Abstract
Heparan sulfate (HS) binds and modulates the transport and activity of a large repertoire of regulatory proteins. The HS phage display antibodies are powerful tools for the analysis of native HS structure in situ; however, their epitopes are not well defined. Analysis of the binding specificities of a set of HS antibodies by competitive binding assays with well defined chemically modified heparins demonstrates that O-sulfates are essential for binding; however, increasing sulfation does not necessarily correlate with increased antibody reactivity. IC50 values for competition with double modified heparins were not predictable from IC50 values with corresponding singly modified heparins. Binding assays and immunohistochemistry revealed that individual antibodies recognize distinct epitopes and that these are not single linear sequences but families of structurally similar motifs in which subtle variations in sulfation and conformation modify the affinity of interaction. Modeling of the antibodies demonstrates that they possess highly basic CDR3 and surrounding surfaces, presenting a number of possible orientations for HS binding. Unexpectedly, there are significant differences between the existence of epitopes in tissue sections and observed in vitro in dot blotted tissue extracts, demonstrating that in vitro specificity does not necessarily correlate with specificity in situ/vivo. The epitopes are therefore more complex than previously considered. Overall, these data have significance for structure-activity relationships of HS, because the model of one antibody recognizing multiple HS structures and the influence of other in situ HS-binding proteins on epitope availability are likely to reflect the selectivity of many HS-protein interactions in vivo.
Introduction
HS2 belongs to the glycosaminoglycan family of polysaccharides and can be considered the most information-rich biopolymer in nature. HS chains consist of repeating β-d-glucuronic acid and GlcNAc units, which are subject to enzymatic post-polymeric modification by N-deacetylation and N-sulfation of GlcNAc, O-sulfation at various positions, and epimerization of d-glucuronic acid to its C-5 epimer, iduronic acid. HS biosynthetic enzymes do not act upon every potential site within a chain, resulting in chains with a high level of structural diversity and a distinct domain structure, with regions of unmodified saccharides (N-acetyl domains) and highly modified saccharides (N-sulfated domains) flanked by regions of alternating modified and unmodified disaccharides (transition or N-acetyl/N-sulfated domains) (1–4). HS chains are usually attached to core proteins, forming HS proteoglycans, and these proteoglycans represent one of the major components of most metazoan cell surfaces and extracellular matrices. Here, they control cell-cell and cell-matrix communication by association with a large repertoire of regulatory proteins (5), including growth factors, morphogens, extracellular matrix components, enzymes, cell adhesion molecules, and cytokines. It is through these interactions with proteins that HS exerts its role in numerous biological processes during embryogenesis, development, and adult homeostasis, largely by regulating transport and/or effector functions of its protein ligands.
The structure of HS is dynamic and is directed by the cellular environment and physiology, i.e. tissue/cell type, state of differentiation and growth, and pathology. Distinct cellular sources have been shown to display a range of HS chain lengths, domain structures, and sulfation (1, 6–10). Native HS is therefore highly diverse, with each cell synthesizing an array of HS chains of different lengths and structures with a variety of modifications and chain domain organization. HS should therefore not be classed as a single entity but rather as a family of structurally related but highly diverse molecules with a range of functional effects. The mechanisms regulating HS biosynthesis and the resulting structure of HS are poorly understood; thus, the precise composition of native chains is difficult to establish. Nucleic acids can be amplified and quantified by PCR and localized by in situ hybridization; there are no such methods for analyzing the minute quantities of HS structures in a tissue. Tissue HS is usually structurally analyzed following extraction and purification; the inherent averaging of this approach, however, limits the information to the overall assessment of mixed populations of HS structures and the loss of all spatial information. Monoclonal antibodies directed toward glycosaminoglycans such as chondroitin sulfate/dermatan sulfate and keratan sulfate are available that enable the localization of specific glycosaminoglycan epitopes to be identified in tissues (11–17). In contrast, HS-specific monoclonal antibodies are relatively limited, with only a few available (18–21). To circumvent the difficulties of analyzing tissue HS in situ, HS-specific single chain variable fragment (scFv) antibodies have been generated by phage display technology, which enables the detection of HS glycosaminoglycan structures in situ (22–24). These antibodies have been used to probe HS in a variety of tissues, including kidney (22, 24, 25), skeletal muscle (23, 26, 27), spleen (28), pancreas (29), fetal lung (30), and adult lung (31), and in disease, such as metastatic malignant melanoma (32). However, their application is complicated by the fact that they are raised against heterogeneous native heparin and HS chains, and consequently the exact structures they recognize within the polysaccharide are unknown. This hinders further application of the antibodies and interpretation of tissue staining patterns. To gain insights into the specificity of these valuable analytical tools, the binding characteristics and epitope structures recognized by a panel of six scFv HS antibodies were examined. Antibody epitopes were probed in situ using immunohistochemistry and dot blotting of embryonic rat lungs, because the development of the lung represents one of the core models of mammalian organogenesis, and its functional transition postpartum is a key event in mammalian physiology. In addition, modeling of the antibody structures and competition binding assays with various heparin analogues were used to determine antibody binding specificities in vitro. The results show that HS phage display antibodies all share a similar, largely basic structure and recognize distinct HS epitopes that are structurally complex. Analysis of antibody binding specificities also provides valuable information on the specificity of HS-protein interactions in general.
EXPERIMENTAL PROCEDURES
Materials
Rabbit anti-VSV-G tag IgG was from Abcam, (Cambridge, UK), mouse anti-His tag IgG was from Sigma-Aldrich, HRP-conjugated sheep anti-mouse IgG was from GE Healthcare, and fluorescein isothiocyanate-conjugated goat anti-rabbit IgG was from Sigma-Aldrich. Bovine serum albumin (BSA) was from Sigma-Aldrich, and nonfat dry milk powder was from Tesco, UK. SuperSignal West Dura Extended Duration Substrate and Hyperfilm ECL were from GE Healthcare. Heparinase enzymes were purchased from IBEX Technologies Inc., porcine mucosal heparan sulfate (PMHS) was a gift from Organon (Oss, Netherlands), and heparin (sodium salt) was purchased from Celsus Laboratories Inc. Protease inhibitor mixture was from Roche Applied Science. Chemically modified heparins were a gift from Dr. E. A. Yates (University of Liverpool) and prepared as described previously (33). Heparin oligosaccharides were a gift from Y. A. Ahmed (University of Liverpool). EZ-LinkTM NHS-LC-biotin was from Pierce, streptavidin was from Promega, and Sephadex G-25 gel filtration column was purchased from Sigma-Aldrich. Maxisorp 96-well microtiter plates were purchased from Nunc (Fisher), and o-phenylenediamine (o-phenylenediamine substrate) was from Dako Ltd. (Ely, UK).
Immunohistochemistry
Lungs from Sprague-Dawley rats (Charles River Ltd, Margate, UK) were harvested at embryonic days (E) 15.5 and 17.5 of gestation and fixed with 4% (w/v) paraformaldehyde in phosphate-buffered saline (PBS), pH 7.4. The lungs were washed in PBS, cryoprotected with 20% (w/v) sucrose in PBS, and gelatin-embedded (7.5% (w/v) gelatin, 15% (w/v) sucrose in PBS) before being covered in Cryo-M-Bed (Bright, Huntingdon, UK) and snap frozen at −40 °C. The lung sections were cut at 8 μm and mounted onto chrome alum gel slides for storage at −40 °C. Tissue sections were blocked with 10% (v/v) goat serum in PBS and incubated overnight at 4 °C with HS scFv antibodies, diluted 1/5 in 1% (v/v) goat serum in PBS. Bound antibody was detected with rabbit VSV-G tag antibody diluted 1/200 in 1% (v/v) goat serum in PBS, followed by fluorescein isothiocyanate-conjugated goat anti-rabbit IgG. The controls were omission of HS antibody or treatment of tissue sections with heparinase III (EC 4.2.2.8) (5 milliunits/ml in 100 mm sodium acetate, 0.1 mm calcium acetate, 50 μg/ml BSA, pH 7.0) for 2 h at 37 °C, prior to antibody incubation to remove HS epitopes.
