Abstract
Protein folding mechanisms have remained elusive mainly because of the transient nature of intermediates. Leech-derived tryptase inhibitor (LDTI) is a Kazal-type serine proteinase inhibitor that is emerging as an attractive model for folding studies. It comprises 46 amino acid residues with three disulfide bonds, with one located inside a small triple-stranded antiparallel β-sheet and with two involved in a cystine-stabilized α-helix, a motif that is widely distributed in bioactive peptides. Here, we analyzed the oxidative folding and reductive unfolding of LDTI by chromatographic and disulfide analyses of acid-trapped intermediates. It folds and unfolds, respectively, via sequential oxidation and reduction of the cysteine residues that give rise to a few 1- and 2-disulfide intermediates. Species containing two native disulfide bonds predominate during LDTI folding (IIa and IIc) and unfolding (IIa and IIb). Stop/go folding experiments demonstrate that only intermediate IIa is productive and oxidizes directly into the native form. The NMR structures of acid-trapped and further isolated IIa, IIb, and IIc reveal global folds similar to that of the native protein, including a native-like canonical inhibitory loop. Enzyme kinetics shows that both IIa and IIc are inhibitory-active, which may substantially reduce proteolysis of LDTI during its folding process. The results reported show that the kinetics of the folding reaction is modulated by the specific structural properties of the intermediates and together provide insights into the interdependence of conformational folding and the assembly of native disulfides during oxidative folding.
Introduction
Studies of the folding of disulfide-containing proteins usually exploit the particular chemistry of disulfide bond formation that permits an efficient trapping of partially oxidized intermediates by alkylation or acidification (1, 2). In the well established method of oxidative folding, fully reduced and unfolded proteins are allowed to gain both their native disulfides and native structure under selected buffer and redox conditions. The heterogeneity of the intermediates that arise during the process and their disulfide connectivity are then used to characterize the folding pathway (3). Various small disulfide-rich proteins have been investigated hitherto using oxidative folding, e.g. proteinase inhibitors such as bovine pancreatic trypsin inhibitor and hirudin (4, 5), as well as enzymes such as RNase A and lysozyme (6, 7). However, these studies have not uncovered any predominant folding scenario but have revealed a great diversity (8, 9). Thus, the structural characterization of disulfide folding intermediates is of fundamental importance to gain insight into the mechanisms that lead to the formation of the native protein state by oxidative folding. So far, such structural studies have largely focused on analogs where one or more disulfide bonds were deleted by mutation of cysteines to alanines or serines, e.g. in the outstanding examples of bovine pancreatic trypsin inhibitor and RNase A (3). Very few genuine intermediates isolated from folding/unfolding reactions have been structurally analyzed, i.e. those of a cyclotide and two metallocarboxypeptidase inhibitors (10–12).
Leech-derived tryptase inhibitor (LDTI)4 is a “nonclassical” Kazal-type inhibitor isolated from the medicinal leech Hirudo medicinalis (13, 14). Besides inhibiting trypsin and chymotrypsin with nanomolar affinity, it is one of two proteins identified so far that tightly inhibit human tryptase β, an oligomeric trypsin-like serine proteinase that is stored in the granules of mast cells and has been implicated in the pathogenesis of allergic and inflammatory disorders such as asthma and rheumatoid arthritis (15–17). LDTI is a small protein of 46 residues and three disulfide bonds with a fold governed by a short central α-helix and a small triple-stranded antiparallel β-sheet (18–20). It comprises a particular cysteine pattern strongly cross-linked by a cystine-stabilized α-helical motif known as CSH that is widely found in bioactive peptides, such as endothelins and toxins from insects and snakes (21). These properties make LDTI an interesting model protein for folding studies. We have recently shown that its oxidative folding is bovine pancreatic trypsin inhibitor-like, i.e. populated by a few native disulfide-bonded intermediates that efficiently funnel this protein toward its native form (22, 23). Here, we report a comprehensive analysis of the pathways of oxidative folding and reductive unfolding of LDTI. Moreover, we have solved the high resolution structures of its major folding and unfolding intermediates to decipher the specific interactions that drive the folding of this molecule. Inhibitory assays on the major folding intermediates complement the work.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Recombinant LDTI (amino acid residues 1–44) was obtained by heterologous overexpression in Escherichia coli and isolated essentially as described previously (13, 14, 24). Protein identity and purity were confirmed by automated Edman degradation and mass spectrometry. The concentration of LDTI in solution was determined by measuring the absorbance at 280 nm, using a calculated absorption coefficient Ε0.1% = 0.35, or a BCA assay (Pierce). Recombinant LDTI was fully active (>95%) as determined by titration with bovine trypsin (see “Trypsin Inhibitory Kinetics”).
Oxidative Folding
2 mg of native protein (N) was incubated in 0.1 m Tris-HCl, pH 8.5, containing 6 m guanidine thiocyanate and 100 mm dithiothreitol for at least 2 h at room temperature. To initiate folding, the fully reduced/unfolded protein (R) was passed through a PD-10 column (Sephadex G-25; GE Healthcare) previously equilibrated with 0.1 m Tris-HCl, pH 7.2. After elution, the protein was immediately diluted to a final concentration of 0.5 mg/ml in the same buffer in the absence (Control −) or presence (Control +) of 0.25 mm 2-mercaptoethanol. In some experiments, increasing amounts of guanidine hydrochloride (GdnHCl) were added to the reaction. To monitor folding, aliquots were removed at various time points, and the reaction quenched with 4% aqueous trifluoroacetic acid (TFA). Acid-trapped intermediates were subsequently analyzed by reversed-phase high performance liquid chromatography (RP-HPLC) using a 4.6-mm Jupiter C4 column (Phenomenex) with a linear gradient from 12–38% acetonitrile with 0.1% TFA at a flow rate of 0.75 ml/min for 50 min.
