an in-frame stop codon 5′ to a consensus polyadenylation signal sequence in a similar location to the human sequence and therefore predicted production of a peptide of almost identical length to human (Fig. 1C). RT-PCR primers were designed to amplify the equivalent mouse sequence, and the presence at the message level of this transcript (mlama3ln1) was confirmed in cDNA derived from cultured mouse keratinocytes (Fig. 2B).
A second EST was identified that also contained the first 9 LAMA3 exons with splicing occurring within exon 10 to a site within exon 18, then continuing to exon 25 (LAMA3LN2). Readthrough of the 5′ splice site of exon 25 incorporated a unique region containing a polyadenylation signal. The sequence corresponding to this mRNA (GenBankTM AK096422) contained a frameshift insertion, which, in turn, led to a termination codon in exon 19, predicting a protein of 483 amino acids (Fig. 1A). RT-PCR primers were designed to test for the presence of this transcript in the various tissues of a multiple tissue cDNA panel. Two PCR products of a larger size than predicted were amplified and sequenced. The larger product consisted of exons 8 to 25 with 5′ splice site readthrough into exon 25e (LAMA3LN2a). The smaller product had exon 10 spliced out but was otherwise identical (LAMA3LN2b) (Fig. 2A). Assuming both these products were spliced normally to the first 7 exons, one would predict products of 1039 amino acids and 994 amino acids, respectively, of which the final 4 amino acids were derived from exon 25e and were therefore unique.
The conserved domain architecture of the various laminin chains, coupled with the expression of fewer isoforms in lower organisms, suggested that gene duplication and rearrangement are responsible for the presence of the multiple chains. With this in mind, the 5′-end of the LN domain containing laminin α chains was analyzed in silico. No potential products were identified in LAMA1 or LAMA2; however, an analogous EST was identified that aligns to LAMA5. This EST (GenBankTM BC015386), had been isolated from a breast adenocarcinoma library and was predicted to encode the first 11 exons of LAMA5 spliced to an alternate 12th exon (exon 12*) including a termination codon (LAMA5LN2) (Fig. 1A). Translation of this mRNA would generate a peptide of 500 amino acids with the final eight being encoded by the alternate 12th exon. The latter would therefore be unique to this transcript. Notably, splice site boundaries at both LAMA3 exon25 and LAMA5 exon 11 also diverged from the consensus sequence (Fig. 1B).
RT-PCR primers were designed to amplify this LAMA5-derived transcript. These yielded two major products (LAMA5LN1 and LAMA5LN2) when tested on normal breast cDNA (Fig. 2A). Sequencing of these products revealed the smaller product to contain the expected exons 1–11 plus 12* and the larger, more abundant, second product to be generated from 5′ splice site readthrough of exon 11 (into exon 11e); therefore, consisting of exons 1–11 plus 11e (Fig. 1A). This additional exon encoded a termination codon 11-bp downstream leading to the generation of a protein of 496 amino acids in size. Both transcripts contained non-consensus polyadenylation signal sequences (38).
RT-PCR Analyses Indicate Tissue-specific Expression Patterns for These Novel Laminin Isoforms
The expression profiles of the newly identified laminin transcripts identified above were studied using multiple tissue cDNA (MTC) panels, which include cDNA from leukocytes and which were supplemented with cDNA from cultured keratinocytes (25–27). Semi-quantitative RT-PCR was performed for each transcript with samples removed at regular intervals, and we scored expression based on signal intensity for each tissue relative to housekeeping genes (Fig. 2A and Table 1).
TABLE 1.
mRNA expression profiling
cDNA from the indicated tissues and cultured cells were processed for sqRT-PCR using primers specific for LAMA3A, LAMA3B, LAMA3LN1, LAMA3LN2, and LAMA5LN1. Amplified products were assigned scores based on signal intensity.