Dot Blotting of Lung Extracts
Lungs from Sprague-Dawley rats (Charles River) were harvested at E15.5, E17.5, E19.5, and E21.5 of gestation and immediately homogenized on ice with 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 2% (w/v) SDS, and protease inhibitor mixture (1 tablet/10 ml buffer). Because earlier gestation stage lungs are smaller, a higher number of E15.5 lungs were used to prepare lung extract samples compared with E21.5 lungs. This ensured similar ratios of tissue weight:volume homogenization buffer. The homogenates were centrifuged at 4 °C at 14,000 rpm for 10 min, and supernatants were collected. Each supernatant was then standardized for protein concentration. Lung extracts (10 μl) were blotted onto nitrocellulose membrane, and endogenous peroxidase was quenched with 0.6% (v/v) hydrogen peroxide in PBS. The membranes were blocked with 5% (w/v) nonfat dried milk in PBS containing 0.2% (v/v) Tween 20 (PBST). HS antibodies were diluted 1/10 in 5% (w/v) milk in PBST, and the membranes were incubated overnight at 4 °C. Bound antibodies were detected with mouse anti-His tag IgG, diluted 1/20,000 followed by HRP-conjugated sheep anti-mouse IgG, diluted 1/2000. After washes in PBST and finally PBS, the membranes were incubation in SuperSignal West Dura extended duration substrate for 5 min and exposed to Hyperfilm ECL. The controls were omission of HS antibody, omission of HS antibody, and anti-His tag antibody or treatment of lung extracts with heparinase I, II, and III (all 5 milliunits/200 μl of extract in 100 mm sodium acetate, 0.1 mm calcium acetate, 50 μg/ml BSA, pH 7.0) for 4 h at 37 °C to remove epitopes prior to blotting lung extract.
Molecular Modeling of Antibody Three-dimensional Structure and Surface Electrostatic Potentials
Antibody light and heavy chain sequences were aligned with known sequences and submitted to web antibody modeling, an online resource hosted by The Centre for Protein Analysis and Design at the University of Bath. This is specifically tailored to construct three-dimensional models of antibody Fv sequences. The resulting models were then viewed in a Swiss-PDB viewer.
Competition Binding Assays and Enzyme-linked Immunosorbent Assay
Biotinylation of Heparan Sulfate
N-Hydroxysuccinimide amino caproate biotin (NHS-LC-biotin) (25 μl of a 50 mm solution in dimethyl sulfoxide) was added to 5 mg of PMHS in 500 μl of distilled water and allowed to react overnight at room temperature. A further 25 μl of NHS-LC-biotin was added after this time. Biotinylated polysaccharide was separated from unconjugated biotin on a Sephadex G-25 gel filtration column, with the absorbance monitored at 280 nm to identify fractions containing biotinylated HS. In this way, PMHS was biotinylated internally within the polysaccharide chain at rare N-unsubstituted glucosamines. PMHS is estimated to contain ∼1.2–7.5% free amines (34), equating to 1–2 biotinylation sites/HS chain; thus, attachment to microtitre plates will be made at a restricted number of sites/chain.
Enzyme-linked Immunosorbent Assay
Three μg/ml streptavidin in 0.1 m Na2CO3HCO3 (pH 9.6) was incubated for 16 h at 4 °C in Maxisorp 96-well microtiter plates. The plates were blocked with 1% (w/v) BSA in PBS for 2 h at room temperature, washed with PBST (0.05% (v/v) Tween 20), and then incubated with biotinylated PMHS (75 μg/ml) in 1% (w/v) BSA, PBST for 2 h at room temperature. The plates were washed with PBST, and 15 μl of antibody was added to wells with 15 μl of competitor saccharide (heparin dp2–dp20 oligosaccharides or chemically modified heparins) and incubated overnight at 4 °C. The antibody dilutions were determined in a titer and were used at final concentrations of; HS3B7V 1/5, HS4E4V 1/15, HS3A8V 1/20, AO4B08V 1/15, EV3C3V 1/15, and EW4G1V 1/2. Competitor saccharides (10 mg/ml in distilled water) were diluted in 2% BSA in PBST (0.1% Tween 20) to the required concentrations. For the positive control, antibody was added to wells with 2% BSA in PBST without competitor saccharide. The plates were washed in PBST and incubated with mouse anti-VSV-G IgG, clone P5D4, diluted 1/100 for 1 h, followed by HRP-conjugated sheep anti-mouse IgG, diluted 1/1000. After washing in PBST, the plates were developed with o-phenylenediamine substrate, and A490 values were measured using a microtiter plate reader.
RESULTS
Six antibodies were chosen on the basis that they had been raised against HS/heparin from a variety of tissue sources and encompassed a broad spectrum of antibody structures. Their selection contained two pairs of antibodies that shared an identical framework structure and therefore only differed in the VH CDR3 sequence (HS4E4V/HS3A8V and EV3C3V/EW4G1V). The two remaining antibodies had distinct framework structures and unique VH CDR3 sequences (Table 1).
TABLE 1.
HS phage display antibodies
Heavy chain variable genes (VH) and gene segments (DP) for each antibody are shown, together with heavy chain CDR-3 sequences, which are considered to be major determinants in antibody specificity. The source of HS against which the antibodies were raised and important references are also shown.
| Clone | VH | DP | CDR3 | Selected against | Ref. |
|---|---|---|---|---|---|
| HS3B7V | 1 | 3 | SRKTRKPFMRK | Bovine kidney HS | 24 |
| AO4BO8V | 3 | 47 | SLRMNGWRAHQ | Mouse skeletal muscle HS | 23 |
| HS4E4V | 3 | 38 | HAPLRNTRTNT | Bovine kidney HS | 23 |
| HS3A8V | 3 | 38 | GMRPRL | Bovine kidney HS | 23 |
| EV3C3V | 3 | 42 | GYRPRF | Human lung HS | 24 |
| EW4G1V | 3 | 42 | GARLKR | Porcine intestine mucosal heparin | 32 |
Anti-HS scFv Antibodies Identify Distinct Epitopes in Fetal Rat Lungs
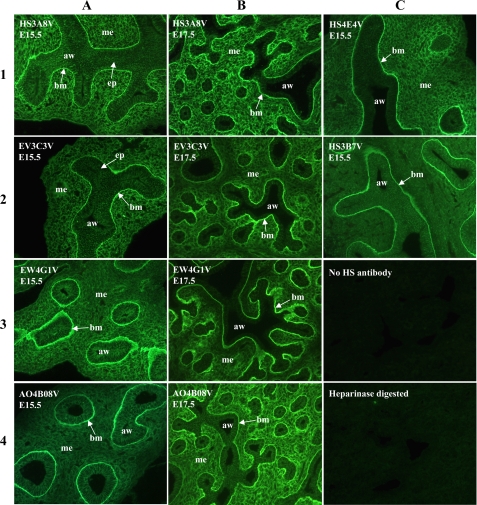
HS phage display antibodies displayed different patterns of immunoreactivity in embryonic rat lungs, indicating distinct HS epitopes (Fig. 1). All of the antibodies showed strong immunoreactivity to airway epithelial basement membranes; one antibody, HS3B7V, exclusively labeled epithelial basement membranes in the lungs and failed to stain mesenchymal or epithelial cells (Fig. 1C, panel 2). HS3A8V and EV3C3V showed similar patterns of immunohistochemistry in E15.5 rat lungs, labeling airway epithelium, underlying basement membranes and the surrounding mesenchyme (Fig. 1A, panels 1 and 2). However, at E17.5, staining by these two antibodies differed, most notably in the lung mesenchyme; HS3A8V staining remained strong throughout the mesenchyme (Fig. 1B, panel 1), whereas a gradient of mesenchymal EV3C3V staining was observed, with the highest levels of immunoreactivity around the smaller developing airways (Fig. 1B, panel 2). EW4G1V and AO4B08V also demonstrated equivalent patterns of staining in E15.5 rat lungs, both showing strong labeling of epithelial basement membranes and weak labeling of the surrounding mesenchyme (Fig. 1A, panels 3 and 4). In E17.5 lungs, however, staining with the two antibodies was distinct; EW4G1V mesenchymal staining remained weak (Fig. 1B, panel 3), whereas AO4B08V mesenchymal staining increased significantly (Fig. 1B, panel 4). The final antibody, HS4E4V showed a unique pattern of immunoreactivity in E15.5 lungs, labeling epithelial basement membranes and areas of mesenchyme surrounding smaller distal airways (Fig. 1C, panel 1). Immunohistochemical analysis of antibody binding specificities thus indicated unique HS epitope expression.