Stop/Go Folding
Acid-trapped intermediates (IIa, IIb, IIc, and I-IIx and I denote species with two and one disulfide bond, respectively) were isolated by RP-HPLC using a linear gradient from 12–32% acetonitrile with 0.1% TFA at a flow rate of 0.75 ml/min for 60 min. Following purification, the intermediates were lyophilized, and folding was reinitiated at room temperature by dissolving samples in 0.1 m Tris-HCl, pH 7.2, at a final protein concentration of 0.5 mg/ml in the absence (Control −) or presence of oxidized glutathione (GSSG) and, in some cases, increasing amounts of GdnHCl. Folding intermediates were then trapped by acidification and analyzed by RP-HPLC as detailed under “Oxidative Folding”.
Reductive Unfolding
Native and intermediate proteins (0.5 mg/ml) were dissolved at room temperature in 0.1 m sodium acetate, pH 4.5, containing selected concentrations of Tris(2-carboxyethyl)phosphine (TCEP, Sigma). To monitor unfolding, time course aliquots of the samples were trapped with 4% aqueous TFA and analyzed by RP-HPLC as detailed in “Oxidative Folding”.
Sample Alkylation
IIa and IIc intermediates were derivatized with 0.1 m 2-aminoethylmethylthiosulfonate (Anatrace) in 0.1 m Tris-HCl, pH 8.0, for 15 min at room temperature. The alkylated intermediates were isolated by RP-HPLC using a linear gradient from 12–24% acetonitrile with 0.1% TFA at a flow rate of 0.5 ml/min for 60 min and subsequently lyophilized. Mass spectrometry served to verify the complete alkylation of free cysteines.
Trypsin Inhibitory Kinetics
The interaction of N and the alkylated intermediates IIaALK and IIcALK with bovine pancreatic trypsin was studied using modifications of methods detailed previously (13, 14, 19, 24). Briefly, a constant concentration of trypsin (see below) was equilibrated with serial dilutions of the inhibitors in buffer containing 50 mm Tris-HCl, 150 mm NaCl, 0.01% Triton, 0.01% Azid, pH 7.6, for 5–300 min. After addition of substrate (5 μm tosyl-Gly-Pro-Arg-AMC or 400 μm carbobenzoxy-Arg-AMC, respectively; Bachem), residual trypsin activity was quantified by following hydrolysis over 10 min using a HTS 7000 Bio Assay Reader (Perkin Elmer). Initial velocities were fitted to the equation describing the binding of “tight-binding” inhibitors (25) with pro Fit (Quantum Soft). Experiments were performed at 20 pm trypsin (i.e. [E] ≪ Ki) to obtain the equilibrium dissociation constants Ki and at 200 nm trypsin ([E] ≫ Ki) to derive the concentrations of inhibitory active proteins and their inactivation over time. In addition, progress curves were obtained by adding the inhibitors at various concentrations to a pre-equilibrated mixture of trypsin (10 pm) and substrate (5 μm tosyl-Gly-Pro-Arg-AMC) in an SFM 25 spectrofluorometer (Kontron) and used to calculate the association and dissociation rate constants according to reaction scheme A as described by Morrison (26).
NMR Samples and Spectroscopy
Samples of N, IIa, IIb, IIc, I, IIaALK, IIbALK, and R were prepared at 1 mm protein concentration by dissolving the lyophilized material in 450 μl of 0.1% aqueous TFA containing 10% D2O, pH 2.0. The LDTI intermediates were analyzed at acidic pH to maintain the free cysteines in a reduced form and prevent their oxidation. For the amide proton exchange experiments, the samples prepared at pH 2.0 were lyophilized and resuspended in 500 μl of 99.98% D2O. NMR experiments were performed at 298 K on Bruker AV 800 and AV 600 spectrometers, both equipped with cryoprobe. For the determination of the sequence-specific polypeptide backbone chemical shift assignments and structure calculations of the native and intermediate forms of LDTI, homonuclear two-dimensional COSY, TOCSY (mixing time of 80 ms) and NOESY (mixing time of 200 and 100 ms) spectra were acquired. NMR data were processed with the program TOPSPIN (Bruker Biospin), whereas the program NMRView (27) was used for interactive spectrum analysis. Comparison of one-dimensional spectra recorded before and after the two-dimensional experiments was used to verify the stability of the native and intermediate forms during the time necessary to perform the NMR measurements. The exchange of amide protons with solvent deuterons was measured at 298 K for the native and intermediate forms of LDTI. NMR data acquisition was started within about 20 min of the initiation of the exchange reaction. A series of TOCSY spectra (60 ms mixing time, 2,048 complex data points, 512 t1 increments, and eight scans per increment) were collected over the course of 3 days. The acquisition time for each experiment was 1 h and 26 min. All the spectra were processed with NMRPipe (28) using the same processing scheme and parameters.