| Transcript | LAMA3A | LAMA3B | LAMA3LN1 | LAMA3LN2a | LAMA3LN2b | LAMA5LN1 | LAMA5LN2 |
|---|---|---|---|---|---|---|---|
| Composition | ex 39–76 | ex 1–38, 40–76 | ex 1–9 + 9e | ex 1–25 + 25e | ex 1–9, 10–25 + 25e | ex 1–11 + 11e | ex 1–11 + 12* |
| Protein | Laminin α3a | Laminin α3b | LaNt α31 | LaNt α32a | LaNt α32b | LaNt α51 | LaNt α52 |
| Heart | + | ++ | ++ | + | |||
| Brain | + | ++ | + | ||||
| Placenta | +++++ | ++++ | +++ | ++ | +++ | + | |
| Lung | ++ | ++++ | ++ | + | |||
| Liver | + | ++ | ++ | + | + | ||
| Skeletal Muscle | + | ||||||
| Kidney | +++ | ++ | +++ | ++ | |||
| Pancreas | +++++ | +++ | + | + | |||
| Spleen | +++ | ++ | |||||
| Thymus | +++++ | + | ++ | + | + | ||
| Prostate | +++++ | +++ | ++ | ++ | +++ | + | |
| Testis | + | + | +++ | + | + | + | + |
| Ovary | + | + | |||||
| Small Intestine | + | ++ | +++ | ++ | |||
| Colon | +++++ | + | ++ | ||||
| Leukocyte | +++ | ++ | |||||
| Keratinocyte | +++++ | +++ | +++ |
For LAMA3LN1, relatively strong expression was detected in heart, placenta, prostate, testis, small intestine, leukocyte, and keratinocyte (Fig. 2A). Lower levels were detected in pancreas, brain, ovary, and fibroblasts (Fig. 2A). No products were present in the colon, liver, skeletal muscle, kidney, thymus, or spleen.
RT-PCR analysis of the expression profile of the other isoforms revealed distinct differences in distribution. LAMA3LN2a was strongly expressed in placenta, liver, thymus, and prostate with weaker expression in testis and colon; LAMA3LN2b was present in liver, testis, and leukocytes; LAMA5LN1 was strongly expressed in heart, brain, placenta, kidney, spleen, prostate, and small intestine and, more weakly, in liver, pancreas, thymus, and colon; LAMA5LN2 was present in tissues similar to LAMA5LN1 albeit at generally lower levels, particularly in kidney and pancreas with minor products in heart, placenta, liver, prostate, and small intestine (Fig. 2A and Table 1).
LAMA3LN1 in Situ Hybridization Shows Epidermal Staining
Of the newly identified isoforms, LAMA3LN1 has the longest region of unique mRNA and protein sequence and therefore was amenable for further study as a paradigm of this new transcript family. Its expression pattern was investigated through ISH using a probe derived from exon 9e, which is specific to the LAMA3LN1 transcript. Positive labeling was obtained in practically all epithelial structures in tongue and skin sections. Staining was seen in the epidermis/interfollicular epidermis from the basal layer to the granular layer with slightly stronger staining in the basal layer (Fig. 2C). Sebaceous glands (not shown), sweat glands, and ducts also stained (Fig. 2D). In the hair follicle, the outer and inner root sheaths were notably labeled with the most prominent label in the hair matrix up to the middle cortex (Fig. 2E). Negligible signal was obtained in connective tissues. These results largely parallel those previously obtained for LAMA3 transcripts in skin and hair (39, 40).
LAMA3LN1 Is Translated and Secreted by Keratinocytes.
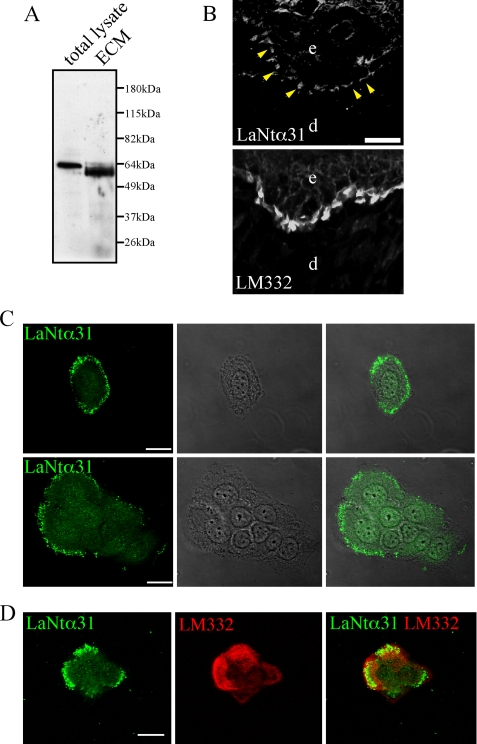
Of all the transcripts identified here, only the LAMA3LN1 transcript encodes a unique sequence of sufficient length (amino acids 423–487) that would allow generation of an antibody probe specific for the putative product of the transcript. This antibody probe would not recognize full length laminin α3a or α3b protein. A rabbit polyclonal antibody raised against this unique region recognizes a protein at the predicted molecular mass of ∼64 kDa in total cell lysate extracts derived from the cultured human keratinocytes prepared for immunoblotting (Fig. 3A). These data indicate that there is a bona fide product translated from the LAMA3LN1 transcript. Based on the structure of the protein, we suggest that it be termed LaNt (Laminin N terminus) α31.
FIGURE 3.