FIGURE 1.
HS phage display antibodies identify distinct HS structures in embryonic rat lung. In E15.5 lungs, HS3B7V exclusively labels epithelial basement membranes and HS4E4V labels basement membranes together with areas of mesenchyme surrounding smaller distal airways. HS3A8V and EV3C3V both label epithelial cells, underlying basement membranes and surrounding mesenchyme in E15.5 lungs; however, at E17.5, staining differs. HS3A8V staining remains strong throughout E17.5 lung mesenchyme, whereas EV3C3V staining becomes more restricted to areas of mesenchyme adjacent to smaller airways. EW4G1V and AO4B08V also show equivalent patterns of staining in E15.5 lungs, strongly labeling epithelial basement membranes and labeling the surrounding mesenchyme more weakly. However, in E17.5 lungs, EW4G1V mesenchymal staining remains weak, whereas AO4B08V mesenchymal staining increases. Immunohistochemistry with individual antibodies is therefore unique, indicating distinct epitopes. E15.5 and E17.5 rat lungs were probed with HS antibodies followed by rabbit VSV-G tag antibody and fluorescein isothiocyanate-conjugated goat anti-rabbit IgG. The negative controls were omission of HS antibody or digestion of HS with heparinase prior to antibody incubation. All of the images are at the same magnification. aw, airway; mes, mesenchyme; epi, epithelium; bm, basement membrane.
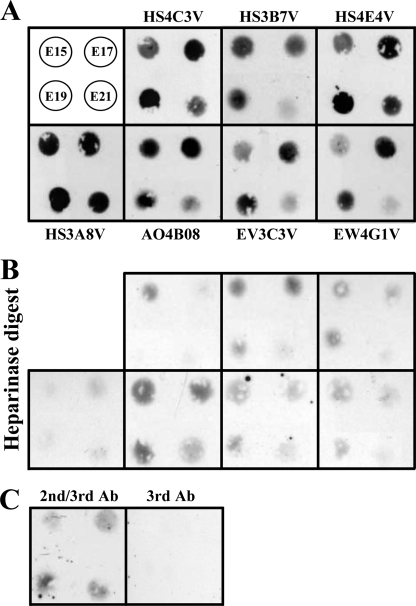
To gain further biochemical insight into the expression of antibody-binding sites, we examined the presence of epitopes biochemically by dot blotting tissue extracts from developing rat lungs and probing with HS antibodies. In many cases the epitopes were identified in lung extracts at varying levels during development that correlated with the relevant immunochemistry data. Surprisingly, however, one antibody, HS4C3V, failed to label developing rat lungs in situ (data not shown), whereas the epitope was clearly identified in vitro in dot blotted lung extracts (Fig. 2). In addition, alongside the major difference in recognition of the HS4C3V epitope, a number of more subtle discrepancies between the in situ and in vitro detection of epitopes were observed. Levels of the AO4B08V epitope were high in E15.5 lung extracts and increased further in E17.5 extracts (Fig. 2), whereas in situ the epitope was present at a low level at E15.5 and significantly increased in E17.5 lungs (Fig. 1, A and B, panels 4). Conversely, EV3C3V epitope levels observed in dot blots were very low at E15.5 and increased significantly at E17.5 (Fig. 2), whereas in situ the epitope was present at a consistently high level throughout development (Fig. 1, A and B, panels 2). These observations are significant, as they highlight an important difference between the presence and availability of epitopes in a native environment in situ compared with an in vitro environment, i.e. in tissue extracts. In the latter, HS chains may be stripped of the majority of their protein partners (e.g. because of detergent in homogenization buffer), whereas in situ, bound proteins could be associated with HS chains and modify antibody binding.
FIGURE 2.
Dot blot analysis of HS epitopes in E15. 5–21.5 fetal rat lung extracts identified differences between in situ and in vitro detection of epitopes. Lung extracts (10 μl) were blotted onto nitrocellulose membrane and probed with HS antibodies as described under “Experimental Procedures.” Bound antibodies were detected via their polyhistidine (His) tag with mouse anti-His-tag IgG, followed by HRP-conjugated sheep anti-mouse IgG. The data are representative of two independent blots. A, the top left panel shows the arrangement of four extracts from E15.5, E17.5, E19.5, and E21.5 fetal lungs. The remaining panels are labeled with the relevant probe HS antibody. B, control data showing background binding after digestion of HS via treatment of lung extracts with a mixture of heparinases I, II, and III (all 5 milliunits/200 of μl extract) prior to blotting onto membrane and probing with antibodies (panels correspond to those in shown in A). C, control probing of blots with secondary anti-His tag and tertiary HRP anti-mouse only (2nd/3rd Ab) or tertiary HRP anti-mouse (3rd Ab) only. Low background immunoreactivity was noted for heparinase-treated lung extract blots; this is likely to reflect reduced enzyme activity in the presence of SDS (data not shown). Ab, antibody.
Three-dimensional Modeling of scFv anti-HS Antibody Structures
The antibodies consisted of a heavy chain (VH) and a light chain variable domain (VL), which are connected by a polypeptide linker. Within both of these domains were three hypervariable regions, termed complementarity determining regions (CDRs), which form the antigen-binding site. All of the antibodies shared an identical VL chain but showed differences in their VH chains.