NMR Assignment and Structure Calculation
An assignment strategy based only on homonuclear two-dimensional NMR spectroscopy (29) was used to assign the 1H NMR resonance of native and intermediate forms of LDTI. Most proton resonance assignments were coincident with the previous ones reported for LDTI variant C (18). The conformational shifts were calculated with the equation, ΔδHα = δHα OBS − δHα RC, where δHα RC values are the shifts corresponding to the random coil, incorporating corrections for the effects of proline and oxidized cysteine on the chemical shifts of peptide backbone nuclei (30). The distance constraints used for the structure calculation of LDTI and its intermediate forms were derived from the homonuclear two-dimensional NOESY spectra. Peak picking of the spectra was carried out manually, and peak volumes were determined using the automatic integration function of NMRView (27). For all X–Pro peptide bonds, the trans conformation was confirmed by intense X(Hα)–Pro(Hδ) sequential NOEs (29). Although the intermediate forms were stable at experimental conditions, pH 2.0, for some residues, double conformations were observed, with one coming from the intermediate and with the other from the native form but with much lower intensity. In these cases, only peaks corresponding to the intermediate form were picked and used for structure calculation. The three-dimensional structures were determined by combined automated NOESY cross-peak assignments (31) and structure calculations with torsion angle dynamics (32), implemented in the program CYANA (33). The standard CYANA protocol of seven iterative cycles of NOE assignment and structure calculation, followed by a final structure calculation, was applied. Stereospecific assignments for some isopropyl methyls and methylene groups were determined by the GLOMSA method (34). This was done before the final structure calculation by analyzing the structures obtained in the preceding seventh NOE assignment/structure calculation cycle. Pseudo atoms with appropriate distance corrections were used for distance restraints involving protons with no stereospecific assignment (34). A set of upper and lower distance limits was introduced for each pair of cysteine residues involved in disulfide bonds (2.1/2.0 Å for Sγ(i)-Sγ(j) and 3.1/3.0 Å for Cβ(i)-Sγ(j) and Sγ(i)-Cβ(j)). The compatibility of the disulfide bond pattern identified in each protein (18) with the structures from the initial rounds of automated calculation was evaluated before introducing these restraints in the subsequent structure calculations. Furthermore, several intercysteine NOE connectivities confirmed the disulfide pairing of the intermediates. Weak constraints on (Φ,ψ) torsion angle pairs and on side chain torsion angles between tetrahedral carbon atoms were used temporarily during the NOE assignment/structure calculation cycles to favor the allowed regions of the Ramachandran plot and staggered rotamer conformations, respectively (36). In each cycle, the structure calculation started from 100 randomized conformers and the standard CYANA-simulated annealing schedule was used with 10,000 torsion angle dynamics steps per conformer. Finally, a 1,000-step of energy minimization with NMR distance constraints using a generalized Born solvent model was applied to the 20 conformers with the lowest values of the final CYANA target function using the AMBER 9.0 program (37). The 20 minimized conformers were used to validate the final structure using the program PROCHECK-NMR (38). The program MOLMOL (39) was used to visualize the structures and prepare the figures.
RESULTS AND DISCUSSION
Folding and Unfolding of LDTI Progresses via the Formation of Native Disulfide-bonded Intermediates
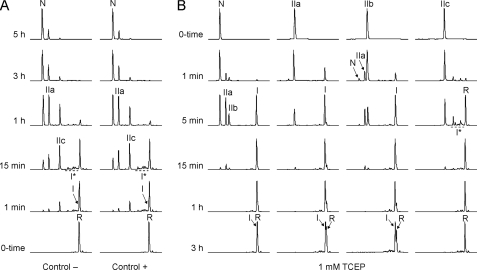
The oxidative folding of LDTI at pH 7.2 undergoes the initial formation of 1-disulfide intermediates (I and I*) that subsequently oxidize, leading to the preferential accumulation of two 2-disulfide species (IIa and IIc) (Fig. 1A). As shown previously, I, IIa, and IIc are intermediates that exclusively contain native disulfide bonds (22). The folding of LDTI is extremely fast compared with that of other small disulfide-rich proteins analyzed under similar conditions (3). Importantly, the chromatographic profiles suggest that folding is little affected by the presence of a reducing agent, implying a low accumulation of non-native scrambled isomers that would otherwise slow down the folding process. Only the presence of large amounts of denaturants affects the efficiency of LDTI folding, diminishing the recovery of native form (see supplemental Fig. S1). Nonetheless, LDTI is able to attain its native state even in the presence of 8 m GdnHCl, illustrating the high conformational stability of the protein. The reductive unfolding of LDTI performed at pH 4.5, which strongly minimizes disulfide reshuffling and favors disulfide reduction, proceeds via the formation of IIa, IIb, and I before reaching the fully reduced/unfolded form (Fig. 1B). Both isolated IIa and IIb reduce one of their two native disulfide bonds (Cys6–Cys25 and Cys4–Cys29, respectively) to form I, which subsequently loses the remaining disulfide (Cys14–Cys40) to render R. Surprisingly, even at acidic pH, IIb is able to reshuffle rapidly into IIa that subsequently regenerates a minor amount of native protein, suggesting that at least one of the two free cysteines in IIb is reactive at this pH and thus has an unusually low pKa. As expected, IIc does not reduce to I but to the I* ensemble, which, in comparison to I, becomes reduced very quickly into R, indicating a low disulfide protection. These results demonstrate that the disulfide stability of LDTI relies on the Cys14–Cys40 disulfide bond and explain that IIc does not accumulate during reductive unfolding due to its low disulfide stability.
FIGURE 1.
Oxidative folding and reductive unfolding of LDTI. RP-HPLC traces of the folding (A) and unfolding (B) intermediates trapped by acidification over time. The folding reactions were performed in the absence (Control −) or presence (Control +) of 0.25 mm 2-mercaptoethanol as thiol catalyst, the unfolding reactions using 1 mm TCEP as reducing agent. Retention times of the native (N) and fully reduced/unfolded (R) forms as well as the major intermediates are labeled. I represents a major intermediate with one native disulfide bond. I* represents a group of native and non-native 1-disulfide intermediates. IIa, IIb, and IIc constitute the major intermediates comprising two native disulfide bonds.
