Abstract
CRIg is a recently discovered complement C3 receptor expressed on a subpopulation of tissue-resident macrophages. The extracellular IgV domain of CRIg (CRIg-ECD) holds considerable promise as a potential therapeutic because it selectively inhibits the alternative pathway of complement by binding to C3b and inhibiting proteolytic activation of C3 and C5. However, CRIg binds weakly to the convertase subunit C3b (KD = 1.1 μm), and thus a relatively high concentration of protein is required to reach nearly complete complement inhibition. To improve therapeutic efficacy while minimizing risk of immunogenicity, we devised a phage display strategy to evolve a high affinity CRIg-ECD variant with a minimal number of mutations. Using the crystal structure of CRIg in complex with C3b as a guide for library design, we isolated a CRIg-ECD double mutant (Q64R/M86Y, CRIg-v27) that showed increased binding affinity and improved complement inhibitory activity relative to CRIg-ECD. In a mouse model of arthritis, treatment with a Fc fusion of CRIg-v27 resulted in a significant reduction in clinical scores compared with treatment with an Fc fusion of CRIg-ECD. This study clearly illustrates how phage display technology and structural information can be combined to generate proteins with nearly natural sequences that act as potent complement inhibitors with greatly improved therapeutic efficacy.
Introduction
Complement consists of serum proteins that, upon target recognition, interact to initiate a cascade of proteolytic reactions leading to coating of pathogens with complement components, formation of a membrane attack complex, and induction of an inflammatory response. As such, the complement system acts as a first line of innate immune defense protecting the host against bacterial and viral infections. Complement activation, initiated by the classical, lectin, or alternative pathways, is tightly controlled by membrane-bound and soluble regulators (1, 2). Under circumstances of complement regulator deficits or mutations that lead to blockade of the regulatory function, complement activation can escalate and cause inflammation and tissue damage (3). There is now accumulating genetic, preclinical, and clinical evidence for complement being a major contributor to the pathophysiology of membranoproliferative glomerulonephritis type II, age-related macular degeneration, and ischemia/reperfusion (4, 5).
Complement component C3, following conversion to C3b, constitutes a central subunit of the alternative pathway convertases that proteolytically converts C3 and C5 to the active products, C3a and C3b or C5a and C5b, respectively (6). The alternative pathway dominates complement activation because it amplifies complement activation initiated through any of the three pathways (7). Therefore, targeting C3b could provide a powerful approach to therapeutic intervention in complement-mediated diseases.
CRIg is a recently discovered complement receptor expressed on a subpopulation of tissue resident macrophages. CRIg binds C3b, iC3b, and C3c but not the parent molecule C3 and efficiently clears complement-coated particles from the circulation (8). Next to its function as a receptor for opsonized particles, the extracellular IgV domain of CRIg (CRIg-ECD)3 is a selective inhibitor of the alternative pathway of complement (9). However, CRIg-ECD binds the convertase subunit C3b with low (KD = 1.1 μm) affinity, which limits potential therapeutic utility.
In order to generate a more potent inhibitor as a therapeutic reagent, information gained from the crystal structure of CRIg in complex with C3b (9) was used in a phage display strategy to improve binding affinity of CRIg-ECD for C3b. Phage-displayed libraries were generated in which codons encoding residues of CRIg-ECD that contact C3b were subjected to limited randomization. Phage-displayed CRIg-ECD variants with improved binding affinity for C3b were selected and sequenced, and recombinant mutant CRIg-ECD proteins were expressed and purified. Two mutations together improved the affinity of CRIg-ECD for C3b by more than an order of magnitude without compromising selectivity for C3b. As a result, in vitro potency of the affinity-matured CRIg-ECD variant was improved by 6-fold relative to the wild type (WT). Finally, recombinant proteins consisting of affinity-matured CRIg-ECD fused to the Fc portion of murine IgG1 showed significantly improved inhibitory potency in the K/BxN serum transfer model of arthritis (10). In sum, we present the first example of structure-based design of a complement inhibitor to improve therapeutic activity.
EXPERIMENTAL PROCEDURES
Construction of Phage-displayed CRIg Libraries
A previously described phagemid (11) was modified by standard molecular biology techniques to construct a phagemid designed to display CRIg-ECD on the surface of M13 bacteriophage as a fusion with the N terminus of a fragment of the gene-3 minor coat protein. The resulting phagemid contained a gene under the control of the Ptac promoter, encoding for the following open reading frame: the maltose-binding protein secretion signal, followed by an epitope tag (gD-tag, amino acid sequence SMADPNRFRGKDLGS), followed by human CRIg-ECD (residues 20–137), followed by the C-terminal domain of the gene-3 minor coat protein. The phagemid was used as the template to construct phage-displayed libraries, as described (12, 13). Briefly, for each library, a “stop template” version of the phagemid (containing TAA stop codons in positions to be mutated) was used as the template for the Kunkel mutagenesis method (14) with a mutagenic oligonucleotide designed to simultaneously repair the stop codons and introduce mutations at the chosen positions using degenerate codons consisting of 70% of the WT nucleotide and 10% each of the other three nucleotides. Each mutagenesis reaction was electroporated into Escherichia coli SS320 to produce a library containing >1010 unique members. After overnight growth at 37 °C, in 2YT medium supplemented with 50 μg/ml carbenicillin and M13-KO7 helper phage (New England Biolabs), library phage were concentrated by precipitation with PEG/NaCl and resuspended in phosphate-buffered saline, 0.5% (w/v) bovine serum albumin (Sigma), 0.1% (v/v) Tween 20 (Sigma) (PBT buffer), as described (12, 13).