LaNt α31 protein expression. In A, HaCaT total protein and ECM extracts were prepared for Western blotting with a rabbit polyclonal antibody against LaNt α31. B shows immunofluorescence images of human foreskin sections stained for LaNt α31 (LaNt31, upper panel) or LM332 (lower panel). e, epidermis; d, dermis. Arrowheads indicate discontinuous staining basal staining. Bar, 20 μm. In C, HaCaT cells were plated onto glass coverslips, fixed, and then processed using antibodies LaNt α31. Cells were imaged by confocal immunofluorescence microscopy. Note punctate distribution of LaNt α31 staining around peripheral regions of both individual (C, upper panels) and groups of cells (lower panels). In D, HaCaT cells were prepared for double labeling using antibodies against LaNt α31 (green) and LM332 (red). Note the area of overlap of stain toward the edge of the cell. Bars, 50 μm.
Although LaNt α31 migrated around 64 kDa in extracts of keratinocytes, it exhibits a difference in mobility in ECM extracts of cultured keratinocytes. The higher mobility suggests the possibility that LaNt α31 undergoes proteolytic processing upon secretion (Fig. 3A).
LaNt α31 antibody generated bright staining along the basement membrane underlying the keratinocytes in the basal layer of the epidermis (Fig. 3B). This staining was not continuous and was distinct from the localization of LM332 along the basement membrane (Fig. 3B). We also evaluated the localization of LaNt α31 by confocal immunofluorescence microscopy in HaCaT cells. The LaNt α31 antibody generated a punctate stain toward the periphery of both individual cells and around the outer edges of groups of cells (Fig. 3C). These puncta were distributed along the substratum-attached surface of the cells and showed partial overlap with staining generated by antibody GB3, which recognizes assembled LM332 trimers (41, 42) (Fig. 3D).
RNAi-mediated Knockdown of LAMA3LN1
LN domains have been implicated in cell adhesion, neurite outgrowth, binding to perlecan, heparin, and heparan sulfate and in laminin self-assembly (43–46). We therefore tested the impact of knockdown of LaNt α31 in HaCaT cells (29), which we have shown to express LaNt α31 by RT-PCR (Fig. 2A), immunoblotting (Fig. 3A), and immunofluorescence (Fig. 3, C and D).
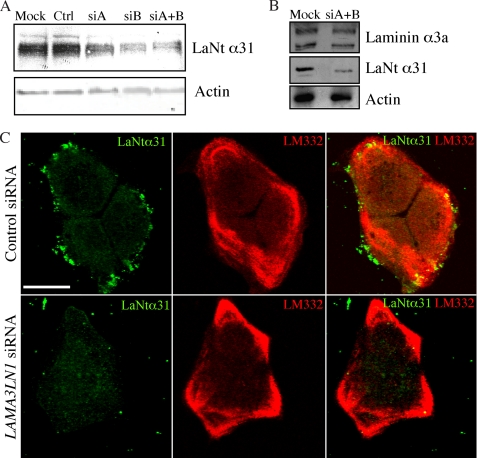
Two siRNA oligos were designed to target regions in LAMA3 exon 9e. These were predicted to be specific for the LAMA3LN1 transcript (33). Effective knockdown of LaNt α31 was observed with both siRNAs individually and to a slightly greater extent when used in combination (Fig. 4A). Most efficient knockdown was obtained at 72-h post-transfection (data not shown). All subsequent experiments were performed at, or spanning, this time point. Following treatment with LAMA3LN1 siRNAs or controls (lamin A/C siRNA, scrambled siRNA) a dye uptake assay was used to measure cellular proliferation rates, and no significant differences were observed between treatments (not shown). Knockdown of LaNt α31 had no obvious effect on expression of laminin α3a or assembly of LM332 matrix, as determined by immunofluorescence (Fig. 4, B and C).
FIGURE 4.
LaNt α31 knockdown using siRNA. In A, HaCaT cells were mock-transfected (Mock), or transfected with lamin A/C siRNA (Ctrl), or LAMA3LN1 siRNAs siA and siB either individually or in combination (siA+B). 72-h post-transfection, total cell extracts were processed for immunoblotting with anti-actin and anti-LaNt α31 antibodies. In B, protein extracts from HaCaT cells, 72 h following transfection with LAMA3LN1 siRNA or mock transfected, were processed for immunoblotting with antibodies against laminin α3a (RG13), LaNt α31, and actin. In C, HaCaT cells were transfected with either LAMA3LN1 or control siRNA, incubated for 48 h, then plated onto glass coverslips. 24 h later, the cells were fixed and prepared for immunofluorescence with antibodies against LaNt α31 (green) and LM332 (red). Bar, 50 μm.