IIa Is the Productive Intermediate That Leads to Native LDTI
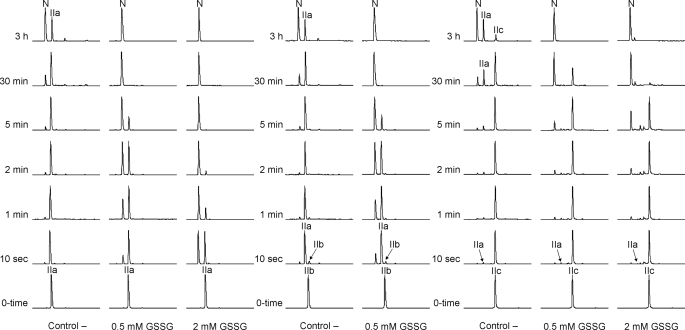
The data obtained by reductive unfolding suggest that IIa and/or IIb are the productive intermediates that convert directly into N. To address this issue, we have isolated the three 2-disulfide species of LDTI (IIa, IIb, and IIc) and reinitiated their folding at pH 7.2 in the absence and presence of oxidizing agents (Fig. 2). IIa converts directly into N in the absence of any redox agent, following a first order reaction with a rate constant of 2.8·10−4 s−1 (supplemental Fig. S2). Increasing concentrations of GSSG, which enhances disulfide formation and prevents disulfide reshuffling, strongly accelerate this conversion and reduce the accumulation of other species; the reaction rate increases proportional to the GSSG concentration with an average slope of 18.4 s−1m−1. The observed reactivity of IIb under acidic conditions suggests that it should be highly unstable at physiological pH. Indeed, in the absence of oxidizing agent isolated IIb reshuffles at a rate of 0.87 s−1 into IIa, which then behaves as described above. This result rules out our previous hypothesis that IIb is the productive intermediate (24) and explains why it does not accumulate during LDTI folding. IIc gives rise to both IIa and N with an overall rate constant of 3.3·10−4 s−1. The amounts of IIa in this reaction can be accurately predicted from the difference between the N-formation and IIc disappearance rates, indicating that IIc reshuffles into IIa and does not directly oxidize into N. In the first 2 min, the generation of N is virtually independent of the presence of GSSG, and only subsequently, the reaction is faster in the presence of higher oxidant concentrations because of the formation of the productive intermediate IIa. The reshuffling of IIc into IIa and the oxidation of IIa into N proceed with very similar reaction rates, providing an explanation for the coexistence of both intermediates in the folding process. On the other hand, stop/go experiments on IIa and IIc intermediates conducted in the presence of denaturant (supplemental Fig. S3) support the hypothesis that these intermediates are highly stable, because both species are detected even in 8 m GdnHCl.
FIGURE 2.
Stop/go folding of the major 2-disulfide intermediates of LDTI. IIa, IIb, and IIc were isolated, lyophilized, and dissolved in the absence (Control −) or presence of oxidizing agent (GSSG) to reinitiate the folding reaction. Folding intermediates were subsequently trapped by acidification over time and analyzed by RP-HPLC.
The Solution Structures of the Major Folding and Unfolding Intermediates of LDTI Reveal Native-like Conformations
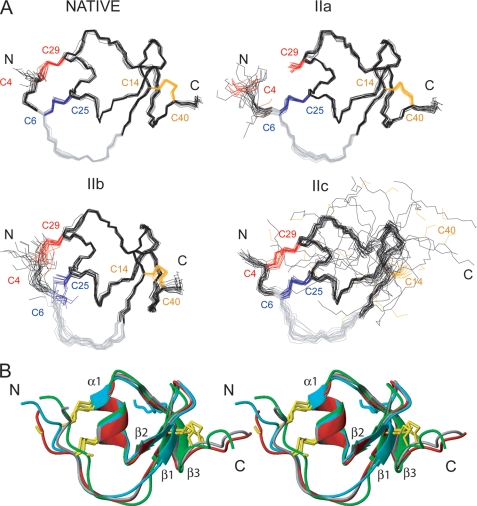
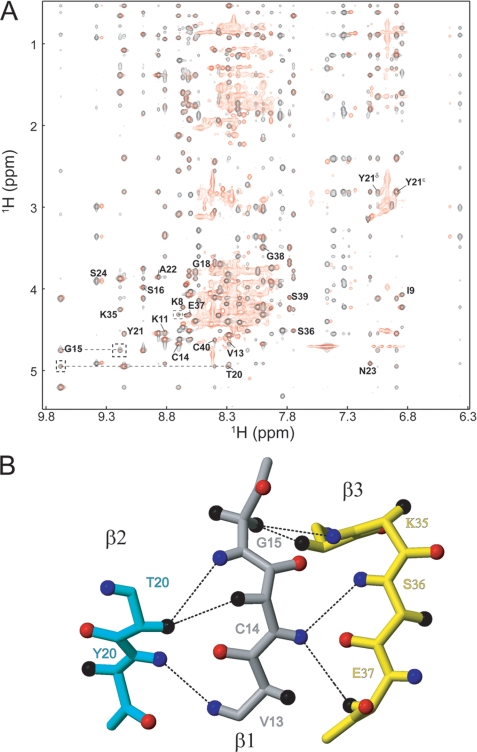
The three 2-disulfide intermediates of LDTI (IIa, IIb, and IIc) are likely to be thermodynamically destabilized relative to LDTI because they lack a covalent bond. The log n rule of Darby and Creighton (40) predicts a destabilization by 3.48, 3.26, and 3.48 kcal mol−1 for IIa, IIb, and IIc, respectively. However, the entropic destabilization does not explain per se the different kinetic behavior of these intermediates, suggesting that their specific conformational properties influence the reaction. To address this issue, we have determined the structures of all three intermediates as well as of native LDTI by NMR spectroscopy at acidic pH, i.e. conditions where they are stable. The solution structures are presented in an ensemble of 20 energy-minimized conformers (Fig. 3A), and quality and precision statistics are summarized in Table 1. All structures are well defined and in excellent agreement with the experimental data. Native LDTI shows the characteristic fold of Kazal-type serine proteinase inhibitors (41) as described previously (18–20). The secondary structure consists of a short α-helix, α1 (Ser24–Asn30), and a small triple-stranded antiparallel β-sheet in this order: β2-β1-β3 (Thr20–Tyr21, Val13–Gly15, and Ile34–Glu37). Hα conformational shifts, sequential and short range NOEs, and amide exchange data completely