Abstract
We previously reported that Dot1a·AF9 complex represses transcription of the epithelial Na+ channel subunit α (α-ENaC) gene in mouse inner medullary collecting duct mIMCD3 cells and mouse kidney. Aldosterone relieves this repression by down-regulating the complex through various mechanisms. Whether these mechanisms are sufficient and conserved in human cells or can be applied to other aldosterone-regulated genes remains largely unknown. Here we demonstrate that human embryonic kidney 293T cells express the three ENaC subunits and all of the ENaC transcriptional regulators examined. These cells respond to aldosterone and display benzamil-sensitive Na+ currents, as measured by whole-cell patch clamping. We also show that AF17 and AF9 competitively bind to the same domain of Dot1a in multiple assays and have antagonistic effects on expression of an α-ENaC promoter-luciferase construct. Overexpression of Dot1a or AF9 decreased mRNA expression of the ENaC subunits and their transcriptional regulators and reduced benzamil-sensitive Na+ currents. AF17 overexpression caused the opposite effects, accompanied by redirection of Dot1a from the nucleus to the cytoplasm and reduction in histone H3 K79 methylation. The nuclear export inhibitor leptomycin B blocked the effect of AF17 overexpression on H3 K79 hypomethylation. RNAi-mediated knockdown of AF17 yielded nuclear enrichment of Dot1a and histone H3 K79 hypermethylation. As with AF9, AF17 displays nuclear and cytoplasmic co-localization with Sgk1. Therefore, AF17 competes with AF9 to bind Dot1a, decreases Dot1a nuclear expression by possibly facilitating its nuclear export, and relieves Dot1a·AF9-mediated repression of α-ENaC and other target genes.
Introduction
Failure of Na+ homeostasis contributes to hypertension, cardiovascular disease, and respiratory diseases such as cystic fibrosis (1). The importance of the epithelial Na+ channel (ENaC)3 in the regulation of salt homeostasis and blood pressure is demonstrated by the association of gain- and loss-of-function mutations in its subunits with genetic hypertensive and hypotensive diseases, such as Liddle syndrome (2) and pseudohypoaldosteronism type 1 (3). ENaC consists of three partially homologous subunits (α, β, and γ), and their expression on the cell surface constitutes the rate-limiting step in active Na+ and fluid absorption in the apical membrane of salt-absorbing epithelia. Aldosterone treatment or hyperaldosteronism caused by Na+ limitation induces α-ENaC transcription in the aldosterone-sensitive distal nephron. In these cells synthesis of α-ENaC is believed to be the rate-limiting step in Na+ channel formation. As a major regulator of epithelial Na+ absorption, aldosterone imposes a tight and complex regulation of ENaC at multiple levels including transcription, trafficking to the cell membrane, and degradation and acts at least partially through α-ENaC induction in the renal collecting duct (4, 5).
We recently identified and characterized a new aldosterone signaling network involving the murine disruptor of telomeric silencing splice variant “a” (Dot1a) (6), putative transcription factor AF9, and serum- and glucocorticoid-inducible kinase isoform 1 (Sgk1). Under basal conditions, Dot1a and AF9 form a repression complex that binds directly or indirectly to the α-ENaC promoter, catalyzes H3 K79 hypermethylation at the promoter, and represses α-ENaC transcription. Aldosterone relieves this repression by inhibiting Dot1a and AF9 expression and by weakening their interaction via Sgk1-mediated AF9 phosphorylation (7–9). Because Dot1a and AF9 are highly conserved and widely expressed and appear to be involved in transcriptional regulation of other genes (7, 8), we hypothesized that this new aldosterone signaling network exists in human cells and that additional protein-protein interactions might regulate the Dot1a·AF9 complex and, thus, the transcription of ENaC and their transcriptional regulators in an aldosterone-dependent or -independent manner.
Here, we report the characterization and use of human embryonic kidney (HEK) 293T cells as a model system to study this new aldosterone-signaling network. We provide evidence showing a novel protein-protein interaction between Dot1a and AF17 that, like AF9, is a mixed lineage leukemia (MLL) fusion partner involved in acute myeloid leukemia (10, 11). We also define AF17 as a new regulator of Dot1a H3 K79 methyltransferase activity and, thus, basal transcription of α-ENaC and other aldosterone-regulated genes.
EXPERIMENTAL PROCEDURES
Reagents
LipofectamineTM 2000 reagent (Invitrogen), Millicell inserts (12 mm in diameter, 0.45 μm pore size, Millipore), and antibodies against dimethyl histone H3 K79, dimethylated histone H3 K9 (Upstate), trimethyl histone H3 K79 (Abcam), GFP, red fluorescence protein (RFP; Clontech), and FLAG (Sigma) were purchased and used according to the manufacturer's instructions. pGL3Zeocin-1.3-ENaC, pDsRedmonomer-V5, and pCDNA3.1 derivatives expressing untagged Dot1a and constructs expressing various Dot1a mutants as GAL4 BD fusions for yeast or mammalian two-hybrid assays as well as those for GFP-Dot1a or glutathione S-transferase (GST)-Dot1a fusions have been described previously (6–9). Human AF17 and its mouse counterpart are equally competent for interacting with Dot1a (see below). For simplicity, AF17 was used to designate these two proteins throughout the manuscript unless otherwise stated. pCDNA-AF17 and pFLAG-AF17 expressing untagged or FLAG-tagged human AF17 were kindly provided by Yoichi Furukawa (12). A 3.2-kb EcoRI/XhoI fragment encoding full-length human AF17 was isolated from pCDNA-AF17 and cloned into pGADT7, pCMV-AD, pGEX6P-1, and pDsRed-monomer-V5 at EcoRI/XhoI sites for expression of AF17 as GAL4 AD, GST, or RFP fusions, respectively. The cDNA insert expressing mouse AF17 aa 635–786 (referred to as AF17 635–786 hereafter) was released from the pGAD10-based isolates and cloned into pGADT7 at the EcoRI site in either forward or reverse orientations. The former was partially digested with EcoRI/XhoI to release the insert. The insert was subsequently cloned into various vectors for expression of AF17 635–786 as FLAG, GST, or GAL4 AD fusions. Two mouse AF17 target sequences (siRNAi#10, CCCGCTGGTCTACTGCGAT, encoding aa 21–26; siRNAi#11, AAGCTTGCTATGGCATCGTCC, encoding aa 36–42) were identical to the corresponding regions of human AF17 except for one mismatch in siRNA#11. They were annealed and cloned into pSilencer-2.1-U6-Hygro (Ambion) at BamHI-HindIII according to the manufacturer's instructions. The Sgk1 coding region was amplified with pCDNA3.1-Sgk1 as template and cloned into pEGFPC2 at EcoRI/SalI to generate pGFP-Sgk1. pGFP-AF17 and pGFP-AF9 were created by replacing the Sgk1 fragment with the EcoRI/XhoI fragments encoding these proteins. The sequences of all inserts in the constructs were verified by DNA sequencing.
Cell Culture, Transient or Stable Transfections, and RNA Interference
HEK 293T cells were maintained with Dulbecco's modified Eagle's medium/F-12 plus 10% fetal bovine serum. Transient transfections were performed using LipofectamineTM 2000 reagent with cells either cultured on tissue culture plates (for immunoprecipitation/immunoblot (IB) experiments (13–16)), on cover slips (for confocal or epifluorescence microscopy (17–19)), or on Millicell inserts (for real-time RT-qPCR experiments). To knock down AF17 mRNA levels by RNA interference, RNA#10, RNA#11, and the parent vector pSilencer-2.1-U6-Hygro, as a negative control, were stably transfected into 293T cells with selection by hygromycin (500 g/ml) as described (8). Briefly, all colonies resistant to the antibiotics were pooled and expanded without the process of clonal selection on the basis of AF17 mRNA expression.
Whole-cell Patch Clamping
Whole-cell patch clamping of HEK 293T cells grown on poly-d-lysine-coated coverslips was performed as described with minor modifications (20–23). To determine the effects of aldosterone, cells were treated with either 1 μm aldosterone or 0.01% ethanol (a vehicle control) for 24 h. For some experiments, 24 h after transfection with GFP as vector control or various GFP fusions, fluorescence-positive cells were selected for recordings. The extracellular solution for whole-cell recordings consisted of 150 mm NaCl, 0.1 mm KCl, 0.8 mm MgCl2, and 10 mm HEPES, adjusted to a pH of 7.4. The pipette solution contained 150 mm NaCl, 6.2 mm MgCl2, 10 mm HEPES, and 10 mm EGTA, adjusted to a pH of 7.2, and electrodes had resistances of 4–6 megaohms after filling. After making whole-cell contact, cells were held at −40 mV, and 1-s voltage ramps from −80 to +80 mV were delivered once every 5 s. Once a stable base line was achieved, standard extracellular solution containing 10 μm benzamil was superfused. ENaC currents are defined as the difference between currents obtained in the absence and presence of 10 μm benzamil as obtained by digital subtraction. Under these recording conditions, the currents showed strong outward rectification, and current density was quantified using currents measured at +80 mV. Quantification of evoked currents was done using Clampfit Version 9.0 (Molecular Devices).