Knockdown of LaNt α31 Impairs Cellular Adhesion
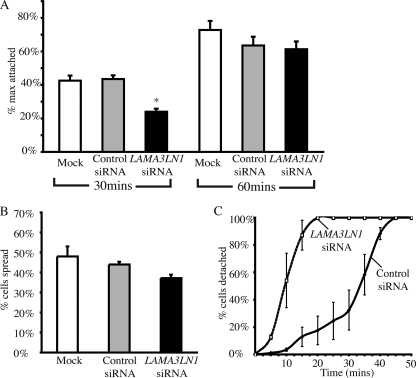
We utilized two different assays to determine if LaNt α31 expression is required for cellular attachment and spreading. For attachment, we used a plate-based assay as described by Yuen et al. (32) to compare the rate of attachment following trypsinization. Compared with controls, LAMA3LN1 siRNA-treated cells initially show a significant delay in adhesion (Fig. 5A; 54% of mock-transfected cell adhesion, p = 0.001 at 30 min). By 60 min, differences between controls and knockdown cells were not significant suggesting that the adhesion defect primarily affected the initial stages of attachment. For cell spreading, cells were imaged and scored by phenotype 2 h after re-seeding onto uncoated tissue culture plates. In comparison to controls, LAMA3LN1 knockdown cells showed a slight reduction in spreading at 2 h after replating that is not seen at later time points (Fig. 5B). However, differences were below significance (p = 0.09).
FIGURE 5.
LaNt α31 knockdown cells display impaired adhesion relative to controls. In A, HaCaT cells, 72-h post-transfection with siRNAs directed against either LAMA3LN1 or lamin a/c, were trypsinized and plated into 96-well plates, allowed to attach for 30, 60, and 360 min, and the percentage of cells attached determined relative to the 360-min time point by staining. Results are plotted as mean ± S.E. from three repetitions, 9 wells per repetition (*, p < 0.001). In B, HaCaT cells, 72-h post-transfection, were plated into 6-well plates, allowed to adhere for 2 h, and their spreading profile scored. Results are plotted as mean percentage cells spread ± S.E. as determined from counting 6 randomly selected fields per treatment, three repetitions (p = 0.09 LAMA3LN1 siRNA relative to control siRNA). In C, HaCaT cells, 72-h post-transfection with LAMA3LN1 siRNA or control siRNA, were exposed to a dilute trypsin (0.005% trypsin, 0.0004% EDTA), and images were collected every 5 min. Cells were scored based on phenotypic appearance, with round cells scored as “detached.” Values represent the mean of three repetitions, 6 fields per repetition (p < 0.01 from 15 to 30 min). p values determined using paired Student's t test.
During routine handling of the siRNA-transfected cells, we became aware that the LAMA3LN1 knockdown cells responded more rapidly to trypsin exposure than control-treated cells. To quantify this, we exposed knockdown and control siRNA-treated keratinocytes to a diluted trypsin (final concentration 0.005% trypsin, 0.0004% EDTA) and scored cell rounding over time. LaNt α31 knockdown cells responded significantly faster than control siRNA-transfected cells (Fig. 5C).
LaNt α31 Is Up-regulated during Wound Healing, and Knockdown Impacts Wound Closure in Vitro
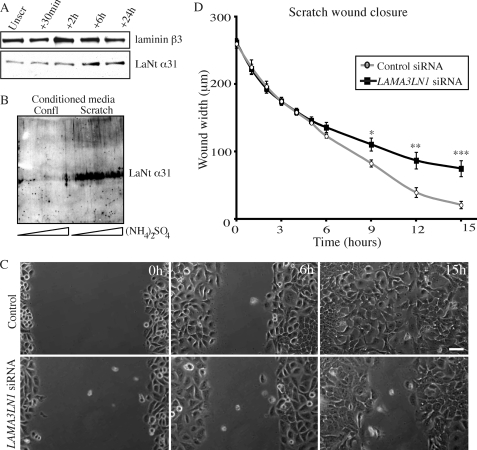
To determine if LaNt α31 expression levels change during wound healing of monolayers of keratinocytes in vitro, ECM extracts were derived from keratinocytes before and 30 min, 2 h, 6 h, and 24 h following introduction of scratch wounds. LaNt α31 protein levels increased following wounding, showing maximal expression around 6-h postwounding, suggesting a potential role in the wound-healing process (Fig. 6A). Moreover, conditioned medium of either confluent or scratch-wounded keratinocytes grown for 24 h in serum-free medium then concentrated with ammonium sulfate revealed a dramatic increase in the level of LaNt α31 secreted into the scratch-wounded culture medium (Fig. 6B).
FIGURE 6.