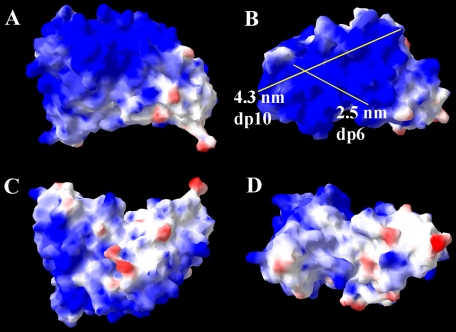
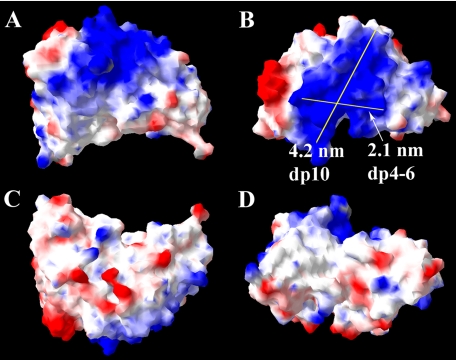
Two pairs of antibodies, HS4E4V/ HS3A8V and EV3C3V/EW4G1V, were encoded by identical gene segments belonging to the VH3 gene family, and consequently, their amino acid sequences were identical apart from their CDR3 (Table 1). AO4B08V was encoded by a different gene segment (DP47) of the VH3 gene family, resulting in minor variations in the heavy chain amino acid sequence, as well as a unique CDR3 (Table 1). HS3B7V was distinct from the other antibodies, originating from a different VH gene family altogether (VH1), resulting in a more diverse heavy chain sequence and more obvious variations in structure and surface electrostatic potential compared with the other five antibodies. The surface electrostatic potentials of the antibodies have not been examined before and were therefore modeled here. Modeling revealed a large basic area on the surface of all antibodies, expected to be a significant mediator of interactions with polyanionic heparin/HS (Figs. 3a and 4 and supplemental Figs. S1–S4). However, the large area of basic residues dominating the surface of the antibodies clearly provided more than one possibility for the binding orientation of HS. Measurement of the potential binding axes for heparin/HS revealed that lengths of between 2 and 4 nm can be accommodated (Figs. 3 and 4 and supplemental Figs. S1–S4), corresponding to a dp4–dp10 heparin/HS sequence, because a disaccharide measured ∼0.8 nm in length (35). Interestingly, on the surface of HS3B7V, however, the basic patch was comparatively smaller than the other antibodies, and there was also a higher proportion of acidic regions on its surface (Fig. 4). The binding axis for heparin/HS would, therefore, be expected to be more restricted in this case than with the other antibodies.
FIGURE 3.
Proposed model and surface electrostatic potential of HS4E4V from four perspectives. A large proportion of basic residues are observed on the surface of the antibody, which will interact with HS and heparin. Two possible HS-binding axes and their lengths are shown. The structure of HS4E4V is extremely similar to HS3A8V, EV3C3V, EW4G1V, and AO4B08V, shown in supplemental Figs. S1–S4. Regions of positive surface charge are represented in blue, and regions of negative charge are in red, whereas neutral areas are white.
FIGURE 4.
Proposed model and surface electrostatic potential of HS3B7V from four perspectives. Two possible HS binding axes and their lengths are shown. Note the smaller proportion of basic residues and higher number of acidic patches on the surface of this antibody compared with the others (Compare with HS4E4V in Fig. 2 and other antibodies described in supplemental Figs. S1–S4). Regions of positive surface charge are represented in blue, and regions of negative charge are in red, whereas neutral areas are white.
Binding Specificities of HS scFv Antibodies
Competition assays were used to evaluate the ability of heparin oligosaccharides of various lengths and with various selective sulfation modifications (Table 2) to compete with immobilized PMHS for binding of six HS antibodies. Chemically modified heparin oligosaccharides were used as model binding competitors, because their sulfation patterns are virtually homogeneous, and they have been structurally characterized (36, 37). This allows systematic “footprinting” of the effects of altered sulfation patterns (41) and allows for interpretation of binding data. HS oligosaccharides, which are generally more heterogeneous, could not be characterized as rigorously or systematically. Sufficient numbers of pure structurally defined HS oligosaccharides are not currently available, making heparin oligosaccharides the most useful model structures available at present to systematically analyze antibody binding specificities. Relative binding affinities of the antibodies and heparin analogues were evaluated by determining IC50 values, defined as the concentration of competitor that inhibits antibody binding to immobilized HS by 50%.
TABLE 2.
Chemically modified heparin structures
The letter I stands for iduronate, and A stands for amino sugar (glucosamine).
| Analogue | Predominant structure | IdoC2 | GlcC6 | GlcC2 |
|---|---|---|---|---|
| Heparin | I2SA6SNS | SO3− | SO3− | SO3− |
| De-N-sulfated | I2SA6SNAc | SO3− | SO3− | COCH3 |
| De-2-O-sulfated | I2OHA6SNS | OH | SO3− | SO3− |
| De-6-O-sulfated | I2SA6OHNS | SO3 | OH | SO3 |
| De-N/de-2-O-sulfated | I2OHA6SNAc | OH | SO3− | COCH3 |
| De-N/de-6-O-sulfated | I2SA6OHNAc | SO3− | OH | COCH3 |
| Only N-sulfated | I2OHA6OHNS | OH | OH | SO3− |
| Fully de-N/O-sulfated | I2OHA6OHNAc | OH | OH | COCH3 |
Minimal Length Heparin-binding Sites
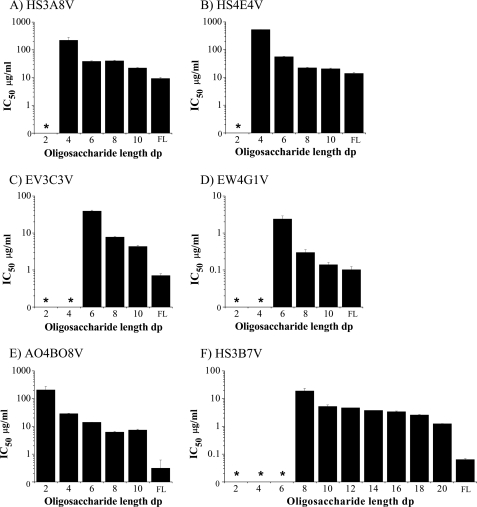
At least a dp4 heparin saccharide was required to compete for antibody-HS binding, with heparin disaccharides (dp2) showing little or no competition. As heparin oligosaccharide length increased from dp4 to dp10, IC50 values progressively decreased, signifying improving competition (Fig. 5 and supplemental Fig. S5). This correlates with the modeling data, which indicated that the basic HS binding patch on the surface of the antibodies can accommodate between a dp4 and dp10 heparin oligosaccharide (Figs. 3 and 4 and supplemental Fig. S5). Binding of HS3A8V, HS4E4V, and AO4B08V antibodies to immobilized HS was competed by relatively high concentrations of dp4 heparin, and substantial competition was observed with heparin oligosaccharides of dp6 and above (Fig. 5, A, B, and E). Conversely, heparin dp4 showed limited ability to compete for EV3C3V and EW4G1V binding; these antibodies required a minimum of heparin dp6, with more significant competition observed with heparin dp8 and longer (Fig. 5, C and D). The HS3B7V antibody showed a more complicated binding profile, with no significant competition with dp2–dp8 heparins (Fig. 5F). Competition was observed with heparin dp10 but remained limited. Thus, oligosaccharide length in the assay was extended to dp12-dp20 for HS3B7V. Heparin dp12-dp18 showed limited variation in competition, with IC50 values between 3 and 5 μg/ml (Fig. 5F). Improved competition was observed with heparin dp20 (IC50 of 1.25 μg/ml); however, full-length heparin, with an IC50 of 0.06 μg/ml, was still a 20-fold stronger competitor for this antibody than dp20 heparin. Intriguingly, molecular modeling of this antibody revealed a relatively smaller basic patch for heparin/HS binding compared with the other antibodies (Fig. 4), suggesting a particularly unusual binding relationship between this antibody and the polysaccharide.
FIGURE 5.
IC50 values for the binding of HS antibodies to immobilized HS in the presence of heparin oligosaccharides of various length and full-length heparin. PMHS was biotinylated and immobilized on streptavidin-coated microtiter plates. Equilibrium binding of HS antibodies in the presence of heparin oligosaccharides, dp2–dp12 and full-length heparin (FL) was quantified at A490 using a VSV-G tag antibody (P5D4) followed by HRP-conjugated anti-mouse antibody and o-phenylenediamine substrate. The range of heparin oligosaccharides was extended to dp12–dp20 for HS3B7V. The IC50 values were calculated as the concentrations of competitor saccharide required for 50% inhibition of antibody binding to immobilized HS. The values are the means of triplicate samples, the error bars represent the S.E., and the data are representative of two separate experiments. * denotes oligosaccharides unable to compete sufficiently to calculate an IC50.