Yeast and Mammalian Two-hybrid Screen, Immunoblot, Immunoprecipitation, GST Pulldown, and Real-time RT-qPCR
These assays were conducted according to our published protocols (6, 9).
Epifluorescence and Confocal Microscopy
HEK 293T cells cultured on coverslips were transfected with various plasmids as indicated in figure legends. 24 h later cells were analyzed either by epifluorescence or confocal microscopy. Cells expressing GFP-Dot1a, RFP-AF17, or both were categorized as cytoplasmic, nuclear, or both depending on the location of the fusion proteins detected by epifluorescence microscopy (24). For confocal microscopy, cells were rinsed briefly in phosphate-buffered saline and fixed with 1% fresh prepared paraformaldehyde for 30 min at room temperature. The nucleus was stained with 300 nm 4′,6-diamidino-2-phenylindole (DAPI, Sigma) for 15 min at room temperature. Coverslips were mounted onto microscope slides with Vectashield mounting medium (Vector Laboratories). A confocal microscope (Olympus FV1000) was used to observe the specimens and take images. Excitation wavelengths for DAPI, GFP, and RFP are 405, 488, and 543 nm, respectively. Sequential scanning mode was used to eliminate the possibility of cross-talk between channels.
Statistical Analysis
All comparisons were conducted using unpaired Student's t test or one-way analysis of variance. p < 0.05 was considered significant.
RESULTS
ENaC Subunits and All of Their Transcriptional Regulators Examined Are Expressed and Regulated by Aldosterone in HEK 293T Cells
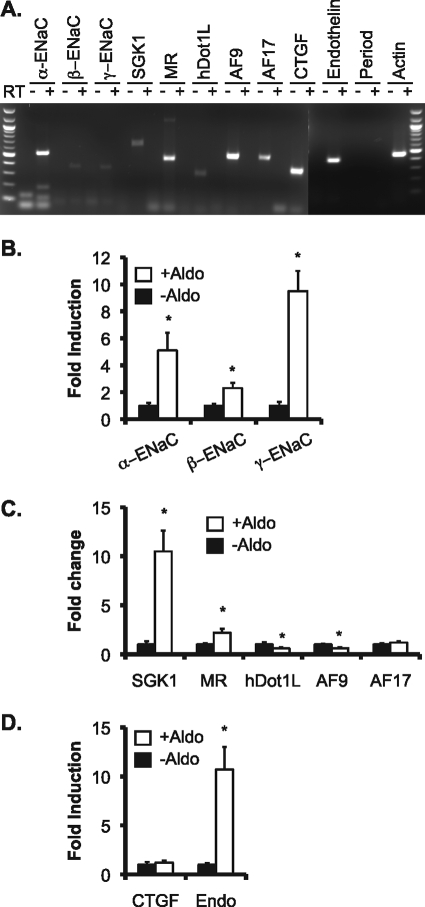
We hypothesized that the mechanisms controlling α-ENaC transcription by the Dot1a·AF9 complex defined in our previous work using mIMCD3 cells and mouse kidney as model systems are applicable to human kidney epithelial cells. To test this hypothesis, we chose 293T cells because they are derived from human embryonic kidney, easily transfected, and widely used in many types of experiments. Several groups have used 293T cells overexpressing ENaC subunits to study the regulation of ENaC cell surface expression, ubiquitination, and activity (13, 19, 25). However, to our knowledge the basal expression of ENaC subunits and their regulatory factors such as SGK1, MR, hDot1L, or AF9 and the effects of aldosterone on their expression have not been documented in 293T cells. As shown in Fig. 1, RT-PCR of total RNAs isolated from 293T cells revealed detectable transcripts of ENaC subunits (α, β, and γ) and their regulators SGK1, MR, hDot1L, AF9, and AF17 (see below). Among the three other aldosterone-regulated genes (CTGF, preproendothelin-1, and period) that were up-regulated by aldosterone in mIMCD3 cells, only period mRNA was not expressed. Real-time RT-qPCR of 293T cells treated with aldosterone (1 μm, for 24 h) led to a 400, 100, and 800% increase in α-, β-, and γ-ENaC mRNA, respectively, compared with control (Fig. 1B). Similarly, aldosterone increased SGK1 and MR mRNA levels to more than 1000 and 200% of control (Fig. 1C). As expected, aldosterone decreased hDot1L and AF9 mRNA expression to about 60% of control and had little effect on AF17 mRNA abundance (Fig. 1C). The hormone also significantly induced mRNA for preproendothelin-1 but not CTGF (Fig. 1D). In brief, 293T cells display expression profiles of ENaC subunits and their positive and negative regulators very similar to the corresponding ones in mIMCD3 cells, suggesting that these cells might share the same or a similar aldosterone signaling network governing the transcription of these genes. Accordingly, 293T cells were used for all subsequent experiments.
FIGURE 1.
ENaC and ENaC transcriptional regulators are expressed and regulated by aldosterone in HEK 293T cells. A, total RNA was isolated from 293T cells cultured in Dulbecco's modified Eagle's medium plus 10% fetal bovine serum and analyzed by RT-PCR in the absence (-) or presence (+) of reverse transcriptase for expression of the genes indicated. B–D, total RNA isolated from 293T cells treated for 24 h with vehicle (−Aldo) or 1 μm aldosterone (+Aldo) was analyzed by real-time RT-qPCR and examined for expression of ENaC subunits (B), ENaC transcriptional regulators (C), or CTGF and preproendothelin-1 (Endo) (D). The mRNA level of each gene was normalized against β-actin mRNA, which was invariant as measured by real-time PCR and set to 1 in the vehicle-treated cells. In all cases *, p < 0.05 versus −Aldo. n = 3.
Dot1a Interacts with AF17 via Its AF9-interacting Domain
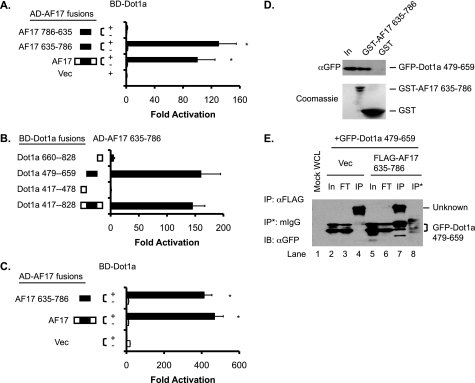
We previously reported a specific interaction between Dot1a and AF9 (8, 9). To identify additional Dot1a-interacting proteins, we screened a yeast two-hybrid cDNA library derived from mouse kidney with GAL4-BD-Dot1a as bait and identified a specific interaction between Dot1a and a peptide corresponding to aa 635–786 of myeloid/lymphoid or mixed lineage-leukemia (trithorax homolog, Drosophila) translocated to 6 (Mllt6, GenBankTM accession number AY050217). The latter shares 92% identity with the corresponding region (aa 648–800) of human AF17 (GenBankTM accession number U07932), which also strongly interacted with Dot1a in the yeast two-hybrid assay (Fig. 2A). These and other observations (see below) suggest that human AF17 and its murine counterpart are equally competent for interaction with Dot1a. Accordingly, AF17 is used to refer to both human and mouse proteins for simplicity. We used full-length human AF17 for transfection studies and mouse AF17 635–786 for GST pulldown and co-immunoprecipitation assays to overcome the technical challenges (see “Discussion”) and assumed that the resulting data were exchangeable between the two proteins.
FIGURE 2.