Keratinocytes up-regulate secretion of LaNt α31 upon wound healing, and LaNt α31 knockdown inhibits their wound-healing ability. In A, ECM extracts derived from confluent wild-type HaCaT cells and 30 min, 2, 6, or 24 h following scratch wounding were probed by Western blotting for LaNt α31 or laminin β3 (B1K) as indicated. In B, either confluent or scratch-wounded HaCaT cells were grown in serum medium for 24 h. Conditioned medium was concentrated with increasing concentrations of ammonium sulfate (1.5-4 m, as indicated by the wedge), and the precipitated proteins were immunoblotted with antibodies against LaNt α31. In C and D, confluent control siRNA or LAMA3LN1 siRNA-transfected cells, 72 h post-transfection, were scratch wounded, and phase contrast images of the wounded area taken every 20 min over 36 h. Representative images at 0 and 6 h and 15 h postwounding are presented in C. Bar, 50 μm. Wound widths were measured at multiple points along the scratch and are plotted in D. Asterisks denote significance; *, p < 0.05; **, p < 0.02; ***, p < 0.01.
To determine if LaNt α31 is required for the closure of wounds, a scratch was introduced into a confluent monolayer of LAMA3LN1 siRNA or control-treated keratinocytes, and images at representative points along the wound margins were taken at 20-min intervals over 36 h (Fig. 6C and supplemental materials). All control keratinocytes had completely closed the introduced wound by 24 h after wounding. In contrast, there were still gaps between the migrating fronts of cells in which LaNt α31 was knocked down (Fig. 6C). Repetition of this experiment in the presence of mitomycin C gave identical results, indicating that the differences in wound closure time was not due to any difference in proliferation rate (not shown).
Closer analyses of the acquired images revealed that the wound closure to be distinctly biphasic (Fig. 6D). During the first phase, cells migrated rapidly over the first ∼6 h to form a distinct line along the edge of the scratch site. Both control and LaNt α31 knockdown cells migrated at similar rates over this period (∼20 μm/h). In the second phase, control cells continued in a slightly slower manner (∼14 μm/h from hours 6–12) during which time they presented numerous forward cellular projections. In contrast, the LaNt α31 knockdown cells projected fewer lamellipodia along their leading edge as they advanced and showed a markedly reduced migration rate relative to controls (8 μm/hr from 6–12 h postscratching). Differences between the populations in terms of wound closure were not significant before 6 h but became increasingly significant thereafter (Fig. 6D, p < 0.05 at 9 h, p < 0.02 at 12 h, p < 0.01 at 15 h).
DISCUSSION
The laminins are an important family of ECM proteins whose functions and interactions have been extensively studied because of their involvement in development, wound healing, cancer, and genetic diseases (14, 47). Here, through in silico analyses and RT-PCR, we have identified multiple novel isoforms derived from the 5′-end of laminin α3 and α5-encoding genes and have mapped their tissue expression profiles. Each of these identified transcripts predicts expression of short polypeptides comprising a laminin N-terminal domain followed by a short stretch of laminin-type epidermal growth factor-like repeats, which therefore structurally resemble the netrin family of axonal guidance cues (11, 48–50). However, all of the predicted proteins of this novel family lack the characteristic C-terminal basic domain found in netrins (48, 50).
Because of the short regions of unique sequence in several of the transcripts we have been unable to generate appropriate tools to confirm their existence at the protein level and to further investigate their functionality. This is not the case with transcript LAMA3LN1. The longer extension to exon 9 has allowed us to generate specific probes, including siRNAs to the LAMA3LN1 mRNA and antibodies to the putative protein product encoded by this transcript. Our antibody confirmed the existence of a secreted protein, which we have termed LaNt α31. The LaNt α31 antibody stains the basement membrane of intact human epithelia although the pattern is distinct from that generated by a LM332 antibody. Likewise, the LaNt α31 antibody localizes along the substratum-associated region of both individual and groups of cultured keratinocytes. Although it fails to perfectly co-localize with LM332 matrix secreted by keratinocytes, its localization partially overlaps with that of LM332. Most important, functional analyses, utilizing RNAi-mediated knockdown, demonstrate a role for this novel protein in cell attachment and cell migration.
One interesting aspect of our study is the identification of tissue-specific expression profiles of the different LaNt encoding transcripts. Of particular note is the differences observed between LAMA3B and LAMA3LN1, which share a common promoter. For example, LAMA3B mRNA is highly expressed in placenta, lung, pancreas, and prostate whereas the LAMA3LN1 message is expressed at a higher level in heart, testis, small intestine, and by leukocytes. These data suggest that transcription of LAMA3LN1 is not purely a consequence of expression of LAMA3B. Rather, this implies that LAMA3LN1 production can be regulated independently of the “full length” transcript. Interestingly, in this regard, our finding that the LAMA3LN1 transcript is conserved between human and mouse implies there has been evolutionary pressure to maintain its expression (1).