agree with these secondary motifs (supplemental Fig. S4). Two of the three disulfide bonds connect the N terminus with the α-helix (Cys4–Cys29 and Cys6–Cys25), whereas the third connects the β1 strand with the C terminus (Cys14–Cys40). Although the intermediates contain only two disulfide bonds, they have essentially the same fold and secondary motifs as native LDTI (Fig. 3B and supplemental Table S1), except for intermediate IIc, which lacks the β3 strand of the β-sheet due to the missing Cys14–Cys40 disulfide bond (Fig. 3 and supplemental Table S2). Importantly, no significant differences are observed in the canonical inhibitory loop, Cys6–Lys11 (Fig. 3B), which is in agreement with the observed inhibitory activity of the intermediates (see below). The three-dimensional structure of intermediate I could not be determined due to severe spectra overlap, with many resonances in the 7.9–8.5 ppm range, indicating that the protein is unstructured. Nevertheless, comparison of its two-dimensional NOESY spectrum with that of native LDTI (Fig. 4A) shows that the residues in the vicinity of the Cys14–Cys40 disulfide bond (i.e. residues 8–24 and 35–40) have approximately the same chemical shift, suggesting the presence of the β-sheet in this intermediate. Furthermore, several interstrand NOEs (β1- β2: 13HN-21HN, 14Hα-20Hα, and 15HN-20-Hα; β1-β3: 14HN-37Hα, 14HN-36HN, 15Hα2–34Hα, and 15Hα2–35HN) (Fig. 4B) confirm the formation of a small triple-stranded antiparallel β-sheet as in native LDTI.
FIGURE 3.
NMR structures of the native and intermediate forms of LDTI. A, representation of 20 minimized structures of the native form and the folding intermediates IIa, IIb, and IIc. The cysteine residues of each protein are colored according to disulfide bonds: Cys4–Cys29 (red), Cys6-Cys25 (blue), and Cys14–Cys40 (orange). The canonical inhibitory loop comprising residues Cys6-Lys11 is colored in gray. The N- and C-terminal ends are labeled. B, comparison of the mean structure of native LDTI (gray), IIa (red), IIb (green), and IIc (blue) in a stereo-view ribbon representation. The secondary structure elements are labeled, and the cysteine residues are shown in yellow.
TABLE 1.
Structural statistics of the 20 best NMR structures of the N and intermediate forms of LDTI
| N | IIa | IIb | IIc | |
|---|---|---|---|---|
| NOE distance constraints | ||||
| Short range distances (i–j) ≥ 1 | 363 | 356 | 304 | 279 |
| Medium range distances (i–j) < 5 | 148 | 133 | 78 | 95 |
| Long range distances (i–j) ≥ 5 | 269 | 236 | 219 | 147 |
| Total | 780 | 725 | 601 | 521 |
| Final CYANA target function valuea (Å2) | 0.72 ± 0.06 | 1.57 ± 0.14 | 1.08 ± 0.06 | 0.48 ± 0.05 |
| AMBER energy (kcal/mol) | −1367 ± 7 | −1360 ± 8 | −1364 ± 8 | −1364 ± 11 |
| Maximal violation (Å) | 0.28 | 0.42 | 0.39 | 0.25 |
| Violations > 0.4 Å | 0/20 | 1/20 | 0/20 | 0/20 |
| Bond lengths (Å) | 0.0102 ± 0.0001 | 0.0101 ± 0.0001 | 0.0102 ± 0.0001 | 0.0108 ± 0.0002 |
| Bond angles (°) | 1.99 ± 0.03 | 2.08 ± 0.04 | 2.02 ± 0.04 | 2.03 ± 0.04 |
| r.m.s.d.c to mean coordinates (Å) (residues 4–40/6–35) | ||||
| Backbone N, Cα, C′ | 0.274/0.248 | 0.436/0.284 | 0.568/0.445 | 2.246/0.655 |
| All heavy atoms | 0.810/0.822 | 0.944/0.880 | 1.003/0.959 | 2.769/1.309 |
| Ramachandran plot statisticsb | ||||
| Most favorable regions (%) | 84.7 | 86.5 | 83.4 | 81.3 |
| Additional allowed regions (%) | 15.3 | 13.2 | 16.3 | 18.1 |
| Generously allowed regions (%) | 0.0 | 0.3 | 0.3 | 0.6 |
| Disallowed regions (%) | 0.0 | 0.0 | 0.0 | 0.0 |
a Average values over the 20 final CYANA conformers.
b Calculated with PROCHECK-NMR (Ref. 38).
c r.m.s.d., root mean square deviation.
FIGURE 4.
NMR characterization of intermediate I. A, overlay of the NOESY spectra of native LDTI (black) and its intermediate I (red). Interstrand NOEs are shown in dashed lines. B, ball-and-stick representation of the three-stranded antiparallel β-sheet (β1, gray; β2, cyan; and β3, yellow), where atoms Hα, HN, and O are represented by a sphere colored in black, blue, and red, respectively. The observed NOEs in the two-dimensional NOESY spectrum of intermediate I are indicated by dashed lines.