Sorting and Analysis of Phage-displayed CRIg Libraries
Maxisorp immunoplates (Nunc) were coated overnight at 4 °C with human C3b protein (5 μg/ml) prepared from purified C3 as described (8) and blocked for 1 h with bovine serum albumin. Phage solutions from each library (1012 phage/ml) were added to the coated immunoplates, incubated for 3 h at 25 °C to allow for binding, and washed 12 times with phosphate-buffered saline, 0.05% Tween 20 (PT buffer). Bound phage particles were eluted with 0.1 m HCl for 10 min, and the eluant was neutralized with 1.0 m Tris base. Eluted phage were amplified in E. coli XL1-blue (Stratagene) with M13-KO7 helper phage and used for further rounds of selection. After five rounds of selection, individual phage clones that bound to C3b were identified by phage ELISA and subjected to DNA sequence analysis.
Purification of CRIg Proteins
Phagemids representing selected CRIg-ECD variants were modified using standard molecular biology techniques to produce expression vectors designed for the secretion of CRIg protein into the E. coli periplasm. The gene encoding for the CRIg-ECD fusion protein was modified as follows. The region encoding the gD-tag was removed, a sequence encoding for a His tag (GGGHHHHH) was fused to the C terminus of CRIg-ECD, and an amber stop codon was inserted between the His-tag and the gene-3 minor coat protein.
A single colony of E. coli BL21(DE3) harboring the expression plasmid was inoculated into 30 ml of LB medium supplemented with 50 μg/ml carbenicillin (LB/carb medium) and was grown overnight at 37 °C. The bacteria were harvested, washed, resuspended, and inoculated into 500 ml of LB/carb medium. The culture was grown at 37 °C to midlog phase (A600 = 0.8). Protein expression was induced with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside, and the culture was grown for 24 h at 30 °C. The bacteria were pelleted by centrifugation, washed twice with phosphate-buffered saline, and frozen for 8 h at −80 °C. The pellet was resuspended in 50 ml of phosphate-buffered saline, and the cells were lysed with an Ultra-Turrax T8 homogenizer (IKA Labortechnik, Staufen, Germany) and an M-110F Microfluidizer® processor (Microfluidics). The CRIg-ECD variant proteins were purified with Ni2+-nitrilotriacetic acid-agarose and gel filtration chromatography.
CRIg-ECD-Fc was generated by subcloning the CRIg-ECD (residues 1–175) into a modified pRK5 expression vector encoding the murine IgG1 Fc region. The CRIg-v27-Fc expression vector was generated using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The Fc fusion proteins were produced by transfection of Chinese hamster ovary cells with the expression vectors followed by purification of secreted proteins by protein A affinity chromatography as described (8).
Fluid Phase Competitive ELISA
Maxisorp plates (384-well, Nunc (Neptune, NJ)) were coated overnight at 4 °C with huCRIg(L)-LFH (2 μg/ml), a construct consisting of the C terminus of the CRIg-ECD fused with a leucine zipper, a FLAG tag, and a His6 tag (9). The plates were washed three times with PT buffer and blocked for 2 h with bovine serum albumin. C3b protein (10 nm) was incubated for 1 h with serial dilutions of CRIg-ECD-Fc in PBT buffer, and the mixture was added to the blocked plates and incubated for 1 h. The plates were washed, and bound C3b was detected by the addition of an anti-human C3b antibody (clone 5F202, U.S. Biological (Swampscott, MA)), horseradish peroxidase-anti-murine IgG antibody conjugate (1:2000 dilution in PBT buffer) (Jackson ImmunoResearch, West Grove, PA), and TMB substrate. Color development was quenched with 1.0 m H3PO4, and the plates were read spectrophotometrically at 450 nm.
Complement Activation Assay
The ability of CRIg-ECD-Fc to inhibit complement activation was evaluated using the WieslabTM complement system alternative pathway kit (Alpco Diagnostics, Salem, NH). Serially diluted CRIg-ECD-Fc (400 to 0.2 nm) and C1q-deficient human serum (5%) (Complement Technology, Tyler, TX) were prepared at twice the final desired concentration, mixed 1:1, and preincubated for 5 min on an orbital shaker at 300 rpm prior to adding to the lipopolysaccharide-coated ELISA plates (100 μl/well). The remainder of the assay was performed according to the manufacturer's instructions.