The AF9-interacting domain in Dot1a is capable of and sufficient for interaction with AF17 in vitro and in vivo. A, shown is a yeast two-hybrid analysis revealing that Dot1a interacts with full-length human AF17 and mouse AF17 635–786 but not with the latter when it is placed in a reverse orientation (AF17 786–635). The bars represent the average -fold activation of the LacZ reporter from three independent experiments (n = 3). *, p < 0.05 versus Vec (pGADT7). B, as in A, mapping by yeast two-hybrid analysis showing that the AF9-interacting domain in Dot1a (aa 479–659) is capable of and sufficient for interacting with AF17; n = 3. C, shown is a mammalian two-hybrid assay confirming that Dot1a interacts with AF17 or AF17 635–786 in 293T cells. The bars represent the average -fold activation of a GAL4-dependent luciferase reporter from three independent experiments (n = 3). *, p < 0.05 versus BD-Dot1a plus Vec (pCMV-AD). D, shown is a GST pulldown assay showing that specific domains in Dot1a and AF17 are responsible for the interaction. GST and GST-AF17 635–786 were purified from E. coli and incubated with whole-cell lysates of 293T cells expressing GFP-Dot1a 479–659. Input (In) of the lysates (5%) and proteins bound to Glutathione-Sepharose 4B beads were examined by immunoblotting with a mouse anti-GFP antibody. The inputs of GST and GST-AF17 were analyzed by Coomassie staining (lower panel). E, co-immunoprecipitation assay demonstrating the interaction between Dot1a and AF17. Whole-cell lysates (WCL) of 293T cells transiently expressing GFP-Dot1a 479–659 with or without FLAG-AF17 635–786 were immunoprecipitated (IP) with a mouse anti-FLAG antibody or equal amount of normal mouse IgG (mIgG). Immunoprecipitated proteins were eluted from the Protein A/G-agarose beads and subjected to IB analysis with the mouse anti-GFP antibody. In, input (2%); FT, flow-through (2%); Vec, vector.
To map the domains of Dot1a responsible for AF17 interaction, we performed additional yeast two-hybrid assays using each of 13 GAL4-BD-Dot1a constructs in combination with the GAL4-AD-AF17 fusion. We detected interactions in all assays involving fragments possessing the previously defined AF9-interacting domain (aa 479–659) of Dot1a (referred to as Dot1a 479–659 hereafter). We observed little or no interaction in all other assays in which Dot1a 479–659 was missing. Similar results were obtained when AF17 was replaced with AF17 635–786 (Fig. 2B and supplemental Fig. S1). Thus, as with AF9, AF17 does not appear to interact with the methyltransferase domain (aa 1–416), the putative leucine zipper domain (aa 576–597), or the C-terminal portion (aa 1112–1540) of Dot1a.
To confirm and extend these findings, we conducted mammalian two-hybrid assays in 293T cells. Co-expression of GAL4-BD-Dot1a with GAL4-AD fusions harboring either AF17 or AF17 635–786 caused a >400-fold activation of a GAL4- dependent luciferase activity construct compared with GAL4-BD-Dot1a alone (Fig. 2C), in agreement with the yeast two-hybrid results.
To verify the interaction biochemically, GST pulldown assays were performed with GST-AF17 635–786 purified from Escherichia coli and 293T cell lysates harboring GFP-Dot1a 479–659. Binding was observed only when the two fusions were combined (lane 2, Fig. 2D). Replacing one or both with the GST and GFP vectors abolished the interaction (lane 3, Fig. 2D and data not shown), confirming the specificity of the interaction. We were unable to express and purify the GST fusion containing full-length AF17 from E. coli, possibly due to its toxicity, insolubility, or both.
Finally, we performed co-immunoprecipitation assays with 293T cell lysates expressing GFP-Dot1a 479–659 with or without FLAG-AF17 635–786. Precipitation of GFP-Dot1a 479–659 by a mouse anti-FLAG antibody was dependent on the presence of the FLAG-AF17 fusion (compare lanes 7 and 4, Fig. 2E). Moreover, it was not precipitated by the same amount of normal mouse IgG in a parallel reaction (lane 8, Fig. 2E). In reciprocal experiments, the FLAG-AF17 fusion was specifically immunoprecipitated by a rabbit anti-GFP antibody in the presence of the GFP-Dot1a fusion but not GFP tag (supplemental Fig. S2). Taken together, the yeast and mammalian two-hybrid, GST pulldown, and co-immunoprecipitation experiments indicate that Dot1a interacts specifically with AF17 in vitro and in vivo. Moreover, the AF9-interacting domain of Dot1a (aa 479–659) is also an AF17 binding domain. Accordingly, this domain is hereafter referred to as the AF9/AF17-interacting domain.
The AF9/AF17-interacting Domain of Dot1a Is Capable of and Sufficient for Mediating Colocalization with AF17 in the Cytoplasm of 293T Cells
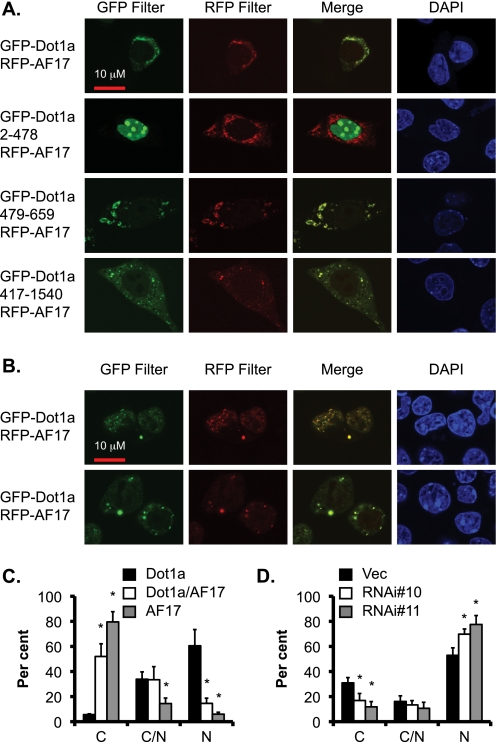
To demonstrate the biological relevance of the Dot1a·AF17 interaction and to further pursue the theory that the AF9-interacting domain in Dot1a is also responsible for interaction with AF17, we co-expressed RFP-tagged AF17 with various GFP-Dot1a fusions in 293T cells and examined their cellular distribution by confocal and epifluorescence microscopy. As shown in supplemental Fig. S3, cells transfected with either GFP-Dot1a or RFP-AF17 alone were detected only by the corresponding filters, confirming the filter specificity. As expected, the three GFP-tagged Dot1a fusions containing the AF9/AF17-interacting domain (full-length, 417–1540, or 479–659) co-localized with RFP-AF17 primarily, if not exclusively, in the cytoplasm (Fig. 3A). In contrast, Dot1a 2–478 apparently failed to co-localize with AF17, although it was highly expressed in the nucleus as a GFP fusion. In these cells most if not all of the RFP-AF17 still resided in the cytoplasm. It should be noted that GFP-Dot1a is apparently easily degraded, as evidenced by multiple small bands in Western blot (8, 9) and progressively diminished fluorescence intensity. These data indicate that 1) Dot1a 479–659 is capable of and sufficient for interacting and co-localizing with AF17, 2) the co-localization of these proteins is not an artifact due to overexpression of the related proteins, and 3) the preferential expression of RFP-AF17 in the cytoplasm is not caused by overexpression of GFP-Dot1a fusions.
FIGURE 3.
AF17 colocalizes with Dot1a and enhances Dot1a cytoplasmic expression at the expense of its nuclear expression. A, representative confocal microscopy images show colocalization of transiently expressed RFP-AF17 with GFP-Dot1a fusions containing full-length, aa 2–478, 479–659, and 417–1540 but not GFP-Dot1a 2–478 in 293T cells. B, as in A, representative images show nuclear (top) and cytoplasmic (bottom) colocalization of the Dot1a and AF17 fusions. C, the bar graph show that AF17 overexpression causes preferential expression of Dot1a in the cytoplasm. As in B, cells expressing GFP-Dot1a, RFP-AF17, or both were categorized as cytoplasmic (C) nuclear (N), or both (C/N) depending on the location of the fusion proteins. The graphed value (%) is the number of cells of each localization type divided by the total number of cells examined. At least 200 cotransfected cells per transfection were examined from three independent experiments (n = 3). Each percentage was compared with control (Dot1a alone) within the category. D, the bar graph shows that AF17 knockdown increased Dot1a nuclear localization, with the observed localization value given as a percentage of the total number of cells surveyed. Results are as in B, except that 293T cells were stably transfected with a control vector (Vec) or one of two RNAi constructs specific for AF17 (see Fig. 5) followed by transient transfection of pGFP-Dot1a. n = 3. *, p < 0.05 versus vector in each category.