The Conformational Properties of the LDTI Intermediates Unveil Their Role in the Folding Reaction
The fact that only IIa oxidizes to native LDTI suggests that this folding intermediate should be highly structured. However, due to the lack of the Cys4–Cys29 disulfide bond the first five N-terminal residues are highly flexible and fully solvent-accessible. In addition, Cys29 at the C terminus of the α-helix is solvent-accessible. Surprisingly, the formation of the α-helix does no require the Cys4–Cys29 link, so IIa has a highly native-like conformation. Although this intermediate converts into N even in the presence of 8 m GdnHCl, under these conditions, the reaction is less efficient and specific and becomes insensitive to GSSG, emphasizing the requirement of the native-like conformation for the direct oxidation into N. The highly unstable IIb intermediate also has a native-like conformation, including a single cross-linked α-helix, in which the free Cys6 and Cys25 residues are accessible to solvent. However, with 8.8 Å, the average distance between Cys6 and Cys25 is large, and the thiols do not face each other in any of the NMR conformers, thus impeding their oxidation to form N because the proximity of two reactive groups (effective concentrations) is determined by the propensity of the backbone to bring the two groups into juxtaposition (7). Moreover, the Cys4–Cys29 disulfide bond is highly flexible, and, in some of the conformers approaches the free thiol of Cys25 as close as 3.2 Å. As discussed above, either Cys6 or Cys25 is highly reactive. The reactivity of an exposed cysteine depends on its local electrostatic environment; in IIb, Cys6 is located in an unstructured context without any side chain in its vicinity, whereas Cys25 is located at the N terminus of the α-helix, where a partial positive charge of the dipole can stabilize the thiolate. The mechanism, which is used by DsbA to lower the pKa of the reactive Cys30 to 3.5 in the catalytic CXXC motif (42), suggests that the conversion of IIb into IIa does not require any major structural rearrangement and is initiated by the fast attack of the Cys4–Cys29 disulfide bond by the Cys25 thiolate.
N, IIa, and IIb share the Cys14–Cys40 disulfide bond. Cys14, located within the β-sheet, is the most buried cysteine residue in the structure of these species, explaining that Cys14–Cys40 initially resists reduction and that intermediate I accumulates in the reductive unfolding reactions. No entropic bias toward the formation of the Cys14–Cys40 disulfide bond from R is expected. Therefore, the folding of the β-sheet likely precedes and promotes the preferential formation of this bond. Subsequently, the formed Cys14–Cys40 disulfide may significantly stabilize the β-sheet and thus promote the accumulation of I during the folding reaction. In proteins of the RNase family, the protection of native disulfide bonds by specific native interactions is shown to play a crucial role during oxidative folding (43). Based on NMR data, no helical conformation is detected in intermediate I suggesting that the two secondary structure elements of LDTI form independently. Because non-natively bonded 2S species are not detected in the folding pathway, the β-sheet constitutes the nucleus of the folding trajectories that leads to the formation of N through the population of I, IIb, and IIa intermediates. Alternative folding routes converge in the formation of the IIc intermediate, with a double cross-linked α-helix, but with a shorter and distorted β-sheet due to the absence of Cys14–Cys40. The lack of this disulfide bond does not restrict the movement of the last 15 residues, moving Cys40 far away from Cys14, which is now exposed to solvent. Thus, the kinetic blockage of IIc does not result from the inaccessibility of its free thiols but rather from a high energy barrier for the formation of this disulfide bond. According to the microscopic reversibility of folding, the transition state for this step coincides with that of the inverse reaction, i.e. the reduction of Cys14–Cys40 that, as discussed above, has a high free energy resulting in a slow disulfide reduction. In general, the formation of a disulfide bond would be always impeded kinetically if, once formed, it becomes buried in a stable folded conformation (1), as in the case of IIc. For the folding of LDTI, it is kinetically advantageous to have a disulfide bond on the surface, such as Cys4–Cys29, which can be directly formed in the final step even in a highly folded context.
The Major Folding Intermediates of LDTI Have Inhibitory Activity
As stated above, the intermediates IIa and IIc are stable only at acidic pH. To avoid their progression along the folding pathway during inhibition kinetic experiments performed at neutral pH, we alkylated the free cysteines of both species. Comparisons of the amide regions of the 1H NMR spectra (supplemental Fig. S5) and fingerprint regions of two-dimensional NOESY spectra (supplemental Fig. S6) of the intermediates without (IIa and IIc) and with (IIaALK and IIcALK) derivatized cysteines show peaks in similar positions and equivalent numbers of NOEs, suggesting that they have analogous folds. Steady-state and pre-steady-state kinetics revealed that the alkylated intermediates have native-like inhibitory activity against trypsin (Table 2). In particular, the inhibitory properties of IIcALK and N toward trypsin are virtually identical. The affinity of IIaALK is reduced ∼25-fold due to both a lower association and higher dissociation rate constant, likely reflecting the higher degree of freedom of the N-terminal part of the reactive site loop that after attack of the scissile peptide bond is connected to the scaffold of the inhibitor only by a single disulfide bond (i.e. Cys6–Cys25). Compared with N both IIaALK and IIcALK are inactivated ∼10-fold more rapidly by trypsin, enhancing the temporary nature of inhibition that has been observed with Kazal-type and many other protein-type serine proteinase inhibitors (44–46). These results are consistent with the native-like structures of the canonical inhibitory loop of IIa and IIc and the higher global flexibility of their scaffolds as compared with native LDTI. They also emphasize the contribution of the Cys4–Cys29 disulfide bond (which is lacking in IIa) to the full inhibitory activity of LDTI, whereas Cys14–Cys40 appears to be dispensable and is indeed missing, e.g. in 13 of 15 domains of the multimeric Kazal-like inhibitor LEKTI (35). Finally, for a proteinase inhibitor such as LDTI, the development of a functional reactive site loop early on during the folding process is physiologically advantageous because it limits degradation and fosters completion of folding even in a proteinase-rich environment.
TABLE 2.