Hemolysis Inhibition Assay
To determine the activity of the alternative complement pathway, a hemolysis assay using rabbit red blood cells (Colorado Serum Company, Denver, CO) was performed as described (9). The extent of hemolysis was determined spectrophotometrically, and the results were expressed as a percentage of hemolysis observed in the absence of inhibitor.
Zymosan Assay for C3 and C5 Convertase
The effect of CRIg-ECD-Fc, CRIg-v27-Fc, and a control Fc fusion protein (anti-gp120) on C3a des-Arg and C5a des-Arg production in mouse serum was determined as described (15).
AlphaScreen Competitive Assay
The potential cross-reactivity of the CRIg-ECD variant to C3 was evaluated using the AlphaScreen® histidine (nickel chelate) detection kit (PerkinElmer Life Sciences). Serially diluted human C3 and C3b (3000 to 0.7 nm), as well as fixed concentrations of biotinylated iC3b (30 nm, Complement Technology), and the wild type or variant CRIg-ECD (15–60 nm) were prepared at three times the final desired concentration, mixed 1:1:1, and preincubated at ambient temperature for 30 min on an orbital shaker at 3000 rpm. A 1:1 mixture of streptavidin donor beads and nickel chelate acceptor beads (0.1 mg/ml each) was prepared at 4 times the final desired concentration and added to the reaction. The reaction plate was protected from light and incubated at ambient temperature for 60 min on an orbital shaker at 3000 rpm. The plate was analyzed on an AlphaQuest®-HTS microplate analyzer (PerkinElmer Life Sciences).
Surface Plasmon Resonance
Affinities of C3b for mutant and wild type CRIg were determined by surface plasmon resonance measurements on a Biacore® A100 instrument (GE Healthcare). An anti-Fc capture format was employed, and the KD was calculated from equilibrium binding measurements. The sensor chip was prepared using the anti-muFc capture kit (catalog number BR-1008-38) following instructions supplied by the manufacturer. Mutant or wild type CRIg was diluted in running buffer (10 mm HEPES, pH 7.4, 150 mm NaCl, 0.01% Tween 20) to 1 μg/ml, and injections of 60 μl were made such that ∼100 response units of fusion protein were captured on one spot of the chip surface. Sensorgrams were recorded for 10-min injections of solutions of varied C3b concentration over the CRIg spot with subtraction of signal for a reference spot containing the capture antibody but no CRIg. Data were obtained for a 2-fold dilution series of C3b ranging in concentration from 4 μm to 15.6 nm with a flow rate at 10 μl/min and at a temperature of 25 °C. The surface was regenerated between binding cycles by a 30-s injection of 10 mm Gly-HCl, pH 1.7. Plateau values obtained at the end of each C3b injection were used to calculate KD using the Affinity algorithm of the Biacore A100 Evaluation software, version 1.1.
K/BxN Serum Transfer Model of Arthritis
All animals were held under sterile pathogen-free conditions, and animal experiments were approved by the Institutional Animal Care and Use Committee of Genentech. Arthritis was induced in BALB/c mice (Jackson Laboratories) by injection of 50 μl of serum obtained from a pooled cohort of K/BxN mice as described (16). Mice received daily subcutaneous injections of 300 μg of fusion protein starting the day prior to serum injection and were scored daily. The extent of disease was scored by visual observation as follows: 0, no evidence of erythema and swelling; 1, erythema and mild swelling confined to the midfoot (tarsal) or ankle; 2, erythema and mild swelling extending from the ankle to the midfoot; 3, erythema and moderate swelling extending from the ankle to the metatarsal joints; 4, erythema and severe swelling encompassing the ankle, foot, and digits. The mean score reflects the sum of the scores over four paws. All mice were sacrificed at the peak of disease 6 days following serum transfer, and paws were processed for histological evaluation as described (15).
Statistical Analysis
All p values were calculated with an unpaired, two-tailed, Student's t test assuming equal variance.