AF17 Competes with AF9 for Interaction with Dot1a
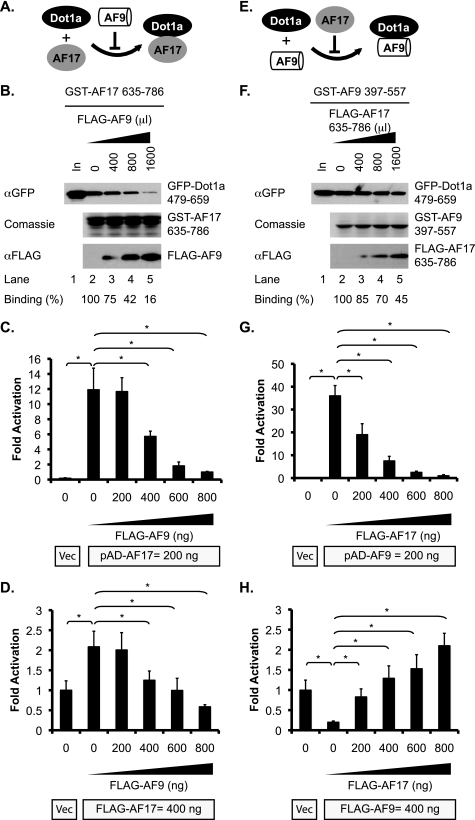
The fact that the same domain of Dot1a can interact with both AF17 and AF9 suggests that these two proteins may compete for binding to Dot1a, as summarized in Fig. 4, A and E. To test this hypothesis, we performed similar GST pulldown assays as described above to investigate competition between AF17 and AF9. As shown in Fig. 4B, increasing the lysate amount containing FLAG-AF9 from 0 to 1600 μl gradually reduced the amount of the GFP-Dot1a fusion retained by a fixed amount of the GST-AF17 fusion. The relative Dot1a·AF17 binding efficiency in the reaction with 1600 μl of FLAG-AF9-containing lysate was ∼16% of the control in which FLAG-AF9 was not added (Fig. 4B), indicating that FLAG-AF9 inhibited the Dot1a·AF17 interaction. To validate this finding in vivo, competitive mammalian two-hybrid assays were performed. 293T cells were transfected with a constant amount of pBD-Dot1a with pAD (as vector control) or with pAD-AF17 in the presence of 0–800 ng of pFLAG-AF9. The total amount of plasmid DNA transfected was kept constant by the addition of an empty vector. The luciferase reporter activity was inversely correlated with the amount of pFLAG-AF9 added; ∼12 times lower reporter expression was observed when 800 versus 0 ng of pFLAG-AF9 was applied (Fig. 4C). Similar experiments were performed to test whether the competition between AF9 and AF17 regulates α-ENaC promoter activity. Transfection of FLAG-AF17 doubled the expression of an α-ENaC promoter-luciferase construct compared with Vec control, indicating that AF17 up-regulates the α-ENaC promoter, possibly by facilitating nuclear export of the endogenous hDot1L (see below). However, this effect was gradually diminished by an increasing amount of FLAG-AF9 that may competitively bind the endogenous hDot1L and repress the promoter (Fig. 4D).
FIGURE 4.
AF17 and AF9 competitively bind Dot1a. A and E, shown are diagrams of AF9 or AF17 preventing its competitor from binding Dot1a. B and F, shown is a GST pulldown assay demonstrating the inhibitory effect of AF9 on Dot1a·AF17 or AF17 on Dot1a·AF9 interactions. GST pulldown assay was performed similar to Fig. 2D except that various amounts of lysates containing FLAG-AF9 (B) or FLAG-AF17 635–786 (F) were premixed with 1 ml of GFP-Dot1a cell lysate as indicated. The total volume of the lysates was equalized by the addition of a mock-transfected lysate. The GFP-Dot1a fusion in the input (5%) or bound to the beads was determined by IB with the anti-GFP antibody. The GST fusions eluted from the columns were examined by Coomassie staining. The FLAG fusions in the flow-through were examined by IB with a mouse anti-FLAG antibody. The amount of bound GFP-Dot1a fusion was normalized to that of the GST-AF17 (B) or the GST-AF9 (F) fusions to calculate the binding efficiency. It was further normalized to the control in which the competitor was omitted. The average of three independent experiments is shown at the bottom. C and G, competitive mammalian two-hybrid assays show that AF9 and AF17 mutually impair their interactions with Dot1a in vivo. Data are similar to Fig. 2C except that varying amounts (ng) of pFLAG-AF9 (C) or pFLAG-AF17 (G) were included in the transfection. The total amount of DNA was normalized by adding an empty vector. In all cases n = 3; *, p < 0.05 versus control, where either no FLAG-AF9 (C) or FLAG-AF17 (G) was added. D and H, AF9 and AF17 mutually antagonize the effect of their competitor on the α-ENaC promoter. 293T cells were transfected with an α-ENaC-promoter luciferase construct with pCDNA3.1 vector (Vec), or pFLAG-AF17 with a range of 0–800 ng of pFLAG-AF9 as competitor (D) or pFLAG-AF9 with 0–800 ng of pFLAG-AF17 as competitor (H) followed by luciferase assay. Luciferase activity was set to 1 in vector-transfected cells. For all cases, n = 3. *, p < 0.05 versus vector. Note: the reporter in D and H was repressed by AF9 and activated by AF17. However, activation of the reporter in C and G is Dot1a·AF17 or Dot1a·AF9 interaction-dependent.
In reciprocal assays the Dot1a·AF9 interaction was progressively inhibited by increasing amounts of FLAG-AF17 635–786 as a competitor in GST pulldown assays (Fig. 4F) or by increasing amounts of FLAG-AF17 in mammalian two-hybrid assays (Fig. 4G). FLAG-AF17 also antagonized FLAG-AF9-mediated repression of α-ENaC promoter-luciferase construct in a dose-dependent manner (Fig. 4H).
AF17 Overexpression Enhances Dot1a Cytoplasmic Expression at the Expense of Its Nuclear Expression
As mentioned above, GFP-Dot1a and RFP-AF17 co-localized in 293T cells. In some of the co-transfected cells, both fusion proteins were primarily in the nucleus (Fig. 3B, top panel). However, in the majority of co-transfected cells, Dot1a and AF17 co-localized in the cytoplasm (Fig. 3B, low panel, and supplemental Fig. S4). The third type of expression pattern displayed substantial signals in both the cytoplasm and the nucleus (supplemental Fig. S4). These data suggest that overexpression of RFP-AF17 with GFP-Dot1a leads to preferential localization of both proteins in the cytoplasm. However, the cellular localization of these proteins might be also regulated in a cell cycle-dependent manner, resulting in variable levels of H3 K79 methylation throughout the cell cycle (26). Moreover, given the non-quantitative nature of the assays, we could not determine whether the expression level of AF17 directly correlated with the degree of cytoplasmic localization in patterns two and three.
To control for any effect of the GFP or RFP tags on the cellular distribution of these fusions, 293T cells were co-transfected with either of three pairs of plasmids: pGFP/pRFP-AF17, pGFP-Dot1a/pRFP, or pGFP-Dot1a/pRFP-AF17. In each transfection we examined only the co-transfected cells and divided them into three types based on the cellular distribution of RFP-AF17, GFP-Dot1a, or both. Without GFP-Dot1a, RFP-AF17 resided in the cytoplasm in almost 80% of cells. Its nuclear expression was detected in only 5% of cells (Fig. 3C). In the absence of RFP-AF17, GFP-Dot1a was present in the nucleus in 62% of cells, with only 5% in the cytoplasm and 33% in both of the compartments. However, when the fusion proteins were co-expressed, the percentage of the cells displaying Dot1a nuclear expression was decreased to about 15% (Fig. 3C), which was accompanied with a dramatic increase (to 52%) of cells with Dot1a located in the cytoplasm. These results suggest that AF17 overexpression enhances cytoplasmic and limits nuclear expression of Dot1a.
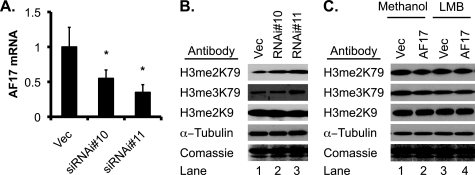
To gain additional evidence, RNAi-mediated depletion of endogenous AF17 was performed. We established three cell lines derived from 293T cells stably carrying an empty vector or one of two AF17-specific siRNA constructs: siRNA#10 and siRNA#11. Real-time RT-qPCR revealed that AF17 mRNA abundance was decreased to 55 and 35% in the cells transfected with these two constructs, respectively, compared with that in the vector-transfected cells (Fig. 5A). Transfection of GFP-Dot1a into these cell lines revealed that the percentage of cells with cytoplasmic GFP-Dot1a was decreased from 31% in vector-transfected cells to 17% in siRNA#10-transfected cells (Fig. 3D), whereas the cells expressing nuclear GFP-Dot1a increased from 53 to 70%. In the cells bearing siRNA#11, which more efficiently knocked down AF17 expression, a more dramatic effect was observed. Only 12% of the cells displayed cytoplasmic GFP-Dot1a expression, whereas 78% showed nuclear expression. In brief, our data are consistent with the notion that AF17 overexpression promotes redistribution of Dot1a from the nucleus to the cytoplasm and that endogenous AF17 is important for Dot1a trafficking.