Inhibition constants of native LDTI and the alkylated folding intermediates IIaALK and IIcALK. The equilibrium dissociation constant Ki of the complex with trypsin, the association and dissociation rate constants kon and koff, and the inactivation rate kinact are given as mean ± S.D.
| Ki | kon | koff | kinact | |
|---|---|---|---|---|
| nm | m−1s−1 | s−1 | s−1 | |
| N | 0.43 ± 0.05 | 7.1 ± 2.4·105 | 5.5 ± 0.5·10−4 | 3 ± 0.6·10−6 |
| IIaALK | 10.48 ± 0.97 | 2.4 ± 0.6·105 | 40.7 ± 9·10−4 | 38 ± 4·10−6 |
| IIcALK | 0.48 ± 0.09 | 10.0 ± 1.1·105 | 7.6 ± 1.6·10−4 | 30 ± 1·10−6 |
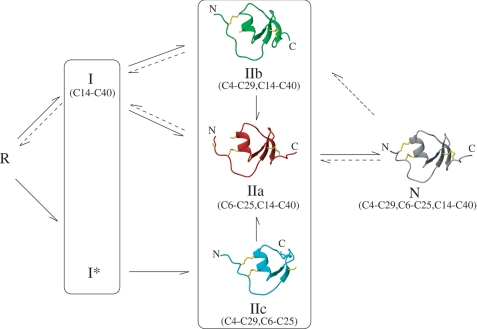
In summary, the findings reported provide an in-depth insight into the mechanisms of oxidative folding and reductive unfolding of LDTI as well as the structures of the major intermediates (Fig. 5). To our knowledge, this work represents the first structural study of a complete collection of genuine folding and unfolding intermediates of a small disulfide-rich protein, which has important implications from theoretical and biomedical perspectives. The results illustrate with unprecedented detail the interdependence of conformational folding and the assembly of native disulfides during oxidative folding. In LDTI folding, the strong bias toward the hierarchical acquisition of the native structure is kinetically modulated by the reactivity and accessibility of thiols and disulfide bonds in its folding intermediates. This interplay of structural and kinetic constraints likely governs the folding of most small disulfide-rich proteins.
FIGURE 5.
Scheme of the oxidative folding and reductive unfolding of LDTI. Solid and dashed arrows outline the predominant pathways of oxidative folding and reductive unfolding of LDTI, respectively. R and N indicate the fully reduced/unfolded and native forms of the protein, respectively, and Ix and IIx intermediates with one or two disulfide bonds. The average NMR structures of IIa, IIb, IIc, and N are represented as ribbon plots with the corresponding disulfide pairings.
Acknowledgments
We thank Sabine Streicher and Stefan Simon for excellent technical assistance. We also thank Ute Marx for performing initial NMR experiments and Sílvia Bronsoms, Werner Machleidt, and Francisco J. Blanco for helpful discussions.
This work was supported by Spanish Ministry for Science and Innovation Grants BIO2007-68046 and BFU2005-01855 and Deutsche Forschungsgemeinschaft Grant SO 249/1-1.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S6.
The atomic coordinates and structure factors (codes 2KMO, 2KMP, 2KMQ, and 2KMR) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- LDTI
- leech-derived tryptase inhibitor
- GSSG
- oxidized glutathione
- N
- native protein
- NOE
- nuclear Overhauser effect
- R
- fully reduced/unfolded protein
- RP-HPLC
- reversed-phase high-performance liquid chromatography
- TCEP
- Tris(2-carboxyethyl)phosphine
- TFA
- trifluoroacetic acid.
REFERENCES
- 1.Creighton T. E. (1997) Biol. Chem. 378, 731–744 [DOI] [PubMed] [Google Scholar]
- 2.Narayan M., Welker E., Wedemeyer W. J., Scheraga H. A. (2000) Acc. Chem. Res. 33, 805–812 [DOI] [PubMed] [Google Scholar]
- 3.Arolas J. L., Aviles F. X., Chang J. Y., Ventura S. (2006) Trends Biochem. Sci 31, 292–301 [DOI] [PubMed] [Google Scholar]
- 4.Darby N. J., Morin P. E., Talbo G., Creighton T. E. (1995) J. Mol. Biol. 249, 463–477 [DOI] [PubMed] [Google Scholar]
- 5.Thannhauser T. W., Rothwarf D. M., Scheraga H. A. (1997) Biochemistry 36, 2154–2165 [DOI] [PubMed] [Google Scholar]
- 6.van den Berg B., Chung E. W., Robinson C. V., Mateo P. L., Dobson C. M. (1999) EMBO J. 18, 4794–4803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Welker E., Narayan M., Wedemeyer W. J., Scheraga H. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 2312–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang J. Y. (2004) Biochemistry 43, 4522–4529 [DOI] [PubMed] [Google Scholar]
- 9.Chang J. Y. (2008) Antioxid. Redox. Signal 10, 171–177 [DOI] [PubMed] [Google Scholar]
- 10.Arolas J. L., D'Silva L., Popowicz G. M., Aviles F. X., Holak T. A., Ventura S. (2005) Structure 13, 1193–1202 [DOI] [PubMed] [Google Scholar]
- 11.Arolas J. L., Pantoja-Uceda D., Ventura S., Blanco F. J., Aviles F. X. (2008) J. Biol. Chem. 283, 27110–27120 [DOI] [PubMed] [Google Scholar]
- 12.Cemazar M., Joshi A., Daly N. L., Mark A. E., Craik D. J. (2008) Structure 16, 842–851 [DOI] [PubMed] [Google Scholar]