Involvement of Particular Structural Moieties in Antibody Binding
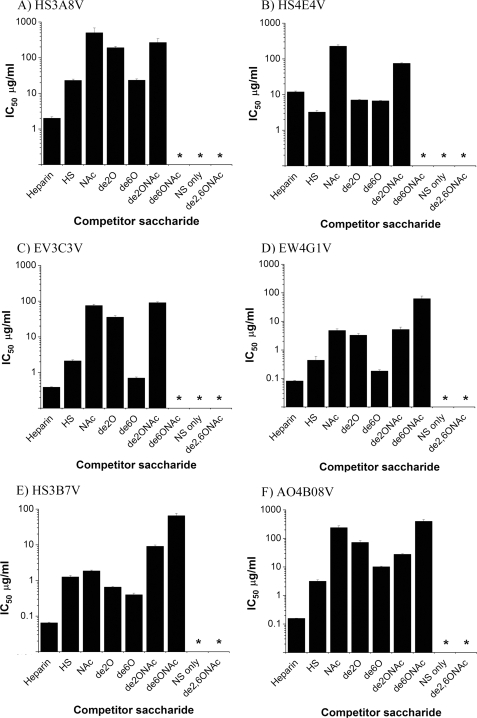
Removal of all sulfate groups from heparin or removal of all O-sulfates abolished competition by the polysaccharide for all antibodies, emphasizing the importance of O-sulfation in binding specificity (Fig. 6). Individual removal of N-, 2-O-, or 6-O-sulfates from heparin reduced competition for five of six antibodies. The exception was HS4E4V; although de-N-sulfation of heparin reduced the competing ability of the polysaccharide for HS4E4V, removal of 2- or 6-O-sulfates from heparin resulted in a structure with 2-fold increased competition for HS4E4V (Fig. 6B). For the other five antibodies, the most significant reduction in competition was observed following removal of N- or 2-O-sulfates, with de-6-O-sulfation having less of an effect. Further information was obtained from competition assays using double modified heparin derivatives, i.e. with sulfates selectively removed from two different positions. This is the first time that these particular modified heparin polysaccharides have been used to systematically explore antibody-HS interactions and provide new and important insights into the binding specificities of the antibodies. De-N-sulfated heparin additionally de-2-O-sulfated or de-6-O-sulfated competed to variable extents and generated intriguing results. First, the double modified de-N/de-6-O-sulfated heparin was, in all cases, a weaker competitor than de-N/de-2-O-sulfated heparin, and for three of the antibodies (HS4E4V, HS3A8V, and EV3C3V), de-N/de-6-O-sulfated heparin did not compete at all (Fig. 6, A–C). This is interesting when compared with singly de-2-O-sulfated or de-6-O-sulfated heparins, where removal of 2-O-sulfates resulted in a weaker competitor than removal of 6-O-sulfates. The exception to this was HS4E4V, for which de-2-O-sulfated and de-6-O-sulfated heparins showed comparable competition (Fig. 6B). Second, competition with the double modified heparins was not simply an additive result of the level of competition achieved with the corresponding singly modified structures. For example, singly modified de-N-sulfated and de-6-O-sulfated heparins were 30- and 6-fold weaker competitors, respectively, than fully sulfated heparin for HS3B7V, whereas removal of both of these sulfate species from heparin resulted in a substantially weaker competitor than might be expected, with 1000-fold reduced competition (Fig. 6F). For a number of antibodies, competition with double modified heparins was comparable with the corresponding singly modified heparins. For example, competition for EV3C3V was similar with de-N/de-2-O-sulfated heparin and singly modified de-N-sulfated heparin with IC50 values of 90 and 75 μg/ml, respectively (Fig. 6C). Similarly, de-N/de-2-O-sulfated heparin competed similarly to singly de-N-sulfated heparin and de-2-O-sulfated heparin for EW4G1V with IC50 values of 5.2, 4.8, and 3.3 μg/ml (Fig. 6D). For a number of antibodies, double modified heparins with two sulfate species removed were actually stronger competitors than the corresponding singly modified polysaccharides. For example, de-N/de-2-O-sulfated heparin was a stronger competitor for HS3A8V (IC50 of 270 μg/ml) than singly de-N-sulfated heparin (IC50 of 500 μg/ml) (Fig. 6A). De-N/de-2-O-sulfated heparin, with an IC50 of 28 μg/ml, was also a stronger competitor for AO4B08V than its corresponding singly modified heparins, which had IC50 values of 240 μg/ml (de-N-sulfated heparin) and 70 μg/ml (de-2-O-sulfated heparin) (Fig. 6F). The binding curves are shown in supplemental Fig. S6.
FIGURE 6.
IC50 values for the binding of HS antibodies to PMHS in the presence of competing chemically modified heparins, unmodified heparin and HS. PMHS was biotinylated and immobilized on streptavidin-coated microtiter plates. Equilibrium binding of HS antibodies in the presence of variable concentrations of heparin, HS, and modified heparin derivatives was quantified at A490 using a VSV-G tag antibody (P5D4) followed by HRP-conjugated anti-mouse IgG and o-phenylenediamine substrate. Modified heparins are: de-N-sulfated/N-acetylated heparin (NAc), de-2-O-sulfated heparin (de2O), de-6-O-sulfated heparin (de6O), de-N-sulfated/N-acetylated and de-2-O-sulfated heparin (de2ONAc), de-N-sulfated/N-acetylated and de-6-O-sulfated heparin (de6ONAc), exclusively N-sulfated heparin (NS only), and completely de-O/N-sulfated/N-acetylated (de2,6ONAc) IC50 values were then calculated as the concentration of competitor saccharide needed for 50% inhibition of antibody binding to immobilized HS. The values are the means of triplicate samples, the error bars represent the S.E., and the data are representative of two separate experiments. * denotes oligosaccharides unable to compete sufficiently to calculate an IC50.
DISCUSSION
The structural heterogeneity of HS renders characterization of native HS chains difficult. HS phage display antibodies allow native HS to be probed in situ, and immunohistochemical analysis of fetal rat lungs indicated that antibodies identify distinct HS epitopes with a developmentally controlled pattern of expression. However, a limitation of the antibodies is that the exact HS structures recognized remain relatively undefined; thus, a characterization of antibody binding specificities and epitope structures was undertaken.
Antibody Surfaces Are Highly Basic, Providing Multiple Possible Binding Orientations
All of the antibodies exhibited a clear trend of improved binding as heparin oligosaccharide length increased. This was not surprising given the observed high basicity of antibody surfaces, which will be increasingly attracted toward the extending length of acidic oligo/polysaccharide. As heparin chain length increased, long range electrostatic forces increased, which acts to maneuver the antibody and polysaccharide toward each other in a favorable orientation for binding, increasing the probability of a productive binding event, a phenomenon described as electrostatic steering (38, 39). The large basic area on the surface of the antibodies provides a number of possible orientations for HS/heparin to bind. The highest affinity binding, however, will result from the interaction that has the largest intermolecular surface area, i.e. the longest oligosaccharide of appropriate structure bound to the longest basic axis of the antibody, with concomitant interactions along the cognate molecular surfaces. The importance of the VH CDR3 in determining binding specificity was evident by distinct binding characteristics of antibodies whose amino acid sequences differed only in the VH CDR3, i.e. HS4E4V/HS3A8V and EV3C3V/EW4G1V (Table 1). However, the large area of basicity of antibody surfaces suggests that regions flanking the VH CDR3 within the antibody framework are also involved in HS interactions, complicating epitope characterization. In competition assays, most antibodies require a minimum of dp4–dp6 heparin, except for HS3B7V, which required a longer polysaccharide. Geometrically, for a heparin oligosaccharide of dp12 or longer to bind HS3B7V, the polysaccharide would have to wrap around the antibody, which is unlikely given the relatively limited flexibility of the heparin glycosidic linkage (40). In addition, the predicted model of HS3B7V revealed a smaller basic patch on its surface compared with the other antibodies, only able to accommodate between a dp4–dp10 heparin (Fig. 4). One possibility is that HS3B7V is not monomeric in solution, perhaps existing as a dimer with a bipartite binding site. Alternatively, HS3B7V may require a rare structural motif in heparin (e.g. an N-unsubstituted amine, 3-O-sulfated glucosamine, or a precise sequence arrangement), the frequency of which would be anticipated to increase as oligosaccharide length increases.