FIGURE 5.
AF17 down-regulates H3 K79 methylation in bulk histones. A, AF17 expression was substantially decreased by transfection of AF17-specific RNAi constructs. 293T cells were stably transfected with pSilencer-2.1-U6-Hygro vector (Vec) or its derivatives bearing AF17-specific siRNA#10 or siRNA#11. Total RNA was isolated and examined by real-time RT-qPCR for AF17, which was normalized to β-actin. n = 3. *, p < 0.05 versus vector. B, AF17 knockdown increases H3 K79 methylation. As in A, acid extracts of 293T cells were analyzed by IB with antibodies recognizing di- and trimethylated H3 K79 (H3me2K79 and H3me3K79), dimethylated H3 K9 (H3 me2K9), or α-tubulin. Coomassie staining was performed with an identical gel. C, AF17 overexpression impairs H3 K79 di- and trimethylation in an LMB-sensitive manner. pGFP-Dot1a was transfected into 293T cells along with pRFP vector or RFP-AF17. 16 h later the cells were treated with vehicle or nuclear export inhibitor LMB (10 nm) for another 16 h followed by IB of the acid extracts as shown in B.
AF17 Decreases H3 K79 Methylation in Bulk Histones, Possibly by Facilitating Dot1a Nuclear Export
If AF17 shifts Dot1a expression from the nucleus to the cytoplasm, Dot1a-catalyzed H3 K79 methylation should be inversely correlated with AF17 expression. To test this hypothesis, we first investigated the effect of AF17 knockdown on H3 K79 methylation. Western blot analysis of acid extracts prepared from the three cell lines mentioned above were analyzed by immunoblotting with antibodies specifically recognizing di- or tri-methylated H3 K79, which were designated as H3 me2K79 and H3 me3K79, respectively. An antibody against dimethylated H3 K9 (H3 me2K9) was included as a control. The levels of H3 me2K79 in the vector-transfected cells was more than doubled or tripled in the 293T cells stably transfected with siRNA#10 and siRNA#11, respectively. Similar results were observed for H3 me3K79 (Fig. 5B). We did not examine the mono-methylated H3 K79 because the available antibody also recognizes the unmethylated H3 K79 isoform (27). However, AF17 knockdown did not obviously affect H3 me2K9 (Fig. 5B).
Because the endogenous levels of H3 me2K79 and H3 me3K79 in 293T cells are low, AF17 overexpression may make their detection and interpretation of experiments difficult. Therefore, we asked whether AF17 overexpression abolishes the Dot1a overexpression-dependent increase of H3 K79 methylation by promoting Dot1a nuclear export. 293T cells were transiently transfected with pGFP-Dot1a along with pRFP as a control or with pRFP-AF17. Cells were treated with methanol vehicle or the nuclear export inhibitor leptomycin B (LMB, 10 nm). This concentration of LMB has been shown to block nucleocytoplasmic shuttling of Id1 in other cells (28). In the vehicle-treated cells, co-expression of RFP-AF17 with GFP-Dot1a reduced the levels of H3 me2K79 and H3 me3K79 to about 40 and 30% that of the control level (compare lane 2 with lane 1 in the corresponding blots, Fig. 5C), respectively. However, this effect was largely abolished by LMB. H3 me2K9 was not measurably affected by RFP-AF17 overexpression regardless of the addition of LMB (Fig. 5C). Together, these data indicate that AF17 limits Dot1a methyltransferase activity, probably by promoting Dot1a nuclear export.
Like AF9, AF17 Displayed Cytoplasmic and Nuclear Colocalization with Sgk1
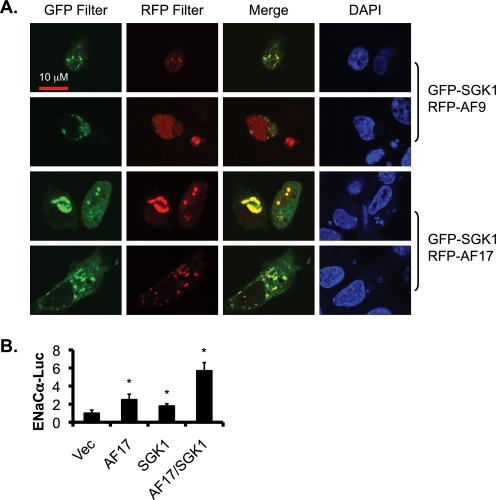
Our previous studies suggest that Sgk1 phosphorylates AF9 in vitro and in vivo in mIMCD3 cells and in mouse kidney and that Sgk1 is associated with the α-ENaC promoter (9). However, whether these two proteins co-localize within cells has not been addressed. Given the fact that AF17 contains three potential consensus Sgk1 phosphorylation sites (see “Discussion”), AF17 might also be a phosphorylation target of Sgk1. As a first step toward addressing this question, we transfected 293T cells with GFP-Sgk1 along with RFP-AF9 as a control or with RFP-AF17 and examined their cellular distribution by confocal microscopy. Sgk1 co-localized with AF9 in the nucleus and cytoplasm. Similar results were obtained when AF9 was replaced with AF17 (Fig. 6A), raising the possibility of a complex regulation of AF9 and AF17 by Sgk1, possibly by phosphorylation.
FIGURE 6.
AF17 and Sgk1 colocalize and synergistically activate the α-ENaC promoter. A, as in Fig. 3A, representative images of confocal microscopy demonstrate colocalization of Sgk1 with AF9 as control or with AF17 in 293T cells. B, luciferase assay indicates a synergistic effect of AF17 and Sgk1 on α-ENaC promoter. 293T cells transfected with an α-ENaC promoter luciferase construct along with pCDNA3.1 vector (Vec) or its derivatives encoding AF17 or Sgk1 were analyzed as in Fig. 4D. *, p < 0.05 versus Vec.
Because Sgk1 and AF17 positively regulate α-ENaC transcription, we sought to determine whether the potential Sgk1-AF17 interaction has a synergistic effect on α-ENaC promoter activity. Luciferase assay revealed that whereas the activity of the α-ENaC was elevated by 150 or 80% versus control in AF17- or Sgk1-transfected cells, respectively, it was increased by 470% when these two proteins were co-expressed, indicating a synergistic effect (Fig. 6B). These observations are consistent with the notion that Sgk1 may regulate AF17 function by phosphorylation. Future studies are required to fully address this question.
AF17 Up-regulates Transcription of ENaC and Several Other Aldosterone-regulated Genes
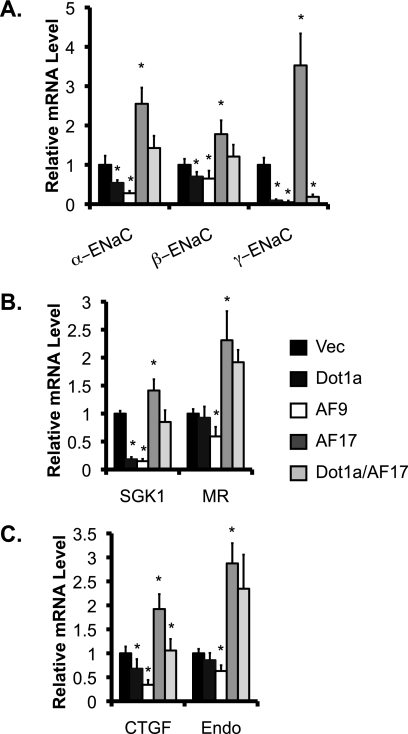
Because AF17 impairs Dot1a nuclear expression and, thus, H3 K79 methylation in the bulk histones, we anticipated that AF17 would relieve Dot1a·AF9-mediated repression. We have reported that overexpression of Dot1a and/or AF9 reduced mRNA expression of α-ENaC and three other aldosterone-regulated genes in mIMCD3 cells (7–9). Whether such regulation is conserved or can be extended to other aldosterone-regulated genes in 293T cells remains unknown. Because AF17 impairs H3 K79 methylation, we anticipated that AF17 would relieve Dot1a·AF9-mediated repression of the α-ENaC promoter. Accordingly, 293T cells were transiently transfected with pcDNA3.1 (Vec) or its derivatives encoding Dot1a, AF9, or AF17 and examined by real-time RT-qPCR. α-ENaC mRNA was observed at ∼54, 28, 250, or 143% of the control levels in cells overexpressing Dot1a, AF9, AF17, or Dot1a/AF17, respectively (Fig. 7A). A similar pattern was obtained for the expression of several other aldosterone-regulated genes examined, including β- and γ-ENaC, SGK1, and CTGF. There were two exceptions to this pattern; Dot1a failed to significantly inhibit mRNA expression of MR (Fig. 7B) and preproendothelin-1 (Fig. 7C). We did not examine the expression pattern of period because its basal level was undetectable in 293T cells (Fig. 1A).