- 13.Sommerhoff C. P., Söllner C., Mentele R., Piechottka G. P., Auerswald E. A., Fritz H. (1994) Biol. Chem. Hoppe Seyler 375, 685–694 [DOI] [PubMed] [Google Scholar]
- 14.Auerswald E. A., Morenweiser R., Sommerhoff C. P., Piechottka G. P., Eckerskorn C., Gürtler L. G., Fritz H. (1994) Biol. Chem. Hoppe Seyler 375, 695–703 [DOI] [PubMed] [Google Scholar]
- 15.Hallgren J., Pejler G. (2006) FEBS. J. 273, 1871–1895 [DOI] [PubMed] [Google Scholar]
- 16.Sommerhoff C. P. (2001) Am. J. Respir. Crit. Care Med. 164, S52–S58 [DOI] [PubMed] [Google Scholar]
- 17.Sommerhoff C. P., Schaschke N. (2007) Curr. Pharm. Des. 13, 313–332 [DOI] [PubMed] [Google Scholar]
- 18.Mühlhahn P., Czisch M., Morenweiser R., Habermann B., Engh R. A., Sommerhoff C. P., Auerswald E. A., Holak T. A. (1994) FEBS Lett. 355, 290–296 [DOI] [PubMed] [Google Scholar]
- 19.Stubbs M. T., Morenweiser R., Stürzebecher J., Bauer M., Bode W., Huber R., Piechottka G. P., Matschiner G., Sommerhoff C. P., Fritz H., Auerswald E. A. (1997) J. Biol. Chem. 272, 19931–19937 [DOI] [PubMed] [Google Scholar]
- 20.Di Marco S., Priestle J. P. (1997) Structure 5, 1465–1474 [DOI] [PubMed] [Google Scholar]
- 21.Hemmi H., Kumazaki T., Yoshizawa-Kumagaye K., Nishiuchi Y., Yoshida T., Ohkubo T., Kobayashi Y. (2005) Biochemistry 44, 9626–9636 [DOI] [PubMed] [Google Scholar]
- 22.Arolas J. L., Bronsoms S., Aviles F. X., Ventura S., Sommerhoff C. P. (2008) Antioxid. Redox. Signal 10, 77–85 [DOI] [PubMed] [Google Scholar]
- 23.Weissman J. S., Kim P. S. (1991) Science 253, 1386–1393 [DOI] [PubMed] [Google Scholar]
- 24.Pohlig G., Fendrich G., Knecht R., Eder B., Piechottka G., Sommerhoff C. P., Heim J. (1996) Eur. J. Biochem. 241, 619–626 [DOI] [PubMed] [Google Scholar]
- 25.Morrison J. F. (1969) Biochim. Biophys. Acta 185, 269–286 [DOI] [PubMed] [Google Scholar]
- 26.Morrison J. F. (1982) Trends Biochem. Sci. 7, 102–105 [Google Scholar]
- 27.Johnson B. A., Blevins R. A. (1994) J. Biomol. NMR 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 28.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 29.Wüthrich K. (1986) NMR of Proteins and Nucleic Acids, Wiley-Interscience Publication, New York [Google Scholar]
- 30.Wishart D. S., Bigam C. G., Holm A., Hodges R. S., Sykes B. D. (1995) J. Biomol. NMR 5, 67–81 [DOI] [PubMed] [Google Scholar]
- 31.Herrmann T., Güntert P., Wüthrich K. (2002) J. Mol. Biol. 319, 209–227 [DOI] [PubMed] [Google Scholar]
- 32.Güntert P., Mumenthaler C., Wüthrich K. (1997) J. Mol. Biol. 273, 283–298 [DOI] [PubMed] [Google Scholar]
- 33.Güntert P. (2004) Methods Mol. Biol. 278, 353–378 [DOI] [PubMed] [Google Scholar]
- 34.Güntert P., Braun W., Wüthrich K. (1991) J. Mol. Biol. 217, 517–530 [DOI] [PubMed] [Google Scholar]
- 35.Mägert H. J., Ständker L., Kreutzmann P., Zucht H. D., Reinecke M., Sommerhoff C. P., Fritz H., Forssmann W. G. (1999) J. Biol. Chem. 274, 21499–21502 [DOI] [PubMed] [Google Scholar]
- 36.Pantoja-Uceda D., López-Méndez B., Koshiba S., Inoue M., Kigawa T., Terada T., Shirouzu M., Tanaka A., Seki M., Shinozaki K., Yokoyama S., Güntert P. (2005) Protein Sci. 14, 224–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Case D. A., Darden T. A., Cheatham T. E., III, Simmerling C. L., Wang J., Duke R. E., Luo R., Merz K. M., Pearlman D. A., Crowley M., Walker R. C., Zhang W., Wang B., Hayik S., Roitberg A., Seabra G., Wong K. F., Paesani F., Wu X., Brozell S., Tsui V., Gohlke H., Yang L., Tan C., Mongan J., Hornak V., Cui G., Beroza P., Mathews D. H., Schafmeister C., Ross W. S., Kollman P. A. (2006) University of California, San Francisco [Google Scholar]
- 38.Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 39.Koradi R., Billeter M., Wuthrich K. (1996) J. Mol. Graph 14, 51–55 [DOI] [PubMed] [Google Scholar]
- 40.Darby N., Creighton T. E. (1995) Methods Mol. Biol. 40, 219–252 [DOI] [PubMed] [Google Scholar]
- 41.Murzin A. G., Brenner S. E., Hubbard T., Chothia C. (1995) J. Mol. Biol. 247, 536–540 [DOI] [PubMed] [Google Scholar]
- 42.Schirra H. J., Renner C., Czisch M., Huber-Wunderlich M., Holak T. A., Glockshuber R. (1998) Biochemistry 37, 6263–6276 [DOI] [PubMed] [Google Scholar]
- 43.Gahl R. F., Scheraga H. A. (2009) Biochemistry 48, 2740–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laskowski M., Wu F. C. (1953) J. Biol. Chem. 204, 797–805 [PubMed] [Google Scholar]
- 45.Sealock R. W., Laskowski M., Jr. (1973) Biochemistry 12, 3139–3146 [DOI] [PubMed] [Google Scholar]
- 46.Tschesche H. (1974) Angew. Chem. Int. Ed. Engl. 13, 10–28 [DOI] [PubMed] [Google Scholar]