RESULTS
Selection and Characterization of CRIg Variants
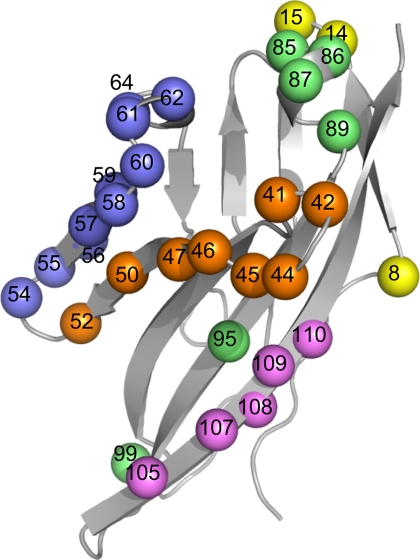
To generate CRIg variants with improved affinity for C3b, we displayed CRIg-ECD on phage and designed libraries targeting the epitope for binding to C3b. Five independent libraries were constructed to cover a total of 32 CRIg residues that make contact with C3b in the crystal structure of the CRIg-C3b complex (Fig. 1) (9). At each randomized position, the library was designed to encode ∼50% of the WT amino acid and ∼50% of a mixture of the other 19 genetically encoded amino acids. Following rounds of selection for binding to immobilized C3b, 24 clones were sequenced from each library and unique sequences were aligned to reveal that 15 of the 32 targeted positions had accumulated mutations (supplemental Fig. S1). For the purposes of affinity maturation, we decided to focus on 11 positions at which most of the sequences were mutated, suggesting that mutations at these positions are well tolerated structurally and may improve affinity for C3b.
FIGURE 1.
CRIg-ECD library design. The CRIg-ECD backbone (Protein Data Bank entry 2ICE) is shown as a gray ribbon, and positions that were diversified in the libraries are shown as spheres. Five libraries were designed to group together residues located close together in the primary sequence, and there was no overlap between the libraries. Spheres colored the same denote positions that were diversified in a single library. The color code and diversified residues for each library are as follows: library 1 (yellow, residues 8, 14, and 15); library 2 (orange, residues 41, 42, 44–47, 50, and 52); library 3 (blue, residues 54–62 and 64); library 4 (green, residues 85–87, 89, 95, and 99); library 5 (magenta, residues 105–110). Structural figures were generated using PyMOL (DeLano Scientific, Palo Alto, CA).
Because our goal was to minimize the number of mutations required to affinity-mature CRIg for binding to C3b, we constructed a panel of CRIg-ECD point mutants with mutations that occurred at the 11 heavily mutated positions described above (Table 1). We relied on two distinct assays to compare these mutants to the WT CRIg-ECD. In the first assay, we estimated the improvement in affinity in a fluid phase competitive ELISA. In the second assay, we assessed the improvement in biological activity using purified proteins in a hemolysis assay that measures the ability of CRIg-ECD to inhibit the lysis of rabbit red blood cells. In total, 21 CRIg-ECD point mutants were tested, and many showed improvements in affinity and/or activity, but notably, improvements in the two assays were not strongly correlated, suggesting that improved affinity does not necessarily translate directly to improved biological activity.
TABLE 1.
Relative C3b affinity and biological activity of CRIg variants
Affinity and activity analysis for CRIg-ECD variants. The -fold improvement in affinity or activity relative to WT was calculated by a fluid phase competitive ELISA or hemolysis assay, respectively. See “Experimental Procedures” for further details.
| Variant | Mutations | IC50 (WT)/IC50 (variant) |
|
|---|---|---|---|
| Affinity | Activity | ||
| v1 | E8Y | 0.9 | 1 |
| v2 | E8W | 2 | 4 |
| v3 | W14F | 0.4 | 2 |
| v4 | G42D | 0.7 | 1 |
| v5 | D44H | 1 | 0.5 |
| v6 | P45F | 0.3 | 1 |
| v7 | Q60I | 0.9 | 5 |
| v8 | Q64R | 10 | 2 |
| v9 | M86Y | 3 | 5 |
| v10 | M86W | 4 | 3 |
| v11 | M86F | 0.5 | 4 |
| v12 | Q99K | 1 | 1 |
| v13 | Q99R | 0.2 | 0.8 |
| v14 | Q99Y | 0.7 | 0.4 |
| v15 | Q99F | 1 | 0.9 |
| v16 | Q99L | 0.2 | 0.8 |
| v17 | Q105R | 0.5 | 0.9 |
| v18 | Q105K | 0.6 | 1 |
| v19 | K110D | 2 | 3 |
| v20 | K110N | 2 | 2 |
| v21 | K110H | 0.3 | 0.5 |
| v22 | E8W/G42D | 1 | 0.3 |
| v23 | E8W/M86W | 6 | 1 |
| v24 | E8W/K110N | 1 | 1 |
| v25 | Q64R/E8Y | 2 | 1 |
| v26 | Q64R/G42D | 1 | 2 |
| v27 | Q64R/M86Y | 18 | 6 |
| v28 | Q64R/M86W | 13 | 3 |
| v29 | Q64R/Q99Y | 1 | 1 |
| v30 | Q64R/Q99F | 1 | 0.8 |
| v31 | Q64R/Q105K | 3 | 2 |
| v32 | M86Y/E8Y | 6 | 2 |
| v33 | M86Y/Q99K | 5 | 4 |
| v34 | M86Y/Q99R | 3 | 2 |
| v35 | M86Y/Q105R | 12 | 2 |
| v36 | M86Y/Q105K | 9 | 4 |
To further improve the affinity and activity of CRIg-ECD, we next constructed a panel of double mutants by combining single mutations selected from different libraries. We chose to combine three mutations that appeared to improve both affinity and activity (E8W, Q64R, and M86Y) with a diverse array of other mutations. In total, we screened 15 CRIg-ECD double mutants and found that CRIg-v27, which contained the mutations Q64R/M86Y, exhibited the highest affinity for C3b and the highest biological activity. Thus, we chose CRIg-v27 for further analysis.