FIGURE 7.
Dot1a, AF9, and AF17 differentially regulate mRNA expression of ENaC and other target genes. As in Fig. 1B, 293T cells were transiently transfected with pCDNA3.1 (Vec) or its derivatives expressing Dot1a, AF9, AF17, or Dot1a/AF17. Total RNA from these cells was analyzed by real-time RT-qPCR for expression of ENaC subunits (A), ENaC regulators (B), or CTGF and preproendothelin (Endo) (C). The relative abundance of mRNA of each gene was set to 1 in vector-transfected cells and used for comparison. n = 3. *, p < 0.05 versus vector for each gene.
Transcriptional Changes in ENaC Genes Are Correlated with Changes in ENaC Activity
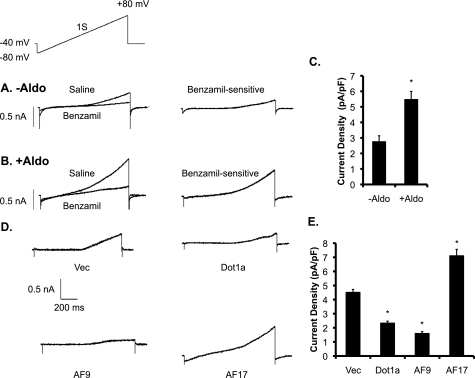
To determine whether the changes in the expression of ENaC genes at the mRNA level are associated with the corresponding changes in ENaC activity, we performed patch clamp experiments first with 293T cells treated with vehicle or aldosterone (1 μm) for 24 h. We then measured currents evoked by ramp voltage commands before and after superfusion of cells with 10 μm benzamil, an inhibitor of ENaC. Benzamil-sensitive cation currents were then obtained by digital subtraction. Under these conditions (symmetrical NaCl in bath and recording pipette), ENaC gives rise to strongly outwardly rectifying currents. Representative examples of recordings from single cells are shown in Fig. 8, A and B, and mean results determined at +80 mV are shown in Fig. 8C. The basal level of benzamil-sensitive Na+ current density was significantly increased by aldosterone treatment (Fig. 8C), a result that is consistent with the increased mRNA expression of all ENaC subunit genes (Fig. 1B).
FIGURE 8.
Aldosterone and ENaC transcriptional regulators regulate ENaC current. A and B, representative traces of whole-cell recordings from cells 24 h after the onset of exposure to equal volume of ethanol as a vehicle control (−Aldo) or 1 μm aldosterone (+Aldo) as indicated. Currents were recorded during a 1-s voltage ramp (−80 to +80 mV) delivered from a holding potential of −40 mV (shown as the inset above the current traces). Currents were obtained in normal external saline and after superfusion of saline containing 10 μm benzamil, as shown in the traces to the left. For each cell, benzamil-sensitive currents were then computed by digital subtraction and are shown in the traces on the right. Under these recording conditions, benzamil-sensitive currents show strong outward rectification. C, mean current density is shown of benzamil-sensitive currents at +80 mV from HEK293 cells treated with vehicle (n = 12) or aldosterone (n = 13). *, p < 0.05 versus −Aldo. pF, picofarads. D–E, effects are shown of transcriptional regulators on ENaC currents in HEK 293T cells. D, representative examples are shown of whole-cell recordings of benzamil-sensitive currents from HEK 293T cells 24 h after transient transfection with expression plasmids encoding GFP (Vec) or one of the GFP-tagged transcriptional regulators (GFP-Dot1a, GFP-AF9, and GFP-AF17) as indicated. Benzamil-sensitive currents recorded from fluorescent cells during application of voltage ramps were obtained by digital subtraction as described above. E, mean densities of benzamil-sensitive currents at +80 mV are compiled from 12 cells in each group. *, p < 0.05 versus vector (Vec).
To determine whether the effects of Dot1a, AF9, or AF17 overexpression on ENaC mRNA expression elicit corresponding changes in ENaC activity, similar experiments were conducted with 293T cells transfected with GFP vector or plasmids encoding the related GFP fusion proteins. With this design, transfected cells can be identified by fluorescence during the recordings. Representative examples of benzamil-sensitive currents are shown in Fig. 8D. The benzamil-sensitive Na+ current density was significantly decreased in Dot1a- or AF9-overexpressing cells compared with GFP controls. However, it was significantly elevated in cells overexpressing AF17 (Fig. 8E). Thus, multiple and independent lines of data indicate that Dot1a, AF9, and AF17 regulate ENaC activity, at least partially through regulation of ENaC gene expression at the mRNA level.
DISCUSSION
In this report we characterized and used HEK 293T cells as a model system to study the aldosterone-signaling network controlling the transcription of ENaC genes, their regulators, and ENaC activity. We demonstrate for the first time the physiological importance of Dot1a and AF9 in suppressing basal ENaC-mediated Na+ transport as measured by patch clamp analysis of transfected HEK 293T cells. We also describe a novel competitive interaction between AF17 with a region of Dot1a known to bind AF9 that antagonizes the epigenetic repressor effects of Dot1a·AF9 on α-ENaC transcription and augments ENaC-mediated Na+ transport under basal conditions to levels comparable with those found with aldosterone induction. The positive regulatory effect of AF17 on α-ENaC transcription appears to involve, at least in part, enhanced nuclear export of Dot1a to the cytoplasm, where we postulate it is degraded. The resulting reduction in nuclear Dot1a expression leads to H3 K79 hypomethylation and de-repression of α-ENaC transcription. Therefore, these studies show that AF17, like two other MLL fusion partners AF9, (8) and AF10 (29), is a Dot1a binding partner and regulator. Dot1a and, thus, H3 K79 methylation is regulated by competition between AF9 and AF17 for binding sites on Dot1a. These data show that AF17 is a regulator of ENaC transcription and ENaC activity, and they support the hypothesis that mistargeting of hDot1L may play a role in some types of MLL fusion-mediated leukemogenesis.
ENaC is expressed in many epithelial tissues including the aldosterone-sensitive distal nephron of kidney, distal colon, lung airway, urinary bladder, and ducts of salivary and sweat glands (30) and is subjected to complex regulation to meet the need for rapid, dynamic changes in salt and water secretion and reabsorption. The channel expression and synthesis, intracellular channel trafficking, maturation and activation, and single-channel properties such as open probability (Po) are controlled by a variety of extrinsic and intrinsic factors (31, 32). Extrinsic factors include mechanical stretch, proteolytic cleavage, and hormones such as aldosterone (33), arginine vasopressin (34), atrial natriuretic peptide (35), insulin, and endothelin (36, 37). Intrinsic regulation may be due to intracellular trafficking, ubiquitination/de-ubiquitination, various kinases, sodium, and metabolic substrates (for review, see Ref. 31). Many proteins have been identified that play a role in these diverse pathways. Among them are Sgk1 (38), Nedd4-2 (39), 14-3-3 (40), ubiquitin-specific protease Usp2–45 (41), glucocorticoid-induced leucine zipper protein (GILZ) (19), small G protein K-Ras2A (42), channel-activating protease CAP1 (43), and furin (44).
For example, Sgk1 is thought to regulate ENaC abundance at the cell surface, in part through phosphorylation of the ubiquitin ligase Nedd4-2 (45). Nedd4-2 phosphorylation reduces its affinity with and, hence, binding to ENaC and induces its interaction with 14-3-3 (40). The concerted actions of Sgk1 and 14-3-3 appear to disrupt Nedd4-2-mediated ubiquitination and subsequent degradation of ENaC, leading to accumulation of ENaC channels at the cell surface.