Because the LaNt proteins are related to the netrins, it is tempting to speculate that they share some common functions. For example, like the netrins, LaNt proteins may regulate adhesion and migration via interaction with cell surface receptors such as the integrins (51–54). Conversely LaNts, may modulate laminin matrix assembly/disassembly through their LN domains because this is a property of the netrins (55). Indeed, in this regard it has already been demonstrated that the mouse α3b laminin N terminus inhibits LM111 polymerization in vitro (45). We are currently investigating such possibilities.
Alternative splicing is clearly an important mechanism via which the diversity of expressed proteins can be expanded. We speculate that LaNt family members may play important roles in regulating laminin matrix functions; this would be consistent with our finding that LaNt protein expression is up-regulated during wound healing and its knockdown modulates keratinocyte adhesion and migration, even though LM332 matrix appears unperturbed, and its secretion is apparently unaffected. How, precisely, LaNt protein members do so should be an interesting avenue of investigation in the future.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AR054184 (to J. C. R. J.), This work was also supported by grants from the Dystrophic Epidermolysis Bullosa Research Association (to W. H. I. M.). Human tissue sections were provided by Northwestern University Skin Disease Research Center, Chicago, IL.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- ECM
- extracellular matrix
- LM
- laminin
- LN
- laminin N-terminal domain
- LaNt
- laminin N terminus
- LE
- laminin-type epidermal growth factor-like repeat
- LCC
- laminin coiled-coil domain
- LG
- laminin globular domain
- ISH
- in situ hybridization
- PBS
- phosphate-buffered saline
- EST
- expressed sequence tag.
REFERENCES
- 1.Kim E., Goren A., Ast G. (2008) Bioessays 30, 38–47 [DOI] [PubMed] [Google Scholar]
- 2.Modrek B., Lee C. J. (2003) Nat. Genet. 34, 177–180 [DOI] [PubMed] [Google Scholar]
- 3.Sakabe N. J., Vibranovski M. D., de Souza S. J. (2004) Genet. Mol. Res. 3, 532–544 [PubMed] [Google Scholar]
- 4.Boyd C. D., Pierce R. A., Schwarzbauer J. E., Doege K., Sandell L. J. (1993) Matrix 13, 457–469 [DOI] [PubMed] [Google Scholar]
- 5.O'Toole E. A. (2001) Clin. Exp. Dermatol. 26, 525–530 [DOI] [PubMed] [Google Scholar]
- 6.Mauch J. C., Sandberg L. B., Roos P. J., Jimenez F., Christiano A. M., Deak S. B., Boyd C. D. (1995) Matrix Biol. 14, 635–641 [DOI] [PubMed] [Google Scholar]
- 7.Sandell L. J. (1996) Connect Tissue Res. 35, 1–6 [DOI] [PubMed] [Google Scholar]
- 8.Kosmehl H., Berndt A., Katenkamp D., Mandel U., Bohle R., Gabler U., Celeda D. (1995) Pathol. Res. Pract. 191, 1105–1113 [DOI] [PubMed] [Google Scholar]
- 9.Timpl R. (1993) Experientia 49, 417–428 [DOI] [PubMed] [Google Scholar]
- 10.White E. S., Baralle F. E., Muro A. F. (2008) J. Pathol. 216, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin Y., Miner J. H., Sanes J. R. (2002) Mol. Cell Neurosci. 19, 344–358 [DOI] [PubMed] [Google Scholar]
- 12.Schor S. L., Ellis I. R., Jones S. J., Baillie R., Seneviratne K., Clausen J., Motegi K., Vojtesek B., Kankova K., Furrie E., Sales M. J., Schor A. M., Kay R. A. (2003) Cancer Res. 63, 8827–8836 [PubMed] [Google Scholar]
- 13.Ferrigno O., Virolle T., Galliano M. F., Chauvin N., Ortonne J. P., Meneguzzi G., Aberdam D. (1997) J. Biol. Chem. 272, 20502–20507 [DOI] [PubMed] [Google Scholar]