Improved Binding Affinity and Complement-inhibitory Activity of CRIg-v27
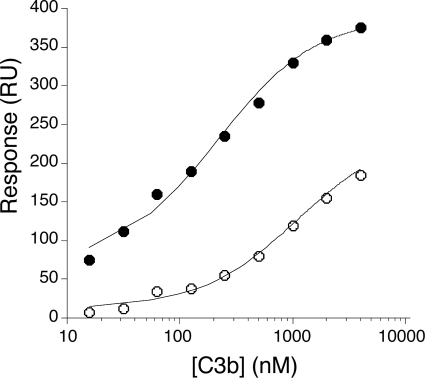
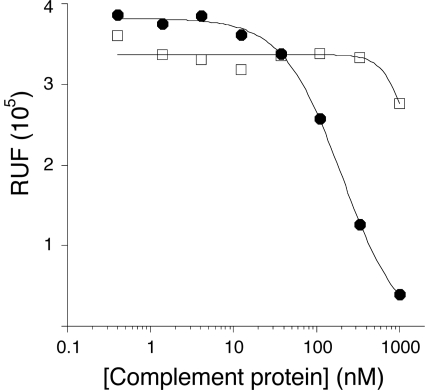
We used surface plasmon resonance analysis to compare binding affinity of WT CRIg-ECD and CRIg-v27 for C3b (Fig. 2). Consistent with the results from the fluid phase phage ELISA, the affinity of CRIg-v27 (KD = 200 nm) as measured by surface plasmon resonance was improved ∼5-fold over the WT (KD = 1100 nm). Previous studies have shown that CRIg selectively binds to C3b but not to native C3 (9). Because mutagenesis may influence this selectivity, we compared the affinity of CRIg-v27 for C3b and C3 in a fluid phase competition assay. CRIg-v27 bound soluble C3b, but not soluble C3, indicating that the mutations did not affect selectivity for the active component C3b (Fig. 3).
FIGURE 2.
CRIg-v27 shows improved affinity for C3b. Binding was measured by surface plasmon resonance by detection of response (y axis) to increasing concentrations of C3b flowed over immobilized WT CRIg-ECD or CRIg-v27 (x axis). Steady state analysis of the binding data was used to derive dissociation constants for C3b binding to WT CRIg-ECD (open circles, KD = 1.1 μm) or CRIg-v27 (closed circles, KD = 0.2 μm). RU, response units.
FIGURE 3.
CRIg-v27 is selective for C3b over C3. AlphaScreen competitive assay shows a dose-dependent decrease in the binding of CRIg-v27 to bead-bound iC3b (y axis) in response to increasing concentrations of solution phase C3b (filled circles) but not C3 (open squares) (x axis). The slight decrease in signal at the highest C3 concentration is also observed for the binding of WT CRIg-ECD (data not shown). RFU, relative units of fluorescence.
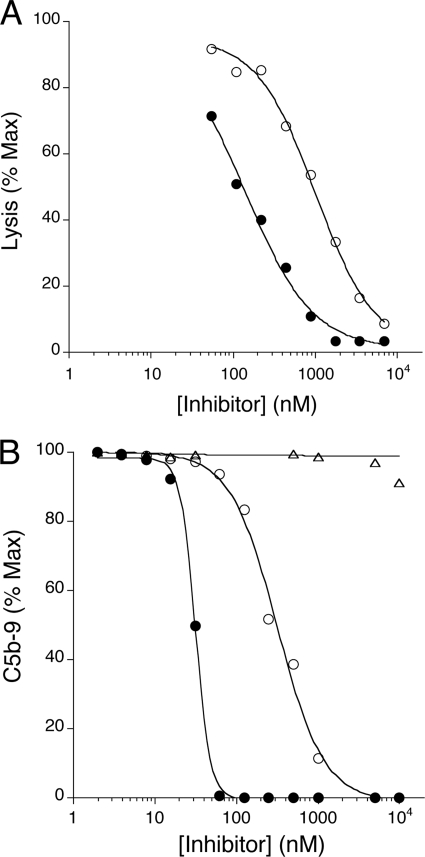
To test whether the improved affinity and conserved selectivity of CRIg-v27 for C3b translates into improved efficacy, we tested the activity of CRIg-v27 compared with WT CRIg-ECD in an erythrocyte-based activity assay selective for the alternative pathway of complement. CRIg-v27 showed a 5-fold improved efficacy in the assay relative to WT CRIg-ECD (Fig. 4A). To further substantiate improved potency of alternative pathway complement inhibition, we compared the inhibitory activities of CRIg-v27 and WT CRIg-ECD in an ELISA selective for the alternative pathway of complement. Consistent with the hemolytic assay results, CRIg-v27 showed ∼6-fold better potency than WT CRIg-ECD (Fig. 4B). Similar to WT CRIg-ECD (9), CRIg-v27 did not inhibit classical pathway complement activation (results not shown). In addition, CRIg-v27 showed superior blockade of alternative pathway activation in mouse serum compared with WT CRIg-ECD, and thus, the variant also exhibits enhanced activity against mouse C3b (supplemental Fig. S2). In summary, two amino acid substitutions in the CRIg binding site for C3b result in improved binding affinity and superior complement-inhibitory activity across species while retaining selectivity for C3b and the alternative pathway of complement.