Our recent work suggests that aldosterone down-regulates the Dot1a·AF9 complex by reducing Dot1a (7) and AF9 expression (8) or by decreasing Dot1a·AF9 interaction via Sgk1-mediated phosphorylation of AF9 (9). Here, we provide evidence showing that AF17 competes with AF9 to bind Dot1a and enhances Dot1a cytoplasmic expression at the expense of its nuclear expression. Although AF17 mRNA expression was not significantly changed by aldosterone treatment, overexpression of AF17 increased the mRNA abundance of all genes examined (α-, β-, and γ-ENaC, SGK1, MR, preproendothelin-1, and CTGF). It can be speculated that this novel epigenetic mechanism most likely regulates the Dot1a·AF9 complex and, thus, H3 K79 methylation in an aldosterone-independent manner. However, the possibility of aldosterone-dependent mechanisms cannot be completely ruled out. Our data do highlight the fact that this epigenetic pathway may govern up to 50% of ENaC-mediated Na+ transport in HEK 293T cells as measured patch clamp analysis (Fig. 8).
Examination of the human AF17 sequence revealed three potential Sgk1 phosphorylation sites with Ser-222, -423, and -662 as the putative phosphorylation acceptors. The first two sites are located outside the Dot1a-interacting domain and are 100% conserved in mouse and rat, whereas Ser-662 is within this domain. The equivalent regions of the mouse and rat homologs share a single amino acid substitution, compared with AF17, which disrupts the consensus sequence motif for Sgk1 phosphorylation. These observations raise the possibility that aldosterone and/or Sgk1 might regulate AF17 function in balancing Dot1a nuclear/cytoplasmic expression and formation of Dot1a·AF17/Dot1a·AF9 complexes. Consistent with this hypothesis, AF17 and Sgk1 exhibit nuclear and cytoplasmic co-localization, indicating that they may be in the same protein complexes and interact either directly or indirectly.
Endogenous AF17 was observed at the mRNA level in 293T cells, as evidenced by RT-PCR (Fig. 1A) and the significant effect of RNAi-mediated AF17 depletion on Dot1a cellular distribution and H3 K79 methylation (Figs. 3D and 5B). Similarly, FLAG- or RFP-AF17 were expressed and detected in immunofluorescence, epifluorescence, and confocal microscopy studies by others (12) and us (Fig. 3). However, all of these proteins remained undetectable by IB analysis with antibodies against FLAG, RFP, or AF17 (data not shown). To our knowledge, no reports exist showing immunoblots with detectable endogenous or overexpressed full-length AF17 in the literature. Therefore, it is very difficult to verify the Dot1a and AF17 interaction or the competitive binding of Dot1a between AF9 and AF17 at the endogenous protein levels or with overexpressed full-length proteins by co-immunoprecipitation/IB assays. This limitation may also prevent the determination of the relative abundance of the endogenous or recombinant full-length AF17 in the cytoplasm and in the nucleus by IB, the confirmation of Dot1a·AF17 co-localization at the endogenous protein level, the definition of AF17 association with α-ENaC promoter or other DNA sequences, and the evaluation of Dot1a preference with AF9 over AF17 under basal conditions and the influence of chromatin context on such preference. Furthermore, the function of Dot1a·AF17 complex in the cytoplasm remains unknown. We repeatedly observed that FLAG-AF17 635–786 was readily detected when expressed alone or with GFP but was barely detectable when co-expressed with GFP-Dot1a 479–659 in the whole-cell lysate by IB (supplemental Fig. S2). Similarly, the biological significance of the cytoplasmic Sgk1-AF17 or Sgk1-AF9 colocalization is elusive. Further studies are needed to address these questions. It is also known that the phenotypic properties of polarized cells may differ depending on whether they are grown on solid substrates or permeable supports. The fact that several of our assays were performed with cells grown on plates or coverslips represents another potential limitation of our studies. Although 293T cells offered practical advantages in the analysis of the effects of overexpression of heterologous genes in a kidney epithelial cell type, they are not specifically derived from the collecting duct (the physiological site of ENaC-mediated Na+ transport in the kidney). Characterization of the renal physiological phenotype of the mutant mice in which Dot1, AF9, or AF17 is disrupted will provide the ultimate proof of the physiological relevance of the current study.
The AF17 gene was originally isolated as a less frequent fusion partner of the MLL gene in t(11;17)(q23;q21) translocations present in some acute myeloid leukemias (10, 11). Limited information exists about its function. AF17 is thought to function as a transcriptional regulator (10) and a downstream target of the β-catenin/T-cell factor pathway and plays a role in G2-M progression (12). Although AF17 can interact with PC2 glutamine/q-rich-associated protein (PCQAP) and CCAAT/enhancer binding protein (C/EBP) in yeast two-hybrid assay (46), the biological relevance and function of these protein-protein interactions remains to be characterized.
Although more than 40 MLL fusion partners have been cloned (47), a common structural motif or biochemical function has not been defined in most of the proteins encoded by these genes. As an exception, similarities between AF9 and ENL and between AF10 and AF17 (48, 49) exist. As mentioned earlier, we and others have reported that AF9 and AF10 interact with Dot1a or its human homolog hDot1L. AF17 and AF10 share significant homology within their respective LAP/PHD finger domains at the NH2 termini and leucine zipper domains toward the COOH termini and differ outside those regions (48). The fragments responsible for interaction with Dot1a or hDot1L (AF17 635-786 and AF10 635-1068) harbor the highly conserved leucine zipper domains. However, the lack of a leucine zipper domain in AF9 and no requirement for the putative leucine zipper domain in Dot1a for its interaction with either AF9 or AF17 argues against a notion that a leucine zipper-dependent dimer configuration is necessary for the interactions of Dot1a or hDot1L with AF9, AF10, or AF17. Because all of these MLL fusion partners (AF9, AF10, and AF17) can interact with Dot1a and the domains responsible for the interactions are retained in the corresponding MLL fusions, our data further support the hypothesis that mistargeting of hDOT1L may play an important role in some types of MLL fusion-mediated leukemogenesis (29).
Acknowledgments
We thank Dr. Yoichi Furukawa for kindly providing the AF17 cDNA constructs. We also thank Dr. Peter Snyder and Dr. Alan Pao for providing technical advice.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DK080236 (to W. Z.), R01 DK082529 (to S. E. D.), and R01 DK075065 (to B. K.). This work was also supported by American Heart Association Beginning Grant-in-aid 0865271F (to W. Z.) and an American Society of Nephrology Carl W. Gottschalk Research Scholar Grant (to W. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- ENaC
- epithelial Na+ channel
- HEK
- human embryonic kidney
- Sgk1
- serum- and glucocorticoid-inducible kinase isoform 1
- GST
- glutathione S-transferase
- RFP
- red fluorescence protein
- LMB
- leptomycin B
- MLL
- mixed lineage leukemia
- IB
- immunoblot
- DAPI
- 4′,6-diamidino-2-phenylindole
- GFP
- green fluorescent protein
- aa
- amino acids
- RT-qPCR
- reverse transcription-quantitative PCR
- BD
- binding domain
- AD
- activation domain
- MR
- mineralocorticoid receptor.