- 14.Aumailley M., Smyth N. (1998) J. Anat. 193, 1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki N., Yokoyama F., Nomizu M. (2005) Connect Tissue Res. 46, 142–152 [DOI] [PubMed] [Google Scholar]
- 16.Aumailley M., Bruckner-Tuderman L., Carter W. G., Deutzmann R., Edgar D., Ekblom P., Engel J., Engvall E., Hohenester E., Jones J. C., Kleinman H. K., Marinkovich M. P., Martin G. R., Mayer U., Meneguzzi G., Miner J. H., Miyazaki K., Patarroyo M., Paulsson M., Quaranta V., Sanes J. R., Sasaki T., Sekiguchi K., Sorokin L. M., Talts J. F., Tryggvason K., Uitto J., Virtanen I., von der Mark K., Wewer U. M., Yamada Y., Yurchenco P. D. (2005) Matrix Biol. 5, 326–332 [DOI] [PubMed] [Google Scholar]
- 17.McLean W. H., Irvine A. D., Hamill K. J., Whittock N. V., Coleman-Campbell C. M., Mellerio J. E., Ashton G. S., Dopping-Hepenstal P. J., Eady R. A., Jamil T., Phillips R. J., Shabbir S. G., Haroon T. S., Khurshid K., Moore J. E., Page B., Darling J., Atherton D. J., Van Steensel M. A., Munro C. S., Smith F. J., McGrath J. A. (2003) Hum. Mol. Genet. 12, 2395–2409 [DOI] [PubMed] [Google Scholar]
- 18.Airenne T., Haakana H., Sainio K., Kallunki T., Kallunki P., Sariola H., Tryggvason K. (1996) Genomics 32, 54–64 [DOI] [PubMed] [Google Scholar]
- 19.Lee G., Kim M. G., Yim J. B., Hong S. H. (2001) Mol. Cells 12, 380–390 [PubMed] [Google Scholar]
- 20.Hayashi Y., Kim K. H., Fujiwara H., Shimono C., Yamashita M., Sanzen N., Futaki S., Sekiguchi K. (2002) Biochem. Biophys. Res. Commun. 299, 498–504 [DOI] [PubMed] [Google Scholar]
- 21.Hao J., McDaniel K., Weyer C., Barrera J., Nagle R. B. (2002) Gene 283, 237–244 [DOI] [PubMed] [Google Scholar]
- 22.Talts J. F., Mann K., Yamada Y., Timpl R. (1998) FEBS Lett. 426, 71–76 [DOI] [PubMed] [Google Scholar]
- 23.Gonzales M., Haan K., Baker S. E., Fitchmun M., Todorov I., Weitzman S., Jones J. C. (1999) Mol. Biol. Cell 10, 259–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langhofer M., Hopkinson S. B., Jones J. C. (1993) J. Cell Sci. 105, 753–764 [DOI] [PubMed] [Google Scholar]
- 25.Airenne T., Lin Y., Olsson M., Ekblom P., Vainio S., Tryggvason K. (2000) Cell Tissue Res. 300, 129–137 [DOI] [PubMed] [Google Scholar]
- 26.Ashton G. H., McLean W. H., South A. P., Oyama N., Smith F. J., Al-Suwaid R., Al-Ismaily A., Atherton D. J., Harwood C. A., Leigh I. M., Moss C., Didona B., Zambruno G., Patrizi A., Eady R. A., McGrath J. A. (2004) J. Invest. Dermatol. 122, 78–83 [DOI] [PubMed] [Google Scholar]
- 27.Cichon S., Anker M., Vogt I. R., Rohleder H., Pützstück M., Hillmer A., Farooq S. A., Al-Dhafri K. S., Ahmad M., Haque S., Rietschel M., Propping P., Kruse R., Nöthen M. M. (1998) Hum. Mol. Genet. 7, 1671–1679 [DOI] [PubMed] [Google Scholar]
- 28.Langbein L., Spring H., Rogers M. A., Praetzel S., Schweizer J. (2004) Methods Cell Biol. 78, 413–451 [PubMed] [Google Scholar]
- 29.Boukamp P., Petrussevska R. T., Breitkreutz D., Hornung J., Markham A., Fusenig N. E. (1988) J. Cell Biol. 106, 761–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones J. C., Goldman A. E., Steinert P. M., Yuspa S., Goldman R. D. (1982) Cell Motil. 2, 197–213 [DOI] [PubMed] [Google Scholar]
- 31.Sehgal B. U., DeBiase P. J., Matzno S., Chew T. L., Claiborne J. N., Hopkinson S. B., Russell A., Marinkovich M. P., Jones J. C. (2006) J. Biol. Chem. 281, 35487–35498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuen H. W., Ziober A. F., Gopal P., Nasrallah I., Falls E. M., Meneguzzi G., Ang H. Q., Ziober B. L. (2005) Exp. Cell Res. 309, 198–210 [DOI] [PubMed] [Google Scholar]