FIGURE 4.
CRIg-v27 shows improved potency for complement inhibition. A, complement inhibition by CRIg-ECD (open circles) or CRIg-v27 (filled circles) measured by an alternative pathway-selective hemolytic assay using rabbit red blood cells and C1q-depleted human serum. B, complement inhibition by CRIg-ECD (open circles), CRIg-v27 (filled circles) or control protein (anti-gp120, triangles) measured by an alternative pathway-selective ELISA using C1q-depleted human serum and plates coated with lipopolysaccharide.
Improved Efficacy of CRIg-v27 in an in Vivo Model of Complement-mediated Inflammation
To determine whether increased binding affinity and potency may translate into improved therapeutic efficacy, we compared the effects of systemically delivered CRIg-v27 or WT CRIg-ECD on complement-mediated inflammation in vivo. Previous studies have shown that CRIg inhibits inflammation and bone destruction in two preclinical models of arthritis (15). Efficacy of WT CRIg-ECD and CRIg-v27 were compared in a third preclinical model of immune complex-mediated arthritis induced by transfer of serum from K/BxN T cell receptor transgenic mice to WT recipients. Serum transfer results in rapid and robust symmetric inflammation of the joints induced by anti-glucose-6-phosphate isomerase autoantibodies (10). Importantly, induction of joint inflammation is fully dependent both on an intact alternative complement pathway and on Fc receptor γ function (16, 17).
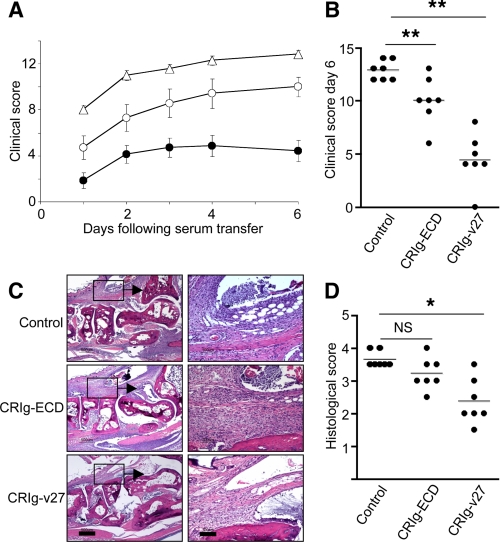
To increase serum half-life in vivo, CRIg-v27 or WT CRIg-ECD was fused to the Fc portion of mouse IgG1. Due to the rapid onset and increased disease severity in the K/BxN model of arthritis as compared with the previously tested anti-collagen antibody-mediated arthritis models, treatment with WT CRIg-ECD-Fc fusion protein reduced arthritis scores by only 22% as compared with mice treated with a control protein (Fig. 5, A and B). In contrast, treatment with CRIg-v27-Fc fusion protein showed a reduction in arthritis scores by 66%. Infiltration of immune cells, consisting primarily of neutrophils and macrophages, was significantly reduced in mice treated with CRIg-v27-Fc relative to those treated with WT CRIg-ECD-Fc or control-Fc (Fig. 5, C and D). Serum concentrations of WT CRIg-ECD-Fc and CRIg-v27-Fc were similar (results not shown), indicating that the improved in vivo efficacy of CRIg-v27 compared with WT CRIg-ECD-Fc was not due to target-mediated differences in the half-lives of the two proteins. Thus, we show that increased binding affinity of CRIg for C3b translates into a significantly improved complement-inhibitory activity in vitro and a significantly better therapeutic efficacy in vivo.
FIGURE 5.