REFERENCES
- 1.Bangel N., Dahlhoff C., Sobczak K., Weber W. M., Kusche-Vihrog K. (2008) J. Cystic Fibrosis 7, 197–205 [DOI] [PubMed] [Google Scholar]
- 2.Shimkets R. A., Warnock D. G., Bositis C. M., Nelson-Williams C., Hansson J. H., Schambelan M., Gill J. R., Jr., Ulick S., Milora R. V., Findling J. W. (1994) Cell 79, 407–414 [DOI] [PubMed] [Google Scholar]
- 3.Chang S. S., Grunder S., Hanukoglu A., Rösler A., Mathew P. M., Hanukoglu I., Schild L., Lu Y., Shimkets R. A., Nelson-Williams C., Rossier B. C., Lifton R. P. (1996) Nat. Genet. 12, 248–253 [DOI] [PubMed] [Google Scholar]
- 4.Eaton D. C., Malik B., Saxena N. C., Al-Khalili O. K., Yue G. (2001) J. Membr. Biol. 184, 313–319 [DOI] [PubMed] [Google Scholar]
- 5.Thomas C. P., Itani O. A. (2004) Curr. Opin. Nephrol. Hypertens. 13, 541–548 [DOI] [PubMed] [Google Scholar]
- 6.Zhang W., Hayashizaki Y., Kone B. C. (2004) Biochem. J. 377, 641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang W., Xia X., Jalal D. I., Kuncewicz T., Xu W., Lesage G. D., Kone B. C. (2006) Am. J. Physiol. Cell Physiol. 290, C936–C946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang W., Xia X., Reisenauer M. R., Hemenway C. S., Kone B. C. (2006) J. Biol. Chem. 281, 18059–18068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang W., Xia X., Reisenauer M. R., Rieg T., Lang F., Kuhl D., Vallon V., Kone B. C. (2007) J. Clin. Invest. 117, 773–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prasad R., Leshkowitz D., Gu Y., Alder H., Nakamura T., Saito H., Huebner K., Berger R., Croce C. M., Canaani E. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 8107–8111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzukawa K., Shimizu S., Nemoto N., Takei N., Taki T., Nagasawa T. (2005) Int. J. Hematol. 82, 38–41 [DOI] [PubMed] [Google Scholar]
- 12.Lin Y. M., Ono K., Satoh S., Ishiguro H., Fujita M., Miwa N., Tanaka T., Tsunoda T., Yang K. C., Nakamura Y., Furukawa Y. (2001) Cancer Res. 61, 6345–6349 [PubMed] [Google Scholar]
- 13.Boulkroun S., Ruffieux-Daidié D., Vitagliano J. J., Poirot O., Charles R. P., Lagnaz D., Firsov D., Kellenberger S., Staub O. (2008) Am. J. Physiol. Renal Physiol. 295, F889–F900 [DOI] [PubMed] [Google Scholar]
- 14.Zhou R., Patel S. V., Snyder P. M. (2007) J. Biol. Chem. 282, 20207–20212 [DOI] [PubMed] [Google Scholar]
- 15.Zhou R., Snyder P. M. (2005) J. Biol. Chem. 280, 4518–4523 [DOI] [PubMed] [Google Scholar]
- 16.Kabra R., Knight K. K., Zhou R., Snyder P. M. (2008) J. Biol. Chem. 283, 6033–6039 [DOI] [PubMed] [Google Scholar]
- 17.Fejes-Tóth G., Pearce D., Náray-Fejes-Tóth A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2973–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soundararajan R., Wang J., Melters D., Pearce D. (2007) J. Biol. Chem. 282, 36303–36313 [DOI] [PubMed] [Google Scholar]
- 19.Soundararajan R., Zhang T. T., Wang J., Vandewalle A., Pearce D. (2005) J. Biol. Chem. 280, 39970–39981 [DOI] [PubMed] [Google Scholar]
- 20.Kim E. Y., Ridgway L. D., Dryer S. E. (2007) Mol. Pharmacol. 72, 622–630 [DOI] [PubMed] [Google Scholar]
- 21.Kim E. Y., Ridgway L. D., Zou S., Chiu Y. H., Dryer S. E. (2007) Neuroscience 146, 1652–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim E. Y., Zou S., Ridgway L. D., Dryer S. E. (2007) J. Neurophysiol. 97, 3508–3516 [DOI] [PubMed] [Google Scholar]
- 23.Ridgway L. D., Kim E. Y., Dryer S. E. (2009) Am. J. Physiol. Cell Physiol. 297, C55–C65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang W., Xia X., Zou L., Xu X., LeSage G. D., Kone B. C. (2004) Am. J. Physiol. Renal Physiol. 286, F1171–FF1177 [DOI] [PubMed] [Google Scholar]
- 25.Rao U. S., Baker J. M., Pluznick J. L., Balachandran P. (2004) Cell Calcium 35, 21–28 [DOI] [PubMed] [Google Scholar]
- 26.Feng Q., Wang H., Ng H. H., Erdjument-Bromage H., Tempst P., Struhl K., Zhang Y. (2002) Curr. Biol. 12, 1052–1058 [DOI] [PubMed] [Google Scholar]
- 27.Jones B., Su H., Bhat A., Lei H., Bajko J., Hevi S., Baltus G. A., Kadam S., Zhai H., Valdez R., Gonzalo S., Zhang Y., Li E., Chen T. (2008) PLoS Genet. 4, e1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishiyama K., Takaji K., Uchijima Y., Kurihara Y., Asano T., Yoshimura M., Ogawa H., Kurihara H. (2007) J. Biol. Chem. 282, 17200–17209 [DOI] [PubMed] [Google Scholar]
- 29.Okada Y., Feng Q., Lin Y., Jiang Q., Li Y., Coffield V. M., Su L., Xu G., Zhang Y. (2005) Cell 121, 167–178 [DOI] [PubMed] [Google Scholar]
- 30.Garty H., Palmer L. G. (1997) Physiol. Rev. 77, 359–396 [DOI] [PubMed] [Google Scholar]
- 31.Bhalla V., Hallows K. R. (2008) J. Am. Soc. Nephrol. 19, 1845–1854 [DOI] [PubMed] [Google Scholar]
- 32.Rossier B. C., Pradervand S., Schild L., Hummler E. (2002) Annu. Rev. Physiol. 64, 877–897 [DOI] [PubMed] [Google Scholar]
- 33.Shigaev A., Asher C., Latter H., Garty H., Reuveny E. (2000) Am. J. Physiol. Renal Physiol 278, F613–F619 [DOI] [PubMed] [Google Scholar]
- 34.Ecelbarger C. A., Kim G. H., Wade J. B., Knepper M. A. (2001) Exp. Neurol. 171, 227–234 [DOI] [PubMed] [Google Scholar]
- 35.Zeidel M. L., Kikeri D., Silva P., Burrowes M., Brenner B. M. (1988) J. Clin. Invest. 82, 1067–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marunaka Y., Hagiwara N., Tohda H. (1992) Am. J. Physiol. 263, F392–F400 [DOI] [PubMed] [Google Scholar]
- 37.Gilmore E. S., Stutts M. J., Milgram S. L. (2001) J. Biol. Chem. 276, 42610–42617 [DOI] [PubMed] [Google Scholar]
- 38.Loffing J., Flores S. Y., Staub O. (2006) Annu. Rev. Physiol. 68, 461–490 [DOI] [PubMed] [Google Scholar]
- 39.Bhalla V., Daidié D., Li H., Pao A. C., LaGrange L. P., Wang J., Vandewalle A., Stockand J. D., Staub O., Pearce D. (2005) Mol. Endocrinol. 19, 3073–3084 [DOI] [PubMed] [Google Scholar]
- 40.Ichimura T., Yamamura H., Sasamoto K., Tominaga Y., Taoka M., Kakiuchi K., Shinkawa T., Takahashi N., Shimada S., Isobe T. (2005) J. Biol. Chem. 280, 13187–13194 [DOI] [PubMed] [Google Scholar]
- 41.Verrey F., Fakitsas P., Adam G., Staub O. (2008) Kidney Int. 73, 691–696 [DOI] [PubMed] [Google Scholar]
- 42.Mastroberardino L., Spindler B., Forster I., Loffing J., Assandri R., May A., Verrey F. (1998) Mol. Biol. Cell 9, 3417–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vallet V., Chraibi A., Gaeggeler H. P., Horisberger J. D., Rossier B. C. (1997) Nature 389, 607–610 [DOI] [PubMed] [Google Scholar]
- 44.Bruns J. B., Carattino M. D., Sheng S., Maarouf A. B., Weisz O. A., Pilewski J. M., Hughey R. P., Kleyman T. R. (2007) J. Biol. Chem. 282, 6153–6160 [DOI] [PubMed] [Google Scholar]
- 45.Debonneville C., Flores S. Y., Kamynina E., Plant P. J., Tauxe C., Thomas M. A., Münster C., Chraibi A., Pratt J. H., Horisberger J. D., Pearce D., Loffing J., Staub O. (2001) EMBO J. 20, 7052–7059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rual J. F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G. F., Gibbons F. D., Dreze M., Ayivi-Guedehoussou N., Klitgord N., Simon C., Boxem M., Milstein S., Rosenberg J., Goldberg D. S., Zhang L. V., Wong S. L., Franklin G., Li S., Albala J. S., Lim J., Fraughton C., Llamosas E., Cevik S., Bex C., Lamesch P., Sikorski R. S., Vandenhaute J., Zoghbi H. Y., Smolyar A., Bosak S., Sequerra R., Doucette-Stamm L., Cusick M. E., Hill D. E., Roth F. P., Vidal M. (2005) Nature 437, 1173–1178 [DOI] [PubMed] [Google Scholar]
- 47.Eguchi M., Eguchi-Ishimae M., Greaves M. (2003) Int. J. Hematol. 78, 390–401 [DOI] [PubMed] [Google Scholar]
- 48.Chaplin T., Ayton P., Bernard O. A., Saha V., Della Valle V., Hillion J., Gregorini A., Lillington D., Berger R., Young B. D. (1995) Blood 85, 1435–1441 [PubMed] [Google Scholar]
- 49.Rubnitz J. E., Morrissey J., Savage P. A., Cleary M. L. (1994) Blood 84, 1747–1752 [PubMed] [Google Scholar]