- 33.Reynolds A., Leake D., Boese Q., Scaringe S., Marshall W. S., Khvorova A. (2004) Nat. Biotechnol. 22, 326–330 [DOI] [PubMed] [Google Scholar]
- 34.Gagnoux-Palacios L., Allegra M., Spirito F., Pommeret O., Romero C., Ortonne J. P., Meneguzzi G. (2001) J. Cell Biol. 153, 835–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gagnoux-Palacios L., Vailly J., Durand-Clement M., Wagner E., Ortonne J. P., Meneguzzi G. (1996) J. Biol. Chem. 271, 18437–18444 [DOI] [PubMed] [Google Scholar]
- 36.Harlow E., Lane D. (1988) Antibodies: A Laboratory Manual, CSHL Press, Cold Spring Harbor, NY [Google Scholar]
- 37.Zhang M. Q. (1998) Hum Mol. Genet. 7, 919–932 [DOI] [PubMed] [Google Scholar]
- 38.Beaudoing E., Freier S., Wyatt J. R., Claverie J. M., Gautheret D. (2000) Genome Res. 10, 1001–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miosge N., Kluge J. G., Studzinski A., Zelent C., Bode C., Sprysch P., Burgeson R. E., Herken R. (2002) Anat. Embryol. 205, 355–363 [DOI] [PubMed] [Google Scholar]
- 40.Sugawara K., Tsuruta D., Kobayashi H., Ikeda K., Hopkinson S. B., Jones J. C., Ishii M. (2007) J. Histochem. Cytochem. 55, 43–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verrando P., Hsi B. L., Yeh C. J., Pisani A., Serieys N., Ortonne J. P. (1987) Exp. Cell Res. 170, 116–128 [DOI] [PubMed] [Google Scholar]
- 42.Matsui C., Nelson C. F., Hernandez G. T., Herron G. S., Bauer E. A., Hoeffler W. K. (1995) J. Invest. Dermatol. 105, 648–652 [DOI] [PubMed] [Google Scholar]
- 43.Nomizu M., Yokoyama F., Suzuki N., Okazaki I., Nishi N., Ponce M. L., Kleinman H. K., Yamamoto Y., Nakagawa S., Mayumi T. (2001) Biochemistry 40, 15310–15317 [DOI] [PubMed] [Google Scholar]
- 44.Nielsen P. K., Yamada Y. (2001) J. Biol. Chem. 276, 10906–10912 [DOI] [PubMed] [Google Scholar]
- 45.Garbe J. H., Göhring W., Mann K., Timpl R., Sasaki T. (2002) Biochem. J. 362, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Odenthal U., Haehn S., Tunggal P., Merkl B., Schomburg D., Frie C., Paulsson M., Smyth N. (2004) J. Biol. Chem. 279, 44504–44512 [DOI] [PubMed] [Google Scholar]
- 47.Lohi J. (2001) Int. J. Cancer 94, 763–767 [DOI] [PubMed] [Google Scholar]
- 48.Barallobre M. J., Pascual M., Del Río J. A., Soriano E. (2005) Brain Res. Brain Res. Rev. 49, 22–47 [DOI] [PubMed] [Google Scholar]
- 49.Aoki-Suzuki M., Yamada K., Meerabux J., Iwayama-Shigeno Y., Ohba H., Iwamoto K., Takao H., Toyota T., Suto Y., Nakatani N., Dean B., Nishimura S., Seki K., Kato T., Itohara S., Nishikawa T., Yoshikawa T. (2005) Biol. Psychiatry 57, 382–393 [DOI] [PubMed] [Google Scholar]
- 50.Yurchenco P. D., Wadsworth W. G. (2004) Curr. Opin. Cell Biol. 16, 572–579 [DOI] [PubMed] [Google Scholar]
- 51.Stanco A., Szekeres C., Patel N., Rao S., Campbell K., Kreidberg J. A., Polleux F., Anton E. S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7595–7600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Staquicini F. I., Dias-Neto E., Li J., Snyder E. Y., Sidman R. L., Pasqualini R., Arap W. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2903–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yebra M., Montgomery A. M., Diaferia G. R., Kaido T., Silletti S., Perez B., Just M. L., Hildbrand S., Hurford R., Florkiewicz E., Tessier-Lavigne M., Cirulli V. (2003) Dev. Cell 5, 695–707 [DOI] [PubMed] [Google Scholar]
- 54.Nikolopoulos S. N., Giancotti F. G. (2005) Cell Cycle 4, e131–e135 [PubMed] [Google Scholar]
- 55.Schneiders F. I., Maertens B., Böse K., Li Y., Brunken W. J., Paulsson M., Smyth N., Koch M. (2007) J. Biol. Chem. 282, 23750–23758 [DOI] [PubMed] [Google Scholar]