CRIg-v27 shows improved efficacy in vivo. A, clinical scores for mice treated with CRIg-ECD-Fc (open circles), CRIg-v27-Fc (filled circles), or a negative control Fc fusion protein (triangles) 1 day prior to injection with K/BxN serum. Data represent mean ± S.E. of 7 mice/group. B, scatter plots of clinical scores for individual mice at day 6 following serum transfer. C, hematoxylin and eosin-stained sections of metacarpo-phalangeal joints. Insets in the left panels are magnified in the right panels to highlight mononuclear and neutrophilic inflammatory infiltrates in the joint. D, scatter plots of histological scores at day 6 following serum transfer. *, p < 0.05; **, p < 0.01; ***, p < 0.001. NS, not significant. Scale bars in C, 500 μm (left) or 100 μm (right).
DISCUSSION
A key role for the complement alternative pathway in a variety of inflammatory and autoimmune diseases has become widely accepted (4), and consequently, the development of therapeutics that selectively target the alternative pathway has become a high priority (18). Along with a renewed interest in the complement pathway, the elucidation of x-ray structures for a number of complement components and regulators has enhanced our understanding of the mechanistic basis for complement activation and inhibition (9, 19–22). As a next step, structural insights into the molecular basis for complement regulation should be applied to the design of more potent inhibitors of the complement cascade. Here, we used the crystal structure of CRIg in complex with C3b as a guide to create a more potent inhibitor of the complement alternative pathway.
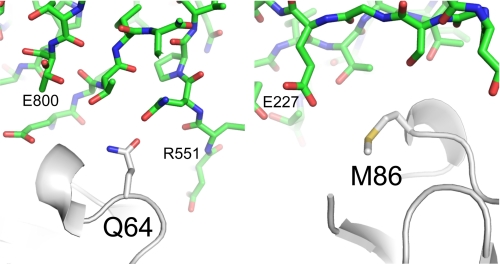
CRIg-v27 contains only two amino acid changes relative to WT CRIg but showed improved potency in vitro and enhanced therapeutic efficacy in vivo, thus illustrating how structural information can be used to create more potent complement inhibitors with only minimal changes to natural proteins. The crystal structure of the complex between CRIg and C3c (9) provides insights into possible molecular mechanisms behind the enhanced affinity of CRIg-v27 for C3b (Fig. 6). Gln-64 in WT CRIg-ECD does not form any hydrogen bonds with C3c, but it is possible that the longer Arg-64 side chain in CRIg-v27 could form a hydrogen bond with the carbonyl oxygen of either Arg-551 or Glu-800 of C3c. Met-86 in WT CRIg-ECD resides on the periphery of binding interface, and the most likely interaction formed by Tyr-86 in CRIg-v27 is a hydrogen bond between the phenol group of the tyrosine side chain and the Glu-227 side chain of C3c.
FIGURE 6.
The environment of CRIg residues Gln-64 and Met-86 in the crystal structure of the complex with C3c. CRIg is shown as a white ribbon presentation with the side chains of Met-86 (right) and Gln-64 (left) shown as sticks. C3c is shown in green as stick representations (Protein Data Bank entry 2ICE).
It is likely that the affinity of CRIg for C3b could be improved further with additional mutations, but our goal was to create a potential therapeutic candidate, and thus, we limited the number of mutations to minimize the risk of immunogenicity. Because affinity and activity were not well correlated, we chose the double mutant that exhibited the largest improvements in both properties. Alternatively, if improved in vitro activity is the dominant factor for improved in vivo efficacy, it is possible that even a single mutation (e.g. CRIg-v9) could provide improved efficacy comparable with the effects of the two mutations incorporated in CRIg-v27. However, testing of this idea would require further in vivo studies with additional CRIg variants.
CRIg-v27 did not bind C3 and retained selectivity for C3 fragments C3b, iC3b, and C3c. From a therapeutic point of view, this selectivity is highly desirable, because the plasma concentration of C3 fragments is ∼200-fold lower than that of native C3 (23). Consequently, targeting of native C3 would require a high dose of therapeutic to achieve favorable pharmacokinetics. In addition to retaining selectivity for C3 fragments, CRIg-v27 specifically inhibited activation of the complement alternative pathway, leaving the classical and mannose-binding lectin pathways intact to mount an appropriate host response to pathogens.
Importantly, although CRIg has the advantage of specificity and selectivity, its critical role in first line host defense against pathogens (8) should be taken into consideration when developing CRIg for therapeutic use. Because systemic treatment with CRIg-ECD proteins could potentially interfere with the function of CRIg as a macrophage receptor, CRIg-ECD variants may prove most valuable as potential therapeutics for disease indications where CRIg can be delivered locally (e.g. intraoculary to treat age-related macular degeneration) or acutely (e.g. in ischemia/reperfusion). Our study should further stimulate the design of potent and selective inhibitors targeting one of three complement pathways, which, subject to appropriate considerations for mode of delivery and indication, should lead to the generation of a new class of complement therapeutics.
Acknowledgments
We thank Micah Steffek and Philip Hass for purification of complement component C3.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- ECD
- extracellular domain
- ELISA
- enzyme-linked immunosorbent assay
- WT
- wild type.