Antibody Epitope Structures Are Complex and Cannot Be Characterized as Simple Linear HS Motifs
Previous attempts to characterize anti-HS scFv antibody epitopes have, in the majority of the cases, used a direct enzyme-linked immunosorbent assay approach, whereby test polysaccharides of various structures are passively adsorbed to a surface and antibody binding is measured directly (24, 31, 41). A weakness of this method is that the structure of the different polysaccharides may affect their immobilization to the surface, and immobilization must mask some structural groups; together these effects will result in variations in surface coverage and exposed binding structures, as suggested previously by Powell et al. (42). In contrast, competition enzyme-linked immunosorbent assay, used here and in two previous studies (24, 43), uses a constant surface of immobilized HS or heparin and is therefore likely to be a more objective measurement of antibody binding specificity. Our binding assays also feature additional competitor polysaccharides to those used in previous analyses, in particular, double modified de-N/de-2-O-sulfated and de-N/de-6-O-sulfated heparins, which have provided more revealing information on antibody binding. Ideally, a large library of structurally defined HS oligosaccharides would be used to evaluate antibody binding specificities. Although the generation of such libraries is underway (44, 45), few HS oligosaccharides have so far been sequenced (46–51) and given the yield of these oligosaccharides, full structural characterization by NMR will only be possible in a very limited number of cases. This approach is therefore not yet feasible. In the interim, simplified model structures generated by the uniform removal of specific sulfate groups from heparin chains provide an invaluable tool, because they are more homogenous and have been structurally characterized (36, 37), allowing a systematic evaluation of antibody binding specificity. The effect of a single alteration in structure, i.e. removal of 6-O-sulfates, on antibody binding can be monitored and interpreted. In the binding assays performed here, sulfation, particularly O-sulfation, was shown to be essential for antibody-HS interactions; however, binding appears to be more intricate than simply relying on sulfates alone, with increasing levels of saccharide sulfation not necessarily correlating with increased antibody reactivity. These data on the effects of selective removal of sulfate groups are clearly not consistent with recognition of single linear epitopes by these antibodies, because if the latter was the case, more dramatic effects on binding would be expected with the loss of one or more sulfate groups. In more than one instance, the IC50 values for competition with double modified heparins was not predictable from the IC50 values of competition with the corresponding singly modified heparins. The most likely explanation is that the conformation of the sugar is particularly significant in determining antibody binding specificity and affinity.
A major area of flexibility within HS chains are iduronic acid residues, which can adopt more than one conformation within HS chains, 1C4 chair, and 2S0 skew boat conformers (52) and may play a role in the generation of antibody epitopes in situ. The balance of iduronic acid conformational equilibrium depends upon both its own substitution with 2-O-sulfate and the substitution of neighboring glucosamines by N-sulfation and 6-O-sulfation (35–37, 52, 53). Sulfate substitutions have also been shown to influence glycosidic linkage geometry (37, 53), adding a further level of structural flexibility to the polysaccharide. Protein binding to HS and the coordination of cations by oxygens in the sugar have also recently been shown to affect the conformational characteristics and flexibility of HS/heparin (53–55), possibly contributing to the formation of epitope structures in vivo. Sulfate substitutions on HS chains coupled with protein binding events and cation coordination will therefore influence the conformation and flexibility of the polysaccharide, ultimately influencing the spatial display of reactive binding groups, principally sulfate, carboxyl, and hydroxyl groups, generating structures in HS that are favorable (or inhibitory) for antibody binding. Previous reports have focused on defining linear epitopes with specific sulfation patterns (24, 28, 41). However, the data presented here suggest that epitopes are more complex than this and are likely to be represented by more than one combination of sulfation pattern and sugar conformation; together these structural characteristics likely create a specific three-dimensional epitope structure within a HS chain that fits optimally into an antibody HS-binding site. It is therefore probable that instead of binding to one definitive HS motif, antibodies bind to a (somewhat restricted) class of structurally related HS epitopes whose sequences present sulfate and carboxyl groups to the antibody in a more (higher affinity) or less (lower affinity) optimal geometry.
Interpretation of in Situ and in Vitro Binding Data
Discrepancies between epitope levels observed in fetal rat lungs in situ and in vitro highlight the need for caution when interpreting antibody binding data and, indeed, data using other heparin-binding proteins. A number of epitopes that were only weakly identified in fetal lungs in situ were measurable at an apparent high level in lung extracts by dot blotting. Most dramatically, the HS4C3V epitope was detected at a high level in lung extracts (Fig. 2) but was not identified at all in lung immunohistochemistry (data not shown). This may be due to occupation of epitopes in situ by HS-binding proteins, i.e. the epitopes are cryptic, making them unavailable for antibody recognition. This may arise directly, whereby endogenous proteins occupy an antibody epitope, or indirectly, whereby endogenous proteins bound to HS modify the conformation of an adjacent epitope. Epitopes that in situ are masked by HS-bound proteins will become available in lung extracts containing the ionic detergent SDS, which will release many of the proteins bound to HS. Conversely, epitopes that appear at a low level in lung extracts but are present at a high level in situ may be generated in HS chains in situ by protein binding and/or cation coordination to HS, which will alter sugar conformation and therefore potentially promote conformations in weak epitopes that are favorable for antibody binding. These epitopes will therefore be diminished in vitro via removal of bound proteins and/or alteration of coordinated cations. These data support the view that the antibody epitopes are highly complex and dependent upon multiple factors. Thus, in situ patterns of antibody binding likely reflect a combination of availability of appropriate underlying HS structures and the overlying presence of other binding proteins, creating a dynamic landscape for functional epitope expression. This notion is supported by in situ studies with other HS-binding proteins using the LACE technique (56). An important implication is that caution must be exercised to avoid overly simplistic interpretation of in situ binding data. This has been noted previously, where immunocytochemical detection of fibroblast growth factor in developing mammary glands suggested the presence of cryptic fibroblast growth factor-binding sites in HS in situ (57). Our data also suggest that in future studies, the exploitation of targeted washing and binding conditions of varying stringency within in situ experiments may permit novel insights into the dynamics of HS epitope expression to be obtained.