REFERENCES
- 1.Kim D. D., Song W. C. (2006) Clin. Immunol. 118, 127–136 [DOI] [PubMed] [Google Scholar]
- 2.Walport M. J. (2001) N. Engl. J. Med. 344, 1058–1066 [DOI] [PubMed] [Google Scholar]
- 3.Walport M. J. (2001) N. Engl. J. Med. 344, 1140–1144 [DOI] [PubMed] [Google Scholar]
- 4.Holers V. M. (2008) Immunol. Rev. 223, 300–316 [DOI] [PubMed] [Google Scholar]
- 5.Jha P., Bora P. S., Bora N. S. (2007) Mol. Immunol. 44, 3901–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Müller-Eberhard H. J., Götze O. (1972) J. Exp. Med. 135, 1003–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Götze O., Müller-Eberhard H. J. (1976) Adv. Immunol. 24, 1–35 [DOI] [PubMed] [Google Scholar]
- 8.Helmy K. Y., Katschke K. J., Jr., Gorgani N. N., Kljavin N. M., Elliott J. M., Diehl L., Scales S. J., Ghilardi N., van Lookeren Campagne M. (2006) Cell 124, 915–927 [DOI] [PubMed] [Google Scholar]
- 9.Wiesmann C., Katschke K. J., Yin J., Helmy K. Y., Steffek M., Fairbrother W. J., McCallum S. A., Embuscado L., DeForge L., Hass P. E., van Lookeren Campagne M. (2006) Nature 444, 217–220 [DOI] [PubMed] [Google Scholar]
- 10.Kouskoff V., Korganow A. S., Duchatelle V., Degott C., Benoist C., Mathis D. (1996) Cell 87, 811–822 [DOI] [PubMed] [Google Scholar]
- 11.Barthelemy P. A., Raab H., Appleton B. A., Bond C. J., Wu P., Wiesmann C., Sidhu S. S. (2008) J. Biol. Chem. 283, 3639–3654 [DOI] [PubMed] [Google Scholar]
- 12.Fellouse F. A., Sidhu S. S. (2007) in Making and Using Antibodies (Howard G. C., Kaser M. R. eds) pp. 157–180, CRC Press, Inc., Boca Raton, FL [Google Scholar]
- 13.Sidhu S. S., Lowman H. B., Cunningham B. C., Wells J. A. (2000) Methods Enzymol. 328, 333–363 [DOI] [PubMed] [Google Scholar]
- 14.Kunkel T. A., Roberts J. D., Zakour R. A. (1987) Methods Enzymol. 154, 367–382 [DOI] [PubMed] [Google Scholar]
- 15.Katschke K. J., Jr., Helmy K. Y., Steffek M., Xi H., Yin J., Lee W. P., Gribling P., Barck K. H., Carano R. A., Taylor R. E., Rangell L., Diehl L., Hass P. E., Wiesmann C., van Lookeren Campagne M. (2007) J. Exp. Med. 204, 1319–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji H., Ohmura K., Mahmood U., Lee D. M., Hofhuis F. M., Boackle S. A., Takahashi K., Holers V. M., Walport M., Gerard C., Ezekowitz A., Carroll M. C., Brenner M., Weissleder R., Verbeek J. S., Duchatelle V., Degott C., Benoist C., Mathis D. (2002) Immunity 16, 157–168 [DOI] [PubMed] [Google Scholar]
- 17.Maccioni M., Zeder-Lutz G., Huang H., Ebel C., Gerber P., Hergueux J., Marchal P., Duchatelle V., Degott C., van Regenmortel M., Benoist C., Mathis D. (2002) J. Exp. Med. 195, 1071–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ricklin D., Lambris J. D. (2007) Nat. Biotechnol. 25, 1265–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gros P., Milder F. J., Janssen B. J. (2008) Nat. Rev. Immunol. 8, 48–58 [DOI] [PubMed] [Google Scholar]
- 20.Janssen B. J., Christodoulidou A., McCarthy A., Lambris J. D., Gros P. (2006) Nature 444, 213–216 [DOI] [PubMed] [Google Scholar]
- 21.Janssen B. J., Halff E. F., Lambris J. D., Gros P. (2007) J. Biol. Chem. 282, 29241–29247 [DOI] [PubMed] [Google Scholar]
- 22.Torreira E., Tortajada A., Montes T., Rodríguez de Córdoba S., Llorca O. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 882–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katschke K. J., Jr., Stawicki S., Yin J., Steffek M., Xi H., Sturgeon L., Hass P. E., Loyet K. M., Deforge L., Wu Y., van Lookeren Campagne M., Wiesmann C. (2009) J. Biol. Chem. 284, 10473–10479 [DOI] [PMC free article] [PubMed] [Google Scholar]