Selectivity of HS-Protein Interactions
HS phage display antibodies essentially represent a panel of HS-binding proteins of similar structure and surface electrostatics, the major difference between them being a 6–11-amino acid VH CDR3 sequence. They are therefore useful model proteins for investigating the selectivity of HS-protein interactions in general. Interestingly, we noted for the first time that, with the exception of HS3B7V, the highly basic antibody structures are virtually identical. Nonetheless, the binding specificities of individual antibodies in immunohistochemistry and competitive binding assays are unique, suggesting that HS-protein interactions are not simply electrostatically driven but rely on a degree of structural specificity in HS creating specific contacts between protein and polysaccharide via hydrogen and van der Waals bonding. This emphasizes the need for caution when affinity selection of the antibodies is used to identify sugar binding partners (41, 43), because elution by increasing ionic strength is likely to bias toward simpler ionic bonding. Importantly, this applies more broadly to HS-protein binding studies, which are often analyzed using this approach, despite significant evidence for the importance of nonionic interactions (58–60). This suggests that an approach examining alternative selection modes could be a productive area of future study. Overall, it is also highly likely that in vivo, a particular HS-binding protein is able to bind more than one HS motif, with subtle alterations in sulfation pattern and sugar conformation influencing the affinity of interaction, thereby regulating the activity of the protein ligand. This view is supported by the vast majority of previous data for this class of glycan-protein interaction, including the prototypic antithrombin III-binding site. Despite the latter often being viewed as a single pentasaccharide sequence, there are a number of variants of this structure that still permit significant binding and activity, indicating a degree of permissible variation around what constitutes a consensus bioactive binding site (61–66). Altering the conformation of heparin, e.g. by coordination of different cations, has been shown to modify polysaccharide biological activity (53), presumably by altered interactions with cellular proteins. Enabling a protein to bind multiple HS structures allows HS to regulate multiple cellular activities of an individual protein, e.g. formation of protein gradients, transport of a protein from the site of production to a target cell, retention of proteins in the extracellular matrix, and formation of active cell surface signaling complexes. Transport of HS-binding proteins through the extracellular matrix to a target cell, for example, is likely to involve sequential binding to different HS chains and movement from low affinity to higher affinity binding structures as suggested previously by analysis of mammary cell-derived HS (67).
In summary, phage display antibodies are useful tools for analyzing native HS in situ, identifying distinct families of HS motifs. In situ, antibody epitope structures will be generated by specific patterns of sulfation and also sugar conformation, which is influenced by sulfate substitutions in and around the epitope, coordination of cations, and protein binding events near the epitope and also at more distant sites along a HS chain.
The HS phage display antibodies provide a model system that generates important insights that likely reflect the specificity of HS-protein interactions in vivo and highlights the complexity of the relationship between proteins and HS-binding motifs. An individual protein is able to bind to multiple HS sequences, with dynamic variations in sulfation and conformation altering the affinity of interaction. This provides a subtle mechanism for regulating the cellular activities of biologically important HS-binding proteins.
This work was supported by the Human Frontiers Science Program, the Birth Defects Foundation, the Biotechnology and Biological Sciences Research Council (United Kingdom), and the North West Cancer Research Fund.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- HS
- heparan sulfate
- scFv
- single chain variable fragment
- VSV-G
- vesicular stomatitis virus glycoprotein
- HRP
- horseradish peroxidase
- PMHS
- porcine mucosal heparan sulfate
- VH
- heavy chain variable domain
- VL
- light chain variable domain
- CDR
- complementarity determining region
- dp
- degree of polymerization
- BSA
- bovine serum albumin
- En
- embryonic day n
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.Turnbull J. E., Gallagher J. T. (1991) Biochem. J. 273, 553–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy K. J., Merry C. L., Lyon M., Thompson J. E., Roberts I. S., Gallagher J. T. (2004) J. Biol. Chem. 279, 27239–27245 [DOI] [PubMed] [Google Scholar]
- 3.Gallagher J. T. (2006) Biochem. Soc. Trans. 34, 438–441 [DOI] [PubMed] [Google Scholar]
- 4.Kreuger J., Spillmann D., Li J. P., Lindahl U. (2006) J. Cell Biol. 174, 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ori A., Wilkinson M. C., Fernig D. G. (2008) Front. Biosci. 13, 4309–4338 [DOI] [PubMed] [Google Scholar]
- 6.Turnbull J. E., Gallagher J. T. (1991) Biochem. J. 277, 297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindblom A., Fransson L. A. (1990) Glycoconj. J. 7, 545–562 [DOI] [PubMed] [Google Scholar]
- 8.Lyon M., Steward W. P., Hampson I. N., Gallagher J. T. (1987) Biochem. J. 242, 493–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turnbull J. E., Gallagher J. T. (1990) Biochem. J. 265, 715–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyon M., Deakin J. A., Gallagher J. T. (1994) J. Biol. Chem. 269, 11208–11215 [PubMed] [Google Scholar]
- 11.Caterson B., Christner J. E., Baker J. R. (1983) J. Biol. Chem. 258, 8848–8854 [PubMed] [Google Scholar]
- 12.Couchman J. R., Caterson B., Christner J. E., Baker J. R. (1984) Nature 307, 650–652 [DOI] [PubMed] [Google Scholar]
- 13.Couchman J. R., Caterson B., Christner J. E., Baker J. R. (1984) Prog. Clin. Biol. Res. 151, 31–46 [PubMed] [Google Scholar]
- 14.Sorrell J. M., Lintala A. M., Mahmoodian F., Caterson B. (1988) J. Immunol. 140, 4263–4270 [PubMed] [Google Scholar]
- 15.Sorrell J. M., Mahmoodian F., Caterson B. (1988) Cell Tissue Res. 252, 523–531 [DOI] [PubMed] [Google Scholar]
- 16.Sorrell J. M., Mahmoodian F., Schafer I. A., Davis B., Caterson B. (1990) J. Histochem. Cytochem. 38, 393–402 [DOI] [PubMed] [Google Scholar]
- 17.Mehmet H., Scudder P., Tang P. W., Hounsell E. F., Caterson B., Feizi T. (1986) Eur. J. Biochem. 157, 385–391 [DOI] [PubMed] [Google Scholar]
- 18.David G., Bai X. M., Van der Schueren B., Cassiman J. J., Van den Berghe H. (1992) J. Cell Biol. 119, 961–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van den Born J., van den Heuvel L. P., Bakker M. A., Veerkamp J. H., Assmann K. J., Berden J. H. (1992) Kidney Int. 41, 115–123 [DOI] [PubMed] [Google Scholar]
- 20.van den Born J., van den Heuvel L. P., Bakker M. A., Veerkamp J. H., Assmann K. J., Berden J. H. (1991) Lab. Invest. 65, 287–297 [PubMed] [Google Scholar]
- 21.Kure S., Yoshie O. (1986) J. Immunol. 137, 3900–3908 [PubMed] [Google Scholar]
- 22.van Kuppevelt T. H., Dennissen M. A., van Venrooij W. J., Hoet R. M., Veerkamp J. H. (1998) J. Biol. Chem. 273, 12960–12966 [DOI] [PubMed] [Google Scholar]
- 23.Jenniskens G. J., Oosterhof A., Brandwijk R., Veerkamp J. H., van Kuppevelt T. H. (2000) J. Neurosci. 20, 4099–4111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dennissen M. A., Jenniskens G. J., Pieffers M., Versteeg E. M., Petitou M., Veerkamp J. H., van Kuppevelt T. H. (2002) J. Biol. Chem. 277, 10982–10986 [DOI] [PubMed] [Google Scholar]
- 25.Lensen J. F., Rops A. L., Wijnhoven T. J., Hafmans T., Feitz W. F., Oosterwijk E., Banas B., Bindels R. J., van den Heuvel L. P., van der Vlag J., Berden J. H., van Kuppevelt T. H. (2005) J. Am. Soc. Nephrol. 16, 1279–1288 [DOI] [PubMed] [Google Scholar]
- 26.Jenniskens G. J., Veerkamp J. H., van Kuppevelt T. H. (2006) J. Cell Physiol. 206, 283–294 [DOI] [PubMed] [Google Scholar]
- 27.Jenniskens G. J., Hafmans T., Veerkamp J. H., van Kuppevelt T. H. (2002) Dev. Dyn. 225, 70–79 [DOI] [PubMed] [Google Scholar]
- 28.ten Dam G. B., Hafmans T., Veerkamp J. H., van Kuppevelt T. H. (2003) J. Histochem. Cytochem. 51, 727–739 [DOI] [PubMed] [Google Scholar]
- 29.van de Westerlo E. M., Smetsers T. F., Dennissen M. A., Linhardt R. J., Veerkamp J. H., van Muijen G. N., van Kuppevelt T. H. (2002) Blood 99, 2427–2433 [DOI] [PubMed] [Google Scholar]
- 30.Thompson S. M., Connell M. G., Fernig D. G., Ten Dam G. B., van Kuppevelt T. H., Turnbull J. E., Jesudason E. C., Losty P. D. (2007) Pediatr. Surg. Int. 23, 411–417 [DOI] [PubMed] [Google Scholar]
- 31.Smits N. C., Robbesom A. A., Versteeg E. M., van de Westerlo E. M., Dekhuijzen P. N., van Kuppevelt T. H. (2004) Am. J. Physiol. Lung Cell. Mol. Physiol. 30, L166–L173 [DOI] [PubMed] [Google Scholar]
- 32.Bernsen M. R., Smetsers T. F., van de Westerlo E., Ruiter D. J., Håkansson L., Gustafsson B., Van Kuppevelt T. H., Krysander L., Rettrup B., Håkansson A. (2003) Cancer Immunol. Immunother. 52, 780–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yates E. A., Guimond S. E., Turnbull J. E. (2004) J. Med. Chem. 47, 277–280 [DOI] [PubMed] [Google Scholar]
- 34.Toida T., Yoshida H., Toyoda H., Koshiishi I., Imanari T., Hileman R. E., Fromm J. R., Linhardt R. J. (1997) Biochem. J. 322, 499–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mulloy B., Forster M. J., Jones C., Davies D. B. (1993) Biochem. J. 293, 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yates E. A., Santini F., Guerrini M., Naggi A., Torri G., Casu B. (1996) Carbohydr. Res. 294, 15–27 [DOI] [PubMed] [Google Scholar]
- 37.Yates E. A., Santini F., De Cristofano B., Payre N., Cosentino C., Guerrini M., Naggi A., Torri G., Hricovini M. (2000) Carbohydr. Res. 329, 239–247 [DOI] [PubMed] [Google Scholar]
- 38.Kozack R. E., d'Mello M. J., Subramaniam S. (1995) Biophys. J. 68, 807–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lookene A., Chevreuil O., Ostergaard P., Olivecrona G. (1996) Biochemistry 35, 12155–12163 [DOI] [PubMed] [Google Scholar]
- 40.Mulloy B., Forster M. J. (2000) Glycobiology 10, 1147–1156 [DOI] [PubMed] [Google Scholar]
- 41.Kurup S., Wijnhoven T. J., Jenniskens G. J., Kimata K., Habuchi H., Li J. P., Lindahl U., van Kuppevelt T. H., Spillmann D. (2007) J. Biol. Chem. 282, 21032–21042 [DOI] [PubMed] [Google Scholar]
- 42.Powell A. K., Yates E. A., Fernig D. G., Turnbull J. E. (2004) Glycobiology 14, 17R–30R [DOI] [PubMed] [Google Scholar]
- 43.Ten Dam G. B., Kurup S., van de Westerlo E. M., Versteeg E. M., Lindahl U., Spillmann D., van Kuppevelt T. H. (2006) J. Biol. Chem. 281, 4654–4662 [DOI] [PubMed] [Google Scholar]
- 44.Powell A. K., Zhi Z. L., Turnbull J. E. (2009) Methods Mol. Biol. 534, 313–329 [DOI] [PubMed] [Google Scholar]
- 45.Guimond S. E., Turnbull J. E. (1999) Curr. Biol. 9, 1343–1346 [DOI] [PubMed] [Google Scholar]
- 46.Shriver Z., Raman R., Venkataraman G., Drummond K., Turnbull J., Toida T., Linhardt R., Biemann K., Sasisekharan R. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10359–10364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turnbull J. E., Hopwood J. J., Gallagher J. T. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 2698–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Venkataraman G., Shriver Z., Raman R., Sasisekharan R. (1999) Science 286, 537–542 [DOI] [PubMed] [Google Scholar]
- 49.Drummond K. J., Yates E. A., Turnbull J. E. (2001) Proteomics 1, 304–310 [DOI] [PubMed] [Google Scholar]
- 50.Merry C. L., Lyon M., Deakin J. A., Hopwood J. J., Gallagher J. T. (1999) J. Biol. Chem. 274, 18455–18462 [DOI] [PubMed] [Google Scholar]
- 51.Kreuger J., Salmivirta M., Sturiale L., Giménez-Gallego G., Lindahl U. (2001) J. Biol. Chem. 276, 30744–30752 [DOI] [PubMed] [Google Scholar]
- 52.Ferro D. R., Provasoli A., Ragazzi M., Casu B., Torri G., Bossennec V., Perly B., Sinaÿ P., Petitou M., Choay J. (1990) Carbohydr. Res. 195, 157–167 [DOI] [PubMed] [Google Scholar]
- 53.Rudd T. R., Guimond S. E., Skidmore M. A., Duchesne L., Guerrini M., Torri G., Cosentino C., Brown A., Clarke D. T., Turnbull J. E., Fernig D. G., Yates E. A. (2007) Glycobiology 17, 983–993 [DOI] [PubMed] [Google Scholar]
- 54.Guerrini M., Guglieri S., Beccati D., Torri G., Viskov C., Mourier P. (2006) Biochem. J. 399, 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hricovíni M., Guerrini M., Bisio A., Torri G., Petitou M., Casu B. (2001) Biochem. J. 359, 265–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Allen B. L., Rapraeger A. C. (2003) J. Cell Biol. 163, 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rudland P. S., Platt-Higgins A. M., Wilkinson M. C., Fernig D. G. (1993) J. Histochem. Cytochem. 41, 887–898 [DOI] [PubMed] [Google Scholar]
- 58.Thompson L. D., Pantoliano M. W., Springer B. A. (1994) Biochemistry 33, 3831–3840 [DOI] [PubMed] [Google Scholar]
- 59.Hjelm R., Schedin-Weiss S. (2007) Biochemistry 46, 3378–3384 [DOI] [PubMed] [Google Scholar]
- 60.Olson S. T., Halvorson H. R., Björk I. (1991) J. Biol. Chem. 266, 6342–6352 [PubMed] [Google Scholar]
- 61.Lindahl U., Bäckström G., Höök M., Thunberg L., Fransson L. A., Linker A. (1979) Proc. Natl. Acad. Sci. U.S.A. 76, 3198–3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Atha D. H., Stephens A. W., Rosenberg R. D. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 1030–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hovingh P., Piepkorn M., Linker A. (1986) Biochem. J. 237, 573–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Petitou M., Lormeau J. C., Choay J. (1988) Eur. J. Biochem. 176, 637–640 [DOI] [PubMed] [Google Scholar]
- 65.Guerrini M., Guglieri S., Casu B., Torri G., Mourier P., Boudier C., Viskov C. (2008) J. Biol. Chem. 283, 26662–26675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen J., Jones C. L., Liu J. (2007) Chem. Biol. 14, 986–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rahmoune H., Chen H. L., Gallagher J. T., Rudland P. S., Fernig D. G. (1998) J. Biol. Chem. 273, 7303–7310 [DOI] [PubMed] [Google Scholar]